Abstract
Background
Glioblastoma multiforme is the most common lethal brain tumor in human adults, with no major therapeutic breakthroughs in recent decades. Research is based mostly on human tumor cell lines deprived of their organotypic environment or inserted into immune-deficient animals required for graft survival. Here, we describe how glioblastoma specimens obtained from surgical biopsy material can be sectioned and transferred into cultures within minutes.
Methods
Slices were kept in 6-well plates, allowing direct observation, application of temozolomide, and irradiation. At the end of experiments, slice cultures were processed for histological analysis including hematoxylin-eosin staining, detection of proliferation (Ki67), apoptosis/cell death (cleaved caspase 3, propidium iodide), DNA double-strand breaks (γH2AX), and neural subpopulations. First clinical trials employed irradiation with the heavy ion carbon for the treatment of glioblastoma patients, but the biological effects and most effective dose regimens remain to be established. Therefore, we developed an approach to expose glioblastoma slice cultures to 12C and X-rays.
Results
We found preservation of the individual histopathology over at least 16 days. Treatments resulted in activation of caspase 3, inhibition of proliferation, and cell loss. Irradiation induced γH2AX. In line with clinical observations, individual tumors differed significantly in their susceptibility to temozolomide (0.4%–2.5% apoptosis and 1%–15% cell loss).
Conclusion
Glioblastoma multiforme slice cultures provide a unique tool to explore susceptibility of individual tumors for specific therapies including heavy ions, thus potentially allowing more personalized treatments plus exploration of mechanisms of (and strategies to overcome) tumor resistance.
Keywords: glioblastoma multiforme, organotypic slice culture, human test system, heavy ions
Glioblastoma multiforme (GBM) is among the most common lethal tumors, with patients having an average life expectancy of <18 months after diagnosis. While substantial progress has been made with regard to understanding tumor pathogenesis,1,2,3,4–7 expansion,8 bystander damage,9 migration,10 altered protein expression, and resistance to cell death,11 none of this information significantly affects life expectancy or quality of life.
Current research often employs immortalized cell lines that are studied in co-culture or after transplantation in immunodeficient mice. While such approaches certainly help identify basic principles of tumor biology and immunology, it is difficult to directly translate observations to the human disease and to develop accurate or even individualized therapeutic strategies based on such experiments. We have previously used organotypic entorhinohippocampal slice cultures in which all neural subpopulations of cells and the basic interneuronal connections are maintained.12–16 These organotypic slice cultures include several advantages, such as (i) the presence of the organotypic matrix, which is increasingly appreciated as providing crucial signaling for site-specific cellular differentiation17–20; (ii) open access allowing treatment and direct observation over extended periods of time; and (iii) collection of supernatants for analysis over time. Using brain slice cultures obtained from epilepsy surgery, we have shown that normal human brain cells, in contrast to the murine system, are susceptible to lysis by tumor necrosis factor–related apoptosis-inducing ligand.21,22 This exemplified the need for the development of human test systems for the evaluation of toxicity and preclinical research, an issue that gained worldwide attention after the “London tragedy,” in which 6 volunteers experienced toxic shocks and cytokine storms in response to an anti-CD28 antibody that had been previously well tolerated by rodents and monkeys.23,24,25 Over the last few years, the use of human slice cultures of tumors in research has begun to emerge in the literature,26,27 underlining an emerging awareness of species differences.
One possible novel approach to treating GBM could be irradiation with high-energy carbon ions. Heavy ion (HI) therapy was developed over 50 years ago at the Lawrence Berkeley Laboratory in the United States and is currently in operation in several centers in Europe and Asia.28 The rationale of using particles heavier than protons for therapy lies in their special radiobiological characteristics, especially in their ability to overcome radioresistance.29 This high relative biological effectiveness makes them attractive for use against tumors resistant to conventional therapy, such as GBM. Clinical results support the rationale of the therapy, and a clinical trial on glioblastoma is currently under way in Heidelberg.30
Here, we tested the survival and suitability of slice cultures derived from GBM as a test system for current and future therapeutic strategies, based on our previous findings.31,32 Tumor tissues obtained directly from neurosurgical operations were immediately transported to the laboratory in media, cut into 350-µm sections, and kept on membranes in 6-well plates for up to 4 weeks. We exposed GBM slice cultures to temozolomide (TMZ), X-rays, and irradiation with the HI carbon (12C) and monitored the effect on proliferation, cell death, and DNA double-strand breaks (DSBs). Our data showed that therapeutic effects can be mimicked well in such slice preparations, which therefore are potentially suitable as an experimental model to better understand mechanisms of tumor resistance, as well as a test system for novel therapies and susceptibility assays for personalized treatment.
Materials and Methods
Tissue Slice Preparation
All patients provided written informed consent according to German law as confirmed by the local committees (144–2008 and 837.211.12-8312-F). Glioblastoma tissue not required for neuropathological diagnostic procedures was obtained after surgical resection at the Department of Neurosurgery (Leipzig or Mainz). An overview of the samples used in this study is given in Table 1. The tissue was transported to the laboratory in minimal essential medium (MEM; Invitrogen). Slice cultures were prepared using a vibratome (Leica VT 1000) or a tissue chopper (McIlwain TC752) at a thickness of 350 µm under sterile conditions. Before preparations, a standard razor blade was wiped with ethanol to remove any oil and then sterilized by autoclaving. Additionally, a normal glass pipette and a pipette with the fine tip broken off were autoclaved. If needed, biopsy specimens were cut into appropriate-size pieces first to obtain evenly shaped slices of ∼5 × 5 mm. When the tissue chopper was used, the tissue was put on a stack of sterile filter membranes, cut, and then transferred carefully into ice-cold MEM by forceps. In most of the preparations, the slices stuck together after cutting, so they were separated under a stereomicroscope with 2 scalpels without cutting into the tissue. Slices were then transferred by the glass pipette with the wide opening onto membrane culture inserts (Millipore) in 6-well plates at a maximum of 3–4 slices per insert depending on size. The cultivation medium consisted of MEM (Gibco), 25% Hank's Balanced Salt Solution (with Ca and Mg; Gibco), 25% N-hydroxysuccinimide (Gibco), 1% L-glutamine (Braun), 1% glucose (stock solution 45%, final concentration 0.45%; Braun), and 1% penicillin/streptomycin (Sigma). Slices were cultivated on a liquid/air interface in a humidified incubator at 37°C and 5% CO2. Medium was changed 3 times a week. After time points ranging from 1 h to 4 weeks, slices were fixed in 4% paraformaldehyde and processed for paraffin embedding. Paraffin sections (8 µm) were cut and stained with hematoxylin and eosin (H&E) for histology. Histology of these sections was compared with the diagnostic histopathology of the same tumor to detect tissue culture–induced changes. For immunocytochemistry, sections were dewaxed in xylene, rehydrated in a decreasing alcohol series, and (if needed) pretreated for antibody staining with citrate buffer (pH 6) in a microwave. Then, sections were washed in phosphate buffered saline (PBS), permeabilized with 1.5% Triton/PBS for 10 min, blocked with 10% normal goat serum in 1.5% Triton/PBS for 1 h, and incubated overnight at 4°C with primary antibodies against Ki67 (rabbit, 1:100; DCS), cleaved caspase 3 (rabbit, 1:400; Cell Signaling), glial fibrillary acidic protein (GFAP; Dako, rabbit, 1:600; Dako), nestin (rabbit, 1:600; Chemicon), vimentin (mouse, 1:100; Dako), neurofilament (mouse, 1:100; Dako), or γH2AX (mouse, 1:100; Millipore). Visualization was achieved by incubation either with appropriate fluorescent-labeled secondary antibodies (goat anti-mouse or anti-rabbit Alexa 488) or with biotinylated anti-rabbit immunoglobulin G followed by streptavidin-conjugated horseradish peroxidase and developing by adding diaminobenzidine for the color reaction. For fluorescent staining, photographs were taken using an Olympus BX51 fluorescent microscope or a Zeiss LSM 510 confocal microscope. In addition to the green fluorescent channel, the red channel was included because some of the samples exhibited a strong autofluorescence, which is normal in the nonjuvenile human brain. Only cells devoid of red fluorescence were used for further analysis. For H&E and diaminobenzidine staining, a Zeiss Axioplan 2 microscope was used.
