Highlights
-
•
Viral polyproteins are cleaved in a spatially and temporally regulated manner.
-
•
Access to the cleavage site is highly regulated through various mechanisms.
-
•
Cleavage can result in structural rearrangement of the termini.
Abstract
Many viruses use a replication strategy involving the translation of a large polyprotein, which is cleaved by viral and/or cellular proteases. Several of these viruses severely impact human health around the globe, including HIV, HCV, Dengue virus, and West Nile virus. This method of genome organization has many benefits to the virus such as condensation of genetic material, as well as temporal and spatial regulation of protein activity depending on polyprotein cleavage state. The study of polyprotein precursors is necessary to fully understand viral infection, and identify possible new drug targets; however, few atomic structures are currently available. Presented here are structures of four recent polyprotein precursors from viruses with a positive sense RNA genome.
Current Opinion in Virology 2013, 3:137–142
This review comes from a themed issue on Virus structure and function
Edited by Joseph Marcotrigiano and Felix Rey
For a complete overview see the Issue and the Editorial
Available online 18th April 2013
1879-6257/$ – see front matter, © 2013 Elsevier B.V. All rights reserved.
Introduction
The regulatory mechanisms of viral gene expression vary throughout the virus world. One widely used mechanism employed by evolutionarily divergent RNA and DNA viruses alike organizes multiple genes into a single open reading frame. Most DNA viruses use extensive alternative splicing to express the viral proteins as needed; although, there are exceptions (ex. African swine fever virus [1]). Almost all retroviruses and RNA viruses use the strategy of translating an open reading frame as a large, precursor polyprotein that is further cleaved by viral or cellular proteases in a highly regulated manner.
Viruses that regulate gene expression by polyprotein processing represent an important group including several important human pathogens such as HIV, poliovirus, rhinovirus, Dengue virus, hepatitis C virus, West Nile virus, Chikungunya virus, and SARS coronavirus. Encoding and translating large viral polyprotein precursors is a strategy with many benefits for the virus lifecycle. First, polyproteins allow for a more compact genome by eliminating additional genetic features, such as promoter or enhancer elements, that would be necessary to express each protein individually. Additionally, differential cleavage site usage is coordinated to regulate activity. Lastly, proteins can sometimes perform alternative functions in their precursor forms versus their mature forms.
Given the wide prevalence of polyprotein processing in many viral families, there are surprisingly few structures of precleavage polyprotein intermediates available. Structure of viral proteins before and after cleavage is essential to provide a greater understanding of the regulation, function, and organization of the viral genome. Here we review four recent atomic structures of polyprotein precursors.
Precursors from viral genome replication proteins
Alphavirus P23
Alphaviruses are enveloped viruses of the Togaviridae family whose genome consists of a positive-sense RNA of ~9–11 kb with a 5′ cap structure and 3′ polyadenosine tail (Figure 1 a). The genome contains two cistrons: the first encodes the viral replication machinery, or nonstructural proteins (nsPs), and the latter contains the structural proteins that form the virion particle. The alphavirus replication machinery is composed of four nonstructural proteins (nsP1 to nsP4), which are expressed as one of two polyproteins (P123 or P1234) caused by read through of an opal codon at the end of nsP3. The nonstructural precursor polyproteins are cleaved in a highly regulated manner by a protease at the C-terminus of nsP2 [2, 3]. Shortly after translation of P1234, cleavage at the P3/4 junction occurs either in cis or trans, followed by the P1/2 junction in cis only [3, 4]. Cleavage intermediates P123 + nsP4 and nsP1 + P23 + nsP4 preferentially use the genomic RNA to synthesize a negative-sense, intermediate RNA [5, 6]. The final cleavage event between P23 yields fully mature nsPs and switches the template to the negative-sense RNA to synthesize positive-sense genomic and subgenomic RNAs.
Figure 1.

Alphavirus genome organization and P23 structure. (a) RNA genome organization of Sindbis virus with 5′ cap and 3′ poly(A) tail. Polyprotein cleavage sites are denoted with an asterisk (nsP2 protease), solid black arrow (Sindbis virus capsid autoprotease), solid black arrowheads (cellular signalase), and hollow arrow (cellular furin). (b) P23 structure (PDB ID: 4GUA) highlighting the nsP2 protease (blue) and methyltransferase-like (cyan) domains and nsP3 macro (yellow) and zinc binding (red) domains. The protease active site is denoted with a green asterisk, zinc ion with a gray sphere, and the P2/3 cleavage site with an arrow. (c) nsP3 macro and zinc binding domains of the P23 structure shown in ribbon format, with coloring similar to panel B. The extended linker between the two nsP3 domains creates a ring-like structure which is fitted around nsP2.
