Abstract
Bacillus cereus G9241 was isolated from a welder suffering from an anthrax-like inhalation illness. B. cereus G9241 encodes two megaplasmids, pBCXO1 and pBC210, which are analogous to the toxin- and capsule-encoding virulence plasmids of B. anthracis. Protein modeling predicted that the pBC210 LF homolog contained an ADP-ribosyltransferase (ADPr) domain. This putative bacterial ADP-ribosyltransferase domain was denoted CerADPr. Iterative modeling showed that CerADPr possessed several conserved ADP-ribosyltransferase features, including an α-3 helix, an ADP-ribosyltransferase turn-turn loop, and a “Gln-XXX-Glu” motif. CerADPr ADP-ribosylated a ~120kDa protein in HeLa cell lysates and intact cells. EGFP-CerADPr rounded HeLa cells, elicited cytoskeletal changes, and yielded a cytotoxic phenotype, indicating that CerADPr disrupts cytoskeletal signaling. CerADPr(E431D) did not possess ADP-ribosyltransferase or NAD glycohydrolase activities and did not elicit a phenotype in HeLa cells, implicating Glu431 as a catalytic residue. These experiments identify CerADPr as a cytotoxic ADP-ribosyltransferase that disrupts the host cytoskeleton.
Bacterial ADP-ribosylating exotoxins (bAREs) catalyze the transfer of ADP-ribose from NAD to a target residue within a eukaryotic protein. This modification is primarily used as a mechanism of virulence by inactivating (or activating) host proteins, often leading to cell death. bAREs are usually categorized into one of four major groups based on their eukaryotic substrates [1]: Cholera toxin (CT)-like toxins modify heterotrimeric G-proteins [2, 3], diphtheria toxin (DT)-like toxins modify Elongation factor 2 [4, 5], C3-like exoenzymes modify small GTPases [6–8], and the vegetative insecticidal protein-like (VIP) binary toxins modify actin [9–12]. C3 and VIP2 proteins share ADPr domain organization and structural conservation, despite different substrate recognition [13–15].
While limited amino acid sequence identity exists between the VIP2 and C3 transferases, there is strict conservation of critical residues required for ADP-ribosyltransferase activity. These residues have been designated the “RSE motif”, and consist of three groups of residues (Arg), (Ser-Thr-Ser), and (Gln/Glu-XXX-Glu) located in discrete regions of the protein [16]. Together, these three motifs form the NAD binding pocket responsible for NAD hydrolysis and transfer of the ADP-ribose moiety onto the substrate acceptor residue. Disruption of these residues, especially the invariant catalytic glutamic acid in the Gln/Glu-XXX-Glu motif results in a non-functional enzyme. The primary structural elements of bARE ADP-ribosyltransferase domains have been designated A–F [17]. Region A is the first N-terminal alpha-helix, and is responsible for recognition of substrate by some bAREs, including ExoT recognition of CrkI/II [17]. Region B, the α-3 helix, is responsible for stabilization of the active site glutamic acid and NAD and positioning of the target residue. Region C, the ADP-ribosyltransferase-turn-turn (ARTT) loop stabilizes the target residue and binds the N-ribose of NAD to create a conformation ideal for hydrolysis. Region E, the phosphate-nicotinamide loop, contains the conserved Ser-Thr-Ser motif and is responsible for positioning the catalytic Glu for proper NAD cleavage. Regions B, C, and E have also been implicated in bARE-substrate interactions [13,14,17,18].
Recently, an in silico analysis was performed on known bacterial protein coding sequences to identify new bAREs through recognition of these conserved protein folding motifs [19]. This approach identified six new putative bAREs from diverse species including Vibrio cholerae, Enterococcus faecalis, Photorhabdus luminescens, and Bacillus cereus. One putative toxin was identified from the genome of a clinical isolate of Bacillus cereus (G9241). B. cereus is a Gram-positive spore-forming organism that is widely distributed in the environment and is the prototype member of the B. cereus group, which includes B. thuringiensis and B. anthracis. The B. cereus group share high genomic similarity; however, the disease phenotypes associated with the different species are widely varied. B. cereus infection is primarily associated with food-borne illness and opportunistic infection of immunocompromised individuals, while B. anthracis is the etiologic agent of anthrax, a disease that can be fatal to humans in cases of pulmonary or gastrointestinal infection.
Anthrax virulence is modulated by both encapsulation of the bacterium within an anti-phagocytic poly-D-glutamate capsule and expression of anthrax toxin, a complex of three proteins: protective antigen (PA), lethal factor (LF), and edema factor (EF) [20]. PA binds to a host receptor [21–23], is processed by proteases [24], and self-associates into heptamers [25, 26] or octamers [27, 28]. Multiple copies of LF, a potent MAPKK zinc metalloprotease [29, 30], or EF, an adenylate cyclase [31, 32], bind PA and enter through clathrin-mediated endocytosis [33]. Upon endosome acidification [34, 35], LF and EF cross the endosome membrane through the oligomeric PA pore and modify cellular signaling [36].
Of recent concern, environmental B. cereus isolates that are responsible for serious illness and death have been isolated from otherwise healthy individuals, primarily welders and metalworkers [37,38]. Several B. cereus strains, including B. cereus G9241, have been implicated in a pulmonary anthrax-like disease, resulting in significant morbidity or death. Many of these B. cereus strains contain a homolog to B. anthracis pXO1, which encodes the anthrax toxin genes [39]. B. cereus G9241 contains two large plasmids, pBCXO1 (191 kb), a pXO1 homolog, and pBC210 (210 kb). pBCXO1 encodes the three subunits of B. anthracis anthrax toxin, consisting of lef (lethal factor; LF-99% identity to B. anthracis LF), cya (edema factor; EF-96% identity), and pag (protective antigen; PA-98% identity), which are expressed in vitro [40]. pBC210 encodes additional copies of pag (60% identity) and lef (36% identity), as well as genes encoding the machinery required to create a polysaccharide capsule. Sequence analysis of pBC210 lef shows the presence of a putative PA binding domain but no coding sequence for the LF MAPKK metalloprotease domain (Supplementary Figure 1). Instead, a “VIP2-like domain” which comprises a putative ADP-ribosyltransferase domain with sequence and structural homology to the binary ADP-ribosylating toxins is present [19]. The pBC210 lef gene product was originally denoted “Certhrax” due to its sequence similarity to anthrax LF; however, we have chosen “Cereus toxin” to describe the full-length protein to remove any confusion with anthrax and lethal factor, while CerADPr will be used to denote the active ADP-ribosyltransferase domain. Iterative modeling of the crystal structure of CerADPr shows remarkable structural similarity to the LF PA binding domain and “VIP2-like” regions, indicating that they may share a conserved structure-function. However, LF contains none of the conserved bacterial ADP-ribosyltransferase residues in the “VIP2-like domain”, which are present in CerADPr (Supplementary Figure 1).
Iterative BLAST analyses with the coding sequence of Cereus toxin (residues 1–476) returned high-scoring matches with multiple members of the VIP2-like and C3-like ADP-ribosyltransferases, including VIP2, Iota toxin, C3bot, and C3Cer. Sequence alignment of these bacterial ADP-ribosyltransferases show very limited conservation of the N-terminal binding domain of Cereus toxin with the binding domains of Iota toxin and VIP2, while the ADP-ribosyltransferase domains of the five proteins show higher levels of conservation, with the “RSE” motif completely conserved (Supplementary Figure 2). Iterative structural modeling of CerADPr using Iota toxin as a template resulted in a model with RMSD of 2.8Å for 170 Cα atoms. However, CerADPr contains an active site Gln-XXX-Glu motif, which is associated with C3 exoenzyme modification of Rho at Asn41 [41]. These similarities prompted the analysis of Cereus toxin as a novel ADP-ribosyltransferase.
Experimental Procedures
Plasmid vectors and mutagenesis
The gene encoding the ADP-ribosyltransferase domain of Cereus toxin (residues 226–476; predicted MW: 29,451 Da, termed CerADPr) was amplified and subcloned into pET15b (pET-CerADPr) (Novagen) and pEGFP-C3 (pEGFP-CerADPr) (Clontech). Site-directed mutagenesis generating an E431D mutation within CerADPr was performed using Quikchange Site-directed Mutagenesis (Agilent Technologies) with the following primers: (+ strand) 5’-GAATATCCAGGGCAATATGACATGTTAATAAATAG-3’and (−strand) 5’-CTATTTATTAACATGTCATATTGCCCTGGATATTC-3’. Plasmids were transformed into Escherichia coli (TG1) and DNA sequence was confirmed.
Expression and purification of recombinant proteins
pET15-CerADPr and pET-CerADPr(E431D) were transformed into E. coli BL21(DE3). Cells were plated overnight at 37°C with 250 µg / ml of ampicillin. Cells were added to 400 ml of LB broth (Miller salts, BD Biosciences) with ampicillin and cultured for 2 hr at 30°C when 1 mM IPTG was added and the cells were cultured overnight at 16°C. Cells were harvested, suspended in binding buffer (20 mM Tris-HCl (pH 7.9), 500 mM NaCl, and 5 mM imidazole) with bacterial protease inhibitor cocktail (Sigma), RNase (20 µg/ml), and DNase (20 µg/ml) and broken by French Press. The lysate was centrifuged (30,000 × g for 20 min) and the soluble fraction passed through a 0.45Sm filter. The filtrate was subjected to Ni2+ affinity resin chromatography (Qiagen). His(6)-fusion proteins were eluted in binding buffer with 250 mM imidazole. The eluate was subjected to size-exclusion fractionation by Sephacryl S200 (Sigma) in 10 mM Tris-HCl (pH 7.6) and 20 mM NaCl. Peak fractions were dialyzed into 10 mM Tris-HCl (pH 7.6), 20mM NaCl, and 40% glycerol, and stored at −80°C. A typical purification yielded ~4 mg CerADPr / L of culture. His(6)-ExoS(78–453) was purified as previously described [42].