Table 1.
Glioblastoma samples used for slice culture experiments in this study
| Sample | Experiment | Data Shown In |
|---|---|---|
| 03082010 | Long-term culture | Fig. 1 |
| 12082010 | Long-term culture | Fig. 1 |
| 25072012 | Long-term culture and X-irradiation | Figs. 1 and 3 |
| 12122011 | TMZ treatment | Figs. 2, 6, and 8 |
| 10092010 | Live imaging and TMZ treatment | Fig. 5 |
| 16092010 | Live imaging and TMZ treatment | Fig. 5 |
| 03012011 | Live imaging and TMZ treatment | Fig. 5 |
| 05042011 | Carbon ion irradiation and TMZ treatment | Figs. 3, 8, 4, and 6 |
| 11102011 | Carbon ion irradiation and TMZ treatment | Figs. 7 and 8 |
| 24062011 | Carbon ion irradiation and TMZ treatment | Figs. 7 and 8 |
| 11052012 | TMZ treatment and X-irradiation | Fig. 8 |
| 15082012 | TMZ treatment and X-irradiation | Fig. 8 |
All patients had a diagnosis of World Health Organization grade IV GBM.
Analysis of Proliferation in Slices
Paraffin sections (8 µm) of slices were dewaxed as previously described here, and proliferating cells were stained with the Ki67 antibody. Nuclei were counterstained with Hoechst 33342. Then, at least 12 images were acquired of 4–6 different sections per group and manually analyzed using ImageJ and the PlugIn CellCounter. The percentage of Ki67-positive cells in relation to the total cell number was referred to as the proliferation index. Statistics were performed using GraphPad Prism 5 (Student's t-test).
Treatment of Glioblastoma Tissue Slices With Temozolomide
Glioblastoma tissue slices maintained on cell culture inserts were incubated with TMZ (dissolved in dimethyl sulfoxide) at a final concentration of 50 or 200 µM of the compound. Control slices were incubated with the corresponding amount of dimethyl sulfoxide (0.2% v/v). After 72 h, tissue slices were removed from the membranes and incubated in 500 µL of medium and 10 µg/mL of Hoechst 33342 (Sigma) at 37°C and 5% CO2 for 1 h to allow for nuclear staining. The tissues were then transferred into 1 mL PBS containing 2 µg/mL propidium iodide (PI; Invitrogen) and gently fixated between 2 glass coverslips.
For confocal imaging and quantification of viable and dead cells within the tissues, an LSM-510 META inverted laser scanning microscope and a 20×/0.8 Plan-Apochromat objective (Carl Zeiss MicroImaging) were used. Hoechst 33342 was excited at 364 nm. PI was excited with the 543-nm laser line. Viable cells with Hoechst-positive and PI-negative nuclei, and dead cells with Hoechst-positive and PI-positive nuclei, were counted in 2 or 3 images of each tissue slice per group. Data were obtained from 3 individual experiments. One-way ANOVA was used, and P < .05 was considered to be statistically significant.
Irradiation of Glioblastoma Slice Cultures
Photon irradiation of slices was performed at the Department for Radiation Therapy and Radio-oncology, University of Leipzig, with a 150-kV X-ray unit (DARPAC 150-MC) with an energy of 13.2 mA and a dose rate of 0.86 Gy/min. Cell culture plates were placed under a specially constructed plate device and irradiated until the desired dose was reached. Alternatively, photon irradiation was performed using the GSI X-ray device (GE Isovolt Titan 320, 250 kV, 16 mA) at a dose rate of 1.4 Gy/min.
HI irradiation with a carbon beam was performed at GSI (Gesellschaft für Schwerionenforschung), Darmstadt, at the former patient irradiation site. The ion beam was generated at the SIS18 synchrotron facility and delivered in a spread-out Bragg peak (SOBP33) as used in carbon ion therapy. The dose applied to the slices was 2 or 4 Gy in a 50-mm-width SOBP corresponding to a linear energy transfer range of 50–70 keV/μm. With this method, the target tissue volume is distributed into voxels in a treatment plan. Then, the ion beam is directed at the 3-dimensional tumor volume, using active energy variation and the raster scanning technique. For experiments with slice cultures, the volume was defined as the area and the height of 1 well. Before and after irradiation, slice cultures were kept in an incubator as previously described here and were removed for only about 15 min for transport and to place them on the irradiation belt.
After irradiation, slices were fixed in 4% paraformaldehyde after one of several time points, washed in PBS, and further processed for paraffin embedding or cryosectioning. Cryosections were cut at 14 µm and stored at –80°C until further use. Paraffin sections were prepared at 8 µm, dried, and stored at room temperature.
For staining of DNA DSBs, cryosections were dried for 20 min at room temperature and then washed twice in PBS and incubated with 1.5% Triton/PBS for 10 min. Then sections were blocked with 10% normal goat serum in 1.5% Triton/PBS for at least 1 h, followed by incubation overnight at 4°C with γH2AX primary antibody (mouse monoclonal, 1:100; Millipore). Then, sections were washed 3 times with PBS and incubated with the secondary antibody (goat anti-mouse 1:1000; Alexa 488, Invitrogen) for 1 h, washed again, counterstained with Hoechst 33342, and mounted with Dako fluorescent mounting medium. Z-stacks were taken using a Zeiss LSM 510 confocal microscope at 400× magnification at intervals of 2 µm. Paraffin sections were stained as previously described here.
