Abstract
Objectives
We hypothesized that microRNA (miR) expression may be involved in memory function because they control local protein translation at synapses and dendritic spines.
Design
Case-control animal study.
Methods
We assessed the miR repertoire in the hippocampus of young, 6-month old (n = 18), mice compared to aged, 26-month old (n = 23), mice and compared miR quantity to memory scores as determined by the novel object recognition task (NORT). We performed a histological brain regional analysis of miR-138, APT1 mRNA, and APT1 protein.
Results
We found that higher miR-138 in the mouse hippocampus is correlated with better memory performance. We also found that acyl protein thioesterase 1 (APT1), a depalmytoylation enzyme expressed at dendritic spines whose translation is controlled by miR-138, mRNA is increased in the mouse hippocampal CA1 and dentate gyrus in aged mice compared to young, but not in mice with memory impairment. We found APT1 protein distribution to be lower in cells with high miR-138 expression.
Conclusions
These results suggest that increased miR-138 is associated with better memory and increased APT1 gene transcription occurs with aging. The role of miR-138 and APT1 protein function in memory and aging warrants further investigation.
Introduction
Controlling localized protein synthesis at the synaptodendritic apparatus may be a molecular mechanism of memory formation (1) and microRNA (miR)s are prime candidate molecules to perform this function (2). MRNAs are transported to dendrites (3) and the components of protein synthesis machinery have been found in dendrites, near synapses (4, 5). There is evidence for activity-mediated synaptic translation and that local protein translation in dendrites allows neurons to selectively rebuild only those synapses that have been activated (6). MiR-138 was identified in a functional screen to affect dendritic spine morphology and activity by modulating the synthesis of acyl protein thioesterase 1 (APT1) (7). De-palmitoylation by APT1 influences protein localization and function at the inner-leaflet of plasma membranes at the synapse (7, 8). A detailed molecular study indicated that receptor or voltage-gated activity lead to proteasomal degradation of a component of RNA-induced silencing complex (RISC), which facilitated release of APT1 mRNA from translation inhibition by miR-138 (9). Here, we planned to determine whether expression of miRs in the brain was correlated with short-term memory performance in older and younger mice. We screened for differential expression of 380 miRs in memory-impaired older mice compared to controls and analyzed the distribution of miR-138, APT1 mRNA, and APT1 protein across a larger cohort of both young and old mice with a range of short-term object recognition performances.
Differential susceptibility of older individuals to developing age-related cognitive decline, including memory impairment, is driven by combinations of factors including genetic, epigenetic, and life-long environmental exposure to stressors (10). The novel object recognition task (NORT) is a one-trial non-matching learning task to study short-term recognition memory in rodents (11) that is suited for preclinical studies of visual learning and memory (12, 13). Older rodents showed deficits in NORT compared to young (14) and hippocampal lesions caused impairments in NORT if the interval between training and testing was greater than 1 min (15, 16). We utilized an animal model of aged mice (22–24 months) compared to young adults (6 month old), a useful tool for studying physiologic, neurobiologic, and cognitive aging in mammals (17). Because of the potential for miR to influence protein synthesis at the synapse, we hypothesized that there would be differentially expressed miRs in the hippocampus of aged mice impaired in NORT. Based on results from our screen, we then analyzed the distribution of miR-138 and its downstream target APT1 mRNA and protein in the brain of mice and compared expression with NORT performance. Higher miR-138 associated with high performance on NORT. In neuronal populations with high miR-138, there was low expression of APT1 protein. Results support a model whereby miR-138 regulates APT1 translation, as found by others (7), which then affects short-term object recognition memory.
Methods
Animals
Young (6 months old, n = 23) male C57BL/6N mice purchased from Charles River Laboratories (Wilmington, MA, USA), and aged (26 months old, n = 18) mice from the National Institute of Aging stock were used for single-trial object recognition testing. Mice were housed two per cage in a temperature-controlled room (21 – 22°C) under a reverse 12 h light/dark cycle. All procedures were approved by the Institutional Animal Care and Use Committee at the University of California, San Diego.
Behavioral testing
The single-trial object recognition test was performed as previously described (18, 19). Specifically, each mouse completed one session of three successive phases. In Phase 1, a 5 min habituation phase, the mouse freely explored an empty open field box. During Phase 2, a 5 min training phase, two identical objects of the same type were placed and the mouse explored the objects. In Phase 3, a 5-min retention phase, one of the objects was replaced with a new object of the other type, the mouse explored the objects, and we assessed novel object exploration. Object exploration was tracked using Ethovision TM Tracking software (Noldus Information Technology, Wageningen, The Netherlands) and scored using Ethovision TM Observe software when the mouse’s nose was within 2 cm of the object. There was a 3-min inter-trial interval; these time periods were chosen to allow for the largest behavioral window to detect performance variance in aged mice.
Behavioral analysis
Assessment of overall locomotor activity and object exploration during the retention phase was performed exactly as previously described (18). The ratio of time spent exploring the novel object over the total amount of time spent exploring both objects during the retention phase of the object recognition test is referred to as the discrimination ratio (DR) (11).
