Abstract
Allelic dropout is a commonly observed source of missing data in microsatellite genotypes, in which one or both allelic copies at a locus fail to be amplified by the polymerase chain reaction. Especially for samples with poor DNA quality, this problem causes a downward bias in estimates of observed heterozygosity and an upward bias in estimates of inbreeding, owing to mistaken classifications of heterozygotes as homozygotes when one of the two copies drops out. One general approach for avoiding allelic dropout involves repeated genotyping of homozygous loci to minimize the effects of experimental error. Existing computational alternatives often require replicate genotyping as well. These approaches, however, are costly and are suitable only when enough DNA is available for repeated genotyping. In this study, we propose a maximum-likelihood approach together with an expectation-maximization algorithm to jointly estimate allelic dropout rates and allele frequencies when only one set of nonreplicated genotypes is available. Our method considers estimates of allelic dropout caused by both sample-specific factors and locus-specific factors, and it allows for deviation from Hardy–Weinberg equilibrium owing to inbreeding. Using the estimated parameters, we correct the bias in the estimation of observed heterozygosity through the use of multiple imputations of alleles in cases where dropout might have occurred. With simulated data, we show that our method can (1) effectively reproduce patterns of missing data and heterozygosity observed in real data; (2) correctly estimate model parameters, including sample-specific dropout rates, locus-specific dropout rates, and the inbreeding coefficient; and (3) successfully correct the downward bias in estimating the observed heterozygosity. We find that our method is fairly robust to violations of model assumptions caused by population structure and by genotyping errors from sources other than allelic dropout. Because the data sets imputed under our model can be investigated in additional subsequent analyses, our method will be useful for preparing data for applications in diverse contexts in population genetics and molecular ecology.
Keywords: expectation-maximization (EM) algorithm, Native Americans, heterozygosity, missing data
MICROSATELLITE markers are widely used in population genetics and molecular ecology. In microsatellite data, distinct alleles at a locus represent DNA fragments of different sizes, typically detected by amplification using the polymerase chain reaction (PCR). Frequently, during microsatellite genotyping in diploid organisms, one or both of an individual’s two copies of a locus fail to amplify with PCR, yielding a spurious homozygote or a spurious occurrence of missing data. This problem is known as “allelic dropout” (e.g., Gagneux et al. 1997; Pompanon et al. 2005). For example, if an individual has genotype AB at a locus, but only allele A successfully amplifies, then only allele A will be detected, and the genotype will be erroneously recorded as AA. If neither allelic copy amplifies, then the genotype will be recorded as missing. Here we follow Miller et al. (2002) by using “copies” to refer to the paternal and maternal variants in an individual and “alleles” to specify the distinct allelic types possible at a locus.
Allelic dropout is common in microsatellite studies and can lead to statistical errors in subsequent analyses (e.g., Bonin et al. 2004; Broquet and Petit 2004; Hoffman and Amos 2005). For example, in estimating population-genetic statistics, because allelic dropout can cause mistaken assignment of heterozygous genotypes as homozygotes, it can lead to underestimation of the observed heterozygosity and overestimation of the inbreeding coefficient (Taberlet et al. 1999). Circumventing allelic dropout is therefore important for microsatellite studies. One general strategy for correcting for allelic dropout involves repeated genotyping, particularly for the apparent homozygotes (e.g., Taberlet et al. 1996; Morin et al. 2001; Wasser et al. 2007). Additionally, computational approaches have been proposed to assess allelic dropout, primarily when replicate genotypes are available (Miller et al. 2002; Wang 2004; Hadfield et al. 2006; Johnson and Haydon 2007; Wright et al. 2009). In practice, however, replicate genotyping is costly and often uninformative or impossible, owing to insufficient DNA or logistical constraints, especially for natural populations with limited DNA samples from noninvasive sources (e.g., Taberlet and Luikart 1999; Taberlet et al. 1999). Therefore, in this study, we develop a maximum-likelihood approach that can correct for allelic dropout without using replicate genotypes.
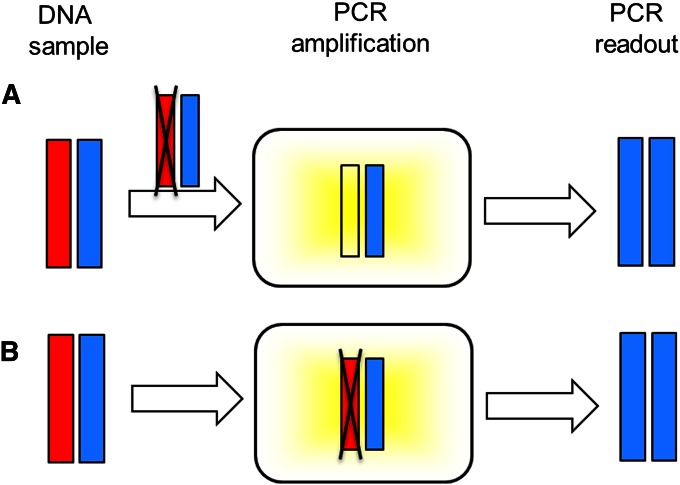
It is believed that the cause of allelic dropout is stochastic sampling of the molecular product, which can occur at two stages of the genotyping process (Figure 1). If DNA concentration is low, then one or both of the allelic copies might not be present in sufficient quantity for successful amplification (e.g., Navidi et al. 1992; Taberlet et al. 1996; Sefc et al. 2003). Poor quality of the template DNA (e.g., high degradation) can also prevent binding by the PCR primers and polymerase, resulting in dropout. An additional problem in the binding step is that some loci might be less likely than others to be bound. Previous studies have found that although different alleles at the same locus have similar probabilities of dropping out, loci with longer alleles tend to have higher dropout rates than those with shorter alleles (e.g., Sefc et al. 2003; Buchan et al. 2005; Broquet et al. 2007); differences in primer annealing efficiency and in template DNA secondary structures might also contribute to different dropout rates across loci (Buchan et al. 2005).
Figure 1 .
Two stages of allelic dropout. The red and blue bars are two allelic copies of a locus in a DNA sample. The black X indicates the location at which allelic dropout occurs. (A) Owing to sample-specific factors such as low DNA concentration or poor DNA quality, one of the two alleles drops out when preparing DNA for PCR amplification. (B) Owing to either locus-specific factors such as low binding affinity between primers or polymerase and the target DNA sequences or sample-specific factors such as poor DNA quality, one of the two alleles fails to amplify with PCR. In both examples shown, allelic dropout results in an erroneous PCR readout of a homozygous genotype.
In this study, we explicitly model the two sources of allelic dropout, using sample-specific dropout rates γi⋅ and locus-specific dropout rates γ⋅ℓ, such that the probability of allelic dropout at locus ℓ of individual i is determined by a function of both γi⋅ and γ⋅ℓ. With a single nonreplicated set of genotypes, we jointly estimate the parameters of the model, including allele frequencies, sample-specific dropout rates, locus-specific dropout rates, and an inbreeding coefficient, thereby correcting for the underestimation of observed heterozygosity and overestimation of inbreeding caused by allelic dropout. We use an expectation-maximization (EM) algorithm to obtain maximum-likelihood estimates (MLEs). With the estimated parameter values, we perform multiple imputation to correct the bias caused by allelic dropout in estimating the observed heterozygosity. We have implemented this method in MicroDrop, which is freely available at http://rosenberglab.stanford.edu.
We first employ the method to analyze a set of human microsatellite genotypes from Native American populations. Using the estimated parameter values, we generate a simulated data set that mimics the Native American data, and we employ this simulated data set to evaluate the performance of our model. First, we compare the patterns of missing data and heterozygosity between the simulated and real data to check whether our model correctly reproduces the observed patterns. Next, we compare estimated and true values of the allelic dropout rates for the simulated data. Finally, we compare the corrected heterozygosity with the “true” heterozygosity calculated from the true genotype data prior to allelic dropout. We further evaluate the robustness of our model, using simulations with different levels of inbreeding, population structure, and genotyping errors from sources other than allelic dropout. We conclude our study by using simulations to argue that our MLEs of dropout rates and the inbreeding coefficient are consistent. That is, we show that as the number of individuals and the number of genotyped loci increase, our estimated values appear to converge to the true values of the parameters.
Data and Preliminary Analysis
The data set on which we focus consists of genotypes for 343 microsatellite markers in 152 Native North Americans collected from 14 populations over many years by the laboratory of D. G. Smith at the University of California (Davis, CA). We identify the populations according to their sampling locations: three populations from the Arctic/Subarctic region, two from the Midwest of the United States (US), two from the Southeast US, two from the Southwest US, three from the Great Basin/California region, and two from Central Mexico. In this data set, the number of distinct alleles per locus has mean 8.0 across loci, with a minimum of 4 and a maximum of 24.
Allelic dropout can generate both spurious homozygotes, when one allelic copy drops out at a heterozygous locus, and missing data, when both copies drop out at either homozygous or heterozygous loci. Thus, under the hypothesis that missing data are caused by allelic dropout, we expect a higher proportion of missing data to be accompanied by a higher proportion of homozygous genotypes. If allelic dropout is caused by low DNA concentration or low quality in certain samples, then a positive correlation will be observed across individuals between missing data and individual homozygosity. Alternatively, if allelic dropout is caused by locus-specific factors such as differences across loci in the binding properties of the primers or polymerase, we instead expect a positive correlation across loci between missing data and locus homozygosity. This type of correlation is also expected if missing data are due to “true missingness”—for example, null alleles segregating in the population at certain loci, as a result of polymorphic deletions in primer regions (e.g., Pemberton et al. 1995; Dakin and Avise 2004). Here, we disregard true missingness and assume that all missing genotypes are attributable to allelic dropout.
