Abstract
Background
Very little is known regarding correlation of micro RNA (miR)–106a with clinical outcomes of patients with glioblastoma multiforme (GBM). This study determined whether miR-106a could be used as an independent prognostic biomarker in those patients.
Methods
A total of 156 GBM patients were divided into 2 cohorts. In the first cohort, matched fresh frozen and formalin-fixed paraffin-embedded (FFPE) samples were collected from 24 GBM patients, while in the second cohort, only FFPE samples were collected from 132 GBM patients. MiR-106a expression levels were examined by quantitative real-time PCR in the 2 cohorts and further validated by in situ hybridization assay in the second cohort. The correlation between miR-106a expression levels and overall survival was evaluated in the second cohort of 114 GBM patients available for follow-up by a log-rank test and a multivariate Cox proportional hazards model.
Results
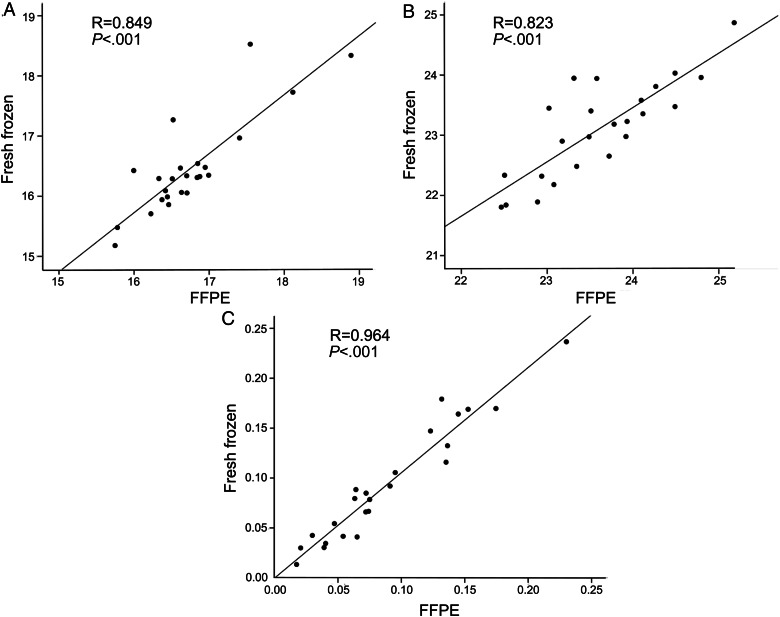
Our data showed a very good correlation of miR-106a or U6 expression between fresh frozen and FFPE GBM specimens, with Pearson's correlation coefficients of 0.849 and 0.823, respectively (P < .001). Their expression levels in archival FFPE samples were quite stable for at least 7 years when stored at room temperature. Multivariate analysis revealed that the expression level of miR-106a was an independent and significant predictor of overall survival in GBM patients (P = .011).
Conclusions
MiR-106a expression was relatively abundant and stable in a large cohort of archival FFPE GBM specimens and could be used as an independent prognostic biomarker in those patients. Thus, miR-106a can be used to predict prognosis and treatment response in individual GBM patients.
Keywords: FFPE, glioblastoma, miR-106a, prognosis, prognostic marker
Glioblastoma multiforme (GBM) is the most frequent primary malignant brain tumor in adults. It is characterized by extremely aggressive invasion and destructive malignancy with a high proliferation rate. Despite aggressive multimodal therapy, patients with GBM have a poor prognosis, with a median survival of about 12–15 months.1,2 Established prognostic factors, such as age at diagnosis, histologic characteristics, and extent of surgical resection, inadequately predict the clinical outcomes of GBM.3 Therefore, identification of novel prognostic indicators for predicting patient outcomes in GBM has a clinical significance. In addition, the investigation of prognostic indicators may elucidate the underlying biological mechanisms involved in the development and progression of GBM. Recently, an important class of small noncoding RNA species, known as micro RNAs (miRNAs), have been shown to regulate critical functions in many cellular and physiological processes, such as cellular proliferation, differentiation, apoptosis, and stem cell maintenance.4 Although the total number of miRNAs remains controversial, characterization of miRNA expression patterns in cancer is thought to have important value for diagnostic and prognostic determinations as well as for target-specific therapy.5 Several miRNAs, such as miR-200c, miR-181a, and miR-210, have recently been reported to act as prognostic biomarkers in ovarian cancer, acute myeloid leukemia, and breast cancer.6–8 However, clinical prognostic miRNA biomarkers in GBM remain unclear.9,10
Formalin-fixed paraffin-embedded (FFPE) samples are an invaluable tool for biomarker discovery and validation because a large number of human FFPE specimens with their detailed clinical annotation have been archived by pathology laboratories and tissue banks, and fresh frozen samples are often difficult to obtain and require special storage conditions. Moreover, recent reports have demonstrated that expression levels of miRNAs are relatively stable in FFPE samples.11,12 This allows us to discover new clinical prognostic miRNA biomarkers of tumors using those archival FFPE samples. In our previous study, we found that miR-106a expression was significantly correlated with the malignant progression of human gliomas.13 Thus, our aim was to investigate whether miR-106a expression in archival FFPE GBM specimens could predict clinical outcomes of GBM patients.
Materials and Methods
Patients With GBM and Their Surgically Excised Specimens
A total of 156 GBM patients enrolled in the Department of Neurosurgery at The First Affiliated Hospital of Harbin Medical University in China from 2005 to 2011 were divided into 2 cohorts. In the first cohort, matched fresh frozen and FFPE samples were collected from 24 GBM patients who underwent surgery in our institute between 2010 and 2011, while in the second cohort, only FFPE samples were collected from 132 GBM patients. The mean age of patients was 48 years (range, 8–80 y); 62% were male. The median preoperative KPS score was 70. Gross total resection (GTR) of the GBM tumor was achieved in 94 patients (71.21%) and subtotal resection (STR) in 29 patients (21.97%) in the second cohort. GTR was defined as no residual enhancement on MRI <48h after surgery, and STR as having residual nodular enhancement.14 All except 8 patients received postoperative radiotherapy. Patients in the current series did not receive chemotherapy. The prognosis was evaluated in all GBM patients with archival FFPE samples in May 2011, and follow-up data were available for 114 cases. Ninety-seven patients (73.48%) died during the follow-up period. The median observation time for overall survival (OS) was 10 months (range, 1–47 mo).