Brain examination
One of the aged mice was found dead before sacrifice. Mice were sacrificed with order counterbalanced for age group to minimize the potential effect of circadian variation in the HPA axis activity. Brains were immediately dissected into two hemi-brains. The frontal lobe of one of the young mice was not available for the current study. One of the young mice was found to have a brain tumor and was excluded. There were 18 aged mice and 23 young mice available, the right hemi-brains of which were included in the histologic study. The left hemi-brain was flash frozen and the hippocampus was extracted for qPCR-based miR expression screening and qPCR quantitation of miR-138 and APT1 mRNA. All of the right hemi-brains were immediately placed in 10% neutral buffered formalin and histologically processed for paraffin embedment. Sagittal sections were taken with Superfrost/Plus microscope slides. One section from each hemi-brain was stained with hematoxylin and eosin for histologic examination.
MiR screening
The four median specimens from each group were chosen as representative samples for screening for differential hippocampal miR expression by young, unimpaired aged, and impaired aged. The mean (standard deviation) DR for representative samples were: young, 0.67 (0.03); unimpaired aged, 0.63 (0.04); and impaired aged, 0.48 (0.04); one-way ANOVA, P < 0.001 comparing all three groups.
RNA was isolated from the frozen hippocampi using Mirvana miR isolation kit (Life Technologies, Carlsbad, CA, USA), quantified using absorbance spectroscopy, and quality-tested by denaturing polyacrylamide gel electrophoresis. To conserve resources, RNA from two samples was pooled so that two screens were performed from each group, each screen representing two specimens, and each screen performed in duplicate. This procedure is efficacious when groups are well-defined with an isolated variable (20). One microgram from each sample was used for reverse transcription using Megaplex Pools for miR Expression Analysis (Life Technologies) and following manufacturer’s protocols for running human Taqman miR Array card A version 2.0 (Life Technologies).
Analysis
Taqman miR array data were analyzed using SDS software (Applied Biosystems, Carlsbad, CA, USA), setting a standard threshold at 0.2 and baseline at 15 cycles. The ΔCT values were calculated using MammU6 as the endogenous control; and ΔCT values exported to JMP software for statistical analysis. A series of Mann-Whitney U tests compared expression between the three groups for each of the 380 miRs on the array card. For illustration, data were calibrated to the average of values for the young group, plotting Log (2−ΔΔCT).
Quantitative PCR
RNA was isolated from the hippocampi of the larger cohort of young (n = 23) and old (n = 19 mice as explained above. We used Taqman assays (Applied Biosystems, Assay ID Mm00493325) for APT1 mRNA, following manufacturer’s protocols. We used β-actin (Assay ID Mm00607939) and GAPDH (Assay ID Mm99999915) as endogenous controls to calculate the ΔCT. The comparative CT method was used to interpret PCR data (21). Statistical analysis was performed on the ΔCT, and for illustration, data were calibrated as mentioned above.
In Situ Hybridization
Ten micrometer - thick paraffin sagittal sections mounted on glass slides were used for in situ hybridization detection of miR-138 and APT1 mRNA using a protocol adapted from (22). Pre-validated digoxigenin (DIG)-conjugated locked nucleic acid (LNA) probes (Exiqon Corporation, Copenhagen, Denmark) at 0.8 μM was prepared in In Situ Hybridization Buffer (Enzo Life Sciences, Farmingdale, NY, USA).
Probes
In situ hybridization probes were purchased from Exiqon Corporation, for miR-138, catalog number 38511–01; for positive control, we used the U6 small nuclear RNA, catalog number 99002–01; for negative control, catalog number 99004–01 on all specimens. For APT1 mRNA, a custom probe was generated using the following sequence: 5′DIG-ATGACAGAGAGGCAATATTAGT-3′.
Immunohistology
Immunohistology was performed exactly as previously described (18). Rabbit anti-APT1 antibody (catalog number TA307163, Origene Technologies Incorporated, Rockville, MD, USA) at 1:500 dilution was used.
Quantification of immunohistochemistry and in situ hybridization
Procedure for quantification of histochemical signal was adapted from previously described methods (18, 24). For immunohistology, DAB-peroxidase reactivity; and for in situ hybridization, NBT/BCIP-alkaline phosphatase reactivity were quantitatively assessed by means of two-dimensional computer-assisted image analysis. The stained slides were digitally scanned using a microscope slide scanner (Aperio ScanScope GL, Vista, CA, USA) equipped with a 20× objective lens (yielding the resolution of 0.5 μm per pixel). The digital slides were viewed on a 24-inch LCD TFT monitor using the ImageScope software (Aperio) the images were exported to tagged image file format (TIFF) and quantitated using Image Pro-Plus version 4.5 software (Media Cybernetics, Bethesda, MD, USA) operating on a Microsoft Windows XP using 640 × 480 pixel full-color image frames.