Results
Slice Cultures From Glioblastoma: Histology and Survival
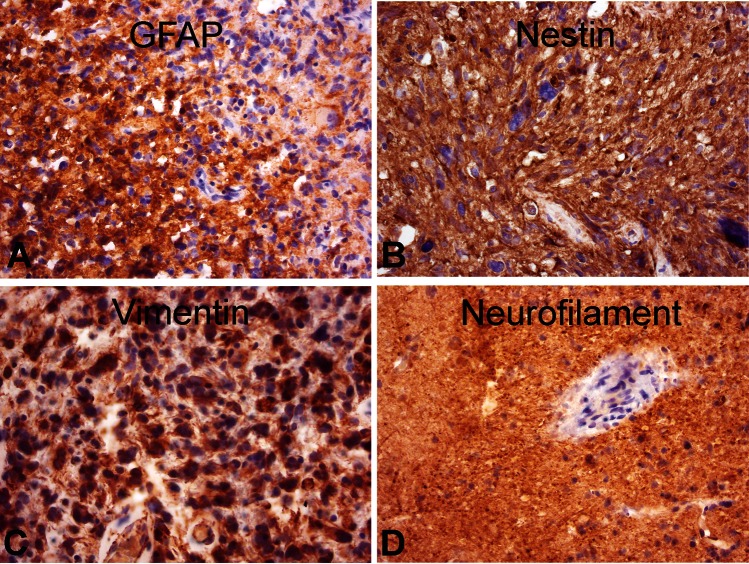
Slices were at first cut with a vibratome and survived well, with histological preservation of the main features of the original tumor for at least 16 days (Fig. 1). At later stages, cell density appeared to decline in some tumor slices, whereas cells in other slices survived longer (Fig. 1E–H and I–L). Some tumors, however, were difficult or even impossible to cut due to their viscous texture, which may have resulted from altered collagen expression.18,34 Using a tissue chopper resolved this problem with equally good histological preservation and maximal survival time. Histological examination of cultured GBM slices and comparison with the original neuropathology used for diagnosis confirmed striking maintenance of the general and individual hallmarks of the tumor (pleomorphic nuclei, palisading, invading vessels/neovascularization, and necrotic areas). As expected, GBM stained positive for GFAP, nestin (intermediate filament protein, GBM “stem” cell marker), and vimentin (mesenchymal intermediate filament protein) and differed strongly in their cell density and content of necrotic areas (Fig. 2).
Fig. 1.
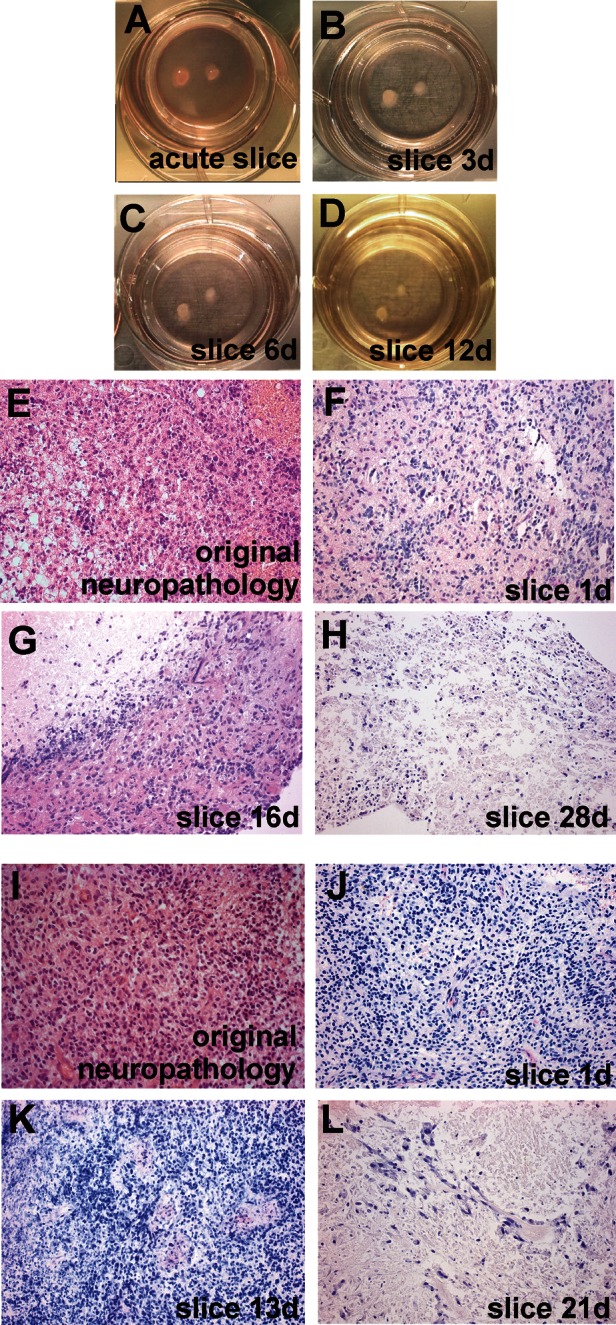
Human GBM slices in culture. Slices were cultured on membrane inserts in six-well plates with no signs of degeneration in acute (A) slices at 1 day or at 3 days (B), 6 days (C), or 12 days (D) in vitro. Original H&E neuropathology (E and I) and H&E-stained paraffin-embedded sections (8 µm; F–H and J–L) prepared from slices after various culture periods. Two different tumors (E–H and I–L) are shown. Note that typical features of individual tumors were maintained at least from 1 to 16 days (F–G) and 1 to 13 days (J–K) in vitro; massive cell loss was observed after 20 days in vitro (H and L). Original magnification: 1× in A–D; 200× in E–L.
Fig. 2.
Typical GBM markers expressed in human GBM slice cultures. Slices were fixed after 7 days in culture and processed for paraffin sectioning (8 µm). Characteristic marker proteins were visualized by antibody staining for GFAP (A), nestin (B), vimentin (C), and neurofilament (D). Counterstaining was performed with hematoxylin. Original magnification: 400×.
Irradiation of Slices: Effect on Proliferation
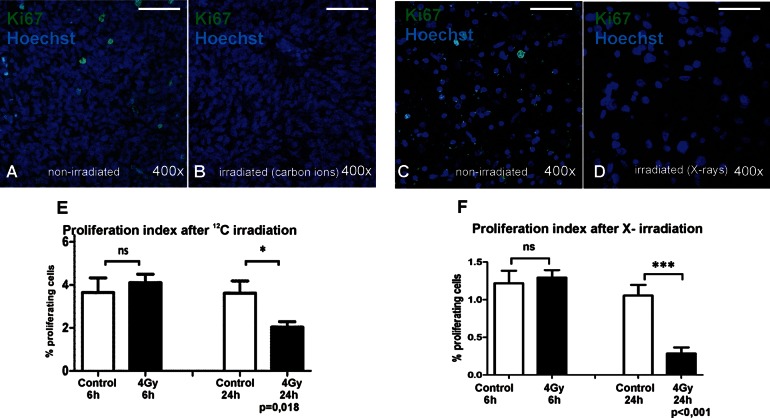
Ionizing radiation is known to cause DNA DSBs. If DNA repair cannot be conducted properly, cell proliferation is arrested at a cell cycle checkpoint and either is inactivated (eg, in postmitotic cells) or proceeds into programmed cell death. Here, we tested the dose- and time-dependent decrease of the proliferation index after SOBP carbon or X-irradiation. GBM slices were fixed 6 or 24 h after irradiation and processed for paraffin sectioning. Proliferating cells were labeled with a Ki67 antibody (Fig. 3A–D), which marks cells in every state of the cell cycle except the G0 resting phase.35 The percentage of Ki67-positive cells in relation to the total number of nuclei was calculated to express the proliferation index. Carbon ions did not significantly diminish proliferation at 6 h, but after 24 h a reduction of ∼40% was found (P = .018; Fig. 3E). This is the first demonstration of specific effects of HI on human GBM tissue ex vivo. Photons also showed the anticipated time-dependent reduction of proliferation (here, ∼50% after 24 h; Fig. 3F). Thus, GBM-derived slice cultures can be irradiated and the biological effects of photons and HI on tumor cells and mechanisms of resistance can be studied in this model.