FFPE tissue samples were routinely collected for histologic diagnosis in strict accordance with World Health Organization criteria, while adjacent specimens were immediately snap-frozen in liquid nitrogen and then stored at −80°C until further analysis. The matched fresh frozen and FFPE specimens were stored from 1 to 2 years, while the archival FFPE blocks ranged in age from 3 to 7 years. Normal brain tissues were obtained from normal adjacent tissues away from tumor tissues or nonneoplastic brain diseases and were histologically confirmed to be free of any pathological lesions. The study was approved by the Institutional Review Board of Harbin Medical University, and subjects gave informed consent.
Sample Preparation
For fresh frozen tissues, a sample of ∼0.5 cm3 in dimension was used for RNA extraction. Total RNA was extracted using a Trizol standard protocol (Invitrogen). For FFPE samples, total RNA was extracted from 5 to 10 sections of 10-μm-thick tissue using the Ambion RecoverAll Kit, and DNA was isolated from tissue sections using the QIAamp DNA FFPE Kit (Qiagen) according to the manufacturer's instructions. Nucleic acid quantitative and qualitative analyses were carried out by a spectrophotometer (Tecan). Synthesis of cDNA was performed with a TaqMan miRNA reverse transcription kit (Applied Biosystems).
Detection of MiRNA Expression
Quantitative real-time PCR
The expression levels of miR-106a, miR-182, miR-196a, and U6 were quantified by quantitative real-time (qRT) PCR using the TaqMan MiRNA Assay Kit (Applied Biosystems) on the Applied Biosystems 7500HT Fast Real-Time PCR System. The expression of miRNAs was defined based on the threshold cycle (Ct), and relative expression levels were calculated using the 2–△△CT method15 after normalization with reference to expression of U6 small nuclear (sn)RNA.
In situ hybridization
In situ hybridization (ISH) was performed with locked nucleic acid probes to detect expression levels of human miR-106a, scramble-miR, or U6 (Exiqon) in FFPE GBM tissues as previously described.16,17 Briefly, after being heated at 75°C for 2h, 5-μm-thick sections of FFPE specimens were deparaffinized in xylene and rehydrated in an ethanol dilution series. Slides were immersed in diethyl pyrocarbonate–treated water at 90°C for 30min, then treated with 30 µg/mL proteinase K (Sigma) at 37°C for 5.5 min and refixed in 4% paraformaldehyde. Slides were prehybridized in a hybridization solution (50% formamide, 5× standard saline citrate [SSC], 0.1% Tween, 9.2 mmol/L citric acid for adjustment to pH 6.0, 50 µg/mL heparin, 500 µg/mL yeast RNA) at 60°C for 5 min, and then at 49.5°C for 1 h. Subsequently, 20 nmol/L of locked nucleic acid–modified 5′ Digoxigenin (DIG)–labeled probe was added to 100 µL of the hybridization solution and hybridized at 49.5°C overnight. Sections were rinsed twice in phosphate buffered saline (PBS)/0.1% NP-40 (nonyl phenoxypolyethoxylethanol) and subsequently blocked in 2× SSC with 2% bovine serum albumin (BSA) at 4°C. An anti-DIG antibody (Roche) at 1:1000 dilution in PBS/1% BSA was applied to the slides at room temperature for 30 min, followed by 3 washes in PBS/0.1% Tween 20. After washing, the sections were incubated with nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine (NBT/BCIP; Roche) at 37°C. The colorimetric reaction was monitored visually and stopped by placing the slides in water when background coloring started to appear on the negative control (scrambled probe), varying 30–60 min.
Scoring of ISH
ISH signals for miR-106a in the GBM specimens were scored by combining the intensity of staining and the proportion of positively stained tumor cells, as previously described.16,18–20 Briefly, the dominant staining intensity in tumor cells was scored as 0 = negative, 1 = weak, 2 = intermediate, and 3 = strong. The proportion of positively stained tumor cells was graded as <10% positive tumor cells, 10%–50% positive tumor cells, and >50% positive tumor cells. Low expression of miR-106a was described as an intensity of 0, 1, 2, or 3 and <10% stained cells or an intensity of 0 or 1 and <50% stained cells, whereas high expression of miR-106a was described as an intensity of 2 or 3 and >10% stained cells or an intensity of 1, 2, or 3 and >50% stained cells. The signals of ISH of tissue sections were scored by 2 independent investigators blinded to patient data.
Sequence Analysis of Isocitrate Dehydrogenase 1 and Tumor Protein p53
The genomic region spanning R132 of isocitrate dehydrogenase (IDH)1 was analyzed by direct sequencing as described previously.21 Mutations of tumor protein p53 (TP53) exons 5–8 were screened as described by Weller et al.22
Fluorescence ISH
Dual-probe fluoresence ISH (FISH) was performed on the FFPE sections with locus-specific probes for epidermal growth factor receptor (EGFR) and centromere of chromosome 7 (Vysis). Standard FISH protocols were followed according to the manufacturer's instructions.
Immunohistochemistry
FFPE sections were immunostained for E2F1 and MIB-1 proteins. Staining was performed with the streptavidin–biotin peroxidase complex method according to the manufacturer's recommendation (Dako). Mouse anti-human E2F1 (1:100; Santa Cruz Biotechnology) or MIB-1 (1:100; Santa Cruz) primary antibody was administered, followed by secondary goat anti-mouse immunoglobulin G (Dako). Negative controls were performed throughout the entire immunohistochemistry procedure. Only nuclear E2F1 or MIB-1 staining was considered positive. Slides were scored on the basis of the percentage of positive tumor cells. All of the immunostained sections were reviewed in blinded fashion by 2 investigators.