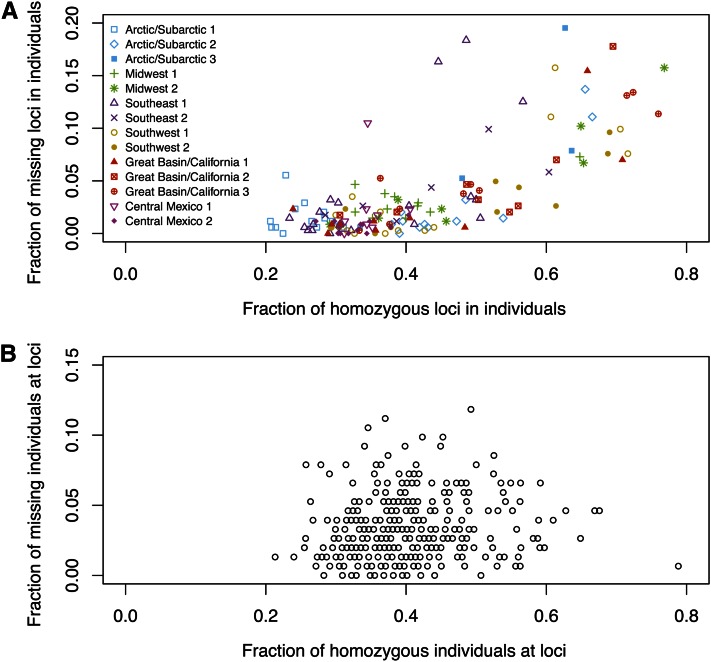
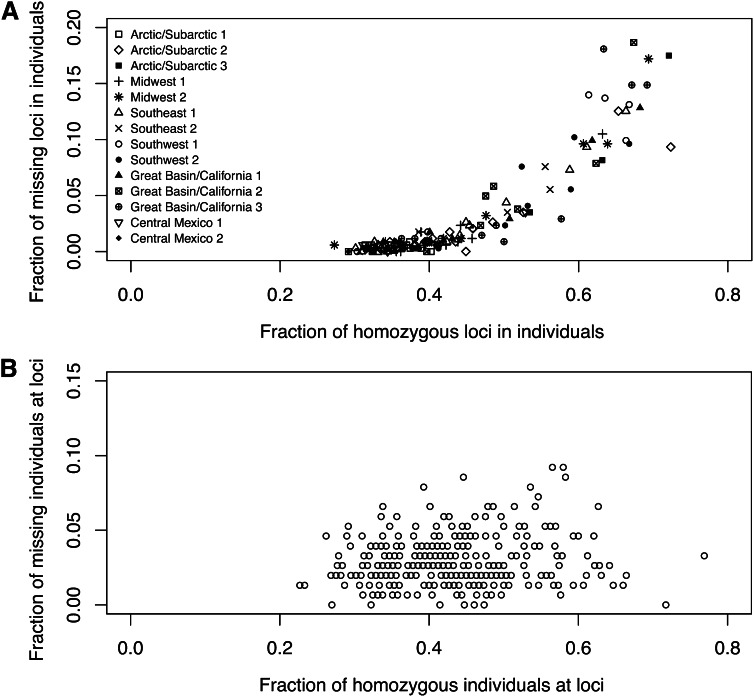
For each individual, we evaluated the proportion of loci at which missing data occurred and the proportion of homozygotes among those loci for which data were not missing. As shown in Figure 2A, missing data and homozygosity have a strong positive correlation: the Pearson correlation is r = 0.729 (P < 0.0001, by 10,000 permutations of the proportions of homozygous loci across individuals). This observation matches the prediction of the hypothesis that missing data result from sample-specific dropout rather than locus-specific dropout or true missingness. By contrast, an analogous computation for each locus rather than for each individual (Figure 2B) finds that the correlation between homozygosity and missing data is much smaller (r = 0.099 and P = 0.0341, by 10,000 permutations of the proportions of homozygous individuals across loci). We therefore suspect that missing genotypes in this data set arise primarily from the allelic dropout caused by low DNA concentration or quality in some samples and that locus-specific factors such as poor binding affinity of primers and polymerase have a smaller effect. In any case, for our subsequent analyses, we continue to consider both sample-specific and locus-specific factors.
Figure 2 .
Fraction of observed missing data vs. fraction of observed homozygotes. (A) Each symbol represents an individual with fraction x of its nonmissing loci observed as homozygous and fraction y of its total loci observed to have both copies missing. The Pearson correlation between X and Y is r = 0.729 (P < 0.0001, by 10,000 permutations of X while fixing Y). (B) Each circle represents a locus at which fraction x of individuals with nonmissing genotypes are observed to be homozygotes and fraction y of all individuals are observed to have both copies missing. r = 0.099 (P = 0.0326).
Model
Consider N individuals and L loci. Denote alleles at locus ℓ by Aℓk with k = 1, 2, … , Kℓ, where Kℓ is the number of distinct alleles at locus ℓ. Denote the observed genotype data by W = {wiℓ: i = 1, 2, … , N; ℓ = 1, 2, … , L}, where genotyping has been attempted for all individuals at all loci. Here, wiℓ is the observed genotype of the ith individual at the ℓth locus. Each entry of W consists of the two observed copies at a locus in a specific individual. If the observed genotype is missing at locus ℓ of individual i, then we specify wiℓ = XX. Otherwise, wiℓ = AℓkAℓh for some k, h ε {1, 2, … , Kℓ}, where k and h are not necessarily distinct. The true genotypes are denoted by G = {giℓ: i = 1, 2, … , N; ℓ = 1, 2, … , L}. A description of the notation appears in Table 1.
Table 1 . Notation used in the article.
| Notation | Meaning | Type |
|---|---|---|
| i | Index of an individual | Basic notation |
| ℓ | Index of a locus | Basic notation |
| k, h | Index of an allele | Basic notation |
| N | No. individuals | Basic notation |
| L | No. loci | Basic notation |
| Kℓ | No. distinct alleles at locus ℓ | Basic notation |
| Aℓk, Aℓh | Allele k (h) at locus ℓ | Basic notation |
| X | Missing data (dropout) | Basic notation |
| γiℓ | Dropout probability at locus ℓ of individual i | Basic notation |
| wiℓ | Observed genotype at locus ℓ of individual i | Observed data point |
| W | Observed genotypes, W = {wiℓ} | Observed data set |
| giℓ | True genotype at locus ℓ of individual i | Latent variable |
| siℓ | IBD state at locus ℓ of individual i | Latent variable |
| ziℓ | Dropout state at locus ℓ of individual i | Latent variable |
| G | True genotypes, G = {giℓ} | Latent variable set |
| S | IBD states, S = {siℓ} | Latent variable set |
| Z | Dropout states, Z = {ziℓ} | Latent variable set |
| ρ | Inreeding coefficient | Parameter |
| φℓk | Frequency of allele Aℓk | Parameter |
| γi⋅ | Sample-specific dropout rate for individual i | Parameter |
| γ⋅ℓ | Locus-specific dropout rate for locus ℓ | Parameter |
| Φ | Allele frequencies, Φ = {φℓk} | Parameter set |
| Γ | Dropout rates, Γ = {γi⋅, γ⋅ℓ} | Parameter set |
| Ψ | Model parameters, Ψ = {ρ, Φ, Γ} | Parameter set |
| nℓk | No. independent copies of allele Aℓk | Summary statistic |
| diℓ | No. dropouts at locus ℓ for individual i | Summary statistic |
| di⋅ | No. sample-specific dropouts for individual i | Summary statistic |
| d⋅ℓ | No. locus-specific dropouts at locus ℓ | Summary statistic |
| s | No. genotypes having two alleles IBD | Summary statistic |
i ε {1, 2, …, N}, ℓ ε {1, 2, … , L}, and k, h ε {1, 2, … , Kℓ}.
To model the dropout mechanism, we specify a set of dropout states Z = {ziℓ: i = 1, 2, … , N; ℓ = 1, 2, … , L} that connects G and W and that indicates which alleles “drop out.” For a heterozygous true genotype giℓ = AℓkAℓh (h ≠ k), supposing allele Aℓk drops out, the dropout state is ziℓ = AℓhX and the observed genotype is wiℓ = AℓhAℓh. For a homozygous true genotype giℓ = AℓkAℓk, the dropout state ziℓ = AℓkX means that exactly one of the two allelic copies drops out.
We make five assumptions in our model:
All distinct alleles are observed at least once in the data set.
All missing and incorrect genotypes are attributable to allelic dropout.
- Both copies at a locus ℓ of an individual i have equal probability γiℓ of dropping out. This probability is a function of a sample-specific dropout rate γi. and a locus-specific dropout rate γ⋅ℓ:
(1) All individuals are unrelated and have the same inbreeding coefficient ρ, such that for any locus of any individual, the two allelic copies are identical by descent (IBD) with probability ρ.
Each pair of loci is independent (i.e., each pair of loci is at linkage equilibrium).
Denote Γ = {γi⋅, γ⋅ℓ: i = 1, 2, … , N; ℓ = 1, 2, … , L} and Φ = {φℓk: ℓ = 1, 2, … , L; k = 1, 2, … , Kℓ}, in which φℓk is the true frequency of allele Aℓk at locus ℓ, γi⋅ is the probability of dropout caused by sample-specific factors for any allelic copy at any locus of individual i, and γ⋅ℓ is the probability of dropout caused by locus-specific factors for any allelic copy at locus ℓ in any individual. Equation 1 arises by noting that the dropout probability for an allelic copy at locus ℓ of individual i, considering the two possible causes as independent, is γiℓ = 1 − (1 − γi⋅)(1 − γ⋅ℓ).
Using assumption 3, the conditional probability ℙ(ziℓ|giℓ, Γ) can be expressed as shown in Table 2. The conditional probability of observing genotype wiℓ given true genotype giℓ and dropout rates γi⋅ and γ⋅ℓ can be calculated as
| (2) |
Here, ℙ(wiℓ|ziℓ, giℓ) is either 0 or 1 because W is fully determined by Z and G, and the summation proceeds over all dropout states ziℓ possible given the observed genotype wiℓ (Table 2).