On each image of the entire parasagittal hemi-brain section, the outline of the area of interest (AOI; i.e., dorsal hippocampal CA3, dorsal hippocampal CA1, and granule cell layer of the dorsal dentate gyrus) was digitally drawn with the Image-Pro Plus software in accordance with neuroantatomic landmarks (24). To measure reactive signals within AOI, histogram-based RGB color segmentation was set to best select the specific signal while ignoring the nonspecific background. Within each probe (miR-138, APT1 mRNA, and APT1 protein), for all slides, the same setting of color segmentation was applied to all the hemi-brains. The values of three Image-Pro Plus measurement statistics included area (= area of object), integrated optical density (= [area] x [average optical density of the object]), and per-area (= [area]/[AOI]) were recorded for each AOI and exported to a spreadsheet for analysis in JMP. The value of AOI (in μm2) was calculated by dividing the mean of area values by the mean of per-area values. The value of reactivity normalized to the AOI (IRn) was calculated by dividing the sum of integrated optical density values by the AOI value was used for subsequent analyses (18).
Statistical Analysis
Unless otherwise indicated, statistical tests were two-sided. For the initial screen, one-way ANOVA was performed to assess differences across three groups: young, aged-unimpaired, and aged-impaired. We report FN, Degrees of Freedom and P - values. For other comparisons, Welch’s t-tests comparing means with unequal variances were performed; comparing mice grouped by age (Aged vs Young) and mice grouped by memory performance (Impaired vs Unimpaired). We report tDegrees of Freedom - statistics and P - values. Statistical analyses were performed and graphs generated using JMP version 9.0 (SAS Institute Inc).
Impairment Grouping
We defined memory impairment as scoring below the lower 95% confidence interval for the group difference from chance performance (chance performance in this task is DR = 0.50). The entire group was significantly different from chance t61 = 4.294, P < 0.0001 with DR = 0.54 as the lower limit of the confidence interval for this test. We thus considered mice that had a DR < 0.54 as impaired, and mice with DR ≥ 0.54 as unimpaired. This statistical classification has been used in the literature to group cognitive performance across aged rodents in various cognitive tasks (26).
Results
Features of the study cohort
In total, 41 mice we used for the study, 18 aged and 23 young. For qPCR of miR-138, one aged mouse was excluded because RNA did not meet quality-control standards. For qPCR of APT1 mRNA, three samples from each group were excluded because of insufficient RNA quantity. For in situ hybridization of miR-138, all specimens were included. For in situ hybridization of APT1 mRNA, one specimen from the young group was excluded from analysis because of quality of the histology section. For immunohistochemistry of APT1 protein, one aged specimen was excluded for the same reason.
For three behavioral measurements, the aged group was significantly different than the young. For the NORT, the aged mice scored, on average, lower DR than the young, t37.6 = −3.29, P = 0.002 (Figure 1A). The locomotor activity or object exploration during the training phase did not differ by age or impairment status, (Distance: t11.83 = −0.12, P = 0.90 by age and t13.52 = 0.997, P = 0.34, Object exploration time: t36.8 = 1.02, P = 0.31 by age; and t11.8 = −0.12, P = 0.91 by impairment status). These data indicate that variations in DR scores during the test phase are not artifacts of differences in overall exploration or object sampling during training.
Figure 1.

Cognitive and behavioral features of the mouse cohort indicating lower object recognition as measured by DR in the aged group compared to the young (a) and no difference in distance traveled (b), center time (c), and sampling time (d) between impairment statuses. The line and heavy lines indicate mean and standard deviation. These data indicate lower memory performance in aging and variation in memory performance is not an artifact of variation in exploratory behavior.
Using the DR = 0.54 as the impairment cut-off, there were ten impaired-aged mice (55.5%) and four impaired-young mice (18%). During the habituation phase of the NORT experiment, the average distance explored (Figure 1B) and the average time spent in the center of the box (Figure 1C) were no different by age or impairment status, suggesting equal activity levels across groups. The time spent sampling the two objects during the habituation (“sampling time”) phase also did not differ across impairment status (Figure 1D). This measure is important because it confirms that significant results are not an effect of variances in exploration behavior. Impairment in the NORT was not associated with impairment in other behavioral tasks (data not shown).
MiR Taqman array
From the 380 miRs measured, 118 had undetermined CT values and were not detected in the hippocampus. Seven miRs were significantly different amongst the three groups as determined by ANOVA and these relative expression levels are shown in Figure 2. MiR-130-b was elevated in the impaired-aged group (F4,2 = 21.64, P = 0.017), but decreased in the unimpaired aged group. All other miRs were decreased in the impaired aged group; they were: miR-138 (F4,2 = 14.92, P = 0.028), miR-222 (F4,2 = 9.13, P = 0.049), miR-29b (F4,2 = 27.32, P = 0.012), miR-339-3p (F4,2 = 13.59, P = 0.031), miR-340-5p (F4,2 = 9.97, P = 0.047), and miR-582-3p (F4,2 = 13.46, P = 0.032) (Figure 2). MiR-138 was chosen for further analysis because it was not significantly changed in the aged group but lower in the aged impaired group and based on these data, we hypothesized that miR-138 would be significantly higher in unimpaired mice as measured by NORT.