Statistical Analysis
A Pearson's correlation coefficient was employed to determine the correlation of miR-106a expression between matched FFPE and fresh frozen samples. An ANOVA and t-test were used for comparison of expression levels of miRNAs between FFPE GBM and normal brain samples or at different storage time points. Fisher's exact test and a Wilcoxon rank-sum test were employed to detect the correlation of miR-106a expression levels with clinical and molecular features of GBM patients. A chi-square test was used to examine the association of miR-106a expression between qRT-PCR and ISH analyses. Survival rates were estimated using the Kaplan–Meier method, and survival curves were compared using a log-rank test. Survival data were evaluated by using univariate and multivariate Cox regression analyses. P < .05 was considered significant.
Results
Expression of MiR-106a Can Be Efficiently Detected in Archival FFPE GBM Specimens by qRT-PCR
Given the compromised integrity of RNAs in FFPE GBM specimens, we examined and compared expression levels of miR-106a or U6 in matched FFPE and fresh frozen samples by qRT-PCR analysis to evaluate whether miR-106a or U6 expression could be efficiently detected in those specimens. U6 is a ubiquitous snRNA, which is commonly used as an endogenous control. Our qRT-PCR results showed highly correlated Ct values between FFPE and fresh frozen specimens for the expression of miR-106a or U6, with a Pearson's correlation coefficient of 0.849 or 0.823, respectively (P < .001; Fig. 1A and B). More importantly, the relative fold change of miR-106a expression after normalization with U6 between archival GBM and normal brain tissues is quite similar to that between their matched fresh frozen specimens, with a Pearson's correlation coefficient of 0.964 (P < .001; Fig. 1C). Our data revealed that miR-106a was relatively abundant and stable in archival specimens, with good correlation of miR-106a expression of the FFPE samples compared with the fresh frozen samples, which is consistent with previous studies.11,16
Fig. 1.
Correlation of miR-106a expression between matched FFPE and fresh frozen GBM samples evaluated by qRT-PCR (n = 24). (A and B) Correlation of Ct values of U6 (A) and miR-106a (B) in FFPE samples compared with corresponding fresh frozen tissues. (C) Correlation of fold changes of miR-106a expression between matched FFPE and fresh frozen samples. The fold changes of miR-106a were calculated as the difference between average expression levels of GBM samples and 4 corresponding normal brain tissues. Abbreviation: FLAIR, fluid attenuated inversion recovery.
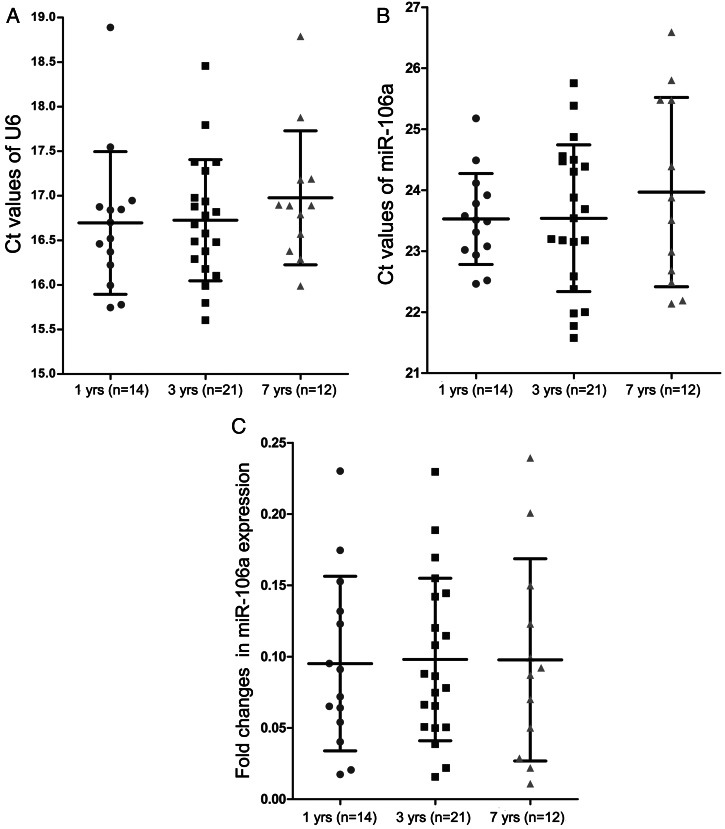
MiR-106a Expression in Archival FFPE Samples Is Quite Stable
Our archival FFPE specimens were stored for 1, 3, or 7 years before RNA extraction. Thus, we compared the expression of miR-106a and U6 in those archival samples at different storage time points to evaluate the effect of prolonged block storage on qRT-PCR analysis. Our qRT-PCR data showed no statistically significant differences in Ct values of miR-106a or U6 in those archival samples stored at different storage time points (Fig. 2A and B). Moreover, the prolonged block storage did not have a significant impact on the relative fold change of miR-106a after normalization with U6 between archival GBM and normal brain tissues (Fig. 2C). Therefore, our results demonstrated that miR-106a expression in those FFPE samples—even those stored for 7 years—is quite stable and that miRNAs extracted from those archival specimens can be used for qRT-PCR analysis.
Fig. 2.
The stability of miR-106a expression in FFPE samples stored at different time points. (A and B) The Ct values of U6 (A) and miR-106a (B) in 1-, 3-, and 7-year-old FFPE GBM samples. (C) The fold changes of miR-106a expression in 1-, 3-, and 7-year-old FFPE GBM samples. The fold changes of miR-106a were calculated as the difference between average expression levels of GBM samples and 4 corresponding normal brain tissues.