The structure of an uncleaved P23 precursor protein from Sindbis virus spanning the nsP2 protease domain through to the central, zinc-binding domain (ZBD) of nsP3 (P23pro-zbd) has been recently determined [7••]. P23pro-zbd shows a solvent exposed but inaccessible P2/3 cleavage site located at the base of an 11–13 Å wide cleft (Figure 1b) [7••]. The nsP2 protease active site is 40 Å away from the P2/3 cleavage site, which supports a trans only cleavage mechanism [4, 7••, 8, 9]. Attempts to model the nsP2 protease onto the P2/3 cleavage site resulted in steric clashes, indicating that accessibility of the cleavage site is highly regulated by an unknown mechanism. P23 has an extensive buried surface area of 3000 Å2 due to a ring-shape structure created by nsP3, which encircles nsP2 (Figure 1c). Several previously described mutations in nsP2 that confer a noncytopathic phenotype [10, 11, 12, 13, 14] map to the surface of nsP2 and make contact with nsP3, implicating this interaction in viral pathogenesis. During the alphavirus lifecycle, nsP2 localizes to the nucleus; however, the mechanism by which nsP2 dissociates from nsP3 post cleavage is unknown. A potential RNA binding surface, which extends over the surface of both nsP2 and nsP3 has been proposed. It is hypothesized that the RNA binding surface may be altered after cleavage, resulting in the RNA template switching that occurs after P23 cleavage.
Poliovirus 3CD
Poliovirus is a nonenveloped virus of the Picornaviridae family with a positive-sense RNA genome of ~7–9 kb. Poliovirus RNA lacks a 5′ cap structure and uses an internal ribosomal entry site (IRES) for translation initiation [15, 16]. The genome contains a single, open reading frame encoding both structural and nonstructural genes as a large polyprotein [17, 18, 19] (Figure 2 a). For simplicity, the proteins are grouped into viral capsid proteins (P1), and proteins involved in viral genome replication and protein processing (P2 and P3). Cleavage of the polyprotein is performed by virus-encoded proteases 2A, 3C, or 3CD precursor. 3CD polyprotein is made up of the 3C protease and the viral RNA polymerase (3D) that is responsible for genome replication. The efficiency of 3C protease and 3D polymerase activity is altered depending on the cleavage state. The 3CD polyprotein preferentially cleaves the P1 precursor compared to mature 3C alone [20, 21] while the 3D polymerase activity is inhibited in the precursor [22]. Furthermore, 3CD promotes RNA synthesis by a redistribution of cellular Arf proteins involved in membrane trafficking, an activity not triggered by the cleaved forms [23, 24•]. These activities highlight the importance of polyprotein cleavage as a mechanism of gene regulation.
Figure 2.
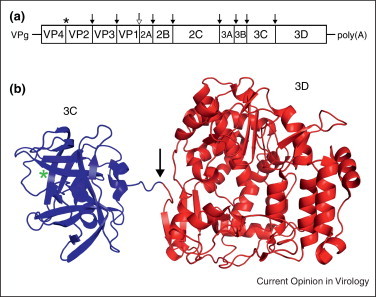
Organization of poliovirus genome and 3CD structure. (a) The RNA genome organization of poliovirus with 5′ covalently linked viral protein VPg and 3′ poly(A) tail. Polyprotein cleavage sites are denoted by an asterisk (autoprotease), solid black arrow (3C or 3CD protease), and a hollow arrow (2A protease). (b) Ribbons representation of the 3CD structure (PDB ID: 2IJD) with a green asterisk indicating the 3C protease active site and an arrow showing the cleavage site.
The atomic structure of 3CD resembles beads-on-a-string with 3C and 3D connected by a flexible, solvent exposed linker region containing the cleavage site [25••] (Figure 2b). Overall, the structures of 3C and 3D alone are very similar to their counterparts in the 3CD precursor. In sharp contrast to alphavirus P23, 3C and 3D have no intramolecular contacts. The 3CD structure supports a trans cleavage mechanism due to the location of the protease active site and the cleavage site [7••, 25••, 26]. In the post cleavage structure of 3D, the N-terminal residues fold inward and insert into a binding pocket where they participate in several hydrogen bonding interactions, while these residues are extended in the 3CD structure. The rearrangement of the N-terminal residues of 3D post cleavage is proposed to be the mechanism for activation of the 3D polymerase activity [25••].