Table 2 . Illustration of the outcomes of allelic dropout using two distinct alleles at locus ℓ, Aℓk and Aℓh.
| True genotype | Genotype frequency | Dropout state | Conditional probability | Observed genotype | Conditional probability |
|---|---|---|---|---|---|
| giℓ | ℙ(giℓ|Φ, ρ) | ziℓ | ℙ(ziℓ|giℓ, Γ) | wiℓ | ℙ(wiℓ|giℓ, Γ) |
| AℓkAℓk | AℓkAℓk | (1 − γiℓ)2 | AℓkAℓk | ||
| AℓkX | 2γiℓ(1 − γiℓ) | ||||
| XX | XX | ||||
| AℓkAℓh | 2(1 − ρ)φℓkφℓh | AℓhAℓk | (1 − γiℓ)2 | AℓkAℓh | (1 − γiℓ)2 |
| AℓhX | γiℓ(1 − γiℓ) | AℓhAℓh | γiℓ(1 − γiℓ) | ||
| AℓkX | γiℓ(1 − γiℓ) | AℓkAℓk | γiℓ(1 − γiℓ) | ||
| XX | XX |
Genotype frequencies are calculated from allele frequencies using Equation 5, where ρ is the inbreeding coefficient, a parameter used to model the total deviation from Hardy–Weinberg equilibrium. Dropout is assumed to happen independently to each copy at locus ℓ of individual i, with probability γiℓ specified by Equation 1. .
We use a set of binary random variables S = {siℓ} to indicate the IBD states of the true genotypes G, such that siℓ = 1 if the two allelic copies in genotype giℓ are IBD, and siℓ = 0 otherwise. Under assumption 4, we have (e.g., Holsinger and Weir 2009)
| (3) |
| (4) |
| (5) |
When ρ = 0, the genotype frequencies in Equation 5 follow Hardy–Weinberg equilibrium (HWE).
With the quantities in Equations 2–5, the probability of observing wiℓ given parameters Ψ is
| (6) |
The summation proceeds over the set of all possible true genotypes giℓ, that is, over all two-allele combinations at locus ℓ. The likelihood function of the parameters Ψ = {Φ, Γ, ρ} is then given by
| (7) |
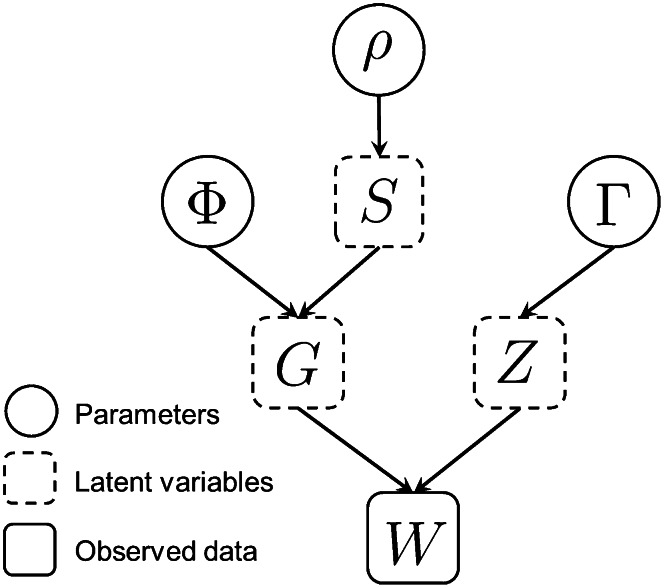
This likelihood assumes that dropout at a locus is independent across individuals, so that each observed diploid genotype of an individual at the locus is a separate trial independent of all others. Further, assumption 5 enables us to take a product across loci, as genotypes at separate loci are independent. A graphical representation of the relationships among the parameters Φ, Γ, and ρ; the latent variables G, S, and Z; and the observation W appears in Figure 3.
Figure 3 .
Graphical representation of the model. Each arrow denotes a dependency between two sets of quantities: Φ allele frequencies; ρ, inbreeding coefficient; Γ, sample-specific and locus-specific dropout rates; G, true genotypes; S, IBD states; Z, dropout states; and W, observed genotypes. W is the only observed data, consisting of N × L independent observations and providing information to infer parameters Φ, ρ, and Γ.
Estimation Procedure
Given the observed genotypes W, we can use an EM algorithm (e.g., Lange 2002) to obtain the MLEs of the allele frequencies Φ, the sample-specific and locus-specific dropout rates Γ, and the inbreeding coefficient ρ. Under the inbreeding assumption (assumption 4), two allelic copies at the same locus need not be independent. If two allelic copies are IBD, then the allelic state of one copy is determined given the allelic state of the other copy, so that the number of independent allelic copies is 1. If two copies at the same locus are not IBD, then the number of independent allelic copies is 2. We introduce a random variable nℓk to represent the number of “independent” copies of allele Aℓk in the whole data set, considering all individuals. We also define a random variable diℓ as the number of copies that drop out at locus ℓ of individual i (diℓ = 0, 1, or 2).
In the E-step of our EM algorithm, we calculate (1) the expectation of the number of independent copies for all alleles, [nℓk|W, Ψ], summing across individuals; (2) for each individual, the total number of dropouts caused by sample-specific factors, ; (3) for each locus, the total number of dropouts caused by locus-specific factors, ; and (4) the expectation of the total number of genotypes that are IBD, summing across the whole data set, . The factors γi⋅/γiℓ and γ⋅ℓ/γiℓ specify the respective probabilities that sample-specific factors and locus-specific factors contribute to the allelic dropouts at locus ℓ of individual i.
To obtain the expectations required for the E-step, we need the posterior probabilities of giℓ, diℓ, and siℓ given the observed genotype wiℓ and the parameters Ψ, for each (i, ℓ) with i = 1, 2, … , N and ℓ = 1, 2, … , L. The posterior joint probabilities of giℓ and siℓ given wiℓ and Ψ are listed in Table 3, and they are calculated from Bayes’ formula:
| (8) |
The second equality holds because the probability of being IBD (siℓ = 1) depends only on the inbreeding coefficient ρ, the true genotype giℓ is independent of ρ and the dropout rate γiℓ given siℓ and the allele frequencies Φ, and the observed genotype wiℓ is independent of Φ and ρ given giℓ and γiℓ.
Table 3 . Posterior joint probabilities of true genotypes giℓ and IBD states siℓ at a single locus ℓ of an individual i.
| Observed genotype | True genotype | IBD state | Joint probability | Posterior probability |
|---|---|---|---|---|
| wiℓ | giℓ | siℓ | ℙ(giℓ, siℓ, wiℓ|Ψ) | ℙ(giℓ, siℓ|wiℓ, Ψ) |
| AℓkAℓh | AℓkAℓh | 1 | 0 | 0 |
| 0 | 2(1 − ρ)φℓkφℓh(1 − γiℓ)2 | 1 | ||
| Others | 1 | 0 | 0 | |
| 0 | 0 | 0 | ||
| AℓkAℓk | AℓkAℓh | 1 | 0 | 0 |
| 0 | 2(1 − ρ)φℓkφℓhγiℓ(1 − γiℓ) | |||
| AℓkAℓk | 1 | |||
| 0 | ||||
| XX | AℓkAℓh | 1 | 0 | 0 |
| 0 | 2(1 − ρ)φℓkφℓh | |||
| AℓkAℓk | 1 | ρφℓk | ||
| 0 |
The calculation of ℙ(giℓ, siℓ | wiℓ, Ψ) is based on Equation 8. h ≠ k.
For example, suppose the observed genotype is wiℓ = AℓkAℓk, and we wish to evaluate ℙ(giℓ = AℓkAℓk, siℓ = 1|wiℓ = AℓkAℓk, Ψ), the posterior joint probability that the true genotype is giℓ = AℓkAℓk and the two allelic copies are IBD. If wiℓ = AℓkAℓk is observed, then the true genotype giℓ can be a homozygote AℓkAℓk or a heterozygote AℓkAℓh, with h ε {1, 2, … , Kℓ} and h ≠ k. Each term in the summation in Equation 8 is a joint probability ℙ(giℓ, siℓ, wiℓ|Ψ). To calculate this quantity, ℙ(siℓ|ρ) and ℙ(giℓ|siℓ, Φ) are obtained using Equations 3 and 4, respectively. The values of ℙ(wiℓ = AℓkAℓk|giℓ, γiℓ) are given by Table 2 and can be obtained using Equation 2. The resulting probabilities ℙ(giℓ, siℓ, wiℓ|Ψ) appear in Table 3. Therefore, for example,
| (9) |
With the values of ℙ(giℓ, siℓ|wiℓ = AℓkAℓk, Ψ), the posterior probabilities of giℓ and siℓ can be easily calculated with Equations 10 and 11, respectively. Results appear in Tables 4 and 5:
Table 4 . Posterior probabilities of true genotypes giℓ at a single locus ℓ of an individual i.
| Observed genotype | True genotype | Posterior probability |
|---|---|---|
| wiℓ | giℓ | ℙ(giℓ|wiℓ, Ψ) |
| AℓkAℓh | AℓkAℓh | 1 |
| Others | 0 | |
| AℓkAℓk | AℓkAℓh | |
| AℓkAℓk | ||
| XX | AℓkAℓh | 2(1 − ρ)φℓkφℓh |
| AℓkAℓk |
The calculation of ℙ(giℓ | wiℓ, Ψ) is based on Equation 10. h ≠ k.
