Abstract
Covalent disulfide bond linkage in a protein represents an important challenge for mass spectrometry (MS)-based top-down protein structure analysis as it reduces the backbone cleavage efficiency for MS/MS dissociation. This study presents a strategy for solving this critical issue via integrating electrochemistry (EC) online with top-down MS approach. In this approach, proteins undergo electrolytic reduction in an electrochemical cell to break disulfide bonds and then online ionized into gaseous ions for analysis by electron-capture dissociation (ECD) and collision-induced dissociation (CID). The electrochemical reduction of proteins allows to remove disulfide bond constraints and also leads to increased charge numbers of the resulting protein ions. As a result, sequence coverage was significantly enhanced, as exemplified by β-lactoglobulin A (24 vs. 73 backbone cleavages before and after electrolytic reduction, respectively) and lysozyme (5 vs. 66 backbone cleavages before and after electrolytic reduction, respectively). This methodology is fast and does not need chemical reductants, which would have an important impact in high-throughput proteomics research.
Top-down mass spectrometry (MS)1–10 has unparalleled strength in protein analysis, which can be used to rapidly identify protein identity1, 5, 11 and to provide extensive molecular connectivity information including labile post-translational modifications (PTMs)12. Despite these striking advantages, it has some challenging issues. For instance, in the case of large proteins, the fragments resulting from electron-capture dissociation (ECD)13 may not be separated from each other due to non-covalent intramolecular interactions of the residues.12 Demonstrated solutions for this issue include prefolding-dissociation (PFD)14 and activated-ion ECD (AI-ECD)15 which unfold proteins and break the intramolecular interactions prior to ECD. Likewise, covalent disulfide bond linkage in a protein represents another challenge for top-down protein analysis. Disulfide bond is one of the most common PTMs (~19% proteins contain multiple disulfide bonds16), which is vital for maintaining the protein structure stability.17 However, the dissociation efficiency of the protein backbone by electron-based ion dissociation is greatly reduced in the presence of disulfide bonds as the latter would be preferentially broken,11, 18 which is exemplified by no fragment ions18 observed for ECD of ribonuclease A ions and a poor sequence coverage for the disulfide-bond protected regions of antibody IgGs19. Thus top-down protein analysis with high sequence coverage is limited to those without disulfide bonds or those with relatively simple disulfide linkages like insulin.18 For proteins containing complicated disulfide bonds such as lysozyme,20 chemical reduction of disulfide bonds using reducing reagents like dithiothreitol (DTT) are often performed prior to top-down analysis. Such a chemical reduction is usually not carried out online with MS analysis of proteins/peptides. The offline reduction therefore could limit the applications of top-down approach to high throughput analysis or rapid identification of protein isoforms.1 Furthermore, the chemical reduction takes minutes to hours, and the removal of the excess amount of reductants is also time-consuming.21 To tackle this critical problem, our strategy in this study is to couple electrochemistry (EC) with top-down MS, in which proteins are rapidly reduced electrochemically followed with online MS and tandem MS analysis. Remarkably, detectable protein backbone cleavages was significantly enhanced by 3–13 folds with the assistance of electrochemical reduction that removes the protein disulfide bond constraints, as revealed by the ECD and collision induced dissociation (CID) data of two chosen proteins, β-lactoglobulin A and lysozyme.
