A streptococcal domain of unknown function 1792 has been crystallized.
Keywords: DUF1792, glycosyltransferase, Streptococcus parasanguinis
Abstract
Serine-rich repeat glycoproteins (SRRPs) belong to a growing family of bacterial adhesins; they play important roles in bacterial virulence. Fap1, the first SRRP protein to be identified, is glycosylated; while the first two steps of its glycosylation have been determined, the remaining glycosylation steps are unknown. In a search for proteins that might be relevant to the glycosylation of Fap1, a putative glycosyltransferase (GalT1) from Streptococcus parasanguinis was identified. GalT1 possesses a domain of unknown function at the N-terminus. This domain is highly conserved in bacteria and is a member of a broad superfamily. However, the structure of this domain has not been determined. Here, the conditions used to produce a recombinant version of this protein domain and to grow protein crystals are reported. The crystals obtained belonged to space group C2, with unit-cell parameters a = 71.0, b = 45.1, c = 78.6 Å, β = 109.6°, and diffracted to 1.55 Å resolution at a synchrotron X-ray source. This domain does not share sequence identity with proteins of known structures above a level of 12%.
1. Introduction
Serine-rich repeat glycoproteins (SRRPs) belong to a growing family of bacterial adhesins found in streptococci, staphylococci and other Gram-positive bacteria (Zhou & Wu, 2009 ▶). Fimbriae-associated protein 1 (Fap1) from Streptococcus parasanguinis was the first identified SRRP; it is important for bacterial adhesion and biofilm formation (Wu et al., 1998 ▶; Wu & Fives-Taylor, 1999 ▶). This family of SRRPs plays an important role in bacterial pathogenesis. Srr-1 and Srr-2, two SRRPs from Group B streptococcus, are involved in the bacterial pathogenesis of meningitis (Van Sorge et al., 2009 ▶) and neonatal infection (Seifert et al., 2006 ▶). The SRRP protein PsrP from S. pneumoniae is associated with frequency of invasive pneumococcal disease (Shivshankar et al., 2009 ▶) and the SRRP protein SraP from Staphylococcus aureus is implicated in the pathogenesis of infective endocarditis (Siboo et al., 2005 ▶).
S. parasanguinis is a primary colonizer of the tooth surface and is important in the formation of dental plaque, the most complicated human biofilm known, and implicated in the development of oral infectious diseases (Drickamer & Taylor, 1998 ▶). Fap1, as the first identified SRRP, has been studied extensively and its biosynthetic pathway has been proposed (Zhu et al., 2011 ▶; Wu & Wu, 2011 ▶; Wu et al., 2007 ▶). Genes flanking fap1 mediate protein glycosylation and secretion (Wu et al., 2007 ▶). They include a gene cluster producing Gly–Gtf3–GalT1–GalT2 located in an upstream region of fap1 and another one producing a SecY2–SecA2–Gap1–Gap2–Gap3–Gtf1–Gtf2 located downstream of fap1. A glycosyltransferase enzyme complex consisting of glycosyltransferase Gtf1 and its chaperone Gtf2 catalyzes the first step of Fap1 glycosylation, transferring GlcNAc residues to the Fap1 polypeptide (Wu & Wu, 2011 ▶; Zhou et al., 2010 ▶; Wu et al., 2007 ▶). Gtf3 mediates the second step by transferring Glc to Gtf1-2-modified Fap1 (Zhu et al., 2011 ▶; Zhou et al., 2010 ▶). The subsequent steps of Fap1 glycosylation are unknown.
Based on amino-acid sequence homology, genes coding for Gly, GalT1 and GalT2 in the fap1 cluster were predicted to encode the remaining glycosyltransferases. One of them might mediate the third step of Fap1 glycosylation. Among these putative glycosyltransferases, GalT1 has a unique conserved domain of unknown function 1972 (DUF1792) and a separate putative GT-A type glycosyltransferase domain. DUF1792 is one of a group of uncharacterized hypothetical protein domains and is number 1792 in the order of addition to the database of hypothetical domains (Bateman et al., 2010 ▶). DUF1792 is located at the N-terminus of GalT1 and is highly conserved in bacteria. DUF1792 does not have any sequence similarity to known glycosyltransferases (lower than 12%). No three-dimensional structure is available for DUF1792. Therefore, structural characterization of DUF1972 by X-ray crystallography may help to define its biochemical function.