Table 5 . Posterior probabilities of the IBD state siℓ at a single locus ℓ of an individual i.
| Observed genotype | IBD state | Posterior probability |
|---|---|---|
| wiℓ | siℓ | ℙ(siℓ|wiℓ,Ψ) |
| AℓkAℓh | 1 | 0 |
| 0 | 1 | |
| AℓkAℓk | 1 | |
| 0 | ||
| XX | 1 | ρ |
| 0 | 1 − ρ |
The calculation of ℙ(siℓ | wiℓ, Ψ) is based on Equation 11. h ≠ k.
| (10) |
| (11) |
The posterior probabilities of diℓ given wiℓ and Ψ appear in Table 6, and they are obtained by
| (12) |
Here,
| (13) |
Therefore, [nℓk|W, Ψ], [di⋅|W, Ψ], [d⋅ℓ|W, Ψ], and [s|W, Ψ] are calculated as
| (14) |
| (15) |
| (16) |
| (17) |
in which f(Aℓk|giℓ, siℓ) indicates the number of independent copies of allele Aℓk in genotype giℓ given the IBD state siℓ, as defined below:
| (18) |
Table 6 . Posterior probabilities of the number of dropouts diℓ at a single locus ℓ of an individual i.
| Observed genotype | No. dropouts | Joint probability | Posterior probability |
|---|---|---|---|
| wiℓ | diℓ | ℙ(diℓ, wiℓ|Ψ) | ℙ(diℓ|wiℓ, Ψ) |
| AℓkAℓh | 0 | 2(1 − ρ)φℓkφℓh(1 − γiℓ)2 | 1 |
| 1 | 0 | 0 | |
| 2 | 0 | 0 | |
| AℓkAℓk | 0 | [ρ + (1 − ρ)φℓk]φℓk(1 − γiℓ)2 | |
| 1 | 2φℓkγiℓ(1 − γiℓ) | ||
| 2 | 0 | 0 | |
| XX | 0 | 0 | 0 |
| 1 | 0 | 0 | |
| 2 | 1 |
The calculations are based on Equations 12 and 13. h ≠ k.
In the M-step of the EM algorithm, we update the estimation of parameters Ψ by
| (19) |
| (20) |
| (21) |
| (22) |
Justification of these expressions appears in Appendix A. With the updated parameter values, we calculate the likelihood ℙ(W|Ψ) using Equation 7 and then repeat the E-step and the M-step. The likelihood is guaranteed to increase after each iteration in this EM process and will converge to a maximum (e.g., Lange 2002); the estimated parameter values are MLEs if this maximum is the global maximum. To lower the chance of convergence only to a local maximum, we repeat our EM algorithm with 100 sets of initial values of Ψ. For each set, the allele frequencies, Φ = {φℓk: k = 1, 2, … , Kℓ; ℓ = 1, 2, … , L}, are sampled independently at different loci from Dirichlet distributions, for locus ℓ; the sample-specific dropout rates γi⋅ (i = 1, 2, … , N), the locus-specific dropout rates γ⋅ℓ (ℓ = 1, 2, … , L), and the inbreeding coefficient ρ are independently sampled from the uniform distribution U(0, 1). An EM replicate is considered to be “converged” if the increase of the log-likelihood log10ℙ(W|Ψ) in one iteration is <10−4; when this condition is met, we terminate the iteration process. The parameter values that generate the highest likelihood among the 100 EM replicates are chosen as our estimates.
Imputation Procedure
To correct the bias caused by allelic dropout in estimating the observed heterozygosity and other quantities, we create 100 imputed data sets by drawing genotypes from the posterior probability , in which , , and are the MLEs of Φ, Γ, and ρ, and ℙ(G|W, Ψ) is specified in Equation 10 and Table 4. In using this strategy, we not only impute the missing genotypes but also replace some of the observed homozygous genotypes with heterozygotes, as it is possible that observed homozygous genotypes represent false homozygotes resulting from allelic dropout. This imputation strategy accounts for the genotype uncertainty that allelic dropout introduces.
Application to Native American Data
We found that in sequential observations of the likelihood of the estimated parameter values, our EM algorithm converged quickly for all 100 sets of initial values for Φ, Γ, and ρ (results not shown). For each of the 100 sets, the EM algorithm reached the convergence criterion within 300 iterations. The difference in the estimated parameter values among the 100 replicates was minimal after convergence, indicating that the method was not sensitive to the initial values (results not shown).
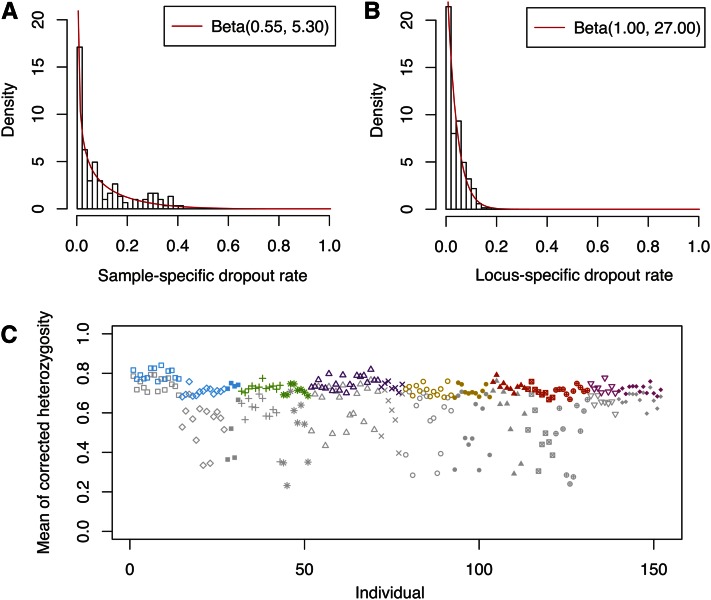
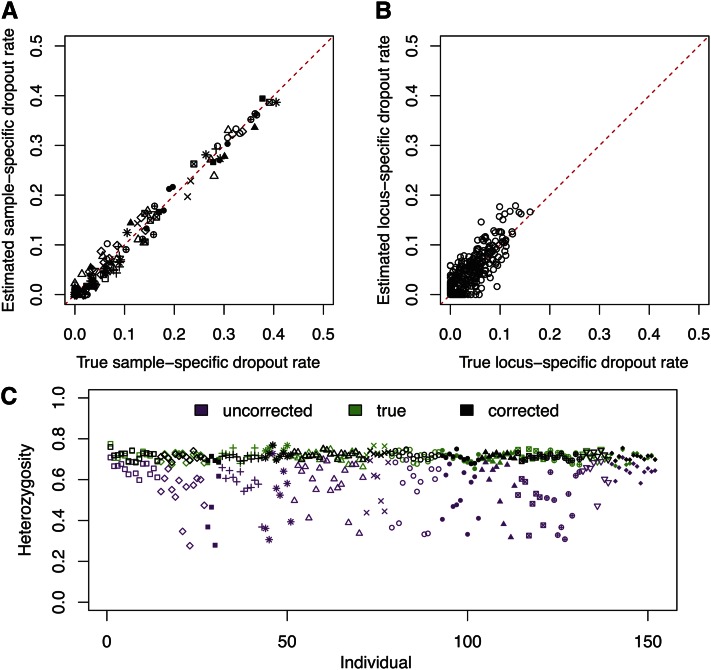
Histograms of the estimated sample-specific dropout rates and the estimated locus-specific dropout rates appear in Figure 4. The mean of the is 0.094, and for most individuals, (Figure 4A). The maximum is 0.405; this high rate indicates that some samples have low quantity or quality and is compatible with the fact that some of the samples are relatively old. Samples from some populations, such as Arctic/Subarctic 1 and Central Mexico 2, have higher overall quality, as reflected in low estimated sample-specific dropout rates.
Figure 4 .
Estimated dropout rates and corrected heterozygosity for the Native American data. (A) Histogram of the estimated sample-specific dropout rates. The histogram is fitted by a beta distribution with parameters estimated using the method of moments. (B) Histogram of the estimated locus-specific dropout rates. The histogram is again fitted by a beta distribution using the method of moments. (C) Corrected individual heterozygosity calculated from data imputed using the estimated parameter values, averaged over 100 imputed data sets. Colors and symbols follow Figure 2. The corresponding uncorrected observed heterozygosity for each individual is indicated in gray.
Compared to the sample-specific dropout rates, the estimated locus-specific dropout rates are much smaller, with mean 0.036 and maximum 0.160 (Figure 4B). The large spread of the compared to the small values of the is consistent with the observation that the positive correlation between missing data and homozygotes is much greater across individuals than across loci (Figure 2).
The estimated inbreeding coefficient is , the minimum possible value, smaller than the positive values typical of human populations. Several explanations could potentially explain the estimate of 0. First, our samples might be close to HWE. Second, our method might systematically underestimate the inbreeding coefficient, a hypothesis that we test below using simulations. Third, genotyping errors other than allelic dropout, such as genotype miscalling, can potentially also contribute to the underestimation. We use simulations to examine this hypothesis as well.
In a given individual, the L loci can be divided into three classes according to the observed genotypes: nhom homozygous loci, nhet heterozygous loci, and L − nhom − nhet loci that have both allelic copies missing. For each individual, we calculated the observed heterozygosity as Ho = nhet/(nhom + nhet), as shown by gray symbols in Figure 4C. High variation exists in Ho for different individuals, and the mean Ho across individuals is 0.590 (standard deviation = 0.137). The observed heterozygosities are negatively correlated with the MLEs of the sample-specific dropout rates (Supporting Information, Figure S1), as is expected from the underestimation of heterozygosity caused by allelic dropout. Averaging the estimated observed heterozygosity over 100 imputed data sets, we see that variation across individuals in estimated heterozygosities is reduced compared to the values estimated directly from the observed genotypes, and the mean heterozygosity increases to 0.730 (standard deviation = 0.035, Figure 4C). The estimated individual heterozygosity does not vary greatly across different imputed data sets (standard deviation = 0.014, averaging across all individuals).
Simulations
We perform three sets of simulations to examine the performance of our method. First, we consider simulations that assume that the model assumptions hold, using as true values the estimated parameter values from the Native American data set (experiment 1). Next, we consider simulations that do not satisfy the model assumptions, by inclusion of population structure (experiment 2) and genotyping errors not resulting from allelic dropout (experiment 3). These latter simulations examine the robustness of the estimation procedure to model violations.
Simulation methods
To generate simulated allelic dropout rates for use in experiments 2 and 3, we first fit the distributions of the estimated sample-specific and locus-specific dropout rates from the Native American data, using beta distributions Beta(α, β). Denote the sample mean and sample variance of the MLEs of the sample-specific (or locus-specific) dropout rates as m and v, respectively. We estimated α and β using the method of moments, with and (Casella and Berger 2001). The estimated sample-specific and locus-specific dropout rates approximately follow Beta(0.55, 5.30) and Beta(1.00, 27.00), respectively (Figure 4, A and B).