Fig. 3.
Proliferation index of human GBM slice cultures irradiated with X-rays or carbon ions. Slices were treated with either 4 Gy of SOBP carbon ions at GSI or 4 Gy of X-rays and fixed 6 or 24 h later. Proliferating cells were then visualized in paraffin-embedded sections (8 µm) using a Ki67 antibody (green) combined with nuclear counterstaining (Hoechst 33342; blue) for quantitative analysis. (A), Non-irradiated; (B), irradiated with 4 Gy carbon ions; (C), non-irradiated; (D), irradiated with 4 Gy X-rays; scale bar = 50 µm). The proliferative fraction in relation to total cell number was determined in at least 12 pictures per group by using ImageJ software. After 6 h, the effect on proliferation was not yet significant after both irradiation types, but after 24 h both treatments resulted in a significant decrease of Ki67-positive cells (E and F).
Exposure of Slices to Carbon Ions and Detection of DNA Damage
After induction of DNA DSBs by ionizing radiation, repair proteins are rapidly recruited to the sites of damage. In mammalian cells, this involves a cascade of proteins that ultimately allows repair of the breaks in the form of rejoining the loose DNA ends.36,37–39 When we established the irradiation setup of GBM slice cultures, we wanted to test whether the slices were evenly hit by the beam, and therefore we used γH2AX as an early DSB marker for the visualization of DNA damage. γH2AX describes a phosphorylation of histone 2AX at serine 139 around the region of the DSB,40 which allows binding of further repair proteins, such as MDC1 and 53BP1. Once the repair process is completed, the repair proteins dissociate and H2AX is dephosphorylated by a distinct phosphatase complex.41 Slices were irradiated with 4 Gy carbon ions in an SOBP to match therapeutic conditions, fixed after 1 h, and embedded in paraffin, and sections were stained with an antibody to γH2AX. γH2AX was mostly absent in controls but appeared in a punctuated nuclear pattern in all sections, confirming induction of DNA damage throughout the irradiated tissues (Fig. 4).
Fig. 4.
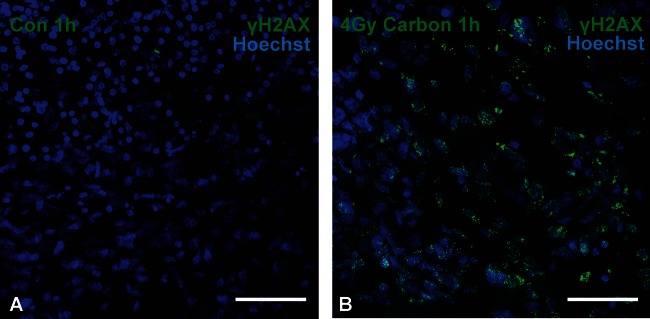
DNA damage in GBM slices after carbon irradiation. Slices were irradiated with 4 Gy of carbon ions in an SOBP and fixed 1 h later. Paraffin sections (8 µm) were assembled and DNA DSBs were visualized by immunocytochemistry with a γH2AX antibody (green) and nuclei with Hoechst (blue). Although γH2AX was rarely detected in non-irradiated controls (A), exposure to therapeutic heavy ions caused massive induction of phosphorylation of H2AX (B). Scale bar = 50 µm. Original magnification: 400×; confocal Z-stacks.
Exposure of Glioblastoma Tissue Slices to Temozolomide: Detection of Cell Death Using Live Imaging
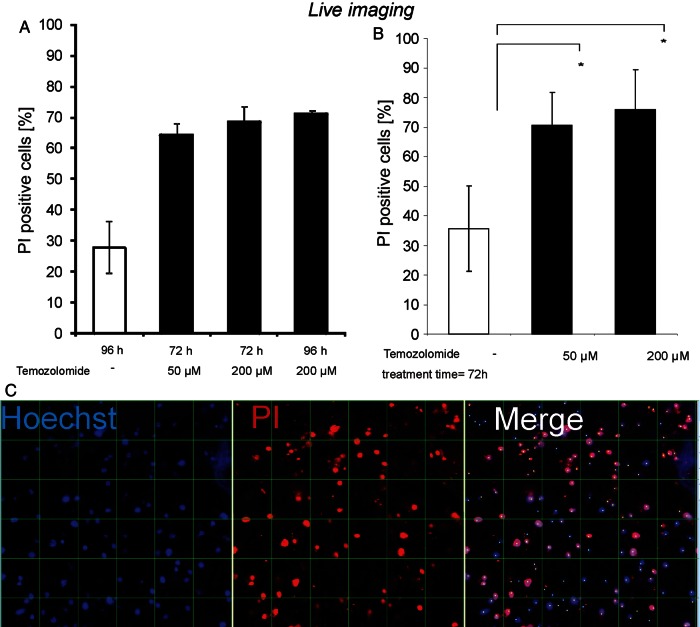
TMZ is the current gold standard in chemotherapy of GBM.42–46 It is an alkylating agent that, in an aqueous solution at physiological pH, dissolves into its bioactive form MTIC (5-(3-methyl-1-triazeno)imidazole-4-carboxamide), which is capable of penetrating the blood–brain barrier.47–49 To test whether GBM tissue in culture responds to TMZ treatment, slices were incubated with vehicle control or TMZ (50 or 200 µM). After 72 or 96 h, Hoechst 33342 and PI were added to the cultures to visualize intact and dying nuclei using confocal live imaging. Microimages were taken in which dying cells (Hoechst/PI double labeled) were counted and their number related to the total number of nuclei (Hoechst single labeled), allowing calculation of a cell death rate. TMZ-induced cell death was highly significant at 72 h, with little or no further increase until 96 h. This effect was more pronounced in slices treated with 200 µM compared with 50 µM of TMZ (Fig. 5). Thus, chemotherapeutic effects can be mimicked in GBM slices and observed over time in situ.
Fig. 5.
Live imaging of cell death in GBM slice cultures treated with TMZ. Slices were treated with either 50 or 200 µM TMZ and then incubated for 72 or 96 h (A). After determination that an incubation of 72 h was sufficient, the experiment was repeated 3 times with 50 or 200 µM TMZ (B). Dying cells were labeled with PI (red), and all nuclei were counterstained with Hoechst 33342 (blue) for quantitative analysis (C). Grid distance= 50 µm. Confocal images were taken and nuclei digitally counted. Student's t-test was performed, and P< .05 was considered statistically significant. Both concentrations showed a significant increase in dying cells compared with controls, with the effect slightly more pronounced at 200 µM. Original magnification: 200× in C; confocal Z-stacks.