Figure 2.
MiR expression levels of 7 miRs from screen that were significantly different (p < 0.05) by two-sided one-way ANOVA comparing Young, Impaired-Aged, and Unimpaired-Aged groups. Illustrated are Log (Fold Change) values calibrated to average of young group, error bars indicate standard deviation (n = 4, each group) and experiment was performed in technical duplicate.
MiR-138 in situ hybridization
MiR-138 reactivity was apparent in most neuronal cells, including pyramidal cells, small interneurons, and medium spiny neurons throughout the brain, Figure 3A. The intensity of the signal was variable from dark to light. Figure 3A shows a representative section from a young, unimpaired specimen, showing the whole section, scanned at 20X, and the CA1-3 regions of the hippocampus (i), the dentate gyrus and subiculum (ii), the motory sensory region of the frontal cortex (iii), and the caudate putamen region (iv). Small interneurons within the CA3 region of Schaffer collaterals also showed intense staining in the young animal (Figure 3A-i). While in a representative sample from the aged-impaired group, the staining was lower throughout the brain, Figure 3B. The insets show Fast Red counterstain to illustrate the presence of neurons and cell bodies in these regions, even though miR-138 reactivity was low or absent, for example within interneurons in the CA3, Figure 3B-i. MiR-138 reactivity showed a wide range of neuron-enriched intensity.
Figure 3.

The distribution of miR-138 in situ hybridization in parasagittal section of mouse brain from a young unimpaired (a) and an aged impaired (b) mouse. Detailed regions are: CA1-CA2-CA3 of hippocampus (i), DG and subiculum of hippocampus (ii), motor cortex (iii), and caudate/putamen (iv). Fast red counterstain (insets) illustrate the presence of cells and nuclei in area of low to absent miR-138 in the aged-impaired specimen (b). For in situ hybridization positive control, probe for U6-small nuclear RNA (c), stains all nuclei, and negative control, scrambled LNA probe (d) illustrates any background coloration. MiR-138 in situ hybridization signal ranged from intense (a) to low (b) and was neuron-enriched in pyramidal neurons, small interneurons, and medium spiny neurons.
The miR-138 signal was quantified and compared by age and impairment status. Because the distributions had unequal variance, Welch’s means comparison test was used. Based on results from the screen, we hypothesized that miR-138 would be lower in the impaired mice, and one-sided testing was performed. MiR-138 was significantly higher in the not-impaired group compared to the impaired group in the DG, t37.72 = 1.82, P = 0.038, and not the CA1, t38.00 = 1.54, P = 0.065; but not significantly different by age, for CA1, t37.60 = −0.25, P = 0.4 and for DG, t33.34 = −1.38, P = 0.08, Figure 4.
Figure 4.

Values for miR-138 in situ hybridization signal in the neuroanatomic regions that were measured (IRn) in the dorsal hippocampal CA1 (a, c) and dorsal DG (b, d). The in situ hybridization signal was not different comparing by age; for CA1, t37.60 = −0.25, P = 0.4 (a) and for DG, t33.34 = −1.38, P = 0.08 (b). MiR-138 signal was significantly higher in CA1 of the unimpaired mice, CA1, t38.00 = 1.55 P = 0.06 (c), and DG, t37.72 = 1.82, P = 0.03 (d). Comparisons were made using one-sided Welch’s test, assuming unequal variances.
APT1 in the mouse brain
We performed in situ hybridization to detect the mRNA of the APT1 gene because it is a target of miR-138. MiR-138 exhibits translational control over APT1 and cellular studies demonstrated this function as a mechanism that controls extension and growth of dendritic spines. In order to explore the possibility of correlations between miR-138 and APT1 on memory functions through physiology of dendritic spine growth, it is necessary to determine whether they were present in the same neural populations of the mouse brain. In Figure 5, the regional expression of APT1 from the same specimens as Figure 3 is illustrated. Within the hippocampus, the APT1 mRNA is more highly expressed in the proximal apical dendrites compared the basal, Figure 5Ai–ii and Figure 5Bi–ii. APT1 mRNA is present in the same neural populations as miR-138, although with more subregional specificity. APT1 mRNA reactivity showed a wide range of neuron-specific intensity.
Figure 5.