Precursors from viral structural proteins
Flavivirus prM-E
Flaviviruses are enveloped viruses with a positive-sense RNA genome of ~11 kb. The flavivirus structural and nonstructural genes are contained within the same open reading fame and are translated as a large polyprotein (Figure 3 a). Both viral and cellular encoded proteases are involved in specific cleavage of the polyprotein. The surface of the virion is composed of the envelope (E) and membrane (M) proteins [27]. E interacts with cellular receptors and contains a loop responsible for membrane fusion [27, 28]. M is expressed as a precursor protein, prM, which prevents premature fusion by E and is cleaved by furin during egress to release the pr fragment, resulting in a mature particle. In addition to its role in virion morphogenesis, the prM precursor may assist in proper folding of the E protein [29, 30].
Figure 3.
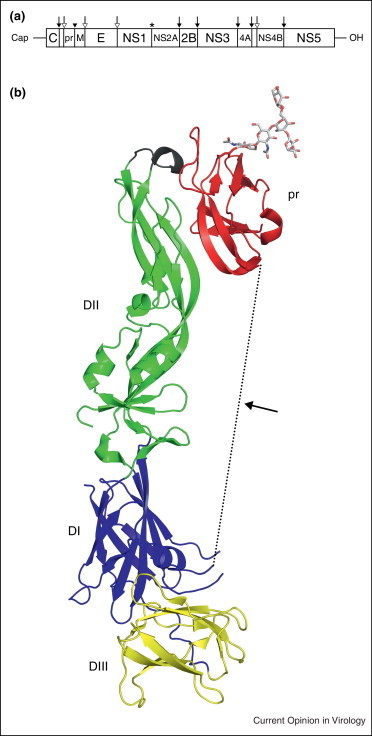
Genomic organization of Flavivirus genome and the structure of prM-E. (a) The genome organization of flavivirus shown with a 5′ cap and 3′ OH. Polyprotein cleavage sites are denoted by an asterisk (unknown protease), solid black arrow (NS2B-NS3 protease), solid black arrowhead (cellular furin), and a hollow arrow (cellular signalase). (b) Structure of prM-E (PDB ID: 3C5X) highlighting pr (red) with E domains DI (blue), DII (green), and DIII (yellow). The fusion loop of E protein is shown in black. The M protein and linker region, which are missing in the structure, are represented as a dotted line.
The structure of a dengue virus prM-E polyprotein was published in 2008 (Figure 3b) [31••]. The construct included prM residues 1 through 91, an eight amino acid linker in place of a transmembrane region, E residues 1 through 394, and a mutated furin cleavage site between pr and M. E contains three distinct domains, termed DI, DII, and DIII, with the fusion loop located at the tip of DII [32, 33, 34, 35, 36, 37]. The pr peptide has a novel beta barrel fold positioned at the distal end of DII, blocking the fusion loop, consistent with its role in preventing membrane fusion [31••, 38]. Similar inaccessibility of the fusion loop in immature virus has also been described in Chikungunya structural precursor polyprotein p62 (discussed by Vaney, Duquerroy, and Ray in current issue of Current Opinion in Virology) [39•]. Cryoelectron microscopy shows that immature flaviviruses at neutral pH have a ‘spiky’ appearance, while mature flaviviruses have a ‘smooth’ surface [40, 41, 42, 43]. These appearances are altered depending on the oligomeric state of immature prM-E or mature M and E proteins. The hinge angle between DI and DII of E alone is very similar to the hinge angle of the prM-E heterodimer [31••, 37]. However, the difference in the angle between DI and DII in immature versus mature virus is 23°, which may determine the oligomeric state of E. Fitting the X-ray structure to cryoEM density shows that immature prM-E at neutral pH would prevent furin cleavage of prM due to steric hindrance [31••]. During egress, immature virions are transported through the trans Golgi Network where exposure to more acidic conditions causes a conformational change in prM-E, allowing furin access to the cleavage site [44].