2. Materials and methods
2.1. Cloning, expression and purification of DUF1792
The gene coding for DUF1792 (amino acids 1–272) from S. parasanguinis FW213 was amplified by PCR using primer set DUF-BamHI-1F (GATCAGGATCCGTGAAGAGATTGTCAGAAATTAG) and DUF-XhoI-272R (GATCACTCGAGTCATCCAATTCTAGCTACTATTTG). The PCR product was gel purified and cloned into a pET-SUMO vector (Lifesensors). The resulting plasmid pET-SUMO-DUF1792 was confirmed by DNA sequencing and then transformed into Escherichia coli BL21 (DE3) for protein expression.
The recombinant strain grown to OD600 = 0.8 in 1 l LB medium was induced with 0.1 mM IPTG at 291 K overnight. The overnight-grown E. coli cells were harvested by centrifugation and lysed by sonication in binding buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 25 mM imidazole). The recombinant protein was purified by passing through a HisTrap column (Ni-affinity column). The N-terminal His-SUMO tag was removed by SUMO protease (ubiquitin-like protein protease) leaving all native and no exogenous residues. The SUMO-protease-digested proteins were then passed through a HisTrap column again to remove cleaved His-SUMO, SUMO protease and uncleaved proteins. The recombinant DUF1792 protein was further purified using a 16/60 Superdex 75 gel-filtration column (GE Healthcare). The DUF1792 protein was maintained in 20 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT.
2.2. Crystallization, data collection and processing
The hanging-drop vapor-diffusion method was used for the crystallization trials. Each drop contains 1 ul protein solution (30 mg ml−1 protein and 10 mM UDP-glucose) and 1 µl reservoir solution. The total volume of solution in each reservoir was 500 µl. Crystallization screening kits (Crystal Screen, Crystal Screen 2, Index and Natrix) from Hampton Research were used to screen for crystallization conditions for the protein. All attempts to grow DUF1792 crystals without the addition of UDP-glucose failed.
The crystals were flash-cooled in a nitrogen-gas stream in cryoprotectant consisting of 15% glycerol in reservoir solution. Diffraction data were collected on the SER-CAT beamline 22-ID at the Argonne National Laboratory (APS). The data were collected using a MAR 300 CCD detector. During data collection, the crystal-to-detector distance was kept at 200 mm. 360 images covering a ϕ range of 360° were collected and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
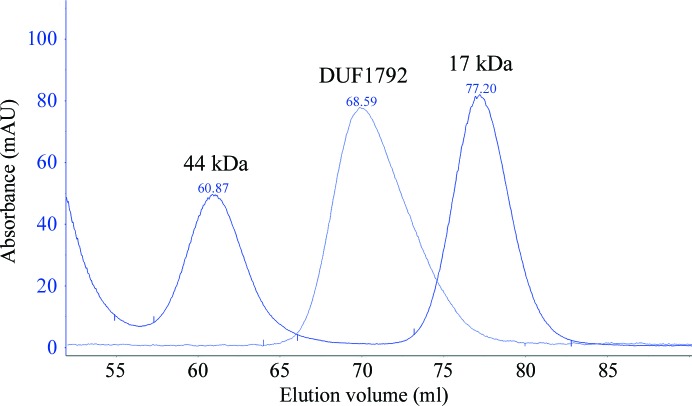
DUF1792 protein was expressed with an N-terminal His-SUMO tag. The His-SUMO tag was completely removed by SUMO protease (Fig. 1 ▶). The DUF1792 protein was further purified by gel filtration (Fig. 2 ▶). From the gel-filtration chromatographic profile, the DUF1792 protein was eluted between ovalbumin (44 kDa) and myoglobin (17 kDa) (Bio-Rad). Since the predicated molecular weight of DUF1792 is at 31.5 kDa, it appears that DUF1792 forms a monomer.
Figure 1.