Exposure of Slices to Carbon Ions and TMZ: Detection of Cell Death Using Activated Caspase 3 Staining
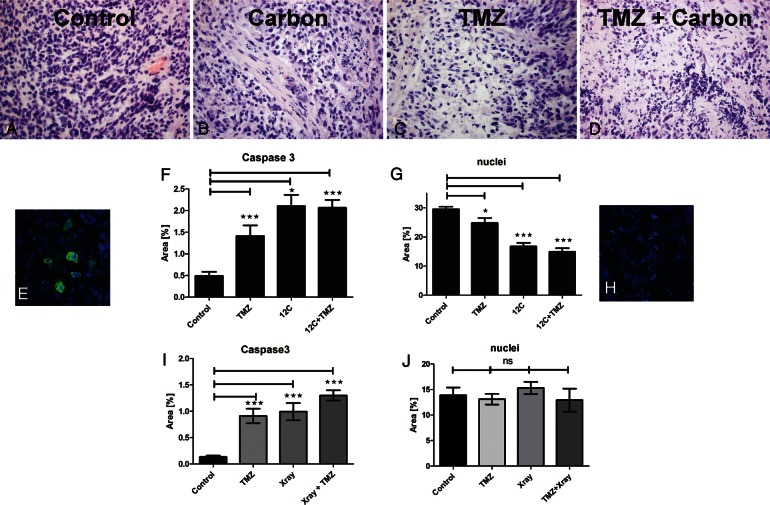
Slices were exposed to either TMZ or carbon ions in an SOBP alone or in combination and fixed 48 h after irradiation. TMZ treatment was started 24 h before irradiation, and a second dose was applied with the regular change of medium 48 h later. At the end of the treatment phase, induction of programmed cell death was determined using cleaved caspase 3 and H&E staining. All treatment regimens caused a decrease in cell density and nuclear alterations (Fig. 6B–D). In some slices, intact nuclei could no longer be identified because only fragments remained. Therefore, instead of relating damaged cells to a total number of cell nuclei, the area coverage of caspase 3–positive (green) and Hoechst-positive (blue) pixels (Fig. 6E, caspase 3–positive cells; 6H, nuclear fragments) was calculated as a measure of treatment-induced damage. After all treatments, the area of caspase 3–positive pixels was significantly increased, whereas Hoechst-positive pixels were decreased (Fig. 6F and G). Both irradiation and TMZ alone induced cell death, but a combination of both treatments did not show a synergistic or additive effect in the cases studied. Irradiation with X-rays, however, also activated caspase 3 but did not cause significant cell loss (Fig. 6I and J). Thus, effects of established treatment options on cell death can be studied in GBM slices.
Fig. 6.
Combined treatment of TMZ and irradiation with carbon ions and X-rays on human GBM slice cultures. GBM slices were treated with TMZ (200 µM), X-ray irradiation (4 Gy), SOBP carbon ions (2 Gy), or irradiation + TMZ. TMZ treatment started 24 h before irradiation and was maintained throughout the entire incubation time. Slices were fixed 2 days after irradiation, and H&E staining was performed for neuropathological assessment (A–D). In addition, activated caspase 3 was labeled (green in E), and fragmented nuclei were visualized using Hoechst 33342 (blue in E and H). Pictures of immunofluorescent stainings (in RGB format) were split into the three single channels of red, green, and blue, resulting in 3 gray-value pictures. The analysis was then performed using ImageJ's Area Measurement function. The area coverage is represented by white pixels in the respective channels and can be fine-tuned by threshold adjusting. In the green channel, the positive area represents the caspase 3–positive cell population, and in the blue channel, the area covered by nuclei is displayed (E–J). Carbon significantly reduced cell numbers (G), whereas X-rays did not (J). Significance was determined using GraphPad Prism 5 (1-way ANOVA, Bonferroni test). Both treatments result in a significant increase in cell death and morphological alterations compared with the vehicle-treated control. Original magnification: 400× in A–F.
TMZ Treatment of Slice Cultures From Tumors With Different MGMT Promoter Methylation Statuses
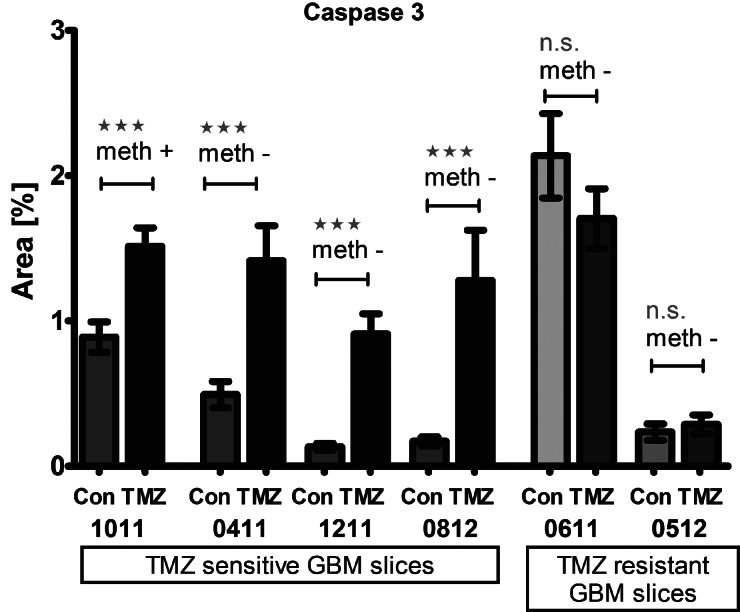
Lack of promoter methylation of O6-methylguanine-DNA methyltransferase (MGMT) has been reported to significantly decrease patients' susceptibility to TMZ and thus survival,50–52 but there are also patients with nonmethylated promoter who benefit from TMZ treatment. In fact, we identified one tumor in which TMZ did not significantly induce cell death and we therefore requested the promoter methylation status, which is assessed by quantitative PCR and sequencing techniques of tumor material obtained from surgery.53–55 This tumor indeed turned out to have a nonmethylated MGMT promoter (Fig. 7). However, in line with the clinical observations that some patients with nonmethylated MGMT respond to TMZ, we subsequently identified other tumors in which TMZ significantly enhanced activation of caspase 3 to levels comparable to those of the methylated tumor (Fig. 8).
Fig. 7.