HIV Gag
The Retroviridae genome consists of two copies of positive-sense RNA, which is reverse-transcribed early during infection. The resulting DNA is inserted into the host genome and used for production of viral RNA and proteins as infection progresses [45]. Retroviruses vary in the number of genes encoded, but they all have at least four open reading frames (gag, pro, pol, and env), which may or may not be in the same reading frame. HIV is a retrovirus whose genome encodes nine (sometimes 10) genes. The gag gene is located at the 5′ end of all retrovirus genomes and is translated as a polyprotein precursor containing three distinct domains: matrix (MA), capsid (CA), and nucleocapsid (NC) (Figure 4 a) [46]. The Gag precursor is associated with the inner surface of the cell's plasma membrane, where it participates in forming the immature virion and selectively packaging viral RNA [45, 46, 47]. Cleavage of precursor protein causes mature CA proteins to organize into cone-like structures and ultimately leads to maturation of the virus [48••].
Figure 4.

HIV Gag polyprotein organization and structure. (a) Schematic representation of the domain organization of HIV-1 Gag polyprotein. Cleavage sites by viral PR protein are denoted with a solid, black arrow. (b) N-terminal 283 residues of the Gag polyprotein (PDB ID: 1L6N) including MA (red) and the N-terminal portion of CA (blue). The post cleavage form of CA (PDB ID: 1GWP) is shown in gray with a box highlighting the N-terminal β hairpin formed after protease digestion. The cleavage site is noted with an arrow.
To discern differences between immature and mature virion structures, the NMR structure for the N-terminal portion of immature HIV-1 Gag, including MA and the N-terminal portion of the CA domain (CAN), was determined (Figure 4b) [48••]. Structures of CAN in the immature polyprotein and mature post cleavage form demonstrate that the N-terminus undergoes a disorder to order transition upon cleavage, resulting in the formation of a β-hairpin, while immature and mature MA structures do not show significant changes. Integral residues located in the CAN hairpin region have previously been shown to prevent proper capsid core formation, leading to formation of noninfectious virus [49, 50, 51]. Therefore, cleavage triggers the formation of the β-hairpin in CAN, and may be responsible for proper capsid core particle assembly.
Conclusions
Presented here are four recent atomic structures of viral polyprotein precursors. Several underlying mechanisms for viral gene regulation can be gleaned from these structures. The first involves accessibility of the polyprotein cleavage site to control the gene's activity, function, and location at specific times post infection. Cleavage of these polyproteins is regulated by different methods. The alphavirus P2/3 cleavage site is solvent exposed but not readily accessible to the protease and the requirement for cleavage is still unknown. Access to the Poliovirus 3CD cleavage site has been postulated to be dependent on the intermolecular scaffolding thought to form between 3CD molecules [25••]. Cleavage of flavivirus prM-E is regulated by the low pH environment found late in viral egress, preventing immature exposure of the fusion loop. Secondly, cleavage events themselves can cause conformational changes in the newly formed termini of the mature, cleaved protein, permitting new functions post cleavage. The 3D polymerase activity is initiated by inserting the N-terminus of the mature protein into a hydrophobic pocket only after cleavage. The N-terminus of HIV CA undergoes the formation a β-hairpin, resulting in maturation of the core particle. Further structures regarding virus particle maturation and the involvement of precursor proteins not discussed here can be found for Chikungunya virus p62 [39•], poliovirus VP0 [52•], influenza HA0 [53•], and human parainfluenza virus 3 fusion protein [54•].
Viral polyproteins represent a largely unexplored structural aspect of the viral lifecycle, especially those involved in viral genome replication. Despite the prevalence of these proteins throughout virus families, very few viral precursor polyprotein structures are currently available. On the basis of the diversity of the four structures and cleavage site regulation presented here, it is likely that many more regulatory mechanisms are yet to be described. Because of their regulatory roles ranging from genome replication to virion maturation, viral polyproteins will be valuable tools for the design of uniquely targeted drugs.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This study was supported by the NIH through a training grant (2T32AI007403-18) to S.A.Y. and an R01 (AI080659) and P50 (GM103368) to J.M.