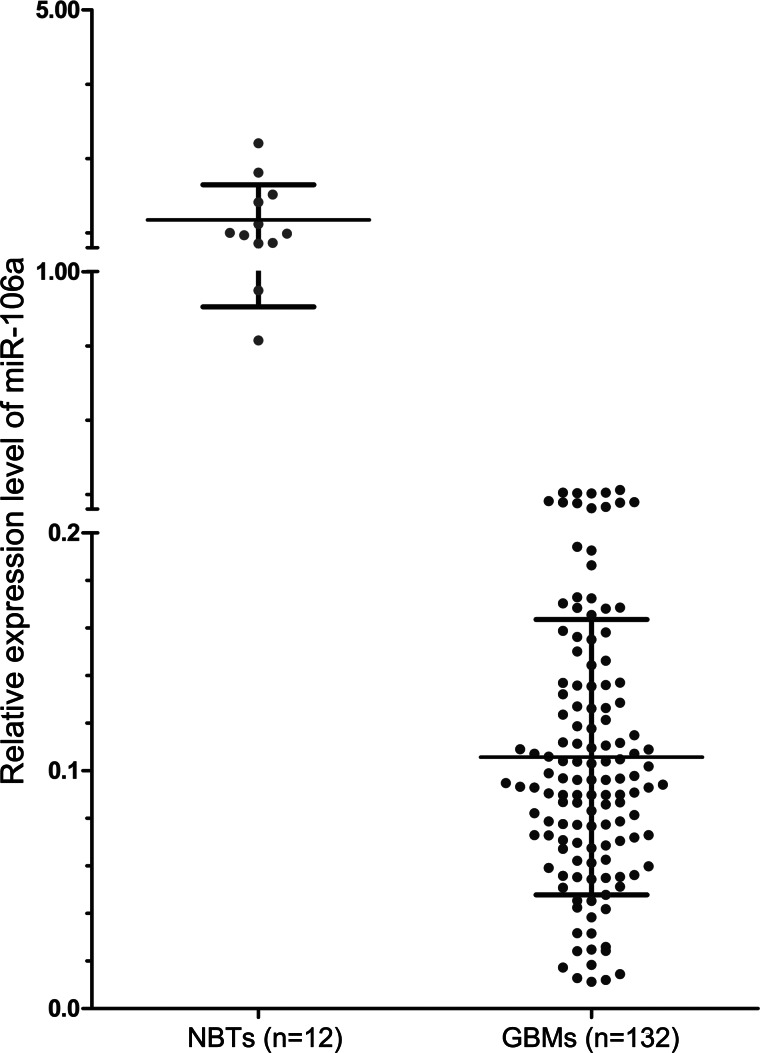
Downregulation of MiR-106a in GBM Specimens
In our previous study, we found that low levels of miR-106a in human glioma specimens were significantly correlated with the malignant progression of gliomas.13 To further detect expression levels of miR-106a in a large cohort of archival GBM specimens, qRT-PCR analysis was employed. Our qRT-PCR data showed that the average relative expression levels of miR-106a in GBM and normal brain tissues were 0.11 ± 0.06 and 1.47 ± 0.59, respectively, and that miR-106a was widely expressed in GBM, with a 56-fold difference between the highest and the lowest value. Expression levels of miR-106a were significantly lower in FFPE GBM samples than in normal brain samples (P < .001; Fig. 3).
Fig. 3.
Scatter plots showing the significant downregulation of miR-106a in GBM patients. The expression levels of miR-106a in FFPE GBM samples and normal brain tissue (NBT) were examined by qRT-PCR. MiR-106a expression was normalized to U6 snRNA.
Correlation of MiR-106a Expression With Other MiRNAs and Molecular Markers in GBM Patients
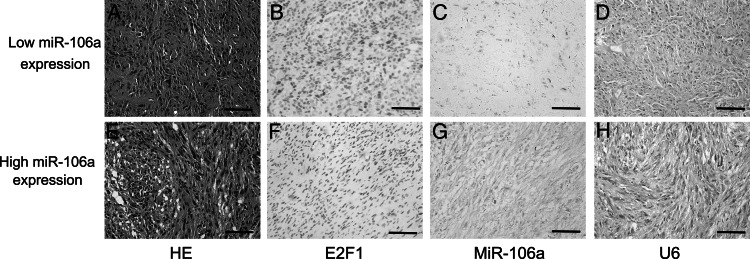
MiR-182 and miR-196a have recently been considered as prognostic miRNAs in glioma patients.9,16,23 Molecular markers, including EGFR, MIB-1, IDH1, and TP53, have been used to evaluate malignant gliomas.24–27 In addition, we previously demonstrated that E2F1 is a direct functional target of miR-106a in patients with gliomas.13 However, the association of miR-106a with those markers in GBM patients remains unclear. To assess correlations among these molecules, we divided GBM patients into 2 groups: a low-expression group containing patients with values less than the average expression levels of miR-106a, and a high-expression group including those with expression values above average. Downregulation of miR-106a was significantly correlated with low levels of miR-182 and with high expression of miR-196a and E2F1. Our immunohistochemistry and ISH results also showed significant negative correlation between miR-106a and E2F1 expression (Fig. 4), with a Pearson's correlation coefficient of −0.691 (P < .001). Nevertheless, miR-106a expression did not correlate statistically with EGFR amplification, MIB-1 expression, and IDH1 and TP53 mutations (Table 1).
Fig. 4.
Negative correlation between miR-106a and E2F1 expression in archival GBM samples. Shown are representative images of hematoxylin/eosin (HE) staining, E2F1 immunoreactivity, and ISH of miR-106a or positive control (U6) from the same tissue area. Increased E2F1 immunoreactivity (B), weak miR-106a ISH staining (C) with corresponding ISH positive control (D) in patients with low levels of miR-106a assessed by qRT-PCR. Decreased E2F1 immunoreactivity (F), strong miR-106a ISH staining (G) with corresponding ISH positive control (H) in GBM patients with high expression levels of miR-106a detected by qRT-PCR. Corresponding HE staining (A and E) to the left. ISH positive signals (miR-106a and U6). Original magnification is 200×, and scale bars are 50 μm.