HeLa cell harvesting and fractionation
HeLa cells (ATCC CCL-2) were cultured in MEM (Life Technologies) supplemented with 10% fetal calf serum, non-essential amino acids, sodium pyruvate, sodium bicarbonate and penicillin/streptomycin and maintained humidified at 37°C in 5% CO2. Confluent plates (150 mm) of HeLa cells were washed (PBS) and scraped into 1 ml HB buffer (200 mM sucrose, 3mM imidazole, mammalian protease inhibitor cocktail (Sigma). Cells were broken by 25–30 passes through a 26 gauge needle, and then centrifuged at 1000 × g to pellet unbroken cells and nuclei. The supernatant (Total lysate) was centrifuged at 100,000 × g in a TLA100.3 rotor (Beckman) for 1.5 hr at 4°C. The supernatant (Soluble) was stored at −80°C. The pelleted membranes were suspended in PBS + 0.1% TritonX-100 (TX-soluble) and stored at −80°C. Protein concentrations in each fraction (Total lysate, Soluble, TX-soluble) were determined by BCA assay (Pierce).
In vitro ADP-ribosylation assay
Reaction mixtures (50µl) contained: 10mM Tris-HCl (pH 7.6), 20 mM NaCl, 50 µg of HeLa lysate, 4 µM 6-biotin-17-NAD (Trevigen), and the indicated amount of CerADPr or ExoS. Reactions were incubated at room temperature (RT) for the indicated time and stopped with Laemmli buffer and boiling. Reactions were subjected to SDS-PAGE and transferred to a polyvinylidene fluoride membrane (PVDF, Millipore) for immunoblot analysis. (Detection of biotin-ADP-ribose) PVDF membranes were blocked for 30 min in PBS + 2% BSA and then incubated with Streptavidin-HRP conjugate (1:80,000 final dilution; Pierce, #21130) for 1 hr at RT. Luminescence from each lane was measured, with the most intense lane (ExoS-50 ng) set to 100%. An unpaired t-test analysis was performed on columns with GraphPad Prism 5 software (GraphPad Software Inc.). * p<0.05, ** p<0.01 (Detection of host proteins in the HeLa cell lysate) PVDF membranes were blocked for 30 min in TBST + 2% milk. Membranes were incubated with α-GFP IgG (mouse, 1:2000; Covance), α-β-actin IgG (mouse, 1:5000; Sigma) or α-tubulin antibody (rabbit, 1:5000; Abcam). Primary antibodies were detected with either goat α-mouse-HRP IgG (1:30000; Pierce) or goat α-rabbit-HRP IgG (1:30000; Pierce). Membranes were probed with Pico Super Signal (Pierce) and luminescence measured with a CCD camera and AlphaView SA software (Cell Biosciences Inc.). Images were cropped in Photoshop (Adobe). Quantification of luminescence was performed with AlphaView software from a minimum of four independent experiments.
NAD glycohydrolase assay
Adapted from a thin-layer chromatography protocol in [43] 1.6 SM recombinant CerADPr, CerADPrE431D, or ExoS was incubated with 2.5 mM NAD (Sigma) for 0–48 hr at 37°C. After the indicated time point, the reaction was spotted onto poly(ethyleneimine)-cellulose (PEI-cellulose) TLC plates (Selecto Scientific) and resolved with 0.4 M LiCl. TLC plates were visualized under UV light and imaged with a CCD camera and AlphaView SA software (Cell Biosciences Inc.). Quantification of ADP-ribose generated from NAD was performed with AlphaView software from three independent experiments and normalized to a standard of ADP-ribose. An unpaired t-test was performed to determine statistical significance. * p<0.05, ** p<0.01
Cell culture and transfection
HeLa cells were cultured to ~70% confluence in 24 well plates and transfected with pEGFP-CerADPr, pEGFP-CerADPr(E431D), or pEGFP using Lipofectamine 2000 (Life Technologies) according to manufacturer protocol. At the indicated times, cells were washed twice with PBS and processed for immunofluorescence as described below. For the analysis of HeLa cell lysates, cells were cultured to ~70% confluence in 6 well plates and transfected with the indicated plasmid in Lipofectamine 2000 for either 5 or 20 hr, scraped into 1mL HB, and lysed by 25–30 passages through a 26 gauge needle. Cell lysates were incubated with 25 µg of TX-soluble protein and 4 µM 6-biotin-17-NAD for 1 hr at RT to assay for CerADPr activity, or the lysates were immediately resolved by SDS-PAGE and transferred to PVDF membranes for immunoblot analysis. Cytotoxic potential of CerADPr was measured in HeLa cells using a trypan blue exclusion assay [44], with ExoS as a positive control, pExoS (RhoGAP−/ADPr+). Cells were transfected according to manufacturer protocol, and at 5 and 20 hr post-transfection (HPT), media was removed, cells were washed and stained for 5 min with 0.4% trypan blue (Life Technologies). Trypan blue was removed, cells were washed in low serum media, and color images were obtained using a 10× objective and Retiga-2000R CCD camera (QImaging) with NIS Elements software (Nikon). Cytotoxicity was determined by counting the number of trypan blue positive HeLa cells / total number of HeLa cells per field. Five random fields were assayed and the results of this determination shown. Three biological replicates were performed for each time point. A 1-way ANOVA analysis was performed in GraphPad Prism 5 software (GraphPad Software Inc.). ** p<0.01
HeLa cell permeabilization and detection of intracellular ADP-ribosylation
Protocol was adapted from [45]. HeLa cells, transfected or non-transfected cells, were washed in 1 ml of ice cold HGI buffer (20 mM PIPES, 2 mM Na-ATP, 4.8 mM magnesium acetate, 0.15 M potassium glutamate, 2 mM EGTA, potassium hydroxide adjusted to pH 7.0), then incubated for 15 min at 4°C with 200 ng/ml of tetanolysin (Santa Cruz Biotechnology) diluted in HGI. Cells were then incubated with HGI containing either 4 µM biotin-NAD (transfected cells) or 3 Sg/ml of recombinant protein (non-transfected cells) for 30 min at 37°C. The buffer was then removed and fresh serum-containing media added to seal membrane pores for two hours. After incubation with biotin-NAD, transfected cells were lysed in RIPA buffer and Laemmli buffer was added. Lysate from cells incubated with recombinant protein was incubated with 50 ng of CerADPr and 4 µM biotin-NAD for an additional hour, followed by addition of Laemmli buffer and boiling. Samples were resolved by SDS-PAGE and subjected to immunoblot analysis.
Immunofluorescence microscopy
HeLa cells were washed twice in PBS + MgCl2 + CaCl2 and then fixed with 4% paraformaldehyde in PBS for 15 min at RT. Cells were permeabilized with PBS + 0.1% Triton-X-100 and 4% formaldehyde for 15 min and then incubated with 150 mM glycine for 15 min. Wells were blocked with PBS + 2.5% BSA, 10% FBS, 0.05% Tween20, and 0.1% Triton-X-100 for 1 hr at RT and then incubated with primary antibody for 1 hr at RT. Alexa Fluor®647-phalloidin (Life Technologies; 1:1000 final dilution) was used to label the actin cytoskeleton, and rabbit α-tubulin IgG (Abcam; 1:2000 final dilution) followed by goat α-rabbit IgG Alexa Fluor®568 (Life Technologies; 1:500 final dilution) labeled microtubules. Antibodies were diluted in PBS + 1% BSA, 5% FBS, 0.05% Tween20, and 0.1% Triton-X-100. After secondary antibody incubation, cells were washed 3 times with PBS and mounted with ProLong Gold antifade reagent (Life Technologies). Cells were viewed with a 60X oil immersion objective, images were taken with a CoolSnap HQ CCD camera (Photometrics) and Metamorph software (Molecular Devices) and processed with ImageJ software (NIH).
Structural Alignment Analysis
CerADPr (residues 226–476) was iteratively modeled on the ADP-ribosyltransferase domain (Ia) of Iota toxin, the top PSI-BLAST match for which a structure has been elucidated (PDB:1GIR), using SWISSMODEL and Swiss-pdb-viewer v4.02 (http://www.expasy.org/ swissmod/SWISS-MODEL.html). ExoS (residues 234–453) was modeled as previously described [17]. Structural alignments of CerADPr, ExoS-ADPr, Iota toxin Ia, VIP2 (1QS2), C3Lim (3BW8 chain A), and C3bot (1G24 chain A) was performed with Visual Molecular Dynamics Multiseq (http://www.ks.uiuc.edu/Research/vmd/; University of Illinois-Urbana/Champaign) [46].