Experiment 1. Native American data:
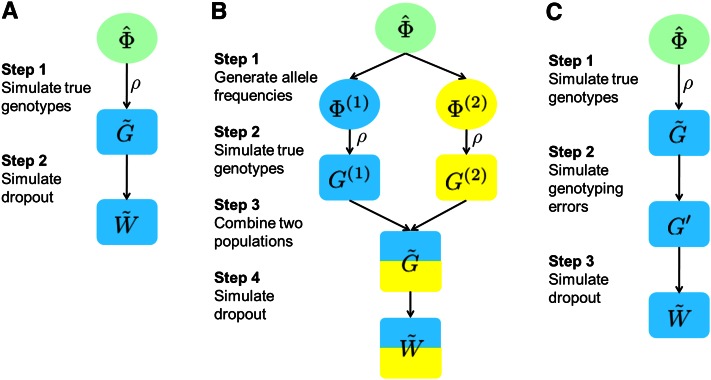
We simulate data under model assumptions 2–5 with parameter values estimated from the actual Native American data (results from Application to Native American Data). The simulation procedure appears in Figure 5A. Suppose , , and are the MLEs of Φ, Γ, and ρ estimated from the data. First, we draw the true genotypes , using probabilities specified by Equation 5, assuming that the allele frequencies are given by and the inbreeding coefficient by . Next, we simulate the dropout state by randomly dropping out copies with probability specified by Equation 1, independently across alleles, loci, and individuals. Using and , we then obtain our simulated observed genotypes . This simulation approach does not guarantee that model assumption 1 will hold, because some alleles might not be observed owing either to allelic dropout or to a stochastic failure to be drawn in the simulation. We simulate one set of genotypes at L = 343 loci for N = 152 individuals.
Figure 5 .
Simulation procedures. In all procedures, represents the allele frequencies estimated from the Native American data, represents the true genotypes generated under the inbreeding assumption, and is the observed genotypes with allelic dropout. (A) Procedure to generate the simulated Native American data (experiment 1). (B) Procedure to generate simulated data with population structure (experiment 2). In step 1, the allele frequencies of two subpopulations are generated using the F model. (C) Procedure to generate simulated data with genotyping errors other than allelic dropout (experiment 3).
Experiment 2. Data with population structure:
To test our method in a setting in which genotypes are taken from a structured population, we simulate data for two subpopulations with equal sample size (N1 = N2 = 76), genotyped at the same set of loci (L = 343). We then apply our method on the combined data set, disregarding the population structure. The procedure appears in Figure 5B.
First, we use the F model (Falush et al. 2003) to generate allele frequencies for two populations that have undergone a specified level of divergence from a common ancestral population. We use the MLEs of the allele frequencies of the 343 loci in the Native American data (results from Application to Native American Data) as the allele frequencies of the ancestral population, . Denote the estimated allele frequencies at locus ℓ by a vector . Under the F model, allele frequencies of locus ℓ for population 1, , and for population 2, , are independently sampled from the Dirichlet distribution , in which F is a parameter constant across loci that describes the divergence of the descendant populations from the ancestral population. F can differ for populations 1 and 2, but for simplicity, we set F to the same value for both populations. Using Equations B1 and B2 in Appendix B and the independence of and , the squared difference of allele frequencies between the two populations satisfies , which is linearly proportional to F. In the limit as F → 0, we get for each ℓ, so that no divergence exists between either descendant population and the ancestral population.
We choose six values of F (0, 0.04, 0.08, 0.12, 0.16, and 0.20) in different simulations. For each value, we first generate allele frequencies, Φ(1) and Φ(2), at all 343 loci for populations 1 and 2. Next, we draw genotypes separately for each population according to the genotype frequencies in Equation 5, with the same value of the inbreeding coefficient ρ. We consider 16 values for ρ, ranging from 0 to 0.15 in increments of 0.01. In total, we generate 6 × 16 = 96 sets of simulated genotypes with different combinations of settings for F and ρ (although for ease of presentation, some plots show only 36 of the 96 cases). Finally, we simulate allelic dropout on each of the simulated genotype data sets using γi⋅ and γ⋅ℓ sampled independently from a Beta(α, β) distribution, in which α = 0.55 and β = 5.30 are estimated from the MLEs of sample-specific dropout rates of the Native American data (Figure 4A). We do not use the estimated α and β from the MLEs of locus-specific dropout rates because these MLEs lie in a relatively small range (Figure 4B) that would not permit simulation of high dropout rates for testing our method. Instead, use of the same beta distribution estimated from the sample-specific dropout rates produces a greater spread in the values of the simulated true locus-specific dropout rates, providing a more complete evaluation.
Experiment 3. Data with other genotyping errors:
In our third experiment, we simulate data with stochastic genotyping errors other than allelic dropout. The simulation procedure appears in Figure 5C. Each simulated data set contains a single population of N = 152 individuals genotyped for L = 343 loci. True genotypes are drawn with probabilities calculated from Equation 5, with allele frequencies Φ chosen as the maximum-likelihood estimated frequencies from the Native American data, and the inbreeding coefficient ρ ranging from 0 to 0.15 incremented in units of 0.01 for different simulated data sets. Next, we simulate genotyping errors using a simple error model, in which at a K-allele locus in the simulated true genotypes, any allele can be mistakenly assigned as any one of the other K − 1 alleles, each with the same probability of e/(K − 1). The parameter e specifies the overall error rate from sources other than allelic dropout, such as genotype miscalling and data entry errors (e.g., Wang 2004; Johnson and Haydon 2007). We consider six values for e (0, 0.02, 0.04, 0.06, 0.08, and 0.10), such that we simulate 96 (= 6 × 16) data sets with different combinations of e and ρ. In the last step, as in experiment 2, we simulate allelic dropout in each data set with both sample-specific and locus-specific dropout rates independently sampled from a Beta(0.55, 5.30) distribution.
Simulation results
Experiment 1. Native American data:
Because we simulate under assumptions 2–5 with parameter values estimated from the real data, we expect that if our model is correctly specified, the simulated data can capture patterns observed in the real data. By comparing plots of the fraction of missing data vs. the fraction of homozygotes in the real and simulated data (Figures 2 and 6), we can see that our simulated data effectively capture the observed positive correlation across individuals and the lack of correlation across loci observed from the real data. For the simulated data set, the Pearson correlation coefficient between the fraction of missing genotypes and the fraction of homozygotes is r = 0.900 (P < 0.0001) across individuals and r = 0.143 (P = 0.0045) across loci. We can also compare the observed heterozygosity for the simulated data (purple symbols in Figure 7C) and the real data (gray symbols in Figure 4C). The simulated data again reproduce the pattern of variation among individual heterozygosities observed in the real data. These two empirical comparisons display the similarity between the real data and the data simulated on the basis of estimates obtained from the real data and thus support the validity of the allelic dropout mechanism specified in our model.
Figure 6 .
Fraction of observed missing data vs. fraction of observed homozygotes for one simulated data set. (A) Each symbol represents an individual with fraction x of its nonmissing loci observed as homozygous and fraction y of its total loci observed to have both copies missing. The Pearson correlation between X and Y is r = 0.900 (P < 0.0001, by 10,000 permutations of X while fixing Y). (B) Each point represents a locus at which fraction x of individuals with nonmissing genotypes are observed to be homozygotes and fraction y of all individuals are observed to have both copies missing. r = 0.143 (P = 0.0045).
Figure 7 .
Estimated dropout rates and corrected heterozygosity for the data simulated on the basis of the Native American data set. (A) Comparison of the estimated sample-specific dropout rates and the assumed true sample-specific dropout rates. (B) Comparison of the estimated locus-specific dropout rates and the assumed true locus-specific dropout rates. (C) Individual heterozygosities in the simulated data. True values of heterozygosity are indicated by green symbols. With allelic dropout applied to true genotypes to generate “observed” data, the uncorrected values of heterozygosity are colored purple. Means of corrected heterozygosities across 100 imputed data sets are colored black. Symbols follow Figure 6.
We can formally compare the estimated dropout rates for the simulation with the true dropout rates specified by the MLEs of the dropout rates for the Native American data. Figure 7A shows that our method accurately estimates the sample-specific dropout rates for all 152 individuals (mean squared error 2.6 × 10−4). The estimated locus-specific dropout rates are also close to their true values, but with a slightly higher mean squared error of 5.2 × 10−4 (Figure 7B). This difference between the estimation of sample-specific dropout rates and that of locus-specific dropout rates can be explained by the fact that the number of loci (L = 343) is more than twice the number of individuals (N = 152). Consequently, more information is available for estimating a sample-specific rather than a locus-specific dropout rate. For the inbreeding coefficient ρ, our estimated value is 1.7 × 10−5, close to the true value of 0 that we used to generate the simulated genotypes.
Finally, in Figure 7C, we can see that our method successfully corrects the bias in estimating heterozygosity from the simulated data. The true observed heterozygosity is calculated using the true genotypes and has mean 0.716, averaging across all individuals. The mean estimated observed heterozygosity, obtained from the observed uncorrected genotypes , is 0.565, lower than the true value. With imputed data sets, we obtain corrected heterozygosities that are close to the true values. The mean and standard deviation of the corrected heterozygosities, evaluated from 100 imputed data sets and averaged across individuals, are 0.715 and 0.014, respectively. The low standard deviation across different imputed data sets indicates that our imputation strategy is relatively robust in correcting the underestimation of observed heterozygosity.