Table 1.
Correlation of miR-106a expression with clinical and molecular characteristics in archival GBM patients
| Clinical Characteristics | High Levels of MiR-106a |
Low Levels of MiR-106a |
P | Molecular Characteristics | High Levels of MiR-106a |
Low Levels of MiR-106a |
P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | ||||
| Age, y | .158 | MiR-196a | .029 | ||||||||
| Median | 46 | 51 | High | 9 | 16.36 | 26 | 33.77 | ||||
| Range | 8–68 | 9–80 | Low | 46 | 83.64 | 51 | 66.23 | ||||
| Sex | .718 | MiR-182 | .034 | ||||||||
| Male | 35 | 63.64 | 46 | 59.74 | High | 22 | 40.00 | 46 | 59.74 | ||
| Female | 20 | 36.36 | 31 | 40.26 | Low | 33 | 60.00 | 31 | 40.26 | ||
| Preoperative KPS | .825 | E2F1 | <.001 | ||||||||
| ≥70 | 45 | 81.82 | 61 | 79.22 | High | 5 | 9.09 | 61 | 79.22 | ||
| <70 | 10 | 18.18 | 16 | 20.78 | Low | 50 | 90.90 | 16 | 20.78 | ||
| Largest diameter, cm | 1.000 | EGFR | .711 | ||||||||
| ≥6 | 36 | 65.45 | 50 | 64.94 | Amplified | 20 | 36.36 | 25 | 32.47 | ||
| <6 | 15 | 27.27 | 22 | 28.57 | Not | 35 | 63.64 | 52 | 67.53 | ||
| Extent of resection | .079 | MIB-1 | .112 | ||||||||
| GTR | 44 | 80.00 | 50 | 64.94 | High | 23 | 41.82 | 44 | 57.14 | ||
| STR | 11 | 20.00 | 27 | 35.06 | Low | 32 | 58.18 | 33 | 42.86 | ||
| Radiotherapy | .719 | IDH1 | .824 | ||||||||
| Yes | 51 | 92.73 | 73 | 94.81 | Mutated | 11 | 20.00 | 14 | 22.08 | ||
| No | 4 | 7.27 | 4 | 5.19 | Wild-type | 44 | 80.00 | 63 | 81.82 | ||
| TP53 | .825 | ||||||||||
| Mutated | 10 | 18.18 | 16 | 20.78 | |||||||
| Wild-type | 45 | 81.82 | 61 | 79.22 | |||||||
MiR-106a Expression Is Correlated With the Prognosis of Patients With GBM
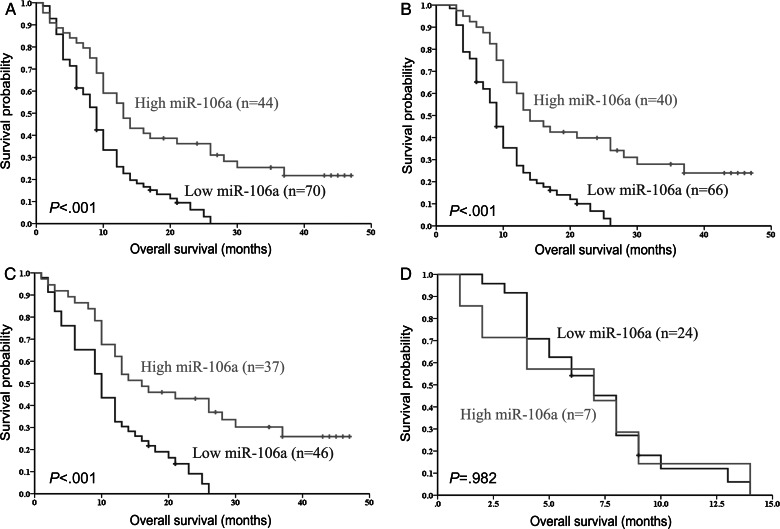
Given the correlation of miR-106a expression with the malignant progression of gliomas, we tested whether miR-106a expression in archival FFPE samples could predict GBM patient outcomes. Patients with miR-106a expression levels below the median showed a shorter OS compared with patients in the high-expression group measured by Kaplan–Meier survival curve analysis with a log-rank comparison (P < .001; Fig. 5A). The median survival time of patients whose tumors had low-level expression of miR-106a was only 9 months (95% confidence interval [CI], 7.55–10.45), whereas the median survival time of those with high expression levels of miR-106a was 13 months (95% CI, 9.29–16.71). More importantly, GBM patients with a median survival of >36 months had a higher than average expression of miR-106a. To eliminate the influence of treatment bias (standard postoperative radiotherapy and extent of resection) on the OS of GBM patients, we reanalyzed our data. Our results demonstrated that low expression levels of miR-106a were significantly correlated with shorter OS in patients with standard postoperative radiotherapy (P < .001; Fig. 5B). In the GTR group, the median survival time of patients with low expression levels of miR-106a was only 10 months (95% CI, 7.36–12.64), whereas the median survival time of those with high expression levels of miR-106a was 16 months (95% CI, 8.41–23.59). There is a significant difference between these 2 groups (P < .001; Fig. 5C). However, in the STR group, our data showed no survival difference between patients regardless of the expression levels of miR-106a (P = .982; Fig. 5D).
Fig. 5.
Kaplan–Meier curves for OS of the second cohort of GBM patients for miR-106a expression. The miR-106a expression levels were converted into discrete variables by discriminating the samples into 2 classes (high and low), under or over median value. Kaplan–Meier survival curves for miR-106a expression detected by qRT-PCR in the second cohort of GBM patients (A), patients with standard postoperative radiotherapy (B), patients in the GTR group (C), and patients in the STR group (D).