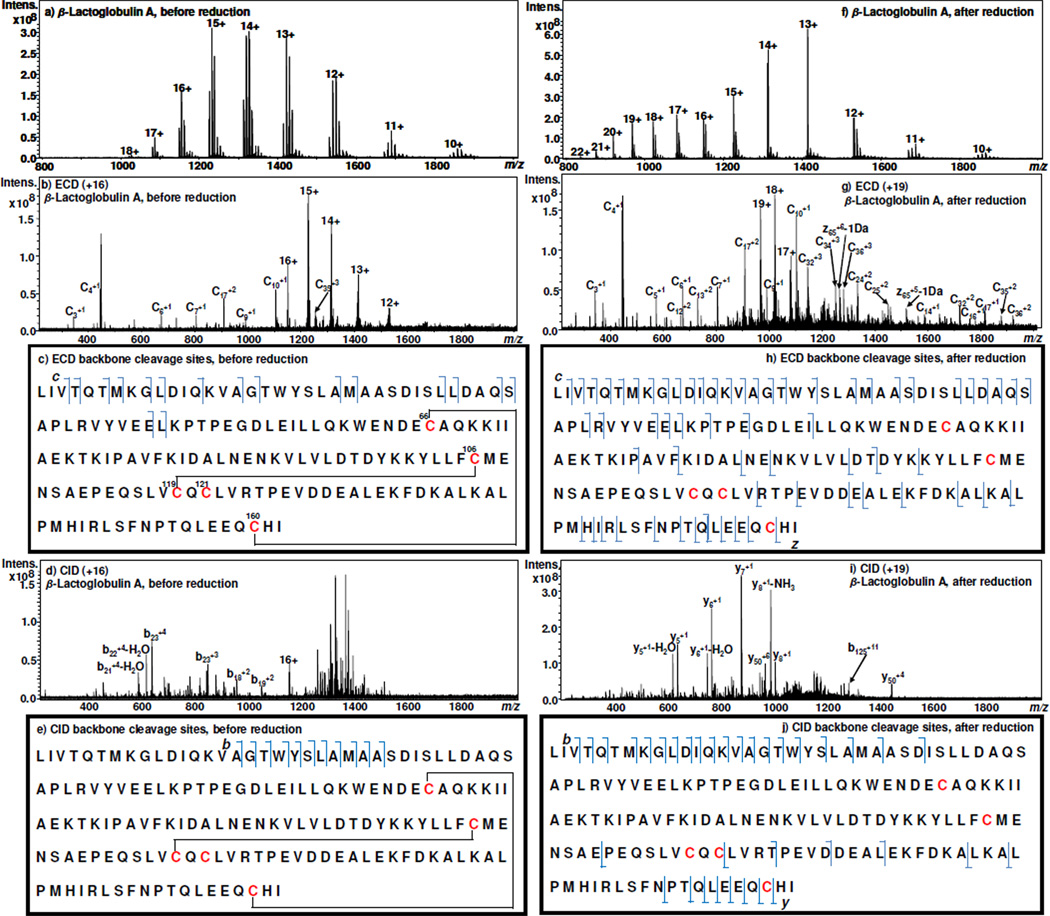
The experimental apparatus of this methodology consists of a thin-layer μ-PrepCell™ electrochemical flow cell (Antec BV, Zoeterwoude, Netherlands) online coupled with a Bruker 12 Tesla SolariX FTICR-MS (Bruker Daltonics, Bremen, Germany) using desorption electrospray ionization (DESI)22 as an interface. In our laboratory, DESI has been shown to be suitable for transfer/ionize electrolyzed compounds from solution to the gas phase for MS detection.23–26 Detailed apparatus and experimental conditions of this study are described in the Supporting Information. β-lactoglobulin A containing two disulfide bridges (Cys66–Cys160 and Cys106–Cys119) and one free Cys121 (refer to Figure 1c for its sequence)27 was first chosen as an example for this study. Figure 1a illustrates the DESI-MS spectrum acquired when a solution of 15 µM β-lactoglobulin A in methanol/water (1:1 by volume) containing 0.5% formic acid flowed through the electrochemical cell with no potential applied to the cell, the multiply charged ions of intact β-lactoglobulin A with a charge state distribution (CSD) of +10~+18 with the most intense peak at +15 were detected. When the +16 protein ion (m/z 1148.0) was selected for ECD, only 17 c ions were observed in the acquired spectrum, arising from the cleavage of the free protein N-terminal where there is no disulfide bond (the major c ions are labeled in the ECD MS/MS spectrum of Figure 1b and all detected fragment ions are marked in Figure 1c). There was no detected fragment ion resulting from broken backbone bonds that are protected by either or both of the disulfide bonds Cys66–Cys160 and Cys106–Cys119. This result can be accounted for by the aforementioned reason that the cleavage of protein backbone bonds by ECD in the presence of disulfide bonds has low efficiency,11, 18, 28 and the observation of fragment ions from those disulfide bond-protected regions requires the cleavage of both backbone and the disulfide bonds. Likewise, in the case of CID of the +16 ion, it is difficult to produce protein fragment ions via backbone cleavages of the residues enclosed by two disulfide bonds because CID in the positive ion mode is known to be unable to effectively cleave disulfide bonds.14, 18, 29 As a result, few fragment ions b16, b17, b18, b19, b20, b21, b22, b23, b24, b25, and b26 were generated from N-terminal cleavages, as illustrated in Figures 1d and e.