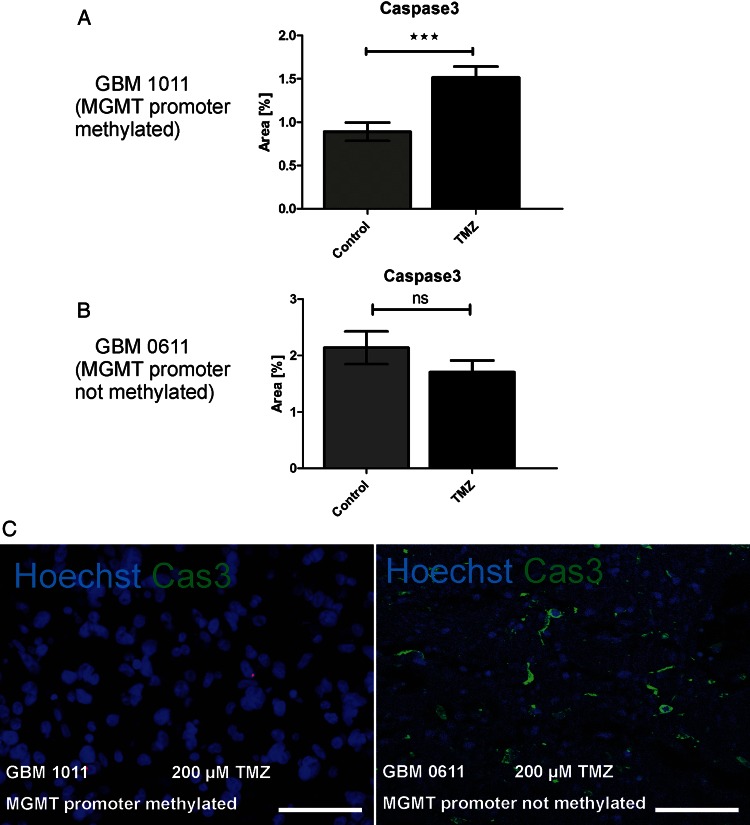
Induction of cell death. GBM slice cultures with different MGMT promoter methylation states were tested for their response to TMZ treatment. In a GBM specimen with methylated promoter sequence (A), TMZ treatment resulted in significant induction of caspase 3 cleavage (GBM 1011), whereas caspase 3 activation was not significantly induced (B) in GBM with an unmethylated promoter sequence (GBM 0611). Immunocytochemistry is shown for the methylated (C, right) and unmethylated (C, left) tumor for cleaved caspase 3 (green) and nuclei (Hoechst, blue). Scale bar= 50 µm. Original magnification: 400× in C; confocal Z-stacks.
Fig. 8.
Caspase 3 activation in GBM slices after TMZ treatment independent of MGMT promoter methylation. After TMZ treatment, caspase 3 activation was detected in some specimens, whereas others seemed to be resistant. This was independent of MGMT promoter methylation. From left to right: GBM 1011 with methylated MGMT promoter (meth +) and significant caspase 3 activation; GBM 0411 with nonmethylated MGMT promoter (meth -) and significant caspase 3 activation; GBM 1211 with nonmethylated MGMT promoter (meth -) and significant caspase 3 activation; GBM 0812 with nonmethylated MGMT promoter (meth -) and significant caspase 3 activation; GBM 0611 with nonmethylated MGMT promoter (meth -) and nonsignificant (ns) caspase 3 activation; GBM 0512 with nonmethylated MGMT promoter (meth -) and nonsignificant (ns) caspase 3 activation. (t-Test, P< .05)
Discussion
Organotypic slice cultures derived from early postnatal rodent brain56 are widely used in neuroscience due to their easy access for pharmacological intervention, electrophysiological studies, and live imaging. We have employed entorhinohippocampal preparations, which, due to the orientation of the trisynaptic pathway (perpendicular to the longitudinal axis of the hippocampus), allow for maintenance of the major connectivity.12,16 In this study, we adjusted cutting and culturing methods to prepare slices from human GBM and demonstrated evidence for their suitability as a test system for novel therapies including irradiation with HI. We irradiated GBM slices with photons and carbon ions and in both instances found that radiation induced DNA damage and strongly affected proliferation. Carbon ion radiation also induced activation of caspase 3, a potent inductor of programmed cell death (Figs. 3–6). In contrast to photon radiation, which delivers energy all the way through the body (eg, in an anterior-posterior direction), the depth of energy deposition of HI can be adjusted and limited to distinct areas of a few millimeters.57,58 In fact, much hope derives from successful intensity-modulated carbon therapy of chondrosarcomas of the skull base, a treatment first established at GSI.59–61 An accelerator specialized for medical applications has been constructed in Heidelberg and opened for patients in 2009. Currently, clinical trials and work with cell lines are aimed at testing the effects of carbon ion radiation in tumors other than chondrosarcomas. Our data support observations in GBM-derived cell lines62,63 and in a small group of patients,64,65 and our approach may lead to a more detailed understanding of the biological effects of HI and additional novel therapies. Moreover, surviving tumor cells can be studied to understand their mode of resistance.
We also applied TMZ to GBM slices and analyzed the effect on cell survival using PI staining and live imaging, as well as labeling of activated caspase 3 in paraffin-embedded sections. TMZ is an alkylating agent widely used to treat GBM that, in combination with radiation therapy, helps prolong patients' survival time. Survival depends on the methylation status of the promoter of the repair enzyme MGMT. Methylation was significantly more frequent in patients who survived longer than 36 months after surgery (P < .05).66 Statistically, methylation status apparently affects susceptibility to TMZ,50,67 but some patients with nonmethylated status also seem to benefit from TMZ. Of note, determining methylation status is not trivial, as only a few of the 109 potential sites have been tested.68 A recent survey among 1053 members of the neuro-oncology community in the United States found that only a small percentage (10.9%) of clinicians regard MGMT status as “always” or “almost always” helpful for their decision making.69 In line with this observation, we identified 2 nonmethylated tumors that were resistant to TMZ (Fig. 7) but also found others that were not (Fig. 8). Thus, the next challenge will be to relate TMZ-induced cell death rates obtained in slices to (progression-free) survival times; we will address this issue with tumor-derived samples during the next 2 years to test the predictive value of this assay.
It may be trivial to state that only in vitro systems allow different therapeutic options to be tested for an individual patient. Our data demonstrate that tumor-derived GBM-slice cultures in principle are suitable for that task as a step on the way to more personalized therapies while also helping unravel basic mechanisms of tumor resistance.
Funding
This work was supported by the Messer Foundation, the Kassel Foundation, and the Dr. Senckenberg Foundation for initial funding, and by BMBF (grant no. 0315498A), ESA (grant no. 50WB0925), and DLR (carbon beam times).
Conflict of interest statement. None declared.
Acknowledgments
The authors thank Friedrich von Metzler, Ekkehardt Sättele, the Messer Foundation, the Kassel Foundation, and the Dr. Senckenberg Foundation for their intellectual and financial support in the initial phase of this project. The authors also thank Prof Ralf-Dieter Kortmann, Dr Annegret Glasow, and Katrin Eisenbruch from the Clinics for Radiation Therapy and Oncology, University Hospital, Leipzig, and Dr Michael Scholz from GSI for help with dosimetry and irradiation setup.