Results
CerADPr is a functional ADP-ribosyltransferase
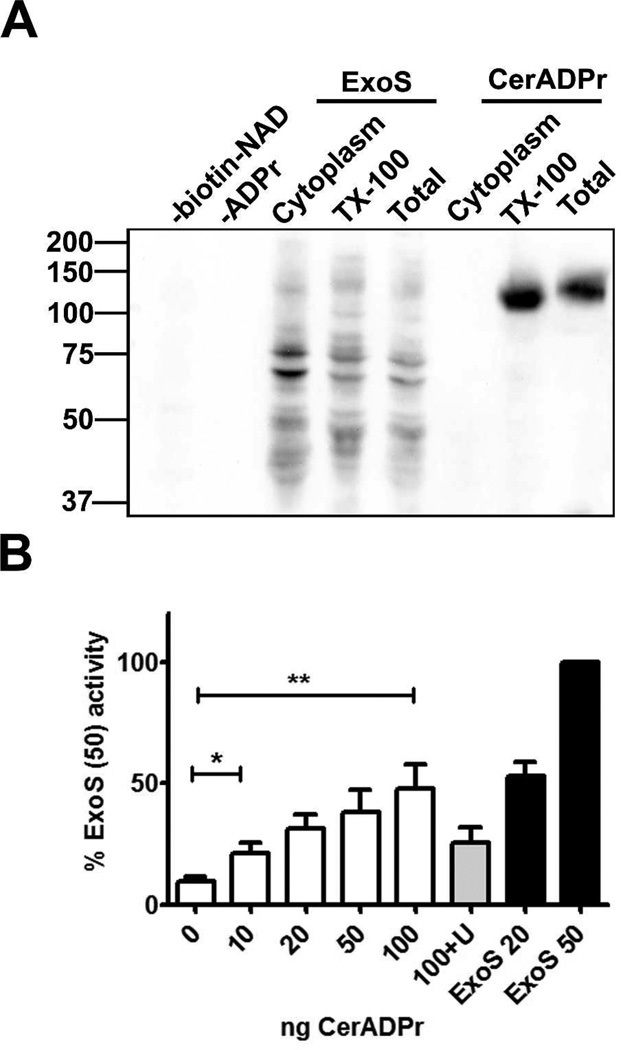
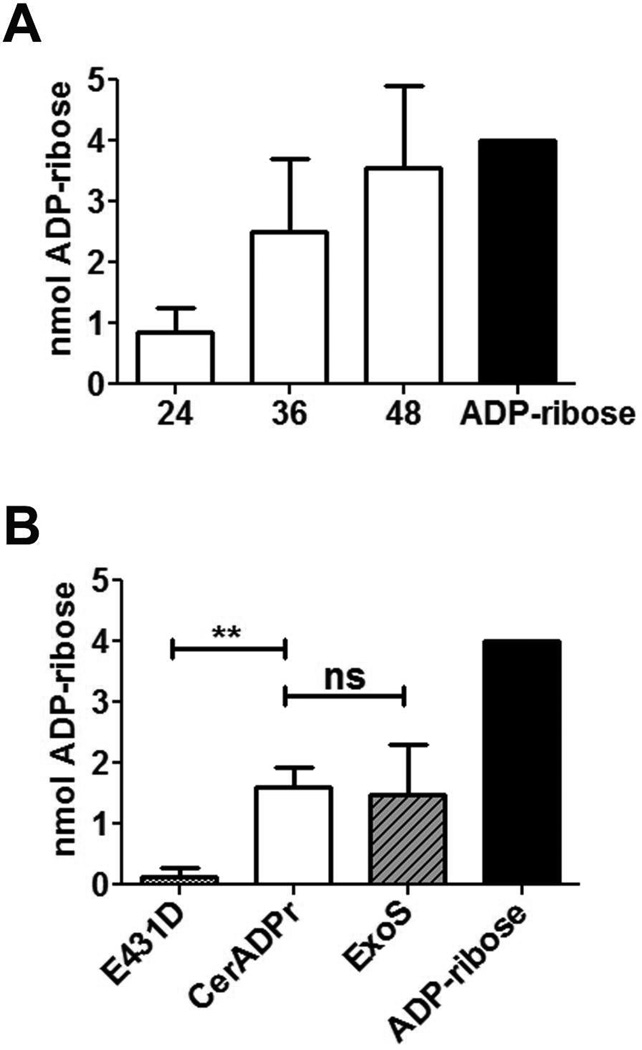
Initial experiments addressed the ADP-ribosyltransferase capacity of the predicted ADP-ribosyltransferase domain of Cereus toxin (residues 226–476, termed CerADPr). Expression of His(6)-CerADPr in E. coli yielded a ~30 kDa protein by SDS-PAGE. Potential host targets for ADP-ribosylation were assessed using a post-nuclear supernatant from HeLa cells, which was fractionated by ultracentrifugation to separate membranes from cytoplasm. CerADPr ADP-ribosylated a 120 kDa protein in the HeLa cell lysate and cell membrane fractions (Figure 1A), but did not ADP-ribosylate host proteins in the cytosol. ADP-ribosylation was dependent upon the addition of biotin-NAD (−biotin) or CerADPr (−ADPr) to the reaction. Pseudomonas aeruginosa ExoS was used as a positive control for ADP-ribosylation; ExoS ADP-ribosylated multiple substrates in the HeLa cell lysate, cytosol, and cell membrane fractions. ADP-ribosylated substrate increased with increased concentrations of CerADPr (Figure 1B). Addition of unlabeled NAD reduced the amount of ADP-ribosylation signal from biotin-NAD, supporting the specificity of CerADPr. NAD glycohydrolase activity of CerADPr was compared to ExoS. CerADPr generated increasing ADP-ribose over time (Figure 2A). Little ADP-ribose was generated when CerADPrE431D was incubated with NAD, supporting a role for E431 in catalysis. Quantification of ADP-ribose generated by the recombinant protein indicated that the glycohydrolase activity of CerADPr was within 2-fold of ExoS (Figure 2B).
Figure 1. CerADPr selectively ADP-ribosylates a high molecular weight host protein in HeLa cell membranes.
(A) A HeLa cell lysate (Total) was centrifuged to generate cytosol (Cytoplasm) and membranes. Membranes were extracted with 0.1% Triton X-100 and centrifuged to remove the insoluble cell matrix (soluble membranes were termed Tx-100). Fractions were normalized for protein and incubated alone (−ADPr) or with 50ng CerADPr or 20ng ExoS for 1 hr at 25°C alone (−biotin-NAD) or with 4 µM biotin-NAD. The reaction was subjected to SDS-PAGE and proteins transferred to a PVDF membrane, which was probed with streptavidin-conjugated HRP. (B) Tx-100 HeLa cell fraction was incubated alone (0) or with the indicated amount of CerADPr (ng) or ExoS (ng) for 1hr at 25°C with 4 µM biotin-NAD or with 10-fold excess unlabeled NAD (100 +U). The reaction mixture was treated as above. The amount of biotin labeling from the 50ng ExoS lane was set to 100% and other signals were normalized, error bars display SEM. An unpaired t-test was performed on selected columns *p<0.05 **p<0.01
Figure 2. NAD glycohydrolase activity of CerADPr.
(A) Recombinant CerADPr was incubated with 5 mM NAD+ for 24, 36, or 48 hr. An aliquot of each reaction was analyzed by PEI-cellulose thin layer chromatography. (B) CerADPr, CerADPr(E431D), or ExoS was incubated with 5 mM NAD+ for 24 hr. An aliquot of each reaction was then subjected to PEI-cellulose TLC. Plates were imaged and the amount of ADP-ribose produced was quantified by densitometry and normalized to an ADP-ribose standard. Quantification is from 3 independent experiments, error bars display SEM. An unpaired t-test was performed on selected columns *p<0.05 **p<0.01
CerADPr contains a conserved bacterial ADP-ribosyltransferase motif and active site glutamic acid
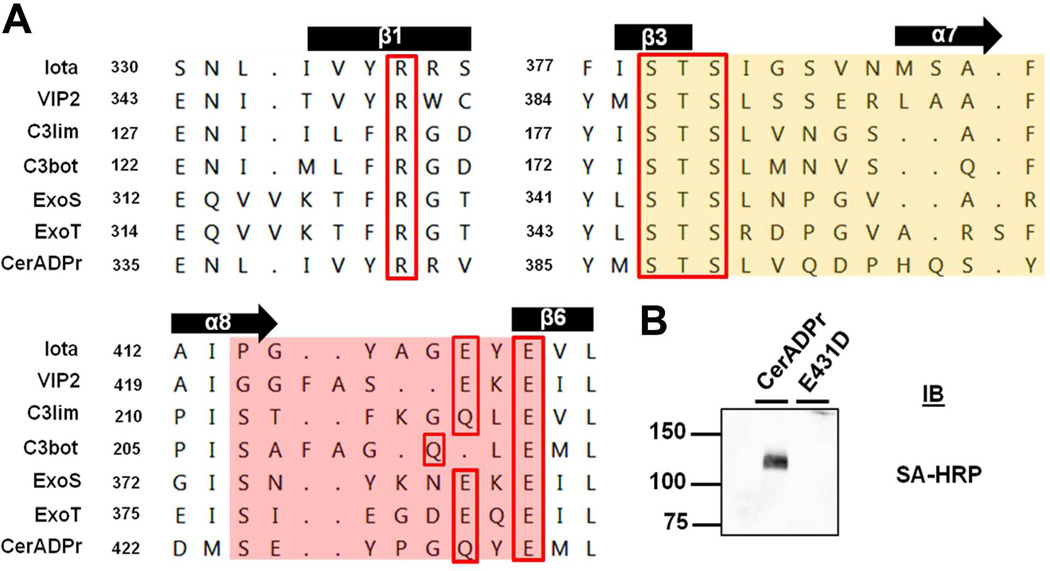
Bacterial ADP-ribosyltransferases possess limited primary amino acid homology, but contain three conserved regions within a conserved structural domain denoted the “RSE” motif [16]. The “RSE” motif forms the active-site pocket and consists of a basic amino acid (Arg), a Ser-Thr-Ser motif present in the phosphate-nicotinamide (PN) loop, and an ADP-ribosyltransferase-turn-turn loop that includes a Gln/Glu-XXX-Glu motif that includes a catalytic glutamic acid (bolded). Structural alignment with several bacterial ADP ribosyltransferases showed that the “RSE” motif was conserved within CerADPr (Figure 3A). The PN loop of CerADPr, highlighted in orange, possesses a Ser-Thr-Ser within β-sheet 3, as well as a conserved aromatic residue forming the tail of the NAD-binding “scorpion” motif [47]. The structure of the ARTT loop (pink) was also conserved, which included α-helix 8 leading into the active site loop and β-sheet 6 containing the catalytic Glu. In addition, a highly charged and lysine-rich loop (Gly303-Glu315) is predicted from the model. This loop, termed the G-loop, is of unknown function and to our knowledge is not present in other bAREs.