Experiment 2. Data with population structure:
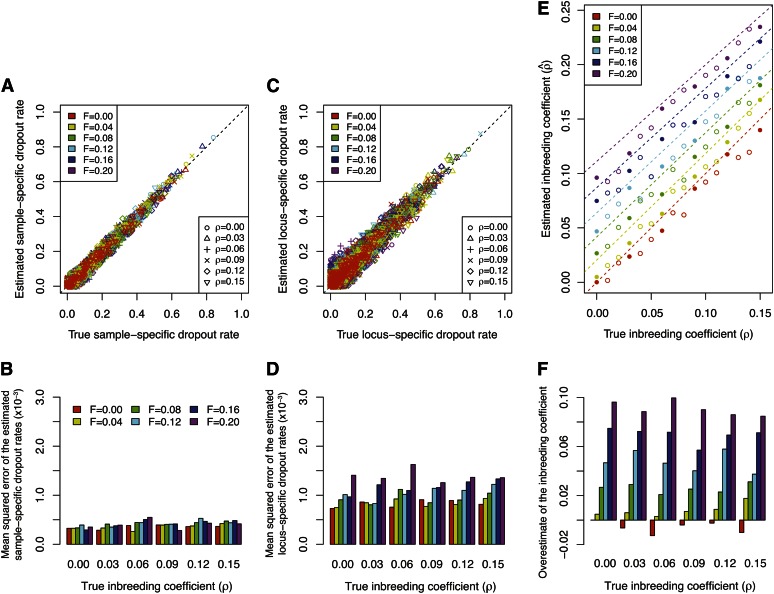
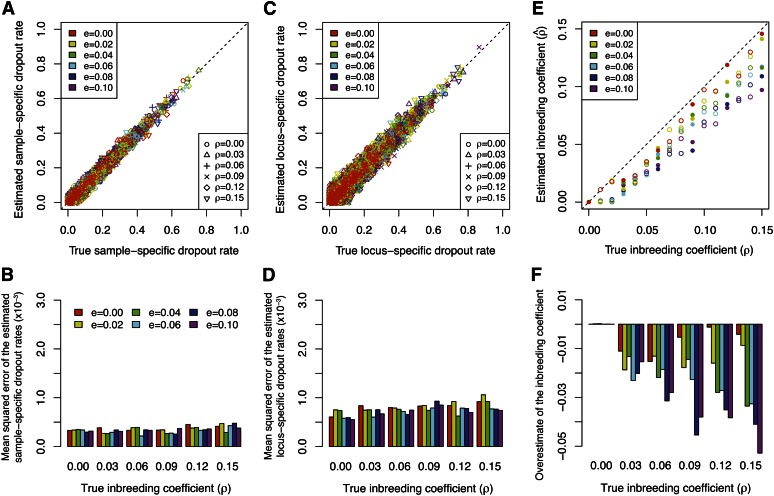
To further test the robustness of our method, we applied our method to 96 simulated data sets with different levels of population structure (parameterized by F) and inbreeding (parameterized by ρ). In Figure 8, A and C, we compare the estimated dropout rates to their true values. Considering the 36 simulated data sets that are displayed, our method accurately estimates both the sample-specific and the locus-specific dropout rates. The accuracy of our estimates is then quantified by mean squared errors for each simulated data set separately, as displayed in Figure 8, B and D. The performance in estimating the sample-specific dropout rate is not greatly affected by either the degree of population structure or the level of inbreeding (Figure 8B). By contrast, while the mean squared error of the estimated locus-specific dropout rates is roughly constant for different levels of inbreeding, it increases with the degree of population structure (Figure 8D).
Figure 8 .
Estimated dropout rates and inbreeding coefficients for simulated data with population structure. (A) Comparison of the estimated sample-specific dropout rates and the assumed true sample-specific dropout rates. (B) Mean squared errors across all the estimated sample-specific dropout rates for each of the 36 data sets shown in A. (C) Comparison of the estimated locus-specific dropout rates and the assumed true locus-specific dropout rates. (D) Mean squared errors across all the estimated locus-specific dropout rates for each of the 36 data sets shown in C. (E) Comparison of the estimated inbreeding coefficient and the assumed true inbreeding coefficient, in which each point corresponds to one of 96 simulated data sets. The 36 solid symbols correspond to the simulated data sets shown in A–D and F. Dashed lines indicate the effective inbreeding coefficients of structured populations under the F model (Equation B11). (F) Overestimation of the inbreeding coefficient, calculated by subtracting the assumed true inbreeding coefficient from the estimated inbreeding coefficient, or .
One possible explanation for this observation is that the accuracy of allelic dropout estimates is closely related to the accuracy of the estimated allele frequencies. This accuracy may decrease as the level of population structure increases, because we do not incorporate population structure in our model for estimation. The estimation of locus-specific dropout rates is more sensitive to inaccurate estimates of allele frequencies because the estimated accuracy of a locus-specific rate relies on the estimation of allele frequencies at that particular locus. By contrast, a sample-specific dropout rate is obtained by averaging the expected number of sample-specific dropouts across all loci in an individual and is less dependent on the accuracy of estimated allele frequencies at any particular locus. Therefore, sample-specific dropout rate estimates are less sensitive to population structure than are locus-specific estimates. When F = 0, with no population structure, the difference between the mean squared error for the sample-specific rates and that for the locus-specific rates arises simply from differences in the numbers of loci and individuals, as discussed for experiment 1.
Figure 8E shows the estimated inbreeding coefficient for all 96 simulated data sets, compared to the simulated true inbreeding coefficient in the subpopulations. With F = 0, a scenario for which no population structure exists and the data are generated under model assumptions 2–5, our method tends to slightly underestimate the inbreeding coefficient. As F increases, the estimate becomes greater than the simulated inbreeding coefficient (Figure 8F). This result is consistent with our expectation, because according to the Wahlund effect (e.g., Hartl and Clark 1997), a pooled population consisting of two subpopulations is expected to have more homozygous genotypes than an unstructured population, resulting in a pattern similar to that caused by a higher level of inbreeding within the unstructured population. Indeed, with no allelic dropout, a structured population under the F model has identical expected allele frequencies and genotype frequencies to those of an unstructured population with a higher inbreeding coefficient ρ* = ρ + (1 − ρ)[F/(2 − F)] (Appendix B). By comparing our estimated inbreeding coefficient with the “effective inbreeding coefficient” ρ* (dashed lines in Figure 8E), we find that most of our estimated inbreeding coefficients are slightly smaller than the corresponding ρ*, indicating that the MLE of ρ is biased downward. It is worth noting that with a single parameter ρ, we capture the deviation of genotype frequencies from HWE introduced by population structure, thereby obtaining accurate estimated allelic dropout rates without explicitly incorporating population structure in our model.
We applied the imputation procedure to correct the bias in estimating heterozygosity for each of the 96 simulated data sets. Similarly to our application in experiment 1, we calculated the uncorrected and true heterozygosities for each individual from the simulated observed genotypes and the simulated true genotypes , respectively. The corrected heterozygosity was averaged across 100 imputed data sets for each simulated data set. Results for 36 simulated data sets appear in Figure S2, in which heterozygosities were averaged across all individuals in each data set. Our results show a significant improvement of the corrected heterozygosity over the uncorrected heterozygosity in all simulations, in that the corrected heterozygosity is considerably closer to the true heterozygosity. This improvement is fairly robust to the presence of population structure.
Experiment 3. Data with other genotyping errors:
This set of simulations tested our method at different levels of genotyping error from sources other than allelic dropout. In all simulated data sets, with genotyping error ranging from 0 to 10% and ρ ranging from 0 to 0.15, our method is successful in estimating both sample-specific and locus-specific dropout rates (Figure 9, A and C). The estimation accuracy of dropout rates is not strongly affected by the genotyping error rate (Figure 9, B and D). We can again see that a smaller number of individuals than loci has led to higher mean squared error for estimated locus-specific rates (Figure 9D) than for sample-specific rates (Figure 9B).
Figure 9 .
Estimated dropout rates and inbreeding coefficients for simulated data with other genotyping errors. (A) Comparison of the estimated sample-specific dropout rates and the assumed true sample-specific dropout rates. (B) Mean squared errors across all the estimated sample-specific dropout rates for each of the 36 data sets shown in A. (C) Comparison of the estimated locus-specific dropout rates and the assumed true locus-specific dropout rates. (D) Mean squared errors across all the estimated locus-specific dropout rates for each of the 36 data sets shown in C. (E) Comparison of the estimated inbreeding coefficient and the assumed true inbreeding coefficient, in which each point corresponds to one of 96 simulated data sets. The 36 solid symbols correspond to the simulated data sets shown in A–D and F. (F) Overestimation of the inbreeding coefficient, calculated by subtracting the assumed true inbreeding coefficient from the estimated inbreeding coefficient, or .
Similar to the F = 0 case in our simulations with population structure, the simulated data sets with no genotyping error (e = 0) are generated under model assumptions 2–5. Consistent with the results for F = 0, our method slightly underestimates the inbreeding coefficient ρ for most simulated data sets with e = 0. As genotyping error increases, the underestimation also increases (Figure 9, E and F). This result can be explained by noting that the simulated genotyping error, which changes the allele frequencies only slightly, tends to create false heterozygotes more frequently than false homozygotes. Therefore, the observed heterozygosity is increased while the expected heterozygosity changes little, leading to a decrease in the estimated inbreeding coefficient. Although our estimation of the inbreeding coefficient ρ becomes less accurate when the genotyping error rate is higher, the underestimation of ρ does not prevent the method from accurately estimating allelic dropout rates.
For the heterozygosity, the corrected values obtained using our imputation strategy are closer to the true values than are the uncorrected values directly obtained from the observed genotypes (Figure S3). However, as the genotyping error rate e increases, our method starts to overcorrect the downward bias in estimating the observed heterozygosity, and the corrected values exceed the true values. Similar to our explanation for the underestimation of the inbreeding coefficient, this overcorrection is introduced by the simulated genotyping error, which creates an excess of false heterozygotes. This excess is in turn incorporated into the corrected estimates of heterozygosity, because we do not model genotyping errors other than those due to allelic dropout.
Discussion
In this study, we have developed a maximum-likelihood approach to jointly estimate sample-specific dropout rates, locus-specific dropout rates, allele frequencies, and the inbreeding coefficient from only one nonreplicated set of microsatellite genotypes. Our algorithm can accurately recover the allelic dropout parameters, and an imputation strategy using the method provides an alternative to ignoring high empirical missing data rates or excluding samples and loci with large amounts of missing data. Investigators can then use the imputed data in subsequent analyses, such as in studies of genetic diversity or population structure, or in software that disallows missing values in the input data. We have demonstrated our approach using extensive analyses of an empirical data set and data sets simulated using parameter values chosen on the basis of the empirical example.