References
- 1.Andres G., Alejo A., Salas J., Salas M.L. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J Virol. 2002;76:12473–12482. doi: 10.1128/JVI.76.24.12473-12482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffin D.E. Alphaviruses. In: David M., Knipe P.M.H., editors. vol 1. Lippincott Williams & Wilkins; 2007. pp. 1023–1067. (Fields Virology). [Google Scholar]
- 3.de Groot R.J., Hardy W.R., Shirako Y., Strauss J.H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasiljeva L., Merits A., Golubtsov A., Sizemskaja V., Kääriäinen L., Ahola T. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem. 2003;278:41636–41645. doi: 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 5.Shirako Y., Strauss J.H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemm J.A., Rümenapf T., Strauss E.G., Strauss J.H., Rice C.M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Shin G., Yost S.A., Miller M.T., Elrod E.J., Grakoui A., Marcotrigiano J. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci U S A. 2012;109:16534–16539. doi: 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference describes the structure of alphavirus nonstructural polyprotein precursor, P23pro-zbd. The importance of regulation in polyprotein cleavage and relation to viral pathogenesis is discussed.
- 8.Shirako Y., Strauss J.H. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology. 1990;177:54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- 9.Hardy W.R., Strauss J.H. Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov I., Agapov E., Hoffman T.A., Prágai B.M., Lippa M., Schlesinger S., Rice C.M. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73:3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dryga S.A., Dryga O.A., Schlesinger S. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology. 1997;228:74–83. doi: 10.1006/viro.1996.8364. [DOI] [PubMed] [Google Scholar]
- 12.Mayuri, Geders T.W., Smith J.L., Kuhn R.J. Role for conserved residues of sindbis virus nonstructural protein 2 methyltransferase-like domain in regulation of minus-strand synthesis and development of cytopathic infection. J Virol. 2008;82:7284–7297. doi: 10.1128/JVI.00224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perri S., Driver D.A., Gardner J.P., Sherrill S., Belli B.A., Dubensky T.W., Polo J.M. Replicon vectors derived from Sindbis virus and Semliki forest virus that establish persistent replication in host cells. J Virol. 2000;74:9802–9807. doi: 10.1128/jvi.74.20.9802-9807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrakova O., Volkova E., Gorchakov R., Paessler S., Kinney R.M., Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewlett M.J., Rose J.K., Baltimore D. 5′-Terminal structure of poliovirus polyribosomal RNA is pUp. Proc Natl Acad Sci U S A. 1976;73:327–330. doi: 10.1073/pnas.73.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nomoto A., Lee Y.F., Wimmer E. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc Natl Acad Sci U S A. 1976;73:375–380. doi: 10.1073/pnas.73.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summers D.F., Maizel J.V., Jr. Evidence for large precursor proteins in poliovirus synthesis. Proc Natl Acad Sci U S A. 1968;59:966–971. doi: 10.1073/pnas.59.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamura N., Semler B.L., Rothberg P.G., Larsen G.R., Adler C.J., Dorner A.J., Emini E.A., Hanecak R., Lee J.J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 19.Racaniello V.R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jore J., De Geus B., Jackson R.J., Pouwels P.H., Enger-Valk B.E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- 21.Ypma-Wong M.F., Dewalt P.G., Johnson V.H., Lamb J.G., Semler B.L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 22.Harris K.S., Reddigari S.R., Nicklin M.J., Hammerle T., Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang W., Harris K.S., Alexander L., Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Belov G.A., Habbersett C., Franco D., Ehrenfeld E. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J Virol. 2007;81:9259–9267. doi: 10.1128/JVI.00840-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference highlights the membrane trafficking role of poliovirus nonstructural polyprotein 3CD and indicates that polyprotein forms can serve as regulators of protein function.
- 25••.Marcotte L., Wass A., Gohara D., Pathak H., Arnold J., Filman D., Cameron C., Hogle J. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81:3583. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structure of poliovirus 3CD precursor protein is described in this reference, giving insight into the regulation of the mature 3D, 3C proteins.
- 26.Crotty S., Saleh M.C., Gitlin L., Beske O., Andino R. The poliovirus replication machinery can escape inhibition by an antiviral drug that targets a host cell protein. J Virol. 2004;78:3378–3386. doi: 10.1128/JVI.78.7.3378-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brett D., Lindenbach H.-J.T., Charles M. Rice: flaviviridae: the viruses and their replication. In: David M, Knipe P.M.H., editors. vol 1. Lippincott Williams & Wilkins; 2007. pp. 1101–1152. (Fields Virology). [Google Scholar]
- 28.Allison S.L., Schalich J., Stiasny K., Mandl C.W., Heinz F.X. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz I.C., Allison S.L., Heinz F.X., Helenius A. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J Virol. 2002;76:5480–5491. doi: 10.1128/JVI.76.11.5480-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konishi E., Mason P.W. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J Virol. 1993;67:1672–1675. doi: 10.1128/jvi.67.3.1672-1675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Li L., Lok S.M., Yu I.M., Zhang Y., Kuhn R.J., Chen J., Rossmann M.G. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]; The structure of flavivirus prM-E structural precursor polyprotein is described in this reference as well as insight into the role of polyproteins in timely viral maturation.