Figure 3. CerADPr domain contains sequence and predicted structural conservation of other bacterial ADP-ribosyltransferases.
(A) A predicted structure-based alignment of the Cereus toxin ADPr domain (CerADPr) was generated with the crystal structure of Iota toxin ADP-ribosyltransferase domain Ia (PDB: 1GIR). Three regions of alignment that are common components of ADP-ribosyltransferase family members, including an Arg, the Ser-Thr-Ser motif, and the Gln/Glu-XXX-Glu sequence (“RSE” motif) are shown. The latter defines an active site glutamic acid that is conserved among ADP-ribosyltransferases. Secondary structure labeling is based off the Iota toxin Ia ADP-ribosyltransferase domain, starting with α-helix1 at residues 214–232. (B) CerADPr(E431D) ADP-ribosyltransferase activity was measured relative to CerADPr by incubation with Tx-100 HeLa cell lysate and 4 µM biotin-NAD for 1 hr at 25°C. The reaction mixture was processed as described in Figure 1 and the ADP-ribosylation of the high-molecular weight substrate is shown in the inset image.
CerADPr contained highest overall similarity with VIP2 and Iota toxins, but highest active site similarity with the C3 toxins that possess a Gln-XXX-Glu motif, rather than a Glu-XXX-Glu found in the binary actin ADP-ribosylating toxins and ExoS and ExoT. Site directed mutagenesis of the predicted catalytic glutamic acid (Glu431) of CerADPr to aspartic acid (E431D) resulted in loss of ADP-ribosylation capacity (Figure 3B), consistent with Glu431 being the catalytic glutamic acid [19].
CerADPr disrupts the host cytoskeleton in HeLa cells
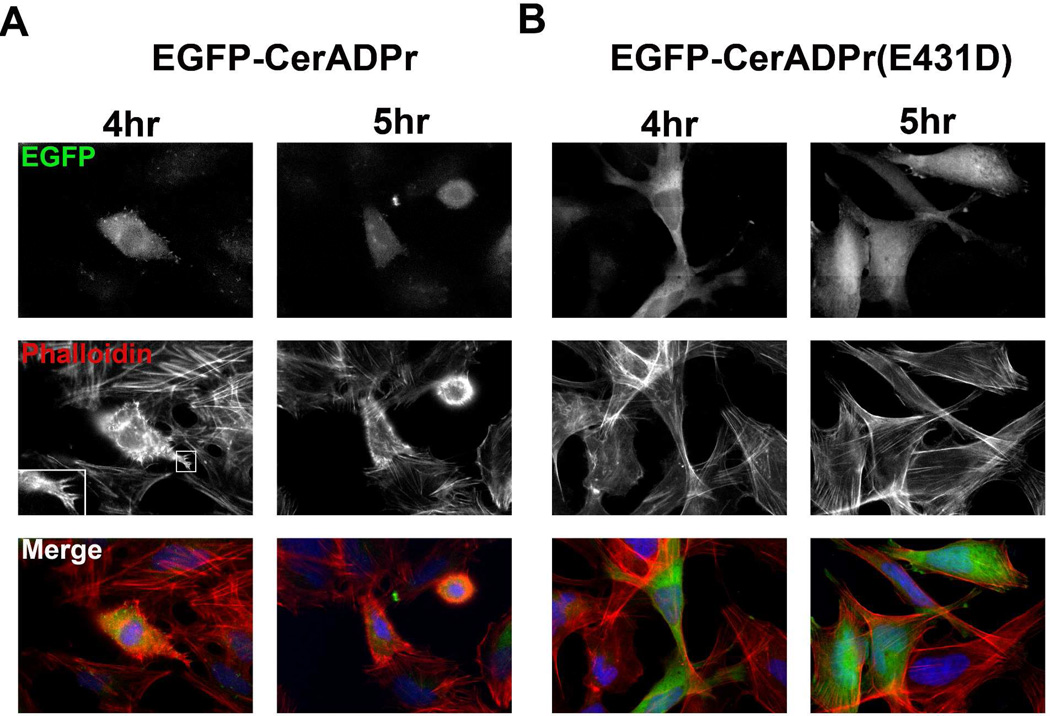
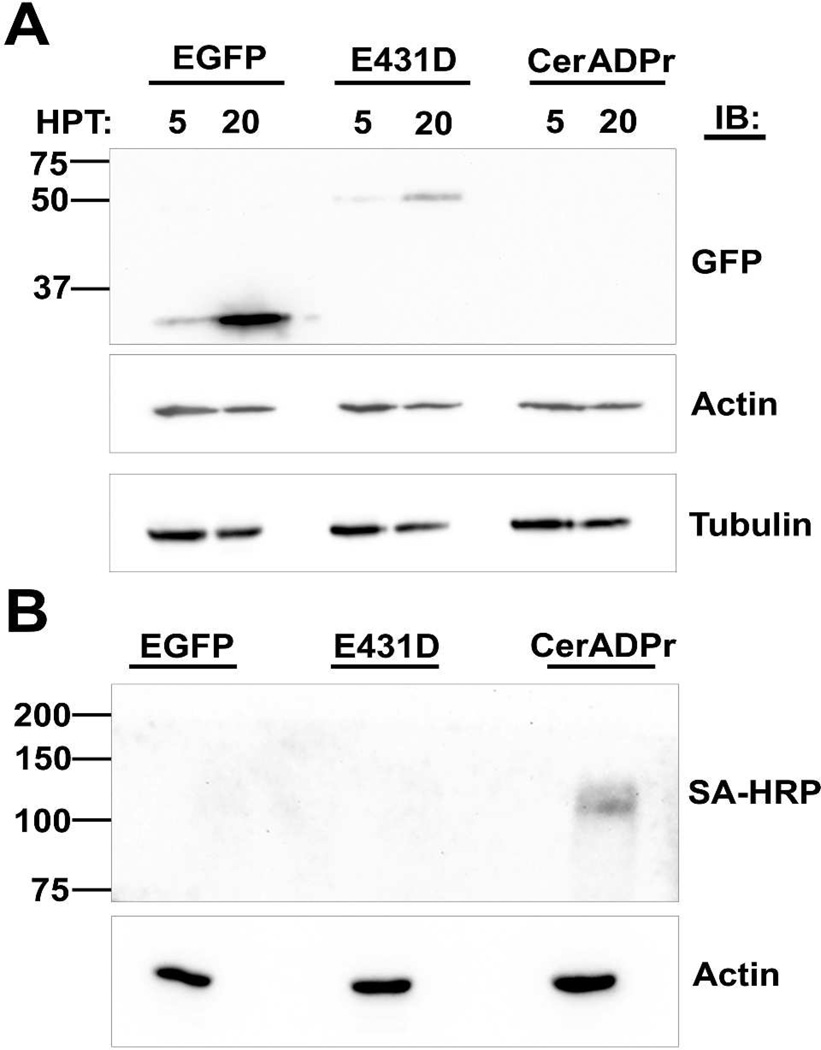
To assess the effect of CerADPr on cell physiology, HeLa cells were transfected with pEGFP-CerADPr, pEGFP-CerADPr(E431D), or pEGFP and stained with phalloidin to examine the cellular actin architecture. By 4 hr post-transfection (HPT), the earliest time that EGFP signal was detectable, EGFP-CerADPr transfected cells displayed an aberrant actin cytoskeleton (Figure 4A). EGFP-CerADPr transfected cells showed a loss of actin stress fibers and formation of actin spikes at the cell periphery (Figure 4A, inset image). At 5 HPT, cells rounded and did not possess defined actin stress fibers, and by 20 HPT, adherent EGFP fluorescent cells were not detected (data not shown). EGFP-CerADPr(E431D) transfected HeLa cells had normal actin cytoskeleton (Figure 4B), supporting a role for ADP-ribosylation in the disruption of the actin cytoskeleton. Since CerADPr disrupted the actin cytoskeleton, the effect of CerADPr expression on the structure of microtubules was also examined by determining microtubule organization with α-tubulin immunostaining. At later incubation times, EGFP-CerADPr transfected HeLa cells displayed disorganized microtubule architecture and appearance of filamentous projections at the cell periphery, which stained positive for tubulin (data not shown). EGFP-CerADPr(E431D) transfected HeLa cells had normal microtubule arrangement.
Figure 4. Expression of EGFP-CerADPr elicits early actin rearrangement in HeLa cells.