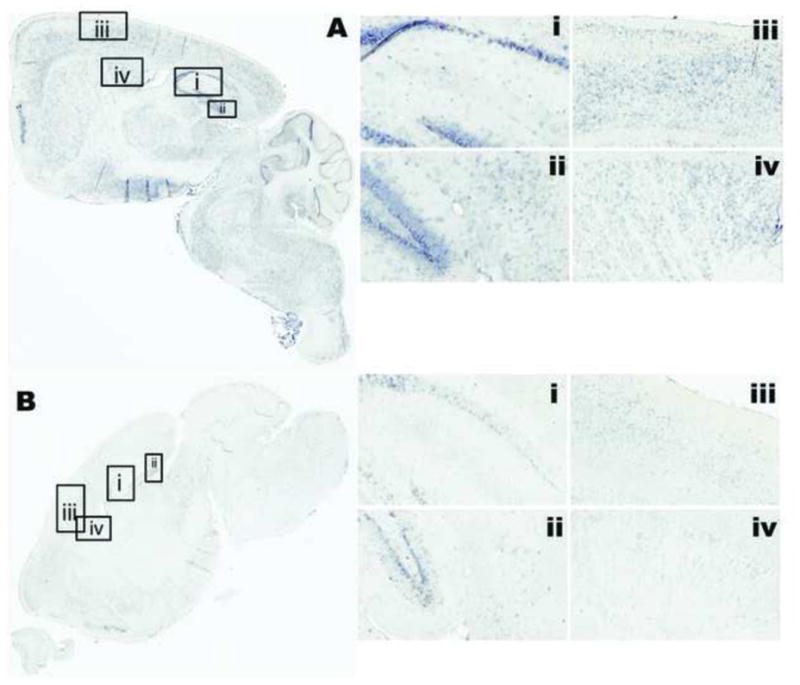
The distribution of APT1 mRNA in situ hybridization in parasagittal section of mouse brain from a young unimpaired (a) and an aged impaired (b) mouse. Detailed regions are: CA1-CA2-CA3 of hippocampus (i), DG and subiculum of hippocampus (ii), motor cortex (iii), and caudate/putamen (iv). In situ hybridization for APT1 mRNA was apparent in apical dendrites of pyramidal neurons. The signal was enriched in basal side of DG granular cells (ii). In the aged mouse, while expression was lower overall, APT1 mRNA appeared to be enriched near mossy fibers of CA1 (b–i).
In order to quantify APT1 mRNA in the brains, we performed quantitative PCR from RNA isolated from all available frozen hippocampi. APT1 mRNA was significantly elevated in the aged animals, t30.69 = 4.21, P = 0.0002, Figure 6A, but was not different when compared by impairment status t14.61 = −0.42, P = 0.68, Figure 6D. We employed histological quantification methods in 40 samples to quantify in situ hybridization signal in the CA1 and DG and did not find any significant differences by age, Figure 6B, t37.00 = −0.08, P = 0.93 for CA1 and t36.89 = −1.33, P = 0.19 for DG. Two-sided hypothesis testing was performed using Welch’s test.
Figure 6.
Values for APT1 mRNA quantification separated by age (a–c) and impairment status (d–f). Quantification by qPCR from RNA isolated from frozen left-hippocampus homogenates, comparing by age (a) and impairment status (d), is illustrated with Log (fold-change) calibrated to average ΔCT value of the young group and using β-actin and GAPDH as endogenous controls. Values for in situ hybridization signal in the neuroanatomic regions that were measured (IRn) in the dorsal hippocampal CA1 (b, e) and dorsal DG (c, f). The line and heavy lines indicate mean and standard deviation. The qPCR signal was significantly different comparing by age, t30.69 = 4.21, P = 0.0002 (a) but not by impairment status, t36.88 = −0.42, P = 0.5 (d). APT1 in situ hybridization signal was not significantly different by age in CA1 (b), t37.00 = −0.08, P = 0.94, or in DG (c), t36.88 = −1.33, P = 0.09. Comparing by impairment status was not significantly different, t36.42 = 1.00, P = 0.48 for CA1 (e) and t26.61 = 0.17, P = 0.2 for DG (f).
We performed immunohistochemical analyses in order to assess the regional location of the APT1 protein product, Figure 7. We found that APT1 is present in the neuropil across the brain and enriched in neurons. Although APT1 protein is found in the same local regions as APT1 mRNA, for example in the apical dendrites of the dentate gyrus, Figure 7Ai and Figure 7Bi, the protein localization is conspicuously lower in cells of high miR-138 intensity. We quantified the expression levels in the CA1 and DG of 40 mice and did not find significant differences comparing APT1 signal by age (Figure 8a–b); for CA1, t35.95 = 0.53, P = 0.60 and for DG, t35.91 = 0.46, P = 0.65; or impairment status (Figure 8c–d); for CA1, t31.2 = 0.53, P = 0.60, and for DG, t31.6 = 1.03, P = 0.31; using two-sided hypothesis testing.
Figure 7.

The distribution of APT1 immunoreactivity in parasagittal section of mouse brain from a young unimpaired (a) and an aged impaired (b) mouse. Detailed regions are: CA1-CA2-CA3 of hippocampus (i), DG and subiculum of hippocampus (ii), motor cortex (iii), and caudate/putamen (iv). Immunoreactivity for APT1 was abundant in neuropil and lower in cell bodies.
Figure 8.

Values for APT1 immunoreactivity in the neuroanatomic regions that were measured (IRn) in the dorsal hippocampal CA1 (a, c) and dorsal DG (b, d). The line and heavy lines indicate mean and standard deviation. The immunoreactivity signal was not different comparing by age, t35.95 =1.45, P = 0.16 in the CA1 (a) and t36.87 = −1.33 P = 0.19 in the DG (b). APT1 immunoreactivity was not significantly different by impairment status, t26.91 = −0.36, P = 0.72 for the CA1 (c) and t36.50 = 1.55, P = 0.13 for DG (d). Comparisons were made using two-sided Welch’s test, assuming unequal variances.