- 32.Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 33.Bressanelli S., Stiasny K., Allison S.L., Stura E.A., Duquerroy S., Lescar J., Heinz F.X., Rey F.A. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modis Y., Ogata S., Clements D., Harrison S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modis Y., Ogata S., Clements D., Harrison S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 36.Kanai R., Kar K., Anthony K., Gould L.H., Ledizet M., Fikrig E., Marasco W.A., Koski R.A., Modis Y. Crystal structure of west Nile virus envelope glycoprotein reveals viral surface epitopes. J Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Zhang W., Ogata S., Clements D., Strauss J.H., Baker T.S., Kuhn R.J., Rossmann M.G. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guirakhoo F., Bolin R.A., Roehrig J.T. The Murray valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology. 1992;191:921–931. doi: 10.1016/0042-6822(92)90267-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Voss J.E., Vaney M.C., Duquerroy S., Vonrhein C., Girard-Blanc C., Crublet E., Thompson A., Bricogne G., Rey F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]; The structures of Chikungunya virus P62-E1 heterodimer and mature E3–E2–E1 complexes are described in this reference. These provide an insight on the role of polyprotein cleavage and viral maturation.
- 40.Zhang Y., Corver J., Chipman P.R., Zhang W., Pletnev S.V., Sedlak D., Baker T.S., Strauss J.H., Kuhn R.J., Rossmann M.G. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Kaufmann B., Chipman P.R., Kuhn R.J., Rossmann M.G. Structure of immature West Nile virus. J Virol. 2007;81:6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W., Chipman P.R., Corver J., Johnson P.R., Zhang Y., Mukhopadhyay S., Baker T.S., Strauss J.H., Rossmann M.G., Kuhn R.J. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu I.M., Zhang W., Holdaway H.A., Li L., Kostyuchenko V.A., Chipman P.R., Kuhn R.J., Rossmann M.G., Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 45.Goff S.P. Retroviridae: the retroviruses and their replication. In: David M., Knipe P.M.H., editors. vol 2. Lippincott Williams & Wilkins; 2007. pp. 1999–2069. (Fields Virology). [Google Scholar]
- 46.Lu K., Heng X., Summers M.F. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogt V.M., Simon M.N. Mass determination of rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Tang C., Ndassa Y., Summers M.F. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]; This reference describes the structure of a fragment of HIV-1 Gag polyprotein and suggests that formation of a β-hairpin at the N-terminus of the CA domain post cleavage is responsible for correct capsid core assembly.
- 49.von Schwedler U.K., Stemmler T.L., Klishko V.Y., Li S., Albertine K.H., Davis D.R., Sundquist W.I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fitzon T., Leschonsky B., Bieler K., Paulus C., Schroder J., Wolf H., Wagner R. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology. 2000;268:294–307. doi: 10.1006/viro.1999.0178. [DOI] [PubMed] [Google Scholar]
- 51.Tang S., Murakami T., Agresta B.E., Campbell S., Freed E.O., Levin J.G. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J Virol. 2001;75:9357–9366. doi: 10.1128/JVI.75.19.9357-9366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Basavappa R., Syed R., Flore O., Icenogle J.P., Filman D.J., Hogle J.M. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 1994;3:1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structure of poliovirus VP0 precursor polyprotein (which yields mature capsid proteins VP4 and VP2) is described in the context of empty viral capsids in this reference.
- 53•.Chen J., Lee K.H., Steinhauer D.A., Stevens D.J., Skehel J.J., Wiley D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]; The structure of influenza hemagglutinin precursor HA0 ectodomain is described in this reference, giving an insight into influenza pathogenicity and regulation of membrane fusion.
- 54•.Yin H.S., Paterson R.G., Wen X., Lamb R.A., Jardetzky T.S. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Uncleaved paramyxovirus fusion (F) protein is described in this reference.