Figure 1.
DESI-MS spectra acquired when a solution of 15 µM β-lactoglobulin A in methanol/water (1:1 by volume) containing 0.5% formic acid flowed through the thin-layer electrochemical cell with an applied potential of (a) 0.0 V and (f) −1.2 V; ECD MS/MS spectra of (b) +16 ion of the intact β-lactoglobulin A (m/z 1148.0) and (g) +19 ion of the reduced β-lactoglobulin A (m/z 967.1); The marked ECD backbone cleavage sites of (c) β-lactoglobulin A and (h) reduced β-lactoglobulin A; CID MS/MS spectra of (d) +16 ion of the intact β-lactoglobulin A (m/z 1148.0) and (i) +19 ion of the reduced β-lactoglobulin A (m/z 967.1); The marked CID backbone cleavage sites of (e) β-lactoglobulin A and (j) reduced β-lactoglobulin A.
By contrast, when a −1.2 V potential was applied to the electrochemical cell to trigger online disulfide bond reduction, the CSD of protein ions was shifted to higher charges with the appearance of a new population of +19~+22 (Figure 1f), indicating a conformational change occurring to the protein due to the reduction.30–32 This phenomenon is in agreement with the early ESI report that increased charges of the protein ions occurred after the cleavage of disulfide bonds31 and our recent observation25. Indeed, the reduction of disulfide bonds in a protein causes the unfolding of the protein, thus allowing the protein to have a greater capacity to accommodate a larger number of charges on its surface.30–32 Thus the newly produced +19 ion was chosen for ECD analysis, under the same experimental conditions used for ECD of +16 unreduced protein ion as described above. As shown in the ECD MS/MS spectrum (Figure 1g) and the labeled sequence map (Figure 1h), 40 c ions and 26 z ions were generated. In addition, 23 b and 12 y ions were observed in the CID of +19 ion, as displayed in Figures 1i and j. These results represent a significant increase the backbone cleavage after electrolytic reduction (totally 73 backbone cleavages from the combined ECD and CID data) in comparison to that before reduction (totally 24 backbone cleavages from the combined ECD and CID data). This change, the approximately 3 fold enhancement in observable backbone cleavages, is ascribed to electrochemical assistance in removing S-S constraints. It might be also contributed to the resulting increased charge states as a result of disulfide bond reduction.6, 33 Although the number of ECD cleavages could be further improved by pre-activation of the precursor ions,14 this was not performed because the purpose in this study is to investigate the effect of electrochemical reduction.
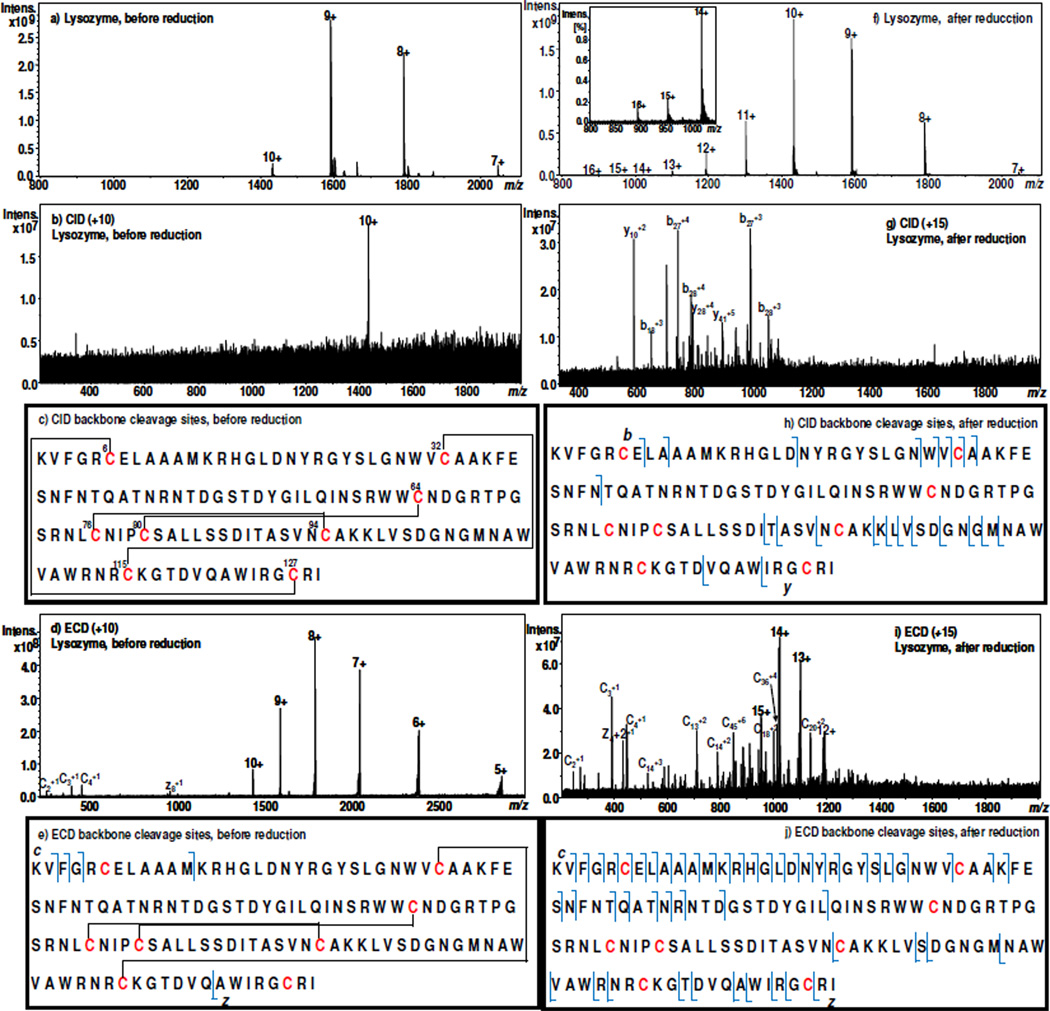