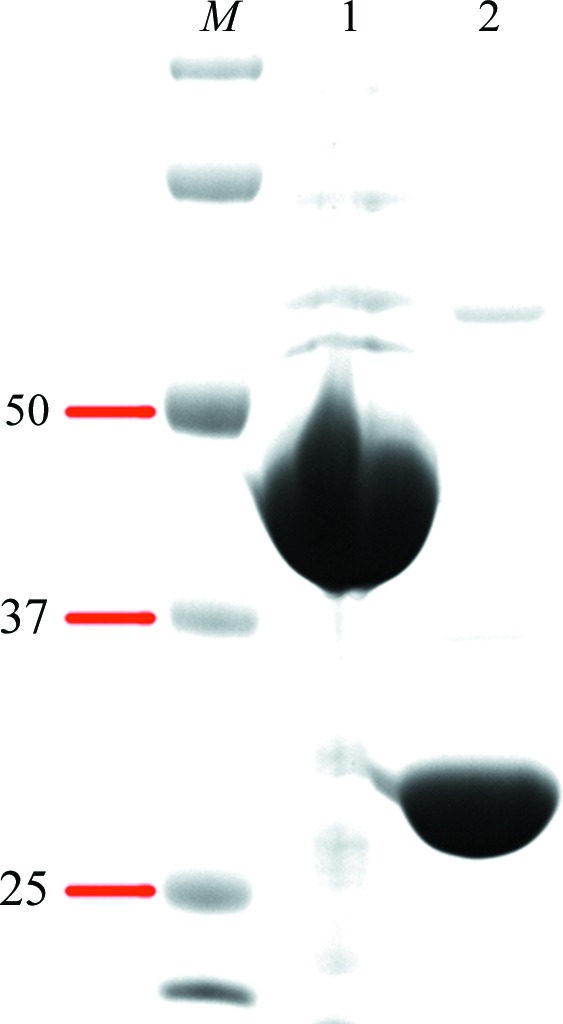
Purification of recombinant DUF1792. DUF1792 with an N-terminal His-SUMO tag (lane 1) and with the SUMO tag removed (lane 2) were stained with Coomassie Blue. The SDS–PAGE analysis revealed that the DUF1792 purity is above 95%. Lane M contains molecular-mass markers (labeled in kDa).
Figure 2.
Gel-filtration chromatographic profile of DUF1792. The DUF1792 and protein standards ovalbumin (chicken, 44 kDa) and myoglobin (horse, 17 kDa) were subjected to gel filtration.
The DUF1792 crystal (0.2 × 0.2 × 0.5 mm) used for data collection was obtained with the following reservoir solution: 100 mM Tris–HCl buffer pH 8.5, 35% PEG 1500, 200 mM sodium acetate (Fig. 3 ▶ a). The DUF1792 crystal grew to full size in 3 d at 293 K. SDS–PAGE analysis of the crystals confirmed that the crystal samples contained the DUF1792 protein (Fig. 3 ▶ b).
Figure 3.
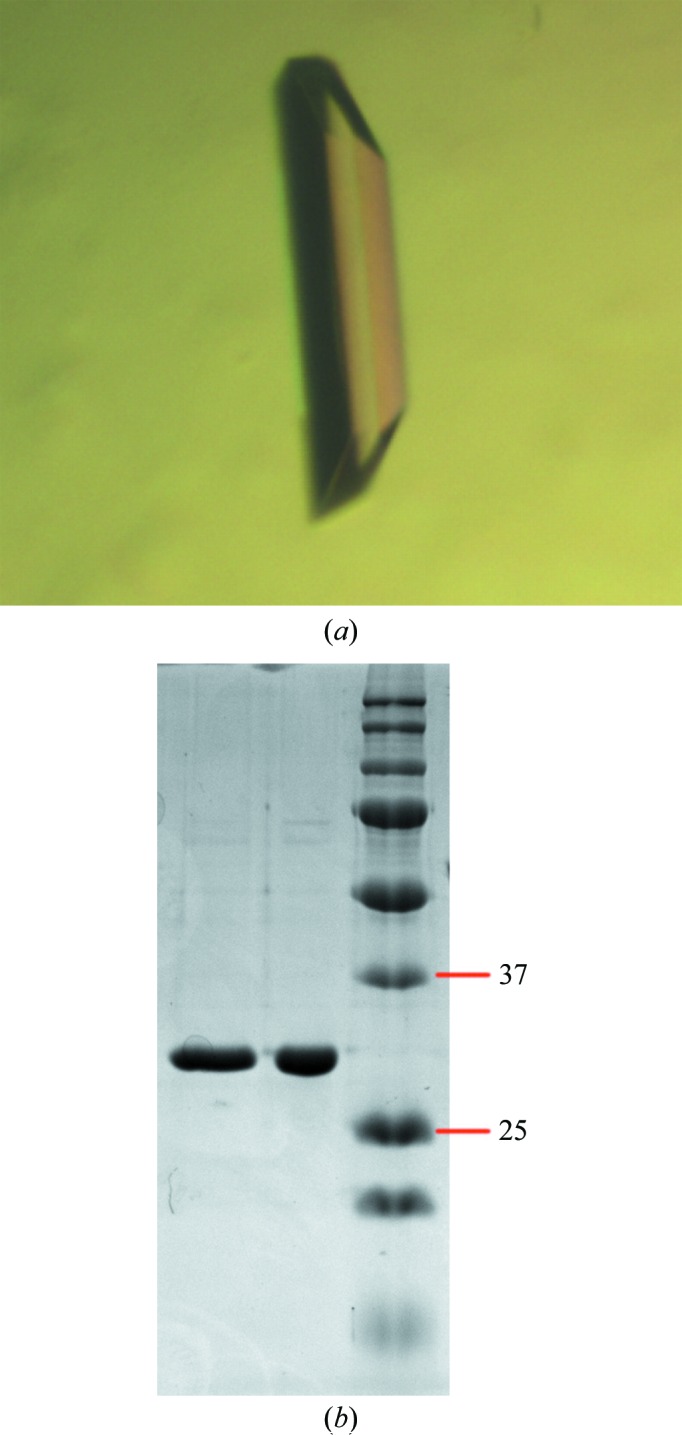
(a) Crystal of DUF1792. (b) SDS–PAGE analysis of two DUF1792 crystals. The right lane contains molecular-mass markers (labeled in kDa).
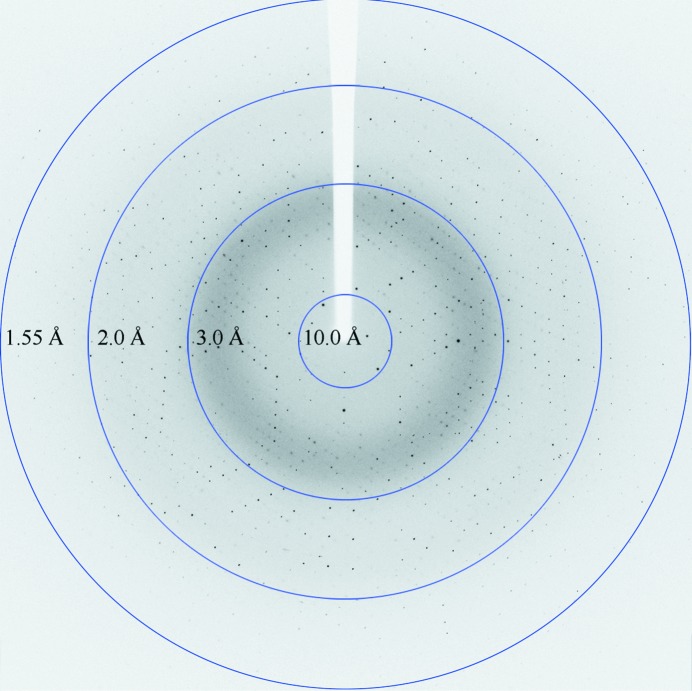
The crystals of DUF1792 diffracted to high resolution, 1.55 Å (Fig. 4 ▶), at the synchrotron source. The DUF1792 crystals bleong to space group C2 with unit-cell parameters a = 71.0, b = 45.1, c = 78.6 Å, β = 109.6°. The R merge of the overall data set was 0.099. Additional data-set statistics and information are given in Table 1 ▶.
Figure 4.
A diffraction pattern of the DUF1792 crystal.
Table 1. Statistics of the data set from the DUF1792 crystal.
Values in parentheses are for the highest-resolution shell.
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 71.0, b = 45.1, c = 78.6, β = 109.6 |
| Wavelength (Å) | 1.0000 |
| Resolution range (Å) | 20.00–1.55 (1.58–1.55) |
| No. of measured reflections | 239255 |
| No. of unique reflections | 34064 |
| R merge † (%) | 9.9 (26.8) |
| Mean I/σ(I) | 58.5 (12.7) |
| Multiplicity | 7.0 (4.9) |
| Completeness (%) | 99.6 (92.9) |
R
merge = 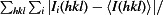
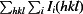 , where I(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the average intensity from multiple observations.