References
- 1.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39(3):193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 3.Jacques TS, Swales A, Brzozowski MJ, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. Embo J. 2010;29(1):222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silver DJ, Steindler DA. Common astrocytic programs during brain development, injury and cancer. Trends Neurosci. 2009;32(6):303–311. doi: 10.1016/j.tins.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 6.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sliwa M, Markovic D, Gabrusiewicz K, et al. The invasion promoting effect of microglia on glioblastoma cells is inhibited by cyclosporin A. Brain. 2007;130(Pt 2):476–489. doi: 10.1093/brain/awl263. [DOI] [PubMed] [Google Scholar]
- 9.Savaskan NE, Heckel A, Hahnen E, et al. Small interfering RNA-mediated xCT silencing in gliomas inhibits neurodegeneration and alleviates brain edema. Nat Med. 2008;14(6):629–632. doi: 10.1038/nm1772. [DOI] [PubMed] [Google Scholar]
- 10.Eke I, Storch K, Kastner I, et al. Three-dimensional invasion of human glioblastoma cells remains unchanged by X-ray and carbon ion irradiation in vitro. Int J Radiat Oncol Biol Phys. 2012;84(4):e515–523. doi: 10.1016/j.ijrobp.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Huang PH, Mukasa A, Bonavia R, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A. 2007;104(31):12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kluge A, Hailer NP, Horvath TL, Bechmann I, Nitsch R. Tracing of the entorhinal-hippocampal pathway in vitro. Hippocampus. 1998;8(1):57–68. doi: 10.1002/(SICI)1098-1063(1998)8:1<57::AID-HIPO6>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Hailer NP, Wirjatijasa F, Roser N, Hischebeth GT, Korf HW, Dehghani F. Astrocytic factors protect neuronal integrity and reduce microglial activation in an in vitro model of N-methyl-D-aspartate-induced excitotoxic injury in organotypic hippocampal slice cultures. Eur J Neurosci. 2001;14(2):315–326. doi: 10.1046/j.0953-816x.2001.01649.x. [DOI] [PubMed] [Google Scholar]
- 14.Dehghani F, Hischebeth GT, Wirjatijasa F, Kohl A, Korf HW, Hailer NP. The immunosuppressant mycophenolate mofetil attenuates neuronal damage after excitotoxic injury in hippocampal slice cultures. Eur J Neurosci. 2003;18(5):1061–1072. doi: 10.1046/j.1460-9568.2003.02821.x. [DOI] [PubMed] [Google Scholar]
- 15.Eyupoglu IY, Savaskan NE, Brauer AU, Nitsch R, Heimrich B. Identification of neuronal cell death in a model of degeneration in the hippocampus. Brain Res Brain Res Protoc. 2003;11(1):1–8. doi: 10.1016/s1385-299x(02)00186-1. [DOI] [PubMed] [Google Scholar]
- 16.Prodinger C, Bunse J, Kruger M, et al. CD11c-expressing cells reside in the juxtavascular parenchyma and extend processes into the glia limitans of the mouse nervous system. Acta Neuropathol. 2011;121(4):445–458. doi: 10.1007/s00401-010-0774-y. [DOI] [PubMed] [Google Scholar]
- 17.Schaefer L, Babelova A, Kiss E, et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J Clin Invest. 2005;115(8):2223–2233. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senner V, Ratzinger S, Mertsch S, Grassel S, Paulus W. Collagen XVI expression is upregulated in glioblastomas and promotes tumor cell adhesion. FEBS Lett. 2008;582(23–24):3293–3300. doi: 10.1016/j.febslet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Moreth K, Brodbeck R, Babelova A, et al. The proteoglycan biglycan regulates expression of the B cell chemoattractant CXCL13 and aggravates murine lupus nephritis. J Clin Invest. 2010;120(12):4251–4272. doi: 10.1172/JCI42213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merline R, Moreth K, Beckmann J, et al. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and microRNA-21. Sci Signal. 2011;4(199):ra75. doi: 10.1126/scisignal.2001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walczak H, Miller RE, Ariail K, et al. Tumoricidal activity of tumor necrosis factor–related apoptosis-inducing ligand in vivo. Nat Med. 1999;5(2):157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 22.Nitsch R, Bechmann I, Deisz RA, et al. Human brain-cell death induced by tumour-necrosis-factor–related apoptosis-inducing ligand (TRAIL) Lancet. 2000;356(9232):827–828. doi: 10.1016/S0140-6736(00)02659-3. [DOI] [PubMed] [Google Scholar]
- 23.Dowsing T, Kendall MJ. The Northwick Park tragedy—protecting healthy volunteers in future first-in-man trials. J Clin Pharm Ther. 2007;32(3):203–207. doi: 10.1111/j.1365-2710.2007.00808.x. [DOI] [PubMed] [Google Scholar]
- 24.Kenter MJ, Cohen AF. Establishing risk of human experimentation with drugs: lessons from TGN1412. Lancet. 2006;368(9544):1387–1391. doi: 10.1016/S0140-6736(06)69562-7. [DOI] [PubMed] [Google Scholar]
- 25.Merz F, Bechmann I. Irradiation of human tumor tissue cultures: optimizing ion radiation therapy. Future Oncol. 2011;7(4):489–491. doi: 10.2217/fon.11.18. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Lang F, Xie X, et al. Efficacy of adenovirally expressed soluble TRAIL in human glioma organotypic slice culture and glioma xenografts. Cell Death Dis. 2011;2:e121. doi: 10.1038/cddis.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaira V, Fedele G, Pyne S, et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci U S A. 2010;107(18):8352–8356. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durante M, Loeffler JS. Charged particles in radiation oncology. Nat Rev Clin Oncol. 2010;7(1):37–43. doi: 10.1038/nrclinonc.2009.183. [DOI] [PubMed] [Google Scholar]
- 29.Pignalosa D, Durante M. Overcoming resistance of cancer stem cells. Lancet Oncol. 2012;13(5):e187–188. doi: 10.1016/S1470-2045(12)70196-1. [DOI] [PubMed] [Google Scholar]
- 30.Combs SE, Jakel O, Haberer T, Debus J. Particle therapy at the Heidelberg Ion Therapy Center (HIT)—integrated research-driven university-hospital–based radiation oncology service in Heidelberg, Germany. Radiother Oncol. 2010;95(1):41–44. doi: 10.1016/j.radonc.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Muller M, Durante M, Stocker H, Merz F, Bechmann I. Modeling radiation effects at the tissue level. Eur Phys J D. 2010;60(1):171–176. [Google Scholar]
- 32.Merz F, Muller M, Taucher-Scholz G, et al. Tissue slice cultures from humans or rodents: a new tool to evaluate biological effects of heavy ions. Radiat Environ Biophys. 2010;49(3):457–462. doi: 10.1007/s00411-010-0293-1. [DOI] [PubMed] [Google Scholar]
- 33.Haberer T, Becher W, Schardt D, Kraft G. Magnetic scanning system for heavy ion therapy. Nucl Instrum Meth Phys Res Sect A. 1993;330(1–2):296–305. [Google Scholar]
- 34.Bauer R, Ratzinger S, Wales L, et al. Inhibition of collagen XVI expression reduces glioma cell invasiveness. Cell Physiol Biochem. 2011;27(3–4):217–226. doi: 10.1159/000327947. [DOI] [PubMed] [Google Scholar]