HeLa cells were transfected with pEGFP-CerADPr (A) or pEGFP-CerADPr(E431D) (B) for 4 or 5 hr when cells were fixed, permeabilized, and incubated with Alexa Fluor®647-phalloidin to label the actin cytoskeleton. Cells were subjected to epifluorescence microscopy. Representative images are shown for each condition. Inset shows close-up of actin spikes seen at cell periphery of EGPF-CerADPr transfected cells.
Since VIP2 family bAREs ADP-ribosylate actin to inhibit polymerization of actin microfilaments, the ability of CerADPr to ADP-ribosylate actin in HeLa cells was determined. Western blotting of transfected cell lysates showed that transfection with EGFP-CerADPr did not elicit a shift in the apparent electrophoretic mobility of either actin or tubulin, suggesting that expression of CerADPr did not directly modify either host protein (Figure 5A). The sequence similarity between CerADPr and C3Cer of B. cereus (Supplementary Figure 2) also prompted the determination of the ability of CerADPr to ADP-ribosylate Rho GTPases. However under conditions where ExoS ADP-ribosylated Rab5, CerADPr did not ADP-ribosylate detectable amounts of Rho GTPases (Supplementary Figure 3).
Figure 5. CerADPr does not covalently modify actin or tubulin.
(A) HeLa cells were transfected with either pEFGP, pEGFP-CerADPr(E431D), or pEGFP-CerADPr. At 5 and 20 hr post-transfection (HPT), cells were harvested and subjected to SDS- PAGE. The gel was transferred to PVDF and probed independently with α-GFP, α-actin and α-tubulin primary antibodies. Membranes were processed as described in Figure 1. (B) Cell lysates were prepared from HeLa cells transfected with either pEFGP, pEGFP-CerADPr(E431D), or pEGFP-CerADPr and incubated with the TX-100-soluble membrane fraction of HeLa cells and biotin-NAD. After 2 hr, the reaction was subjected to SDS-PAGE and proteins were transferred to PVDF membranes and probed for the ADP-ribosylation of the high-molecular weight substrate of CerADPr and actin reactivity measured to normalize the cell lysates.
Western blotting showed that EGFP-CerADPr(E431D) transfected HeLa cell lysate possessed a GFP reactive protein of the expected molecular weight of an EGFP-CerADPr fusion Figure 5A). A GFP reactive protein was not detected by immunoblot in the lysates of EGFP-CerADPr transfected HeLa cells, although intracellular expression of bAREs reduces host protein expression [44]. Additionally, the lysate from cells transfected with pEGFP-CerADPr ADP-ribosylated the previously observed high-molecular weight host protein in HeLa cell lysates (Figure 5B), confirming that EGFP-CerADPr was expressed in the transfected HeLa cells.
CerADPr ADP-ribosylates a 120kDa protein in HeLa cells
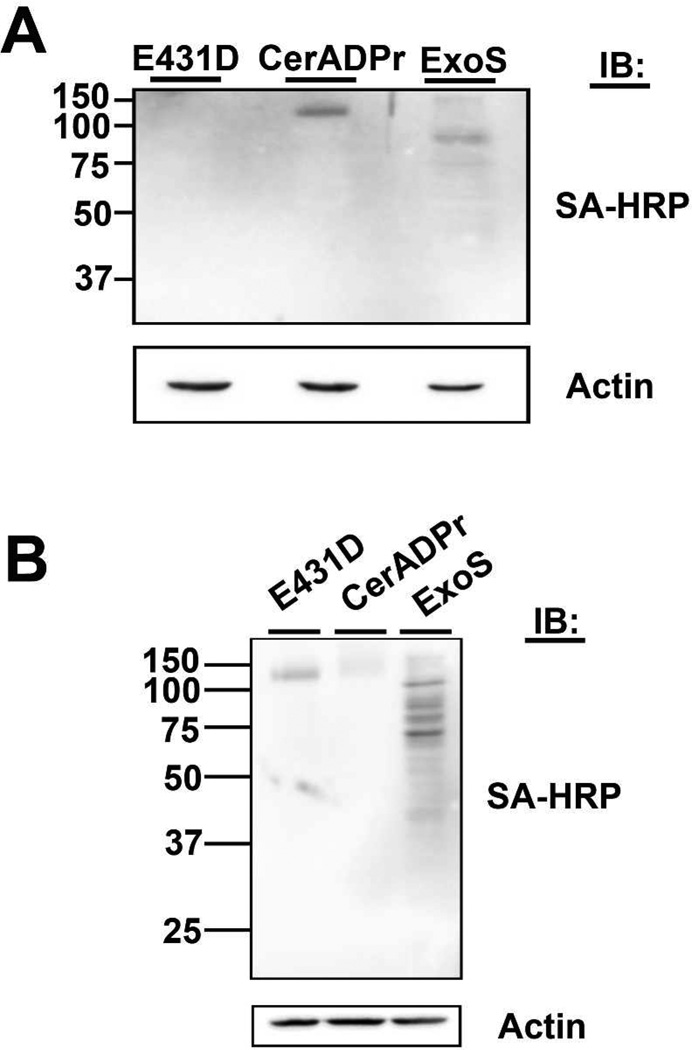
To determine if CerADPr ADP-ribosylates the 120kDa protein within intact HeLa cells, tetanolysin was used to reversibly generate pores in the plasma membrane to allow diffusion of externally added molecules into the cell [48–50]. HeLa cells transfected with EGFP-CerADPr, EGFP-CerADPr(E431D), or ExoS were incubated with tetanolysin and biotin-NAD. Lysate from cells transfected with EGFP-CerADPr showed the ADP-ribosylation of the 120kDa protein, while lysates from cells transfected with EGFP-CerADPr(E431D) did not contain detectable ADP-ribosylated proteins Figure 6A). Lysate from cells transfected with ExoS showed multiple ADP-ribosylated proteins, consistent with the ADP-ribosyltransferase specificity of this protein.
Figure 6. CerADPr ADP-ribosylates the 120KDa target within HeLa cells.
(A) HeLa cells were transfected with pEGFP-CerADPr, pEGFP-CerADPr(E431D), or pExoS for 2 hr. Cells were treated with 200 ng/ml of tetanolysin and then incubated with biotin-NAD for 2.5 hr. Cell lysates were resolved by SDS-PAGE and biotin-ADP-ribose incorporation was visualized by immunoblotting with streptavidin-HRP. Actin was used as a loading control. (B) HeLa cells were treated with 200 ng/ml of tetanolysin, then incubated with 3 µg/ml recombinant CerADPr, CerADPr(E431D), or ExoS for 2 hr. Cell lysates prepared and then incubated with biotin-NAD and recombinant CerADPr for an hour. Lysates were resolved by SDS-PAGE, transferred to PVDF membranes, where biotin-ADP-ribose incorporation was visualized with streptavidin-HRP. Actin was used as a loading control.
To corroborate the ability of CerADPr to ADP-ribosylate the 120 kDa protein in HeLa cells, a back ADP-ribosylation experiment was performed. HeLa cells were incubated with tetanolysin, then incubated with recombinant CerADPr or CerADPr(E431D). Cell lysates from these cells were then incubated with recombinant CerADPr and biotin-NAD to ADP- ribosylate accessible 120 kDa protein. Lysates from cells incubated with CerADPr(E431D) possessed accessible 120kDa protein for ADP-ribosylation, while lysates from cells incubated with CerADPr possessed a reduced amount of accessible 120 kDa protein for ADP-ribosylation (Figure 6B). This indicated that CerADPr had ADP-ribosylated the 120 kDa protein within HeLa cells.
CerADPr is cytotoxic for HeLa cells
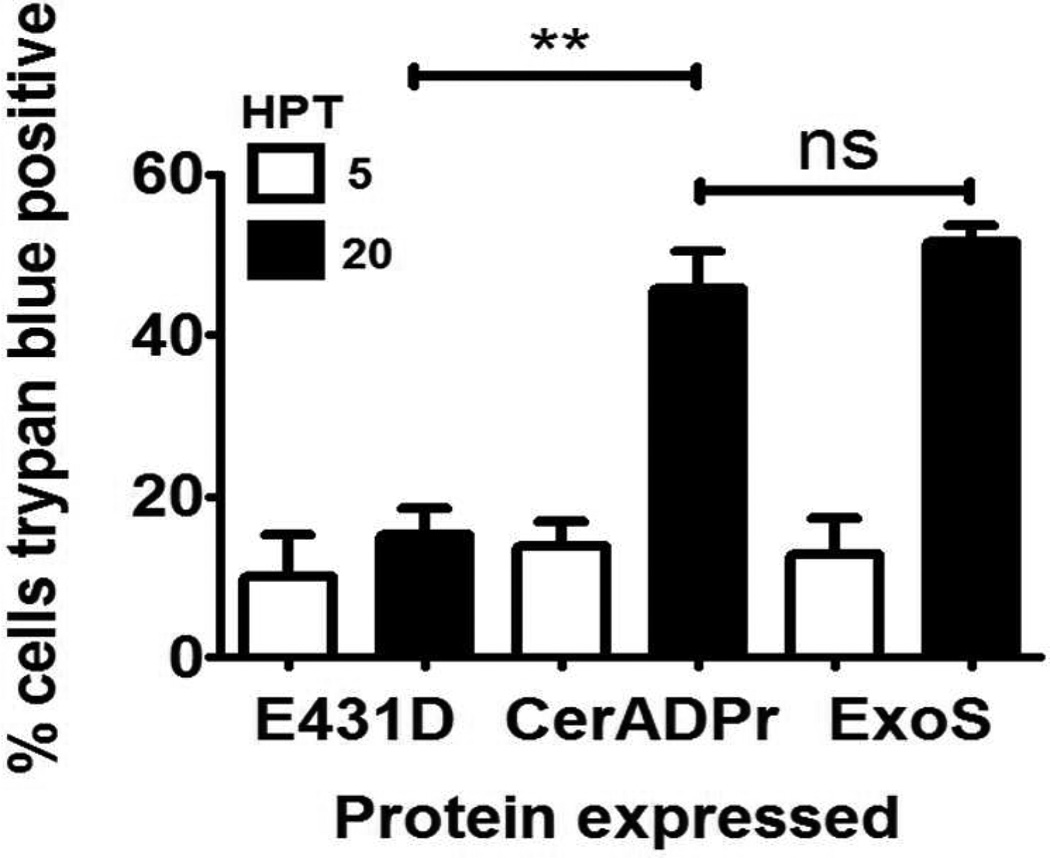
To determine if CerADPr was cytotoxic, HeLa cells were transfected with pEGFP-CerADPr, pEGFP-CerADPr(E431D) or pExoS(R146K), a RhoGAP−/ADPr+ ExoS derivative, and scored for trypan blue exclusion. ExoS was used as a positive control for cytotoxicity [44]. Trypan blue is excluded from viable cells, but enters necrotic or apoptotic cells. At 5 HPT, ~10% of cells transfected with pExoS were trypan blue positive, while at 20 HPT, ~50 % of transfected cells were trypan blue positive (Figure 7). At 5 HPT, ~15% of pEGFP-CerADPr transfected cells were trypan blue positive and at 20 HPT, ~45% of transfected cells were trypan blue positive. Cells transfected with pEGFP-CerADPr(E431D), showed limited numbers of trypan blue positive cells (~15% at 20 HPT). This indicated that expression of CerADPr elicits a cytotoxic response in HeLa cells.