Subsequently, we determined the correlation of miR-106a expression with clinical and molecular variables using a Cox proportional hazards regression model (Table 2). Univariate analysis showed a highly statistically significant correlation between OS and expression levels of miR-106a (P < .001; hazard ratio [HR] = 0.430). Multivariate analysis further revealed that expression level of miR-106a was an independent and significant predictor of OS in GBM patients (P = .011; HR = 0.504). Additionally, univariate and multivariate analyses demonstrated that preoperative KPS, EGFR amplification, and miR-196a and MIB-1 expressions were also independent and significant predictors of OS in those GBM patients but that miR-182 expression and IDH1 and TP53 mutations were not (Table 2).
Table 2.
Univariate and multivariate Cox regression analyses of OS in archival GBM patients
| Variables |
RT-PCR |
ISH |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Analysis |
Multivariate Analysis |
Multivariate Analysis |
||||||||
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | ||
| Gender | Female vs male | 1.020 | 0.681–1.527 | .924 | 1.071 | 0.682–1.682 | .767 | 1.084 | 0.678–1.736 | .735 |
| Age, y | 60 vs ≤60 | 1.705 | 1.083–2.685 | .021 | 1.298 | 0.745–2.260 | .357 | 1.135 | 0.629–2.049 | .673 |
| KPS | ≥70 vs <70 | 0.583 | 0.357–0.952 | .031 | 0.484 | 0.279–0.841 | .010 | 0.494 | 0.278–0.878 | .016 |
| Tumor size, cm | ≥6 vs <6 | 1.088 | 0.690–1.715 | .717 | 1.165 | 0.704–1.928 | .553 | 1.335 | 0.782–2.280 | .290 |
| Extent of resection | GTR vs STR | 0.328 | 0.203–0.529 | <.001 | 0.561 | 0.316–0.996 | .049 | 0.582 | 0.323–1.047 | .071 |
| MiR-106a expression | High vs low | 0.430 | 0.273–0.677 | <.001 | 0.504 | 0.297–0.854 | .011 | 0.452 | 0.255–0.800 | .006 |
| MiR-196a expression | High vs low | 2.877 | 1.822–4.542 | <.001 | 2.252 | 1.321–3.841 | .003 | 1.906 | 1.108–3.281 | .020 |
| MiR-182 expression | High vs low | 1.303 | 0.872–1.949 | .197 | 0.974 | 0.611–1.554 | .912 | 1.032 | 0.630–1.693 | .900 |
| EGFR | Amplified vs not | 1.672 | 1.102–2.536 | .016 | 1.613 | 1.004–2.593 | .048 | 1.785 | 1.067–2.987 | .027 |
| MIB-1 | High vs low | 2.128 | 1.409–3.213 | <.001 | 1.610 | 1.008–2.572 | .046 | 1.715 | 1.048–2.806 | .032 |
| IDH1 | Mutation vs wild-type | 0.720 | 0.426–1.216 | .219 | 0.712 | 0.403–1.256 | .241 | 0.709 | 0.386–1.301 | .267 |
| TP53 | Mutation vs wild-type | 1.357 | 0.835–2.204 | .218 | 1.234 | 0.709–2.147 | .457 | 1.278 | 0.706–2.315 | .418 |
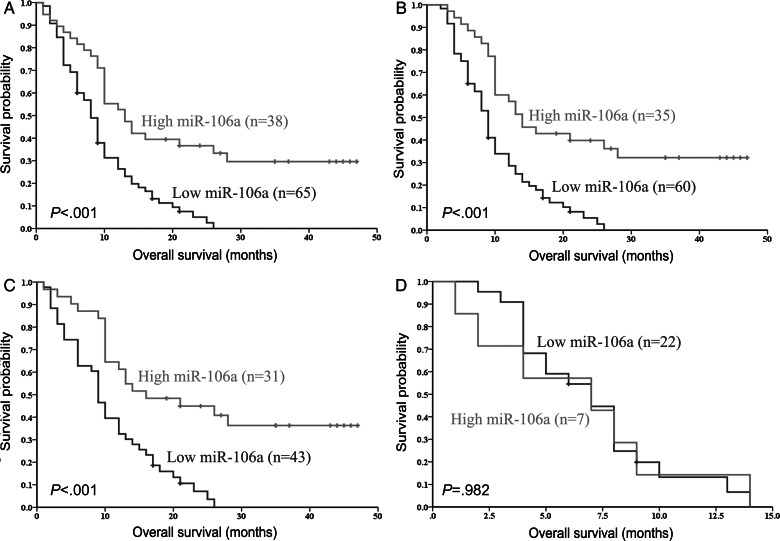
Independent Prognostic Biomarker in GBM Patients Further Validated via ISH
MiR-106a expression levels were again examined by ISH to further validate whether miR-106a could indeed be an independent prognostic biomarker in GBM patients. Our ISH analysis first demonstrated that miR-106a was widely expressed in GBM specimens, which coincided with the results we obtained from qRT-PCR assay (Fig. 4). MiR-106a expression was detected in 120 (91%) of 132 GBM patients assigned to the second cohort, while no staining was observed with negative control probes in any sample. A chi-square test revealed good correlation of miR-106a expression levels in the same specimen detected by 2 different methods (ISH and qRT-PCR analyses) (P < .001; McNemar = 1.000). Subsequently, we sought to verify the correlation of miR-106a expression levels with GBM patient outcomes via ISH technology, and our ISH data showed that miR-106a expression levels strongly correlated with OS of GBM patients (Fig. 6A). Multivariate analysis showed that the expression level of miR-106a detected by ISH was an independent predictor of OS in GBM patients, consistent with qRT-PCR results (Table 2). Again, to eliminate the influence of treatment bias on the OS of GBM patients, we reanalyzed our data. Our ISH data confirmed that low expression levels of miR-106a were significantly correlated with shorter OS in patients with standard postoperative radiotherapy (Fig. 6B). The patients in the GTR group also showed similar correlation between miR-106a expression and OS, but those in the STR group did not (Fig. 6B–D). These findings acquired by ISH (Fig. 6) were consistent with those results obtained by qRT-PCR (Fig. 5). Our results showed that either ISH or qRT-PCR could be used to detect miR-106a expression in those archival FFPE samples. Taken together, our results suggest that miR-106a expression, as demonstrated in both ISH and qRT-PCR analyses, is correlated with OS of patients with GBM and can be used as a novel and potentially significant independent biomarker for the prognosis of patients with GBM.