We further examined chicken lysozyme, a 14 kDa protein having 129 amino acid residues and containing four disulfide bonds (Cys6–Cys127, Cys30–Cys115, Cys64–Cys80, and Cys76–Cys94). The latter two disulfide bonds are intertwined and further enclosed by the former two bonds (Figures 2c). Figure 2a shows the DESI-MS spectrum acquired when a solution of 15 µM lysozyme in methanol/water (1:1 by volume) containing 0.5% formic acid flowed through the electrochemical cell, the intact lysozyme exhibits the CSD of +7~+10 with the most abundant peak located at +9. Interestingly, no fragments were observed upon CID of +10 ion of m/z 1430.5 (Figures 2b and c). This result is not surprising as the protein has 4 cross-linked disulfide bonds that protect most of the protein sequence. Again, ECD of +10 ion (m/z 1430.5) gives rise to few fragment ions of c2, c3, c4, c12, and z8 with low abundances, as displayed in Figures 2d and e. This is in agreement with the previous report about the ECD of intact lysozyme ions.20 In this case, although the backbone dissociation efficiency is very low, the cleavage of one disulfide bond (Cys6–Cys127) by ECD occurred, evidenced by the formation of c12, and z8. When a −1.5 V potential was applied to the cell to reduce the protein, the CSD of the protein ions was broadened and shifted to the higher charge states (+7~+16, Figure 2f). It appears that the electrochemical reduction can serve as a stimulant to trigger protein conformational change by reducing S-S linkages to eliminate the protein tertiary structure, which can be monitored online with DESI-MS detection. As shown in the CID mass spectrum of +15 ion (m/z 954.7) of the reduced lysozyme (Figure 2g) and the labeled backbone cleavage sites (Figure 2h), b7, b9, b18, b27, b28, b29, b30, b31, b39, y6, y10, y24, y25, y26, y28, y30, y31, y32, y33, y37, y40 and y41 were generated after electrolytic reduction of lysozyme, which is in stark contrast with the CID of intact protein ion (Figure 2b). In addition, much more backbone cleavages took place with ECD. As shown in Figures 2i and j, 36 c ions and 14 z ions were observed upon ECD of the newly produced +15 ion (this ECD result is similar to ECD of chemically reduced mononitrated lysozyme which shows a total of 59 fragment ions20). Thus, it can be seen that, with electrochemical assistance to remove disulfide bond linkages, the sequence information for lysozyme was remarkably enhanced. Based on the combined CID and ECD data, totally 5 and 66 unique backbone cleavages were resulted before and after electrolysis, respectively). Other methodologies have been reported for enhancing the top-down dissociation efficiency, including CID of low charged lysozyme ions,11 which showed a maximum of 13 backbone cleavages. In contrast, our method is more effective.