Figure 9 shows a qualitative (non-quantitative) assessment of serial sections at 400x magnification from example young/normal and aged/impaired mice for both miR-138 in situ hybridization and APT1 immunostaining in two serial section with nuclear counterlabels. For reference, a schematic of a sagittal mouse brain section shows the locations of the fields that are presented (Figure 9F) (25). APT1 immunoreactivity was positive in interneurons of Schaffer collaterals (Figure 9A, top and bottom). MiR-138 and APT1 was present in pyramidal neurons (Figure 9B). Focal APT1 was present in oligodendrocytes as well as neurons, examples are indicated with white arrowheads (Figure 9B). We observed diffuse, concentric staining the cell bodies of neurons and neuropil of the caudate/putamen (Figure 9C). In the cell bodies of pyramidal cells of the CA3, we observed APT1 staining (black arrowheads) that lacked miR-138. In this region and aged/impaired specimen, there was APT1 staining (white arrowheads) in oligodendrocytes that was not present in the young/normal example.
Figure 9.
MiR-138 and APT1 in two serial sections each from a young/normal and aged/impaired specimen, showing 400X original magnification of dentate gyrus (a), pyramidal layer of the somato-motor cortex (b), caudate-putamen (c), CA3 of hippocampus (d), and thalamus (e). A schematic shows the location of the fields imaged fields (f), reproduced from Allen Brain Atlas (25). The sections labeled with in situ hybridization for miR-138 were developed with NBT/BCIP and counterstained using Fast Red (Left, all panels), while the sections immunolabeled for APT1 were developed with DAB and counterstained with haematoxylin (right, all panels). The 400X magnification shows APT1 immunoreactivity in some cell bodies and neuropil (right, all panels). In (a) the dentate gyrus, there was focal staining in oligodendrocytes of APT1 in cells lacking miR-138 (white arrowheads, top), and a lack of APT1 in the granular layer with abundant miR-138 (top). In the granular layer of the age/impaired specimen (bottom), there was some APT1 staining. In the pyramidal neurons of the frontal cortex (b), the young/normal specimen specimen had intense miR-138 and little in the aged/impaired mouse. The focal APT1 staining in oligodendrocytes was apparent in the FC as with the DG (white arrowheads). There was also a notable concentric staining in cell bodies (black arrowheads). In the CP, there was notably more miR-138 in the young/normal specimen compared to the aged/normal, with miR-138 in the medium spiny neurons, and there was less APT1 in these specific cells (white arrowhead, top). In cells with little miR-138, there was more APT1 (black arrowheads) in the neuropil and conversely, intense miR-138, there was less APT1 (white arrowheads, top). The APT1 staining in the CA3 granular layer seemed to mainly be concentric in cell bodies (black arrowheads, top and bottom). In this aged/impaired specimen, one APT1-positive oligodendrocyte was observed (white arrowhead, bottom) as in the DG and FC, but was not observed in this example of young/normal. In the thalamus (e), both miR-138 and APT1 were observed. The APT1 was both in the concentric, cell body staining (black arrowheads, top and bottom) and oligodendrocytes (white arrowheads, bottom). For miR-138, there was staining in many cell bodies of the young, normal specimen (top). Abbreviations: DG - dentate gyrus, FC - frontal cortex, CP - caudate-putamen, CA3 - CA3 field of the hippocampus, Thal - thalamus.
Discussion
Recent studies focused on the function of miR-138 and APT1 in dendritic spine growth in vitro. Here, we identified miR-138 as a candidate miR in a screen measuring the quantity 380 miRs from the hippocampus in young, aged, and aged-memory-impaired mice. Our main findings were as follows: miR-138 was significantly lower in the CA1 and DG of the memory-impaired mice, APT1 mRNA was elevated in hippocampus in the aged mice as measured by qPCR. We observed through qualitative (non-quantitative) assessment of serial sections that intensity of APT1 immunostaining was lower in cells of intense miR-138 (Figure 9).
We analyzed the distribution of miR-138 in the brains of 42 mice (n = 16 impaired and n = 26 not impaired), and found that higher levels of miR-138 in the DG of the hippocampus predicted higher object recognition memory. This observation is not consistent with a previously published report that indicated knockdown of general miR expression, through Dicer1 knockout, led to improved learning and memory performance in mice (27). We report here significant results based on object recognition memory with a three-minute interval between training and retention testing; the previous report used methods to detect long-term spatial learning and memory, while we analyzed visual recognition. The mechanisms by which miRs affect these different types of memory may vary. The previous study also knocked out many other miRs, including miR-132, whose overexpression caused increased density of dendritic spines in hippocampal pyramidal neurons and impaired recognition memory, as measured by NORT (28). The cumulative effect of knocking out all miRs that function through Dicer1 might overwhelm the effects of what would occur if specifically knocking out miR-138 only. Aside from highlighting the importance of miRs to dendritic spine morphology and memory function, it is difficult to discern a mechanism from a low resolution approach. The nature and results of our study support the notion that higher steady-state levels of miR-138 in the hippocampus may improve capacity to create and recall object recognition. Future interventional studies would elucidate an effect-size and mechanism.