, where I(hkl) is the intensity of reflection hkl and 〈I(hkl)〉 is the average intensity from multiple observations.
DUF1792 is highly conserved in Gram-positive and Gram-negative bacteria. Its structure and function are unknown. Interestingly, DUF1792 is associated with many putative glycosyltransferases in Gram-positive bacteria as an independent domain. It is often annotated as a glycosyltransferase. DUF1792 shows little sequence identity to proteins of known structures (maximum 12%), so that one might expect that solving its structure might provide hints to the function of the domain. In this study, we grew the crystals of DUF1792 in the presence of UDP-glucose and found that its presence is critical for producing DUF1792 crystals. This finding suggests that UDP-glucose may interact with DUF1792. The fact that many glycosyltransferases bind to their active sugar donors including UDP-glucose (Ramakrishnan et al., 2006 ▶; Zhu et al., 2011 ▶) lends credence to the supposition that DUF1792 may function as a glycosyltransferase. While we obtained a 1.55 Å resolution data set for the native DUF1792 protein, we could not solve the structure by molecular-replacement methods as no high-similarity structural homologs exist in databases. We continue to work on determining the DUF1792 structure with the help of selenomethionine-substituted DUF1792 crystals by MAD (multi-wavelength anomalous dispersion) methods. Structural analysis of DUF1792 should provide new insights into the biochemical properties of DUF1792.
Acknowledgments
We are grateful to the staff scientists at the APS SER-CAT beamline for their help in the data collection. We also thank the editor for extraordinary editorial help in producing the final draft of this article. This study was supported by NIH/NIDCR grant No. DE017954 (HW).
References
- Bateman, A., Coggill, P. & Finn, R. D. (2010). Acta Cryst. F66, 1148–1152. [DOI] [PMC free article] [PubMed]
- Drickamer, K. & Taylor, M. E. (1998). Trends Biochem. Sci. 23, 321–324. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ramakrishnan, B., Ramasamy, V. & Qasba, P. K. (2006). J. Mol. Biol. 357, 1619–1633. [DOI] [PubMed]
- Seifert, K. N., Adderson, E. E., Whiting, A. A., Bohnsack, J. F., Crowley, P. J. & Brady, L. J. (2006). Microbiology, 152, 1029–1040. [DOI] [PubMed]
- Shivshankar, P., Sanchez, C., Rose, L. F. & Orihuela, C. J. (2009). Mol. Microbiol. 73, 663–679. [DOI] [PMC free article] [PubMed]
- Siboo, I. R., Chambers, H. F. & Sullam, P. M. (2005). Infect. Immun. 73, 2273–2280. [DOI] [PMC free article] [PubMed]
- Sorge, N. M. van, Quach, D., Gurney, M. A., Sullam, P. M., Nizet, V. & Doran, K. S. (2009). J. Infect. Dis. 199, 1479–1487. [DOI] [PMC free article] [PubMed]
- Wu, H. & Fives-Taylor, P. M. (1999). Mol. Microbiol. 34, 1070–1081. [DOI] [PubMed]
- Wu, H., Mintz, K. P., Ladha, M. & Fives-Taylor, P. M. (1998). Mol. Microbiol. 28, 487–500. [DOI] [PubMed]
- Wu, R. & Wu, H. (2011). J. Biol. Chem. 286, 34923–34931. [DOI] [PMC free article] [PubMed]
- Wu, H., Zeng, M. & Fives-Taylor, P. (2007). Infect. Immun. 75, 2181–2188. [DOI] [PMC free article] [PubMed]
- Zhou, M. & Wu, H. (2009). Microbiology, 155, 317–327. [DOI] [PubMed]
- Zhou, M., Zhu, F., Dong, S., Pritchard, D. G. & Wu, H. (2010). J. Biol. Chem. 285, 12140–12148. [DOI] [PMC free article] [PubMed]
- Zhu, F., Erlandsen, H., Ding, L., Li, J., Huang, Y., Zhou, M., Liang, X., Ma, J. & Wu, H. (2011). J. Biol. Chem. 286, 27048–27057. [DOI] [PMC free article] [PubMed]