Figure 2.
DESI-MS spectra acquired when a solution of 15 µM lysozyme in methanol/water (1:1 by volume) containing 0.5% formic acid flowed through the thin-layer electrochemical cell with an applied potential of (a) 0.0 V and (f) −1.5 V (the zoomed-in mass range for +14~+16 for the reduced lysozyme is shown in the inset); CID MS/MS spectra of (b) +10 ion of the intact lysozyme (m/z 1430.5) and (g) +15 ion of the reduced lysozyme (m/z 954.7); The marked CID backbone cleavage sites of (c) lysozyme and (h) reduced lysozyme; ECD MS/MS spectra of (d) +10 ion of the intact lysozyme (m/z 1430.5) and (i) +15 ion of the reduced lysozyme (m/z 954.7); The marked ECD backbone cleavage sites of (e) lysozyme and (j) reduced lysozyme.
It is evident that substantially increased backbone cleavage by ECD and CID (3–13 fold increments as shown with the two chosen proteins in this study) occurs to the electrolytic reduced protein in comparison to intact protein, producing many more structurally informative fragment ions. The electrolytic reduction for the disulfide bonds is fast and takes place instantaneously when the reduction potential is applied. Equally importantly, it is a clean and “green” approach as it does not involve the use of toxic chemical reductants. Comparison to DTT reduction was also made, using lysozyme as an example. The protein did not show any reduction at pH of 3 with DTT in 100 fold excess amount even when the lysozyme was incubated with DTT for overnight (Figure S-2b, Supporting Information). At pH 8, the protein can be reduction and the complete reduction takes ca. 90 min (see Figures S-2c to S-2f, Supporting Information). The pH of the sample solution needs to be adjusted to be acidic prior to MS analysis. The reduction of lysozyme by DTT for 90 min (Figure S-2f, Supporting Information) does show higher reduction efficiency than the electrolytic reduction shown in Figure 2f (as indicated by the higher relative abundances of newly produced protein ion peaks in Figure S-2f). However, the electrolytic reduction efficiency could be significantly improved, using a different electrode (Figure S-3, Supporting Information). Overall, the online electrochemistry employed is well suited for the top-down protein analysis, which would be particularly valuable in high-throughput proteomics applications. It is also an addition to the study of proteins by combined electrochemistry and mass spectrometry.34, 35
Supplementary Material
ACKNOWLEDGMENT
Financial support from NSF (CHE-0911160), instrument support from Prof. Michael L. Gross to access the NIH/NCRR Mass Spectrometry Resources at Washington University in St. Louis (Grant number 2P41RR000954) and High-End Instrument Program of the NCRR (Grant No. 1S10 025101), and research support from Antec BV Company, Dr. Agnieszka Kraj and Mr. Martin Eysberg are gratefully acknowledged.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental apparatus, procedure description and additional supporting data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SMM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelleher NL, Lin HY, Valaskovic GA, Aaseruud DJ, Fridriksson EK, McLafferty FW. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- 3.Kelleher NL. Anal. Chem. 2004;76:196A–203A. [PubMed] [Google Scholar]
