Abstract
Purpose: The PathVisio-Faceted Search plugin helps users explore and understand complex pathways by overlaying experimental data and data from webservices, such as Ensembl BioMart, onto diagrams drawn using formalized notations in PathVisio. The plugin then provides a filtering mechanism, known as a faceted search, to find and highlight diagram nodes (e.g. genes and proteins) of interest based on imported data. The tool additionally provides a flexible scripting mechanism to handle complex queries.
Availability: The PathVisio-Faceted Search plugin is compatible with PathVisio 3.0 and above. PathVisio is compatible with Windows, Mac OS X and Linux. The plugin, documentation, example diagrams and Groovy scripts are available at http://PathVisio.org/wiki/PathVisioFacetedSearchHelp. The plugin is free, open-source and licensed by the Apache 2.0 License.
Contact: augustin@mail.nih.gov or jakeyfried@gmail.com
1 Introduction
Pathway diagrams are important tools that facilitate understanding biological systems by aggregating many disparate observations into concise representations. There is a growing trend to use formal graphical notations, such as the Molecular Interaction Map (MIM) notation and the Systems Biology Graphical Notation (SBGN), and formats, such as BioPAX, to relate greater mechanistic detail about pathway interactions than is typical in protein–protein interaction networks (Demir et al., 2010; Le Novère et al., 2009; Luna et al., 2011). The well-known tool Cytoscape possesses powerful annotation and search capabilities, but it is focused on network analysis, not diagram rendering (Smoot et al., 2011). This tool lacks the capabilities necessary to render diagrams that are fully compliant with MIM or SBGN. The CySBGN plugin is the best current attempt, but has some rendering problems, such as z-ordering issues (Goncalves et al., 2013).
PathVisio (http://pathvisio.org/) is a free open-source tool built to display and edit diagrams using formalized notations (van Iersel et al., 2008). PathVisio is capable of visualizing microarray and metabolomics data, similar to GenMAPP, but lacks other search features including the visualization and search of nominal data, which limits analysis, especially on the growing number of pathways in the related WikiPathways database (Dahlquist et al., 2002; Pico et al., 2008).
Here, we present a robust search plugin for PathVisio using the faceted search method that is found on many e-commerce websites for product search using discrete attributes, known as facets. Facet entries can be selected to filter down to content of interest, and faceted searching is found in bioinformatics tools, including the Protein Data Bank (http://www.rcsb.org/). Facets allow users to attach data to pathways using biological identifiers. Any piece of data that can be looked up in a database based on identifiers, such as ‘HGNC: TP53’, can be used as a facet. For example, a user might wish to know which pathway proteins are involved in cell division, are targets of cancer drugs and are over-expressed in an experiment. This plugin allows users to easily ask complex queries of biological pathways and identify biological entities of interest through the powerful search capability on user data, webservice data and data processed using its scripting capability.
2 Implementation
The plugin interface and dataservice options are described below:
Plugin interface: The plugin’s interface is found in the ‘Facets’ pane of the PathVisio sidebar. Figure 1 shows a search example for pathway components that are transcribed in lymphomas and encoded by genes on chromosomes 9 and 17. The main area of the sidebar is where facets appear. At the bottom, are buttons for managing dataservices and adding facets. There are two types of facets: string and numeric. String facets are for nominal data (e.g. cellular location). Entries for all the nodes are displayed initially. For example, clicking on ‘nucleus’ in a cellular location facet will highlight only nodes annotated as existing in the nucleus and removes entries from other facets for nodes not in the nucleus. Numeric facets allow users to find values in a range. This facet type returns a Boolean response or error for missing values. All facets entries have appended counts with the number of nodes satisfying a particular condition. When using multiple facets, a selection in one facet restricts entries across all the facets, as fewer nodes are able to satisfy each additional condition. Active facet data and highlighted nodes can be exported for use in future sessions or in other software.
Compatible pathways: This plugin can operate on any pathway loaded into PathVisio, but it only functions on nodes with database identifiers. This is a key requirement for connecting imported data to the nodes on the diagram. Many database providers use unique identifier schemas for the same biological entities, thus requiring ID mapping; this plugin uses BridgeDb as its mapping service (van Iersel et al., 2010). BridgeDb works either through a local database loaded in PathVisio or the BridgeREST API.
Dataservices: The plugin can use (i) local files formatted as tab-separated values (TSV) or comma-separated values (CSV) files, (ii) Ensembl BioMart webservice and (iii) Groovy scripts.