Figure 7. CerADPr is cytotoxic for HeLa cells.
HeLa cells were grown to ~70 % confluence in 6-well dishes and transfected with 500 ng of either pExoS, pEGFP-CerADPr(E431D), or pEGFP-CerADPr. At 5 and 20 hr post-transfection (HPT), media was removed, cells were washed and stained for 5 min with trypan blue. Trypan blue was removed, cells were washed, and color images were obtained as described in Methods. Cytotoxicity was determined by counting the number of trypan blue positive HeLa cells / total number of HeLa cells per field. Five random fields were assayed from three biological replicates, and the results of this determination shown. 1-way ANOVA was performed on all columns ** p<0.01.
Discussion
In silico structural analyses are powerful tools to discover candidate bacterial virulence factors by predicting conservation of enzymatic folding motifs or catalytic residues within open reading frames from newly sequenced bacterial genomes. In this study, the predicted ORF of Cereus toxin (Certhrax) from Bacillus cereus G9241 is shown to be a functional ADP ribosyltransferase. CerADPr contained a conserved bacterial ADP-ribosyltransferase “RSE” motif where mutation of the predicted conserved, invariant Glu resulted in loss of ADP-ribosyltransferase activity. CerADPr targeted actin polymerization at early expression time points. However, these effects appeared to be off-target, since direct modification or loss of either actin or tubulin was not detected. CerADPr displayed a cytotoxic phenotype with similar temporal properties to ExoS, a known cytotoxic effector [44].
Bacterial ADP-ribosyltransferase exotoxins (bAREs) have limited amino-acid sequence identity, with conservation of structure and sequence active site motifs. The canonical bARE domain contains a “RSE” motif, consisting of Arg, Ser-Thr-Ser, and Gln/Glu-XXX-Glu motifs, except in the diphtheria toxin/Exotoxin A subfamily [16]. bAREs within the VIP2 or C3 subfamilies contain two additional structures, the phosphate-nicotinamide loop and α-3 helix, that are absent in the cholera toxin-like bAREs [51]. Sequence and structural alignment with other bAREs showed that CerADPr had a conserved “RSE” motif, as well as the PN loop and α-3 helix, as in members of the VIP2-like or C3-like bAREs. Unexpectedly, CerADPr also contains an additional 12 residue loop (Gly303-Glu315), which, to our knowledge, is not present in other bAREs. This loop, termed the G-loop, is of unknown function and highly charged with lysine residues.
Alignment of CerADPr with previously characterized bAREs yielded several dichotomies. The protein organization shows homology to the VIP2 subfamily of binary toxins, which ADP-ribosylate actin [12]. CerADPr had the highest scoring PSI-BLAST match with Iota toxin from Clostridium perfringens and showed similarity to the binary actin-ADP-ribosylating VIP2. However, presence of a Gln-XXX-Glu motif, rather than Glu-XXX-Glu, in the ARTT loop implied that CerADPr may ADP-ribosylate targets in a similar manner to the C3 exoenzymes. A C3-like mechanism cannot be ruled out, as TccC5, a bARE from Photorhabdus luminescens contains a Gln-XXX-Glu motif and ADP-ribosylates Gln61/63 of host Rho proteins, despite limited homology to the C3 exoenzymes [52]. However, in vitro ADP-ribosylation assays showed that CerADPr does not ADP-ribosylate the Rho GTPases (Supplementary Figure 3). Since the target of CerADPr is detergent solubilized from cellular membranes, the substrate of CerADPr may be an integral membrane protein, or a part of a membrane associated complex. The sequence and structural duality suggests that CerADPr may belong to a new subfamily of bAREs with unique substrate specificity, since CerADPr ADP-ribosylates a 120 kDa target in eukaryotic cell lysate. Further, despite sequence and structural similarities between CerADPr and the VIP2-like toxin subfamily members, CerADPr does not appear to directly modify actin either in vitro (Figure 1A), or in HeLa cells (Figure 5A). In other bAREs, mutation of the conserved, invariant Glu correlates with a >10,000-fold reduction in ADP-ribosyltransferase activity [53, 54]. While precise kinetic calculations are difficult without the identity of the substrate, CerADPr(E431D) displayed limited cytotoxicity for HeLa cells and no ADP-ribosylation of the 120 kDa substrate in vitro, as well as a loss of detectable NAD glycohydrolase activity.
Most bAREs target the cytoskeleton, either directly, through actin ADP-ribosylation, or indirectly, by interfering with cytoskeletal signaling pathways. VIP2-like toxins and C3 exoenzymes disrupt actin filament polymerization, resulting in rounding and cell death, while other bAREs like ExoS or ExoT disrupt signaling by ADP-ribosylating ezrin/radixin/moesin proteins or CrkI/II, respectively [13, 55–58]. These modifications alter cytoskeleton dynamics and result in cell rounding. CerADPr appears to share a similar ability to disrupt the cytoskeleton, causing stress fiber rearrangement and formation of actin spikes at the cell periphery early, followed by cell rounding and detachment from the growth surface with longer incubations. Formation of actin spikes is somewhat similar to effects induced by the E. coli effector Map, a Cdc42 Guanine-nucleotide exchange factor, which causes filapodia formation in intoxicated cells [59, 60]. Current studies are directed towards resolving the identity of the 120 kDa host target that is ADP-ribosylated by Cereus toxin.
CerADPr disrupted microtubule structure, with filamentous projections extending from the periphery of cells displaying disrupted actin architecture, although microtubule disruption was only observed at longer incubation times. This phenotype has previously been observed in cells intoxicated with actin ADP-ribosylating binary toxin CDTa from Clostridium difficile, and is thought to be a result of depolymerization of the F-actin cytoskeleton [61]. Therefore, it is likely the tubulin projections elicited by CerADPr result from actin depolymerization, since the extensions were observed upon extended incubations, after actin rearrangements were observed. Many bAREs, including the VIP2-like and C3-like bAREs, also stimulate a cytotoxic phenotype following cytoskeletal disruption, with ExoT and heat-labile enterotoxin notable exceptions [17,44, 62–64]. The CerADPr phenotype, then, appears similar to that of the VIP2 or C3 bAREs, despite an inability of CerADPr to directly ADP-ribosylate actin or the Rho proteins in vitro or in cell culture.
B. cereus pathogenesis is typically characterized by expression of various enterotoxins and hemolysins, resulting in mild to moderate gastroenteritis, and emetic or diarrheal illness. However, several recently isolated B. cereus strains, including G9241, contain homologs of B. anthracis plasmid pXO1 that encode the three anthrax toxin components [39]. G9241 produces these proteins in vitro, and their expression is required for full virulence, as is the production of both a hyaluronic acid capsule [40] and a polysaccharide capsule [40, 65]. Pulmonary anthrax-like disease observed in individuals infected with B. cereus G9241 may be due to the effects of the anthrax toxin homologs, acting in a similar mechanism to B. anthracis anthrax toxin. However, G9241 also contains a second large plasmid, encoding a novel ADP-ribosyltransferase, Cereus toxin (Certhrax). Other B. cereus strains encode bAREs, such as C3Cer or VIP2, but the pathogenic contributions of these proteins have not been well elucidated in mammals. Cereustoxin may contribute to B. cereus G9241 pathogenesis in an antiphagocytic capacity, eitherthrough disruption of actin polymerization, or through its cytotoxic activity. Other bAREs studied in mammalian models, such as Salmonella SpvB, induce cytotoxicity in macrophages within 24 hours of bacterium phagocytosis [66]. Alternatively, Cereus toxin could play a limitedrole in mammalian pathogenesis, and be more directed towards environmental targets, like the crystal toxins expressed by B. thuringiensis, which primarily target insects. However, whileconsidered an insect pathogen, B. thuringiensis has been associated with isolated cases of wound infection, food poisoning, and catheter-associated infections in immunocompromised humans [69].