A significant factor in our study is that we were interested in the natural history of age-related brain changes and we sought to identify biological factors that correlated with successful and non-successful cognitive aging. In our model, successful cognitive aging is to have NORT memory scores above chance, or DR ≥ 0.540. Ability to form object recognition memory is multifactorial and we present evidence that higher expression of miR-138 may be one factor. MiR-138 inhibits synthesis of APT1 protein at the post-synapse, which prevents depalmitoylation of membrane-bound proteins and functionally results in stability, rather than growth, of new dendritic spines (9). Pertinent to the geriatric population, physiologic factors that may influence executive processing speed, like efficient synaptodendritic contacts, correlate with cognitive performance (29). We would speculate that retaining higher levels of miR-138 would inhibit new protein synthesis and depalmitoylation in dendrites which may in-turn inhibit production of new dendritic spines. In our model, retaining short-term recognition memory may be a matter of synaptic stability and preservation of neuronal energy stores by reducing rates of protein turnover, a mechanism supported by evidence suggesting that the fillipodia stability, not density, promotes synapse formation (30). The results presented here justify future experiments with transgenic miR-138 and assessing hippocampal neurophysiology and memory function. Examination of miR-138 in other systems of aging and cognition, for example retrospective autopsy studies, is warranted.
The DR for the young mice in the cohort was 0.67 ± 0.14, which is quite consistent with previously published studies on NORT in rodents (31). Researchers debate the importance of the hippocampus to recognition memory (32), however, studies consistently indicate that the ability to remember a single item across a delay of more than a few minutes depends substantially on the hippocampus (33). There is compelling evidence that the perirhinal cortex plays a significant role in NORT performance (16), in our study, available sagittal sections of the mouse brains were too medial to sample the perirhinal cortex. Sampling the perirhinal cortex would have required sections at lateral coordinates 3.0 – 4.0 mm and these sections were lateral 1.25 mm to 1.75 mm (25). Future studies using this model of aging should focus on sample acquisition to quantify miR-138 and APT1 in the perirhinal cortex and hippocampus in order to determine effects of this molecular system on neurons of these regions relative to one another.
Our findings are limited to correlational and cross-sectional study relating to natural history of aging and memory function. We cannot determine from our studies the mechanisms driving differential expression of either miR-138 or APT1 across the various brain regions.
In formulating our original hypothesis, we hypothesized a three-group model of Young, Unimpaired Aged, and Impaired Aged. The screening procedure utilized mice fitting within this framework by definition and choosing animals representing the median performers within these groups, as determined by the NORT score. However, in the larger cohort of 42 animals, the data did not fit this model. We observed an apparent disconnect between the levels of miR-138, APT1 mRNA, and protein levels with age and impairment status. Instead, miR-138 expression correlated with impairment status, regardless of age. APT1 mRNA expression correlated with age, regardless of impairment status. Though there is a clear relationship between age and memory impairment (Figure 1), our data lead to a new hypothesis that APT1 mRNA increases with age and higher levels of miR-138 may be protective from short term memory impairment, possibility functioning through the effect of APT1 activity on dendritic spine growth and development. These mouse brain specimens were formalin-fixed and paraffin embedded, which precluded dye-based labeling for dendritic spine quantification; however this is an aim for future studies. Finally, the other six miRs identified to be dysregulated from the initial screen, shown in Figure 1, warrant further investigation.
To conclude, we found that miR-138 was higher in the hippocampus of mice without memory impairment as measured by the NORT than mice with impairment. We found that APT1 mRNA was higher in the hippocampus of the aged mice, but APT1 mRNA levels did not differ by impairment status. We present an illustration of brain distribution of miR-138 and find that it is neuron-enriched with wide inter-individual variation. We present also brain distribution of APT1 mRNA, which is neuron-specific but not present in all neurons, and less variable than miR-138 across individuals. APT1 protein was found broadly in the neuropil and in oligodendrocytes. Our results support a model whereby steady state level of miR-138 is one factor to a mechanism for preservation of short term object recognition memory in aging.