We have found that our method works well on simulated data. In particular, it performs well in estimating the sample-specific dropout rates γi⋅ and locus-specific dropout rates γ⋅ℓ. Further, in the examples we have considered, it is reasonably robust to violations of the model assumptions owing to the existence of population structure or to sources of genotyping error other than allelic dropout. This robustness arises partly from the inclusion of the inbreeding coefficient ρ in our model, which enables us to capture the deviation from HWE caused by multiple factors, such as true inbreeding, population structure, and genotyping errors. Because the various sources of deviation from HWE are incorporated into the single parameter ρ, the estimation of ρ itself is more sensitive to violation of model assumptions; therefore, it is important to be careful when interpreting the estimated value of ρ, as it may reflect phenomena other than inbreeding. When data are simulated under our model, such as in the cases of F = 0 and e = 0, our method tends to slightly underestimate ρ (Figures 8E and 9E), indicating that our MLEs are biased, at least for the inbreeding coefficient.
We can use simulation approaches to further explore the statistical properties of our estimates. To examine the consistency of the estimators, we performed two additional sets of simulations, in which we generated genotype data under our model with either different numbers of individuals N or different numbers of loci L (Appendix C). When L is fixed, although estimates of the sample-specific dropout rates γi⋅ are not affected by the value of N, our estimates of the locus-specific dropout rates γ⋅ℓ and the inbreeding coefficient ρ become closer to the true values as N increases (Figure S4). When N is sufficiently large (e.g., N = 1600), the estimates of γ⋅ℓ and ρ are almost identical to the true values. If we instead fix N and increase L, then the estimates of γi⋅ and ρ eventually approach the true values, while the estimates of γ⋅ℓ remain unaffected (Figure S5). These results suggest, without a strict analytical proof, that our MLEs of the dropout rates and inbreeding coefficient are likely to be consistent.
For the Native American data, we can compare the estimated heterozygosities under our model with other data on similar populations. Wang et al. (2007) studied microsatellites in 29 Native American populations, including 8 populations from regions that overlap those considered in our data. We reanalyzed these populations, 3 from Canada and 5 from Mexico, by calculating observed heterozygosity Ho from the same 343 loci as were genotyped in our data. We obtained a mean Ho of 0.670 with standard deviation 0.051 across 176 individuals in the pooled set of 8 populations. In comparison, mean Ho across our 152 Native American samples is 0.590 (standard deviation = 0.137) before correcting for allelic dropout, substantially lower than in Wang et al. (2007), and it is 0.730 (standard deviation = 0.035) after correcting for allelic dropout, higher than in Wang et al. (2007). Several possible reasons can explain the imperfect agreement between our corrected heterozygosity and the estimate based on the Wang et al. (2007) data. First, the sets of populations might differ in such factors as the extent of European admixture, so that they might truly differ in underlying heterozygosity. Second, the Wang et al. (2007) data might have some allelic dropout as well, so that our Ho estimates from those data underestimate the true values. Third, our method might have overcorrected the underestimation of Ho; our simulations show that because we do not model genotyping errors from sources other than allelic dropout, the existence of such errors can lead to overestimation of Ho (Figure S3). It is also possible that missing genotypes caused by factors other than allelic dropout could have been erroneously attributed to allelic dropout, leading to overestimation of dropout rates and hence to overcorrection of Ho.
Our model assumes that all individuals are sampled from the same population with one set of allele frequencies and that inbreeding is constant across individuals and loci. We applied this assumption to the whole Native American data set as an approximation. However, evidence of population structure can be found by applying multidimensional scaling analysis to the Native American samples. As shown in Figure S6, individuals from different populations tend to form different clusters, indicating that underlying allele frequencies and levels of inbreeding differ among populations. Although our simulations have found that estimation of allelic dropout rates is robust to the existence of population structure, estimation of allele frequencies and the inbreeding coefficient can become less accurate in structured populations. It would therefore have been preferable in our analysis to apply our method on each population instead of on the pooled data set; however, such an approach was impractical owing to the small sample sizes in individual populations. To address this problem, it might be possible to directly incorporate population structure into our model (e.g., Falush et al. 2003), thereby enabling allele frequencies and inbreeding coefficients to differ across the subpopulations in a structured data set. Further, because samples from the same population are typically collected and genotyped as a group, full modeling of the population structure might allow for a correlation in dropout rates across individuals within a population.
An additional limitation of our approach is that during data analysis, we do not take into account the uncertainty inherent in estimating parameters. We first obtain the MLEs of allele frequencies , allelic dropout rates , and the inbreeding coefficient and then create imputed data sets by drawing genotypes using , , and . This procedure is “improper” because it does not propagate the uncertainty inherent in parameter estimation (Little and Rubin 2002). To obtain “proper” estimates, instead of using an EM algorithm to find the MLEs of the parameters, we could potentially use a Gibbs sampler or other Bayesian sampling methods to sample parameter values and then create imputed data sets using these sampled parameter sets. In such approaches, parameters sampled from their underlying distributions would be used for different imputations, instead of using the same MLEs for all imputations.
Finally, we have not compared our approach with methods that rely on replicate genotypes. While we expect that replicate genotypes will usually lead to more accurate estimates of model parameters, our method provides a general approach that is relatively flexible and accurate in the case that replicates cannot be obtained. Compared with existing models that assume HWE (e.g., Miller et al. 2002; Johnson and Haydon 2007), our model uses a more general assumption of inbreeding, and we also incorporate both sample-specific and locus-specific dropout rates. The general model increases the applicability of our method for analyzing diverse genotype data sets, such as those that have significant dropout caused by locus-specific factors (e.g., Buchan et al. 2005). It is worth noting that HWE is the special case of ρ = 0 in our inbreeding model; when it is sensible to assume HWE, we can simply initiate the EM algorithm with a value of ρ = 0. This choice restricts the search for MLEs to the ρ = 0 parameter subspace, because Equation 22 stays fixed at 0 in each EM iteration. Similarly, if we prefer to consider only sample-specific dropout rates (or only locus-specific dropout rates), then we can simply set the initial values of γ⋅ℓ to 0 for all loci (or initial values of γi⋅ to 0 for all individuals). These choices also restrict the search to subspaces of the full parameter space. We have implemented these options in our software program MicroDrop, which provides flexibility for users to analyze their data with a variety of different assumptions.
Supplementary Material
Acknowledgments
We thank Roderick Little for helpful advice, Michael DeGiorgio for comments on a draft of the manuscript, and Zachary Szpiech for help in evaluating and testing the MicroDrop software. We also thank two anonymous reviewers for constructive comments that have led to substantial improvement of this article. This work was supported by National Institutes of Health grants R01-GM081441 and R01-HG005855, by the Burroughs Wellcome Fund, and by a Howard Hughes Medical Institute International Student Research Fellowship.
Appendix A: The EM Algorithm
The main text describes an EM algorithm for estimating parameters in our model. Here, we provide the derivation of Equations 19–22 for parameter updates in each EM iteration. We start from a general description of the EM algorithm (e.g., Casella and Berger 2001; Lange 2002).
To obtain the MLEs, our goal is to maximize the likelihood ℒ = ℙ(W|Ψ). Because ℒ is difficult to maximize directly, we use an EM algorithm to replace the maximization of ℒ with a series of simpler maximizations. We introduce three sets of latent variables: the true genotypes G, IBD states S, and dropout states Z, each representing an N × L matrix. Instead of directly working on likelihood ℒ, the EM algorithm starts with a set of initial values arbitrarily chosen for Ψ and, in each of a series of iterations, maximizes the Q function defined by Equation A1. This iterative maximization is easier and sequentially increases the value of ℒ (e.g., Lange 2002), so that the parameters eventually converge to values at a maximum of ℒ.
In the E-step of iteration t + 1, we want to calculate the following expectation:
| (A1) |
This computation is equivalent to calculating [G|W, Ψ(t)], [S|W, Ψ(t)], and [Z|W, Ψ(t)] and then inserting these quantities into the expression for ln ℙ(W, G, S, Z|Ψ), such that Equation A1 is a function of parameters Ψ = {Φ, Γ, ρ}. In the M-step, the parameters are updated with values Ψ(t+1) that maximize Equation A1. The explicit expression for Equation A1 is cumbersome, but given the dependency described in Figure 3, we can greatly simplify our EM algorithm by a decomposition of ℙ(W, G, S, Z|Ψ):
| (A2) |
Equation A2 implies that we can maximize and separately to maximize Q(Ψ|Ψ(t)) (Equation A1). Further, it can be shown that nℓk, di⋅, d⋅ℓ, and s are sufficient statistics for φℓk, γi⋅, γ⋅ℓ, and ρ, respectively. Therefore, in the E-step, we can simply calculate the expectations of these four sets of statistics (Equations 14–17) rather than evaluating the full matrices [G|W, Ψ], [S|W, Ψ], [Z|W, Ψ].
In the M-step, the dropout rates Γ are updated by maximizing , resulting in Equations 20 and 21, quantities that can be obtained intuitively by considering each dropout as an independent Bernoulli trial. The allele frequencies Φ and the inbreeding coefficient ρ are updated by maximizing , resulting in Equations 19 and 22 after some algebra. As an example, we show the derivation of Equations 19 and 22 for a single biallelic locus (L = 1, Kℓ = 2).