The sequence homology with the PA binding domain of lethal factor (Supplementary Figure 1) suggests that Cereus toxin could utilize a clathrin-dependent cell entry mechanism, similar to that employed by LF, although whether the entry mechanism and host receptor are identical or divergent needs exploration [22, 33]. Alternatively, the similarities with the VIP2-like binary toxins could implicate an entry mechanism similar to either Iota or C2, which utilize a clathrin-independent pathway, or a clathrin-dependent pathway involving the Rho proteins respectively [70, 71].
Dissecting the mechanisms behind novel bARE activity may lead to an effective strategy to neutralize the virulence of a wide range of bacterial pathogens, while avoiding off-target effects on the host microbiome and development of resistance often observed with extended antibiotic therapy [72–74]. Unlike B. anthracis, B. cereus is naturally resistant to β-lactams including cephalosporins, but currently responds well to other available antibiotics like vancomycin [75, 76]. However, given the rise of multi-drug resistance in other human pathogens, directed targeting of virulence factors is likely a safer long-term strategy to combat these infections. Determination of the substrate and mechanism of Cereus toxin action may also lead to its use as a molecular tool to elucidate mammalian signaling pathways. Finally, understanding the activity of Cereus toxin will broaden our understanding of the pathogenesis of B. cereus G9241 and may lead to therapies to address the severe morbidities and mortality associated with emerging pathogenic strains of B.cereus.
Supplementary Material
Acknowledgements
We acknowledge the assistance of Amanda Przedpelski for protein production. The opinions expressed here are those of the authors and do not reflect the views of the United States Navy or the Department of Defense.
Funding Source: JTB acknowledges membership in the GLRCE and support by 1-U54-AI-057153 from Region V Great Lakes Regional Center of Excellence, the NIAID, and National Institutes of Health Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research Program. NCS was supported by NIAID AI-AI30162. ADO acknowledges support from the Biological Defense Research Directorate of the Naval Medical Research Center.
Abbreviations and Textural Footnotes
- ADPr
ADP-ribosyltransferase
- bAREs
bacterial ADP-ribosylating exotoxins
- CT
cholera toxin
- DT
diphtheria toxin
- VIP
vegetative insecticidal protein
- PA
protective antigen
- LF
lethal factor
- EF
edema factor
- CerADPr
cereus toxin ADP-ribosyltransferase domain
- PVDF
polyvinylidene fluoride membrane
Footnotes
Supplemental Information
Supplementary figures include an amino acid homology alignment of CerADPr/Lethal factor protein domains and a homology alignment of CerADPr with other bacterial ADP-ribosyltransferases, as well as data showing the inability of CerADPr to ADP-ribosylate Rho proteins. These figures may be accessed free of charge online at http://pubs.acs.org
References
- 1.Krueger KM, Barbieri JT. The family of bacterial ADP-ribosylating exotoxins. Clinical Microbiology Reviews. 1995;8(1):34–47. doi: 10.1128/cmr.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill DMaRM. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: basis of the activation of adenylate cyclase. Proc Natl Acad Sci. 1978;75(7):3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moss J, et al. NAD-dependent ADP-ribosylation of arginine and proteins by Escherichia coli heat-labile enterotoxin. Journal of Biological Chemistry. 1979;254(14):6270–6272. [PubMed] [Google Scholar]
- 4.Collier RJ. Effect of diphtheria toxin on protein synthesis: Inactivation of one of the transfer factors. Journal of Molecular Biology. 1967;25(1):83–98. doi: 10.1016/0022-2836(67)90280-x. [DOI] [PubMed] [Google Scholar]
- 5.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. Journal of Biological Chemistry. 1980;255(22):10717–10720. [PubMed] [Google Scholar]
- 6.Braun U, et al. Purification of the 22 kDa protein substrate of botulinum ADP-ribosyltransferase C3 from porcine brain cytosol and its characterization as a GTP-binding protein highly homologous to the rho gene product. FEBS Letters. 1989;243(1):70–76. doi: 10.1016/0014-5793(89)81220-7. [DOI] [PubMed] [Google Scholar]
- 7.Coburn J, et al. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. Journal of Biological Chemistry. 1989;264(15):9004–9008. [PubMed] [Google Scholar]
- 8.Aktories K, et al. The rho gene product expressed in E. Coli is a substrate of botulinum ADP-ribosyltransferase C3. Biochemical and Biophysical Research Communications. 1989;158(1):209–213. doi: 10.1016/s0006-291x(89)80199-8. [DOI] [PubMed] [Google Scholar]
- 9.Han S, et al. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Mol Biol. 1999;6(10):932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 10.Aktories K, et al. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322(6077):390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 11.Schering B, et al. ADP-ribosylation of skeletal muscle and non-muscle actin by Clostridium perfringens iota toxin. European Journal of Biochemistry. 1988;171(1–2):225–229. doi: 10.1111/j.1432-1033.1988.tb13780.x. [DOI] [PubMed] [Google Scholar]
- 12.Vandekerckhove J, et al. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Letters. 1987;225(1.2):48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- 13.Tsuge H, et al. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens ι-toxin. Proceedings of the National Academy of Sciences. 2008;105(21):7399–7404. doi: 10.1073/pnas.0801215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han S, et al. Crystal structure and novel recognition motif of Rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. Journal of Molecular Biology. 2001;305(1):95–107. doi: 10.1006/jmbi.2000.4292. [DOI] [PubMed] [Google Scholar]
- 15.Tsuge H, et al. Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin. Journal of Molecular Biology. 2003;325(3):471–483. doi: 10.1016/s0022-2836(02)01247-0. [DOI] [PubMed] [Google Scholar]
- 16.Domenighini M, Rappuoli R. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Molecular Microbiology. 1996;21(4):667–674. doi: 10.1046/j.1365-2958.1996.321396.x. [DOI] [PubMed] [Google Scholar]
- 17.Sun J, et al. How bacterial ADP-ribosylating toxins recognize substrates. Nat Struct Mol Biol. 2004;11(9):868–876. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- 18.Wilde C, et al. Recognition of RhoA by Clostridium botulinum C3 Exoenzyme. Journal of Biological Chemistry. 2000;275(22):16478–16483. doi: 10.1074/jbc.M910362199. [DOI] [PubMed] [Google Scholar]
- 19.Fieldhouse RJ, et al. Cholera- and Anthrax-Like Toxins Are among Several New ADP-Ribosyltransferases. PLoS Comput Biol. 2010;6(12):e1001029. doi: 10.1371/journal.pcbi.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little SF, Ivins BE. Molecular pathogenesis of Bacillus anthracis infection. Microbes and Infection. 1999;1(2):131–139. doi: 10.1016/s1286-4579(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 21.Escuyer V, Collier RJ. Anthrax protective antigen interacts with a specific receptor on the surface of CHO-K1 cells. Infection and Immunity. 1991;59(10):3381–3386. doi: 10.1128/iai.59.10.3381-3386.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradley KA, et al. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414(6860):225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 23.Scobie HM, et al. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proceedings of the National Academy of Sciences. 2003;100(9):5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh Y, et al. The chymotrypsin-sensitive site, FFD315, in anthrax toxin protective antigen is required for translocation of lethal factor. Journal of Biological Chemistry. 1994;269(46):29039–29046. [PubMed] [Google Scholar]
- 25.Milne JC, et al. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. Journal of Biological Chemistry. 1994;269(32):20607–20612. [PubMed] [Google Scholar]
- 26.Petosa C, et al. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385(6619):833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 27.Kintzer AF, et al. The Protective Antigen Component of Anthrax Toxin Forms Functional Octameric Complexes. Journal of Molecular Biology. 2009;392(3):614–629. doi: 10.1016/j.jmb.2009.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kintzer AF, et al. Role of the Protective Antigen Octamer in the Molecular Mechanism of Anthrax Lethal Toxin Stabilization in Plasma. Journal of Molecular Biology. 2010;399(5):741–758. doi: 10.1016/j.jmb.2010.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duesbery NSWC, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 30.Duesbery NS, Woude GFV. Anthrax lethal factor causes proteolytic inactivation of mitogen-activated protein kinase kinase. Journal of Applied Microbiology. 1999;87(2):289–293. doi: 10.1046/j.1365-2672.1999.00892.x. [DOI] [PubMed] [Google Scholar]
- 31.Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proceedings of the National Academy of Sciences. 1982;79(10):3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SH L. Bacillus anthracis calmodulin-dependent adenylate cyclase: chemical and enzymatic properties and interactions with eucaryotic cells. Adv Cyclic Nucleotide Protein Phosphorylation Res. 1984;17:189–198. [PubMed] [Google Scholar]
- 33.Abrami L, et al. Anthrax toxin triggers endocytosis of its receptor via a lipid raft–mediated clathrin-dependent process. The Journal of Cell Biology. 2003;160(3):321–328. doi: 10.1083/jcb.200211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedlander AM. Macrophages are sensitive to anthrax lethal toxin through an acid-dependent process. Journal of Biological Chemistry. 1986;261(16):7123–7126. [PubMed] [Google Scholar]