Local files: TSV/CSV files can be added as dataservices. Each file row maps to a node and each column to a facet. The first file row should be headers describing the column contents (e.g. HGNC Symbol, Molecular Weight, etc.) and are used as the facet titles. At least one column must contain identifiers; the identifier column and type is specified in the ‘Manage Dataservices’ dialogue.
BioMart webservice: The BioMart (http://www.biomart.org/) webservice provides the ability to query multiple biological databases through a single interface (Guberman et al., 2011). This dataservice is built-in to the plugin, and can be enabled or disabled. A subset of ∼70 Ensembl attributes is available through this dataservice; other attributes are available through the plugin’s scripting functionality. Turning on the dataservice makes available all these attributes in the ‘Add Facet’ dialogue. Adding a facet from BioMart may take some time as data must be downloaded. Currently, data are not cached between PathVisio sessions, and performance information is provided on the project website.
Groovy scripting: This dataservice provides additional flexibility by incorporating Groovy (http://groovy.codehaus.org), a scripting language compatible with Java. Groovy may be used to perform additional logic, incorporate and pre-process other data formats and access other webservices. These are added by clicking ‘Add Groovy’ in the ‘Manage Dataservices’ dialogue. There are two fields in the ‘Add Groovy’ dialogue: one for the facet name and the other for Groovy code. Identifiers may be converted to a specific format (e.g. for custom webservices) before being passed to the Groovy script; this is specified in the ‘Manage Dataservices’ dialogue, and the default setting is to perform no ID conversion. The Groovy script will be run once for each diagram node with an identifier and expects a return string or an array of strings to be returned as facet entries. The online documentation goes into detail on how to use the Groovy scripting features and provides in-depth examples of its use. The first example illustrates how users may connect histone binding data to diagram nodes by using Enseml BioMart to produce a facet. Other included examples demonstrate retrieving information from ChEMBL (http://www.ebi.ac.uk/chembl/) and from QuickGO (http://www.ebi.ac.uk/QuickGO/).
Fig. 1.
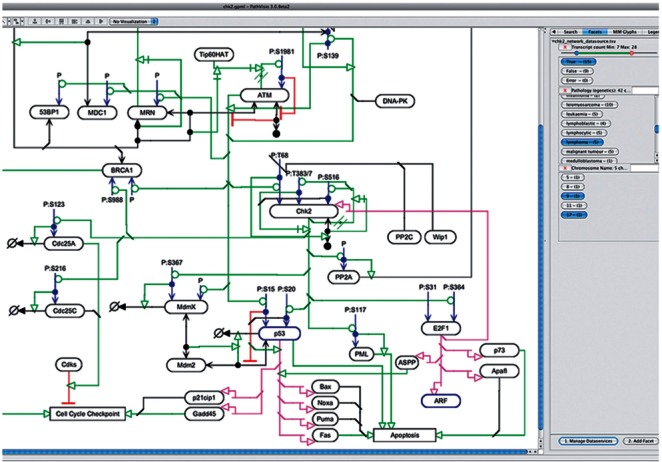
The Faceted Search plugin used to identify genes with a select number of variants, related to lymphomas, and located on specified chromosomes resulting in two highlighted nodes
3 Conclusion
The PathVisio-Faceted Search plugin was designed to help users understand complex pathways by making it easier to cross-reference diagrams with data from multiple sources, both local experimental data and via webservices. Faceted search is a widely used search method that overcomes the limitations of more conventional methods to easily find relevant items. In the future, we plan to introduce a visual programming interface to simplify the Groovy scripting component of the plugin and make the flexibility of this scripting support accessible to more users.
Acknowledgements
We thank the PathVisio community, Margot Sunshine and Alexander Pico for their help with this project.
Funding: Project was funded by Google Summer of Code and the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute.
Conflict of Interest: none declared
References
- Dahlquist KD, et al. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Demir E, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves EJ, et al. CySBGN: a cytoscape plug-in to integrate SBGN maps. BMC Bioinformatics. 2013;14:17. doi: 10.1186/1471-2105-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guberman JM, et al. BioMart Central Portal: an open database network for the biological community. Database (Oxford) 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N, et al. The systems biology graphical notation. Nat. Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- Luna A, et al. A formal MIM specification and tools for the common exchange of MIM diagrams: an XML-based format, an API, and a validation method. BMC Bioinformatics. 2011;12:167. doi: 10.1186/1471-2105-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico AR, et al. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:4. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iersel MP, et al. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iersel MP, et al. The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services. BMC Bioinformatics. 2010;11:5. doi: 10.1186/1471-2105-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