Acknowledgments
Sources of Support
Stein Institute for Research on Aging (ET and DJ). National Institute on Drug Abuse and National Institute of Mental Health research grants DA031591 (ET), MH062512 (CA), DA026306 (CA), MH074697 (VR), MH019934 (DJ), and MH080002 (DJ). The UC San Diego Center for AIDS Research (AI36214) supported work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008;89:293–311. doi: 10.1016/j.nlm.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashraf SI, Kunes S. A trace of silence: memory and microRNA at the synapse. Curr Opin Neurobiol. 2006;16:535–539. doi: 10.1016/j.conb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 4.Martin KC, Barad M, Kandel ER. Local protein synthesis and its role in synapse-specific plasticity. Curr Opin Neurobiol. 2000;10:587–592. doi: 10.1016/s0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 5.Ostroff LE, Fiala JC, Allwardt B, et al. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35:535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- 6.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Siegel G, Obernosterer G, Fiore R, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalva MB. Neuronal activity moves protein palmitoylation into the synapse. J Cell Biol. 2009;186:7–9. doi: 10.1083/jcb.200906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 10.Goosens KA, Sapolsky RM. Stress and Glucocorticoid Contributions to Normal and Pathological Aging. 2007 2011/01/05. [PubMed] [Google Scholar]
- 11.Bevins RA, Besheer J. Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc. 2006;1:1306–1311. doi: 10.1038/nprot.2006.205. [DOI] [PubMed] [Google Scholar]
- 12.Dere E, Huston JP, De Souza Silva MA. The pharmacology, neuroanatomy and neurogenetics of one-trial object recognition in rodents. Neurosci Biobehav Rev. 2007;31:673–704. doi: 10.1016/j.neubiorev.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Young JW, Powell SB, Risbrough V, et al. Using the MATRICS to guide development of a preclinical cognitive test battery for research in schizophrenia. Pharmacol Ther. 2009;122:150–202. doi: 10.1016/j.pharmthera.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scali C, Giovannini MG, Prosperi C, et al. Tacrine administration enhances extracellular acetylcholine in vivo and restores the cognitive impairment in aged rats. Pharmacol Res. 1997;36:463–469. doi: 10.1006/phrs.1997.0252. [DOI] [PubMed] [Google Scholar]
- 15.Hammond RS, Tull LE, Stackman RW. On the delay-dependent involvement of the hippocampus in object recognition memory. Neurobiol Learn Mem. 2004;82:26–34. doi: 10.1016/j.nlm.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Brown MW, Warburton EC, Aggleton JP. Recognition memory: material, processes, and substrates. Hippocampus. 2010;20:1228–1244. doi: 10.1002/hipo.20858. [DOI] [PubMed] [Google Scholar]
- 17.Murphy GG, Shah V, Hell JW, et al. Investigation of age-related cognitive decline using mice as a model system: neurophysiological correlates. Am J Geriatr Psychiatry. 2006;14:1012–1021. doi: 10.1097/01.JGP.0000209404.54310.b3. [DOI] [PubMed] [Google Scholar]
- 18.Soontornniyomkij V, Risbrough VB, Young JW, et al. Short-term recognition memory impairment is associated with decreased expression of FK506 binding protein 51 in the aged mouse brain. Age (Dordr) 2010;32:309–322. doi: 10.1007/s11357-010-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young JW, Powell SB, Geyer MA, et al. The mouse attentional-set-shifting task: a method for assaying successful cognitive aging? Cogn Affect Behav Neurosci. 2010;10:243–251. doi: 10.3758/CABN.10.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng X, Wood CL, Blalock EM, et al. Statistical implications of pooling RNA samples for microarray experiments. BMC Bioinformatics. 2003;4:26. doi: 10.1186/1471-2105-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods. 2010;52:307–315. doi: 10.1016/j.ymeth.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Soontornniyomkij V, Risbrough VB, Young JW, et al. Increased hippocampal accumulation of autophagosomes predicts short-term recognition memory impairment in aged mice. Age. 2011;32:309–321. doi: 10.1007/s11357-011-9234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valverde F. Golgi atlas of the postnatal mouse brain. New York, NY: Springer/Wein; 1998. [Google Scholar]
- 25.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 26.Schoenbaum G, Setlow B, Saddoris MP, et al. Encoding changes in orbitofrontal cortex in reversal-impaired aged rats. J Neurophysiol. 2006;95:1509–1517. doi: 10.1152/jn.01052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konopka W, Kiryk A, Novak M, et al. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen KF, Sakamoto K, Wayman GA, et al. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown PJ, Liu X, Sneed JR, et al. Speed of Processing and Depression Affect Function in Older Adults With Mild Cognitive Impairment. Am J Geriatr Psychiatry. 2012 doi: 10.1016/j.jagp.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arstikaitis P, Gauthier-Campbell C, Huang K, et al. Proteins that promote filopodia stability, but not number, lead to more axonal-dendritic contacts. PLoS ONE. 2011;6:e16998. doi: 10.1371/journal.pone.0016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reger ML, Hovda DA, Giza CC. Ontogeny of Rat Recognition Memory measured by the novel object recognition task. Dev Psychobiol. 2009;51:672–678. doi: 10.1002/dev.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemanic S, Alvarado MC, Bachevalier J. The hippocampal/parahippocampal regions and recognition memory: insights from visual paired comparison versus object-delayed nonmatching in monkeys. J Neurosci. 2004;24:2013–2026. doi: 10.1523/JNEUROSCI.3763-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Squire LR, Wixted JT, Clark RE. Recognition memory and the medial temporal lobe: a new perspective. Nat Rev Neurosci. 2007;8:872–883. doi: 10.1038/nrn2154. [DOI] [PMC free article] [PubMed] [Google Scholar]