Fig. 6.
A strong correlation of miR-106a expression levels with OS of GBM patients was further confirmed by ISH. Kaplan–Meier survival curves for miR-106a expression detected by ISH in the second cohort of GBM patients (A), patients with standard postoperative radiotherapy (B), patients in the GTR group (C), and patients in the STR group (D).
Discussion
Clinically relevant tissue-based biomarkers of gliomas may help neuro-oncologists to further classify gliomas with different biologies, improve prognosis prediction, and predict a response to therapy.28 Some important molecular markers with clinical implications have recently been identified in GBM patients, such as deletions of chromosome arms 1p and 19q, mutations of the p53 gene, promotor hypermethylation of O6-methylguanine-DNA methyltransferase, and mutations of EGFR or IDH genes.29 Although these genetic and epigenetic markers of GBM have some clinical implications, none of the molecular markers can be used in daily decision making.22,30,31
MiRNAs are a class of endogenous small noncoding RNA molecules that regulate cell growth, differentiation, and apoptosis.32,33 MiRNAs are aberrantly expressed in a wide variety of human cancers, and certain miRNA expression patterns have had a good correlation with specific clinical features of cancers.34–36 The unique miRNA signatures have been observed to be correlated with prognostic factors and disease progression in chronic lymphocytic leukemia, lung cancers, colon cancers, and breast cancers.37–40 MiR-182 has been considered to be a biological marker of glioma progression, and high miR-182 expression is associated with poor OS in glioma patients.16 Expression of 2 other miRNAs, miR-487b and miR-410, also demonstrated predictive value in neuroblastoma.41 MiR-196 was reported to play a role in malignant progression and may have been a prognostic predictor in 74 GBM patients.9 Therefore, miRNA expression may be a clinically useful biological marker to predict patient outcomes. We previously presented evidence that the low expression of miR-106a was significantly correlated with the malignant progression of human gliomas.13 In addition, low expression of miR-106a was recently observed to be associated with poor patient survival in 84 astrocytoma samples.42 However, very little is known regarding its role in GBM and its correlation with clinical outcomes of patients with GBM. MiR-106a is located at Xq26.2, and mature miR-106a is 23 nucleotides in length. Downregulation of miR-106a has been observed to predict shortened disease-free survival and OS in human colon cancer patients.43 In the present study, we revealed that miR-106a showed 13-fold lower mean expression levels in GBM samples than in normal brain tissues. During a median follow-up period of 10 months (range, 1–47 mo), the expression level of miR-106a was an independent and significant predictor of OS in GBM patients. Our results also showed that downregulation of miR-106a was significantly correlated with low levels of miR-182 and high expression of miR-196a in GBM patients. Multivariate analysis also revealed that miR-196a expression level was an independent predictor of OS in GBM patients, consistent with the previous findings.9 Although little is known about the function of miR-106a in tumor genesis and progression, our previous study has shown that miR-106a provides a tumor-suppressive effect via suppressing proliferation of and inducing apoptosis in human glioma cells, and the suppressive effect of miR-106a on gliomas may result from inhibition of E2F1 via posttranscriptional regulation.13 In this study, we further revealed the significant negative correlation between miR-106a and E2F1 expression in a large cohort of GBM patients.
Although well-annotated FFPE samples present extremely valuable resources for clinical cancer research, the formalin preservation process significantly decreases the yield and integrity of RNAs via enzymatic and chemical degradation, extensive cross-linking with proteins, and various chemical modifications.44,45 Thus, those archival specimens were not routinely employed for the analysis of gene expression. Optimal yield and quality of RNAs could be generated from only fresh frozen specimens suitable for the analysis of mRNA expression via qRT-PCR and microarray technologies.11,46 However, the availability of fresh frozen tissues is very limited, and they are not suitable for retrospective studies. In contrast, it has been generally believed that miRNAs in archival FFPE specimens may not be susceptible to RNA degradation because they are about 20–22 nucleotides in length and protein protected.12,47 Therefore, studies of miRNA stability and expression in those archival specimens are clinically significant because they have been seen as potential biomarkers for diagnosis, prognosis, and response to therapy. Recent studies have demonstrated a very good correlation of miRNA expression between matched fresh frozen and FFPE samples.48,49 In the present study, we detected and compared miR-106a expression levels in FFPE samples with those in corresponding fresh frozen specimens, and our results showed that their expression levels were highly correlated between FFPE specimens and matched fresh frozen tissues. Moreover, no significant differences in expression levels of miR-106a were observed among the different storage times. These data demonstrated that miR-106a was relatively stable and easily recovered in FFPE samples when stored for at least 7 years.
In summary, our study provided the first evidence that miR-106a expression was relatively abundant and stable in a large cohort of archival FFPE GBM specimens obtained at a single institute, and there was a good correlation of miR-106a expression of FFPE samples compared with matched fresh frozen samples. The long-term storage of FFPE samples seems to have no significant negative effect on the qRT-PCR analysis. Expression level of miR-106a was significantly lower in FFPE GBM samples and was an independent and significant predictor of OS in GBM patients. Thus, miR-106a could be used to predict prognosis and treatment response in individual GBM patients.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81272788, 30973078, 30901533, 81172388) and the Special Prophase Project of the National Basic Research Program of China (grant no. 2009cb526404).
Acknowledgments
We thank Drs. Hongyu Yang, Jinxing Li, and Yuandong Dong, Department of Neurosurgery, for data collection.