- 4.Chait BT. Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 5.Cui W, Rohrs HW, Gross ML. Analyst. 2011;136:3854–3864. doi: 10.1039/c1an15286f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, Zhang J, Yin S, Loo JA. J. Am. Chem. Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 7.Sohn CH, Chung CK, Yin S, Ramachandran P, Loo JA, Beauchamp JL. J. Am. Chem. Soc. 2009;131:5444–5459. doi: 10.1021/ja806534r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reid GE, McLuckey SA. J. Mass Spectrom. 2002;37:663–675. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- 9.Breuker K, McLafferty FW. Angew. Chem. Int. Ed. 2005;44:4911–4914. doi: 10.1002/anie.200500668. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Ge Y. Circ. Cardiovasc. Genet. 2011;4:711–711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Shiyanov P, Zhang L, Schlager JJ, Green-Church KB. Anal. Chem. 2010;82:6079–6089. doi: 10.1021/ac1006766. [DOI] [PubMed] [Google Scholar]
- 12.McLafferty FW, Breuker K, Jin M, Han X, Infusini G, Jiang H, Kong X, Begley TP. FEBS J. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 13.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 14.Han X, Jin M, Breuker K, McLafferty FW. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 15.Horn DM, Ge Y, McLafferty FW. Anal. Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 16.Alegre-Cebollada J, Kosuri P, Rivas-Pardo JA, Fernandez JM. Nature Chemistry. 2011;3:882–887. doi: 10.1038/nchem.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sevier CS, Kaiser CA. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 18.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. J. Am. Chem. Soc. 1999;121:2857–2862. [Google Scholar]
- 19.Tsybin YO, Fornelli L, Stoermer C, Luebeck M, Parra J, Nallet S, Wurm FM, Hartmer R. Anal. Chem. 2011;83:8919–8927. doi: 10.1021/ac201293m. [DOI] [PubMed] [Google Scholar]
- 20.Mikhailov VA, Iniesta J, Cooper HJ. Anal. Chem. 2010;82:7283–7292. doi: 10.1021/ac101177r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bilusich D, Bowie JH. Mass Spectrom. Rev. 2009;28:20–34. doi: 10.1002/mas.20206. [DOI] [PubMed] [Google Scholar]
- 22.Takats Z, Wiseman JM, Gologan B, Cooks RG. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Dewald HD, Chen H. Anal. Chem. 2009;81:9716–9722. doi: 10.1021/ac901975j. [DOI] [PubMed] [Google Scholar]
- 24.Miao Z, Chen H. J. Am. Soc. Mass Spectrom. 2009;20:10–19. doi: 10.1016/j.jasms.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Dewald HD, Chen H. J. Proteome Res. 2011;10:1293–1304. doi: 10.1021/pr101053q. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Yuan Z, Dewald HD, Chen H. Chem. Commun. 2011;47:4171–4173. doi: 10.1039/c0cc05736c. [DOI] [PubMed] [Google Scholar]
- 27.Surroca Y, Haverkamp J, Heck AJR. J. Chromatogr. A. 2002;970:275–285. doi: 10.1016/s0021-9673(02)00884-1. [DOI] [PubMed] [Google Scholar]
- 28.Zubarev RA, Haselmann KF, Budnik B, Kjeldsen F, Jensen F. Eur.J. Mass Spectrom. 2002;8:337–349. [Google Scholar]
- 29.Ryan CM, Souda P, Halgand F, Wong DT, Loo JA, Faull KF, Whitelegge JP. J. Am. Soc. Mass Spectrom. 2010;21:908–917. doi: 10.1016/j.jasms.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katta V, Chait BT. J. Am. Chem. Soc. 1991;113:8534–8535. [Google Scholar]
- 31.Loo JA, Edmonds CG, Udseth HR, Smith RD. Anal. Chem. 1990;62:693–698. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 32.Kaltashov IA, Abzalimov RR. J. Am. Soc. Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Vasicek L, Brodbelt JS. Anal. Chem. 2009;81:7876–7884. doi: 10.1021/ac901482s. [DOI] [PubMed] [Google Scholar]
- 34.Permentier HP, Jurva U, Barroso B, Bruins AP. Rapid Commun. Mass Spectrom. 2003;17:1585–1592. doi: 10.1002/rcm.1090. [DOI] [PubMed] [Google Scholar]
- 35.Permentier HP, Bruins AP. J. Am. Soc. Mass Spectrom. 2004;15:1707–1716. doi: 10.1016/j.jasms.2004.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.