Denote the alleles by A1 and A2 and the corresponding allele frequencies by φ1 and φ2, with φ1 + φ2 = 1. Suppose that in the whole data set, xhk,u individuals have true genotype AhAk (1 ≤ h ≤ k ≤ 2) and IBD state u (u = 0 or 1). Then ℙ(G, S|Φ, ρ) can be written as
| (A3) |
in which s is the total number of genotypes that are IBD (u = 1), and n1 and n2 are the numbers of independent copies for alleles A1 and A2, respectively. We can see from Equation A3 that s is a sufficient statistic for ρ, and n1 and n2 are sufficient statistics for Φ. Following Equation A3, can be expressed as
| (A4) |
in which c = [x12,0|W, Ψ(t)]ln 2 is a constant with respect to parameters ρ and Φ. To maximize , we can solve the following equations:
| (A5) |
| (A6) |
The solutions for the case of L = 1 and Kℓ = 2 agree with Equations 19 and 22:
| (A7) |
| (A8) |
Appendix B: Inbreeding and the F Model
In the presence of population structure, the proportion of homozygotes in the pooled population exceeds that of an unstructured population, leading to a deviation from Hardy–Weinberg equilibrium similar to inbreeding. Therefore, we expect our algorithm to overestimate the inbreeding coefficient when population structure in the genotype data is not taken into account for the estimation. In this section, we derive an expression for this overestimation in a structured population under the F model (Falush et al. 2003). We show that a structured population with two subpopulations, whose inbreeding coefficients are ρ1 and ρ2, has expected allele and genotype frequencies identical to those of an unstructured population with a certain inbreeding coefficient ρ* higher than ρ1 and ρ2.
Consider a structured population with N1 = c1N and N2 = c2N = (1 − c1)N individuals sampled from subpopulations 1 and 2, respectively (Figure S7). Without loss of generality, we examine only a single locus with K alleles. Under the F model, the allele frequencies of subpopulation j (j = 1, 2), Φj = {φj1, … , φjK}, follow a Dirichlet distribution , in which ΦA = {φA1, … , φAK} denotes the allele frequencies of a common ancestral population of the two subpopulations and Fj measures the divergence of subpopulation j from the ancestral population. We need the first and second moments of the allele frequencies Φj, quantities that can be obtained from the mean, variance, and covariance of a Dirichlet distribution. For h ≠ k,
| (B1) |
| (B2) |
| (B3) |
Suppose the two subpopulations have inbreeding coefficients ρ1 and ρ2, respectively. Under the inbreeding model (e.g., Holsinger and Weir 2009), the frequency of genotype AkAh in subpopulation j can be written as
| (B4) |
Using Equations B1–B4, in the structured population, homozygote AkAk has expected genotype frequency
| (B5) |
Similarly, the expected genotype frequency of heterozygote AkAh (h ≠ k) is
| (B6) |
We now search for the value of ρ* at which genotype frequencies in an unstructured population satisfy Equations B5 and B6. If we are unaware of the population structure, then the allele frequencies in the pooled population are
| (B7) |
Our goal is to derive an inbreeding coefficient ρ* for an unstructured population with allele frequencies Φ*, such that expected genotype frequencies of an unstructured population with inbreeding are identical to those of the structured population (Equations B5 and B6).
The expected genotype frequency of a homozygote AkAk in an unstructured population with an inbreeding coefficient ρ* can be written as
| (B8) |
For a heterozygote AkAh (h ≠ k), the expected genotype frequency is
| (B9) |
Comparing Equations B5 and B6 and Equations B8 and B9, the genotype frequencies in the two scenarios agree if
| (B10) |
In summary, under the F model, for both homozygotes and heterozygotes, the expected genotype frequencies in a structured population are identical to those in an unstructured population with allele frequencies Φ* (Equation B7) and inbreeding coefficient ρ* (Equation B10). For testing the robustness of our method for allelic dropout, we simulated genotype data with population structure using c1 = c2 = 0.5, F1 = F2 = F, and ρ1 = ρ2 = ρ (experiment 2). In this setting, Equation B10 reduces to
| (B11) |
The values of Equation B11 for our simulated data sets are indicated by dashed lines in Figure 8.
Appendix C: Additional Simulation Procedures
To assess the performance of our method as a function of the size of the data set, we performed two additional sets of simulations. In one, we fixed the number of loci and modified the number of individuals, and in the other, we fixed the number of individuals and modified the number of loci.
Experiment C1. Simulating data with different numbers of individuals
We used a similar procedure to that shown in Figure 5A, following assumptions 2–5 of our model. We fixed the number of loci at L = 250. This value is chosen to be between 152 (the number of individuals in the Native American data) and 343 (the number of loci in the data). The numbers of individuals were chosen to be N = 50, 100, 200, 400, 800, and 1600. For each pair consisting of a choice of N and L, we simulated data sets with the inbreeding coefficient ρ ranging from 0 to 0.15 in increments of 0.01. Therefore, we generated 6 × 16 = 96 simulated data sets.
For each simulated data set, the allele frequencies Φ at L loci were independently sampled (with replacement) from the estimated allele frequencies of the 343 loci in the Native American data (results from Application to Native American Data). Given the allele frequencies Φ and the inbreeding coefficient ρ, true genotypes were drawn according to the inbreeding assumption. Next, the observed genotype data were created by adding allelic dropout. The sample-specific dropout rates γi⋅ and the locus-specific dropout rates γ⋅ℓ were both independently sampled from Beta(0.55, 5.30), as in experiments 2 and 3 in the main text.
Experiment C2. Simulating data with different numbers of loci
The procedure we used to simulate data with different numbers of loci was similar to that in experiment C1, except that we fixed the number of individuals at N = 250 and varied the number of loci (L = 50, 100, 200, 400, 800, and 1600). Therefore, we generated 96 simulated data sets, each of which has the same amount of data as a corresponding data set generated by experiment C1.
Footnotes
Communicating editor: Y. S. Song
Literature Cited
- Bonin A., Bellemain E., Eidesen P. B., Pompanon F., Brochmann C., et al. , 2004. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 13: 3261–3273 [DOI] [PubMed] [Google Scholar]
- Broquet T., Petit E., 2004. Quantifying genotyping errors in noninvasive population genetics. Mol. Ecol. 13: 3601–3608 [DOI] [PubMed] [Google Scholar]
- Broquet T., Ménard N., Petit E., 2007. Noninvasive population genetics: a review of sample source, diet, fragment length and microsatellite motif effects on amplification success and genotyping error rates. Conserv. Genet. 8: 249–260 [Google Scholar]
- Buchan J. C., Archie E. A., van Horn R. C., Moss C. J., Alberts S. C., 2005. Locus effects and sources of error in noninvasive genotyping. Mol. Ecol. Notes 5: 680–683 [Google Scholar]
- Casella G., Berger R. L., 2001. Statistical Inference, Ed. 2 Duxbury, Pacific Grove, CA [Google Scholar]
- Dakin E., Avise J. C., 2004. Microsatellite null alleles in parentage analysis. Heredity 93: 504–509 [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P., Boesch C., Woodruff D. S., 1997. Microsatellite scoring errors associated with noninvasive genotyping based on nuclear DNA amplified from shed hair. Mol. Ecol. 6: 861–868 [DOI] [PubMed] [Google Scholar]
- Hadfield J. D., Richardson D. S., Burke T., 2006. Towards unbiased parentage assignment: combining genetic, behavioural and spatial data in a Bayesian framework. Mol. Ecol. 15: 3715–3730 [DOI] [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., 1997. Principles of Population Genetics, Ed. 3 Sinauer Associates, Sunderland, MA [Google Scholar]
- Hoffman J. I., Amos W., 2005. Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol. Ecol. 14: 599–612 [DOI] [PubMed] [Google Scholar]
- Holsinger K. E., Weir B. S., 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. C. D., Haydon D. T., 2007. Maximum-likelihood estimation of allelic dropout and false allele error rates from microsatellite genotypes in the absence of reference data. Genetics 175: 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K., 2002. Mathematical and Statistical Methods for Genetic Analysis, Ed. 2 Springer-Verlag, New York [Google Scholar]
- Little R. J. A., Rubin D. B., 2002. Statistical Analysis with Missing Data, Ed. 2 John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Miller C. R., Joyce P., Waits L. P., 2002. Assessing allelic dropout and genotype reliability using maximum likelihood. Genetics 160: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P. A., Chambers K. E., Boesch C., Vigilant L., 2001. Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Mol. Ecol. 10: 1835–1844 [DOI] [PubMed] [Google Scholar]
- Navidi W., Arnheim N., Waterman M. S., 1992. A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am. J. Hum. Genet. 50: 347–359 [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M., Slate J., Bancroft D. R., Barrett J. A., 1995. Nonamplifying alleles at microsatellite loci: a caution for parentage and population studies. Mol. Ecol. 4: 249–252 [DOI] [PubMed] [Google Scholar]
- Pompanon F., Bonin A., Bellemain E., Taberlet P., 2005. Genotyping errors: causes, consequences and solutions. Nat. Rev. Genet. 6: 847–859 [DOI] [PubMed] [Google Scholar]
- Sefc K. M., Payne R. B., Sorenson M. D., 2003. Microsatellite amplification from museum feather samples: effects of fragment size and template concentration on genotyping errors. Auk 120: 982–989 [Google Scholar]
- Taberlet P., Luikart G., 1999. Non-invasive genetic sampling and individual identification. Biol. J. Linn. Soc. 68: 41–55 [Google Scholar]
- Taberlet P., Griffin S., Goossens B., Questiau S., Manceau V., et al. , 1996. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 24: 3189–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P., Waits L. P., Luikart G., 1999. Noninvasive genetic sampling: look before you leap. Trends Ecol. Evol. 14: 323–327 [DOI] [PubMed] [Google Scholar]
- Wang J., 2004. Sibship reconstruction from genetic data with typing errors. Genetics 166: 1963–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Lewis C. M., Jr, Jakobsson M., Ramachandran S., Ray N., et al. , 2007. Genetic variation and population structure in Native Americans. PLoS Genet. 3: 2049–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasser S. K., Mailand C., Booth R., Mutayoba B., Kisamo E., et al. , 2007. Using DNA to track the origin of the largest ivory seizure since the 1989 trade ban. Proc. Natl. Acad. Sci. USA 104: 4228–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. A., Barker R. J., Schofield M. R., Frantz A. C., Byrom A. E., et al. , 2009. Incorporating genotype uncertainty into mark-recapture-type models for estimating abundance using DNA samples. Biometrics 65: 833–840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.