Conflict of interest statement. None declared.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Neyns B, et al. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28(16):2712–2718. doi: 10.1200/JCO.2009.26.6650. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Wei Q, Wang LE, et al. Survival prediction in patients with glioblastoma multiforme by human telomerase genetic variation. J Clin Oncol. 2006;24(10):1627–1632. doi: 10.1200/JCO.2005.04.0402. [DOI] [PubMed] [Google Scholar]
- 4.Sumazin P, Yang X, Chiu HS, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147(2):370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camps C, Buffa FM, Colella S, et al. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 7.Schwind S, Maharry K, Radmacher MD, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(36):5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchini S, Cavalieri D, Fruscio R, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12(3):273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 9.Guan Y, Mizoguchi M, Yoshimoto K, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16(16):4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 10.Roth P, Wischhusen J, Happold C, et al. A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118(3):449–457. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- 11.Szafranska AE, Davison TS, Shingara J, et al. Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling. J Mol Diagn. 2008;10(5):415–423. doi: 10.2353/jmoldx.2008.080018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng L, Wu X, Gao H, et al. MicroRNA profiling of clear cell renal cell carcinoma by whole-genome small RNA deep sequencing of paired frozen and formalin-fixed, paraffin-embedded tissue specimens. J Pathol. 2010;222(1):41–51. doi: 10.1002/path.2736. [DOI] [PubMed] [Google Scholar]
- 13.Yang G, Zhang R, Chen X, et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J Mol Med (Berl). 2011;89(10):1037–1050. doi: 10.1007/s00109-011-0775-x. [DOI] [PubMed] [Google Scholar]
- 14.McGirt MJ, Chaichana KL, Gathinji M, et al. Independent association of extent of resection with survival in patients with malignant brain astrocytoma. J Neurosurg. 2009;110(1):156–162. doi: 10.3171/2008.4.17536. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(t)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L, Mao P, Song L, et al. MiR-182 as a prognostic marker for glioma progression and patient survival. Am J Pathol. 2010;177(1):29–38. doi: 10.2353/ajpath.2010.090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnem T, Eklo K, Berg T, et al. Prognostic impact of miR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali-Fehmi R, Che M, Khalifeh I, et al. The effect of cyclooxygenase-2 expression on tumor vascularity in advanced stage ovarian serous carcinoma. Cancer. 2003;98(7):1423–1429. doi: 10.1002/cncr.11650. [DOI] [PubMed] [Google Scholar]
- 19.Donnem T, Al-Saad S, Al-Shibli K, Busund LT, Bremnes RM. Co-expression of PDGF-B and VEGFR-3 strongly correlates with lymph node metastasis and poor survival in non-small-cell lung cancer. Ann Oncol. 2010;21(2):223–231. doi: 10.1093/annonc/mdp296. [DOI] [PubMed] [Google Scholar]
- 20.Semaan A, Qazi AM, Seward S, et al. MicroRNA-101 inhibits growth of epithelial ovarian cancer by relieving chromatin-mediated transcriptional repression of p21(waf(1)/cip(1)) Pharma Res. 2011;28(12):3079–3090. doi: 10.1007/s11095-011-0547-x. [DOI] [PubMed] [Google Scholar]
- 21.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 22.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 23.Ma X, Yoshimoto K, Guan Y, et al. Associations between microRNA expression and mesenchymal marker gene expression in glioblastoma. Neuro Oncol. 2012;14(9):1153–1162. doi: 10.1093/neuonc/nos145. doi:10.1093/neuonc/nos145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faria MH, Goncalves BP, do Patrocinio RM, de Moraes-Filho MO, Rabenhorst SH. Expression of Ki-67, topoisomerase II alpha and c-MYC in astrocytic tumors: correlation with the histopathological grade and proliferative status. Neuropathology. 2006;26(6):519–527. doi: 10.1111/j.1440-1789.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 25.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purow B, Schiff D. Advances in the genetics of glioblastoma: are we reaching critical mass? Nat Rev Neurol. 2009;5(8):419–426. doi: 10.1038/nrneurol.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 28.Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G. Molecular diagnostics of gliomas: state of the art. Acta Neuropathol. 2010;120(5):567–584. doi: 10.1007/s00401-010-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9(7):717–726. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krex D, Klink B, Hartmann C, et al. Long-term survival with glioblastoma multiforme. Brain. 2007;130(Pt 10):2596–2606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 31.Tabatabai G, Stupp R, van den Bent MJ, et al. Molecular diagnostics of gliomas: the clinical perspective. Acta Neuropathol. 2010;120(5):585–592. doi: 10.1007/s00401-010-0750-6. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3’ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16(2):144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tricoli JV, Jacobson JW. MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67(10):4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 35.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki HI, Miyazono K. Dynamics of microRNA biogenesis: crosstalk between p53 network and microRNA processing pathway. J Mol Med (Berl). 2010;88(11):1085–1094. doi: 10.1007/s00109-010-0650-1. [DOI] [PubMed] [Google Scholar]
- 37.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 38.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 39.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gattolliat CH, Thomas L, Ciafre SA, et al. Expression of miR-487b and miR-410 encoded by 14q32.31 locus is a prognostic marker in neuroblastoma. Br J Cancer. 2011;105(9):1352–1361. doi: 10.1038/bjc.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhi F, Chen X, Wang S, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur J Cancer. 2010;46(9):1640–1649. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Diaz R, Silva J, Garcia JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47(9):794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 44.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27(22):4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive—gene expression in paraffin-embedded tissue. J Pathol. 2001;195(1):66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 46.Huang WY, Sheehy TM, Moore LE, Hsing AW, Purdue MP. Simultaneous recovery of DNA and RNA from formalin-fixed paraffin-embedded tissue and application in epidemiologic studies. Cancer Epidemiol Biomarkers Prev. 2010;19(4):973–977. doi: 10.1158/1055-9965.EPI-10-0091. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Li J, Smyth P, Flavin R, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13(10):1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10(3):203–211. doi: 10.2353/jmoldx.2008.070153. [DOI] [PMC free article] [PubMed] [Google Scholar]