- 35.Gordon VM, Leppla SH, Hewlett EL. Inhibitors of receptor-mediated endocytosis block the entry of Bacillus anthracis adenylate cyclase toxin but not that of Bordetella pertussis adenylate cyclase toxin. Infection and Immunity. 1988;56(5):1066–1069. doi: 10.1128/iai.56.5.1066-1069.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller CJ, Elliott JL, Collier RJ. Anthrax Protective Antigen: Prepore-to-Pore Conversion†. Biochemistry. 1999;38(32):10432–10441. doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- 37.Hoffmaster AR, et al. Characterization of Bacillus cereus Isolates Associated with Fatal Pneumonias: Strains Are Closely Related to Bacillus anthracis and Harbor B. anthracis Virulence Genes. J. Clin. Microbiol. 2006;44(9):3352–3360. doi: 10.1128/JCM.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller JM, et al. Fulminating bacteremia and pneumonia due to Bacillus cereus. Journal of Clinical Microbiology. 1997;35(2):504–507. doi: 10.1128/jcm.35.2.504-507.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmaster AR, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson MK, et al. Bacillus cereus G9241 Makes Anthrax Toxin and Capsule like Highly Virulent B. anthracis Ames but Behaves like Attenuated Toxigenic Nonencapsulated B. anthracis Sterne in Rabbits and Mice. Infect. Immun. 2011;79(8):3012–3019. doi: 10.1128/IAI.00205-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekine A, Fujiwara M, Narumiya S. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. Journal of Biological Chemistry. 1989;264(15):8602–8605. [PubMed] [Google Scholar]
- 42.Knight DA, Barbieri JT. Ecto-ADP-ribosyltransferase activity of Pseudomonas aeruginosa exoenzyme S. Infection and Immunity. 1997;65(8):3304–3309. doi: 10.1128/iai.65.8.3304-3309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Randerath K, Randerath E. Ion-exchange chromatography of nucleotides on poly-(ethyleneimine)-cellulose thin layers. Journal of Chromatography A. 1964;16(0):111–125. doi: 10.1016/s0021-9673(01)82445-6. [DOI] [PubMed] [Google Scholar]
- 44.Pederson KJ, Barbieri JT. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Molecular Microbiology. 1998;30(4):751–759. doi: 10.1046/j.1365-2958.1998.01106.x. [DOI] [PubMed] [Google Scholar]
- 45.Ahnert-Hilger G, et al. Exocytosis from permeabilized bovine adrenal chromaffin cells is differently modulated by guanosine 5'-[gamma-thio]triphosphate and guanosine 5'-[beta gamma-imido]triphosphate. Evidence for the involvement of various guanine nucleotide-binding proteins. Biochem. J. 1992;284(2):321–326. doi: 10.1042/bj2840321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y-M, et al. Conserved Structural Motif for Recognizing Nicotinamide Adenine Dinucleotide in Poly(ADP-ribose) Polymerases and ADP-Ribosylating Toxins: Implications for Structure-Based Drug Design. Journal of Medicinal Chemistry. 2010;53(10):4038–4049. doi: 10.1021/jm1001106. [DOI] [PubMed] [Google Scholar]
- 48.Riese MJ, et al. Auto-ADP-ribosylation of Pseudomonas aeruginosaExoS. Journal of Biological Chemistry. 2002;277(14):12082–12088. doi: 10.1074/jbc.M109039200. [DOI] [PubMed] [Google Scholar]
- 49.Riese MJ, Barbieri JT. Membrane Localization Contributes to the In Vivo ADP-Ribosylation of Ras by Pseudomonas aeruginosa ExoS. Infection and Immunity. 2002;70(4):2230–2232. doi: 10.1128/IAI.70.4.2230-2232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walev I, et al. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proceedings of the National Academy of Sciences. 2001;98(6):3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. FEBS Journal. 2006;273(20):4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 52.Lang AE, et al. Photorhabdus luminescens Toxins ADP-Ribosylate Actin and RhoA to Force Actin Clustering. Science. 2010;327(5969):1139–1142. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- 53.Visschedyk DD, et al. Photox, a Novel Actin-targeting Mono-ADP-ribosyltransferase from Photorhabdus luminescens. Journal of Biological Chemistry. 2010;285(18):13525–13534. doi: 10.1074/jbc.M109.077339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radke J, Pederson KJ, Barbieri JT. Pseudomonas aeruginosa Exoenzyme S Is a Biglutamic Acid ADP-Ribosyltransferase. Infection and Immunity. 1999;67(3):1508–1510. doi: 10.1128/iai.67.3.1508-1510.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maresso AW, Baldwin MR, Barbieri JT. Ezrin/Radixin/Moesin Proteins Are High Affinity Targets for ADP-ribosylation by Pseudomonas aeruginosa ExoS. Journal of Biological Chemistry. 2004;279(37):38402–38408. doi: 10.1074/jbc.M405707200. [DOI] [PubMed] [Google Scholar]
- 56.Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 Regulator of Kinase (Crk) Proteins. Journal of Biological Chemistry. 2003;278(35):32794–32800. doi: 10.1074/jbc.M304290200. [DOI] [PubMed] [Google Scholar]
- 57.Barth H, et al. Binary Bacterial Toxins: Biochemistry, Biology, and Applications of Common Clostridium and Bacillus Proteins. Microbiol. Mol. Biol. Rev. 2004;68(3):373–402. doi: 10.1128/MMBR.68.3.373-402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogelsgesang M, Pautsch A, Aktories K. C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn-Schmiedeberg's Archives of Pharmacology. 2007;374(5):347–360. doi: 10.1007/s00210-006-0113-y. [DOI] [PubMed] [Google Scholar]
- 59.Bulgin R, et al. The bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infect. Immun. :IAI.01250-09. doi: 10.1128/IAI.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kenny B, et al. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Molecular Microbiology. 2002;44(4):1095–1107. doi: 10.1046/j.1365-2958.2002.02952.x. [DOI] [PubMed] [Google Scholar]
- 61.Schwan C, et al. Clostridium difficile Toxin CDT Induces Formation of Microtubule-Based Protrusions and Increases Adherence of Bacteria. PLoS Pathog. 2009;5(10):e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hilger H, et al. The Long-Lived Nature of Clostridium perfringens Iota Toxin in Mammalian Cells Induces Delayed Apoptosis. Infection and Immunity. 2009;77(12):5593–5601. doi: 10.1128/IAI.00710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant CC, Messer RJ, Cieplak W. Role of trypsin-like cleavage at arginine 192 in the enzymatic and cytotonic activities of Escherichia coli heat-labile enterotoxin. Infection and Immunity. 1994;62(10):4270–4278. doi: 10.1128/iai.62.10.4270-4278.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heine K, et al. ADP-Ribosylation of Actin by the Clostridium botulinum C2 Toxin in Mammalian Cells Results in Delayed Caspase-Dependent Apoptotic Cell Death. Infection and Immunity. 2008;76(10):4600–4608. doi: 10.1128/IAI.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh S-Y, et al. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Molecular Microbiology. 2011;80(2):455–470. doi: 10.1111/j.1365-2958.2011.07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Libby SJ, et al. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cellular Microbiology. 2000;2(1):49–58. doi: 10.1046/j.1462-5822.2000.00030.x. [DOI] [PubMed] [Google Scholar]
- 67.Hernandez E, et al. Bacillus thuringiensis subsp.konkukian (Serotype H34) Superinfection: Case Report and Experimental Evidence of Pathogenicity in Immunosuppressed Mice. Journal of Clinical Microbiology. 1998;36(7):2138–2139. doi: 10.1128/jcm.36.7.2138-2139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McIntyre L, et al. Identification of Bacillus cereus Group Species Associated with Food Poisoning Outbreaks in British Columbia, Canada. Applied and Environmental Microbiology. 2008;74(23):7451–7453. doi: 10.1128/AEM.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuroki R, et al. Nosocomial Bacteremia Caused by Biofilm-Forming<i>Bacillus cereus</i>and <i/>Bacillus thuringiensis</i>. Internal Medicine. 2009;48(10):791–796. doi: 10.2169/internalmedicine.48.1885. [DOI] [PubMed] [Google Scholar]
- 70.Gibert M, et al. Endocytosis and toxicity of clostridial binary toxins depend on a clathrin-independent pathway regulated by Rho-GDI. Cellular Microbiology. 2011;13(1):154–170. doi: 10.1111/j.1462-5822.2010.01527.x. [DOI] [PubMed] [Google Scholar]
- 71.Pust S, Barth H, Sandvig K. Clostridium botulinum C2 toxin is internalized by clathrin- and Rho-dependent mechanisms. Cellular Microbiology. 2010;12(12):1809–1820. doi: 10.1111/j.1462-5822.2010.01512.x. [DOI] [PubMed] [Google Scholar]
- 72.Nicasio AM, Kuti JL, Nicolau DP. The Current State of Multidrug-Resistant Gram-Negative Bacilli in North America. Pharmacotherapy. 2008;28(2):235–249. doi: 10.1592/phco.28.2.235. [DOI] [PubMed] [Google Scholar]
- 73.Davies J, Davies D. Origins and Evolution of Antibiotic Resistance. Microbiology and Molecular Biology Reviews. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Micro. 2009;7(12):887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luna VA, et al. Susceptibility of Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides and Bacillus thuringiensis to 24 antimicrobials using Sensititre® automated microbroth dilution and Etest® agar gradient diffusion methods. Journal of Antimicrobial Chemotherapy. 2007;60(3):555–567. doi: 10.1093/jac/dkm213. [DOI] [PubMed] [Google Scholar]
- 76.Turnbull PCB, et al. MICs of Selected Antibiotics for Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, and Bacillus mycoides from a Range of Clinical and Environmental Sources as Determined by the Etest. Journal of Clinical Microbiology. 2004;42(8):3626–3634. doi: 10.1128/JCM.42.8.3626-3634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.