- 35.Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31(1):13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 36.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol. 2003;81(3):123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 37.Prise KM, Schettino G, Folkard M, Held KD. New insights on cell death from radiation exposure. Lancet Oncol. 2005;6(7):520–528. doi: 10.1016/S1470-2045(05)70246-1. [DOI] [PubMed] [Google Scholar]
- 38.Jeggo PA, Lobrich M. DNA double-strand breaks: their cellular and clinical impact? Oncogene. 2007;26(56):7717–7719. doi: 10.1038/sj.onc.1210868. [DOI] [PubMed] [Google Scholar]
- 39.Tobias F, Durante M, Taucher-Scholz G, Jakob B. Spatiotemporal analysis of DNA repair using charged particle radiation. Mutat Res. 2010;704(1–3):54–60. doi: 10.1016/j.mrrev.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 41.Keogh MC, Kim JA, Downey M, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439(7075):497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 42.Fukushima T, Takeshima H, Kataoka H. Anti-glioma therapy with temozolomide and status of the DNA-repair gene MGMT. Anticancer Res. 2009;29(11):4845–4854. [PubMed] [Google Scholar]
- 43.Bei R, Marzocchella L, Turriziani M. The use of temozolomide for the treatment of malignant tumors: clinical evidence and molecular mechanisms of action. Recent Pat Anticancer Drug Discov. 2010;5(3):172–187. doi: 10.2174/157489210791760526. [DOI] [PubMed] [Google Scholar]
- 44.Becker KP, Yu J. Status quo—standard-of-care medical and radiation therapy for glioblastoma. Cancer J. 2012;18(1):12–19. doi: 10.1097/PPO.0b013e318244d7eb. [DOI] [PubMed] [Google Scholar]
- 45.Johnson DR, Chang SM. Recent medical management of glioblastoma. Adv Exp Med Biol. 2012;746:26–40. doi: 10.1007/978-1-4614-3146-6_3. [DOI] [PubMed] [Google Scholar]
- 46.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 47.Tisdale MJ. Antitumor imidazotetrazines—XV. Role of guanine O6 alkylation in the mechanism of cytotoxicity of imidazotetrazinones. Biochem Pharmacol. 1987;36(4):457–462. doi: 10.1016/0006-2952(87)90351-0. [DOI] [PubMed] [Google Scholar]
- 48.Denny BJ, Wheelhouse RT, Stevens MF, Tsang LL, Slack JA. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33(31):9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 49.Danson SJ, Middleton MR. Temozolomide: a novel oral alkylating agent. Expert Rev Anticancer Ther. 2001;1(1):13–19. doi: 10.1586/14737140.1.1.13. [DOI] [PubMed] [Google Scholar]
- 50.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 51.Kim YS, Kim SH, Cho J, et al. MGMT gene promoter methylation as a potent prognostic factor in glioblastoma treated with temozolomide-based chemoradiotherapy: a single-institution study. Int J Radiat Oncol Biol Phys. 2012;84(3):661–667. doi: 10.1016/j.ijrobp.2011.12.086. [DOI] [PubMed] [Google Scholar]
- 52.Christians A, Hartmann C, Benner A, et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS One. 2012;7(3):e33449. doi: 10.1371/journal.pone.0033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah N, Lin B, Sibenaller Z, et al. Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PLoS One. 2011;6(1):e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Nifterik KA, van den Berg J, van der Meide WF, et al. Absence of the MGMT protein as well as methylation of the MGMT promoter predict the sensitivity for temozolomide. Br J Cancer. 2010;103(1):29–35. doi: 10.1038/sj.bjc.6605712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitange GJ, Carlson BL, Mladek AC, et al. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009;92(1):23–31. doi: 10.1007/s11060-008-9737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gahwiler BH. Slice cultures of cerebellar, hippocampal and hypothalamic tissue. Experientia. 1984;40(3):235–243. doi: 10.1007/BF01947561. [DOI] [PubMed] [Google Scholar]
- 57.Tobias CA, Blakely EA, Alpen EL, et al. Molecular and cellular radiobiology of heavy ions. Int J Radiat Oncol Biol Phys. 1982;8(12):2109–2120. doi: 10.1016/0360-3016(82)90554-5. [DOI] [PubMed] [Google Scholar]
- 58.Chatterjee A, Takada E, Torikoshi M, Kanazawa M. Diagnostic imaging by energetic radioactive particle beams: applications in Bragg peak cancer therapy. Nucl Phys A. 1997;A616(1–2):478c–489c. doi: 10.1016/s0375-9474(97)00119-x. [DOI] [PubMed] [Google Scholar]
- 59.Eickhoff H, Haberer T, Kraft G, et al. The GSI Cancer Therapy Project. Strahlenther Onkol. 1999;175(suppl 2):21–24. doi: 10.1007/BF03038880. [DOI] [PubMed] [Google Scholar]
- 60.Gademann G, Hartmann GH, Kraft G, Lorenz WJ, Wannenmacher M. The medical heavy ion therapy project at the Gesellschaft fur Schwerionenforschung facility in Darmstadt. Strahlenther Onkol. 1990;166(1):34–39. [PubMed] [Google Scholar]
- 61.Schulz-Ertner D, Tsujii H. Particle radiation therapy using proton and heavier ion beams. J Clin Oncol. 2007;25(8):953–964. doi: 10.1200/JCO.2006.09.7816. [DOI] [PubMed] [Google Scholar]
- 62.Oishi T, Sasaki A, Hamada N, et al. Proliferation and cell death of human glioblastoma cells after carbon-ion beam exposure: morphologic and morphometric analyses. Neuropathology. 2008;28(4):408–416. doi: 10.1111/j.1440-1789.2008.00899.x. [DOI] [PubMed] [Google Scholar]
- 63.Combs SE, Bohl J, Elsasser T, et al. Radiobiological evaluation and correlation with the local effect model (LEM) of carbon ion radiation therapy and temozolomide in glioblastoma cell lines. Int J Radiat Biol. 2009;85(2):126–137. doi: 10.1080/09553000802641151. [DOI] [PubMed] [Google Scholar]
- 64.Mizoe JE, Tsujii H, Hasegawa A, et al. Phase I/II clinical trial of carbon ion radiotherapy for malignant gliomas: combined X-ray radiotherapy, chemotherapy, and carbon ion radiotherapy. Int J Radiat Oncol Biol Phys. 2007;69(2):390–396. doi: 10.1016/j.ijrobp.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Combs SE, Kieser M, Rieken S, et al. Randomized phase II study evaluating a carbon ion boost applied after combined radiochemotherapy with temozolomide versus a proton boost after radiochemotherapy with temozolomide in patients with primary glioblastoma: the CLEOPATRA trial. BMC Cancer. 2010;10:478. doi: 10.1186/1471-2407-10-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skiriute D, Vaitkiene P, Saferis V, et al. MGMT, GATA6, CD81, DR4, and CASP8 gene promoter methylation in glioblastoma. BMC Cancer. 2012;12:218. doi: 10.1186/1471-2407-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakada M, Furuta T, Hayashi Y, Minamoto T, Hamada J. The strategy for enhancing temozolomide against malignant glioma. Front Oncol. 2012;2:98. doi: 10.3389/fonc.2012.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Everhard S, Tost J, El Abdalaoui H, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11(4):348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holdhoff M, Ye X, Blakeley JO, et al. Use of personalized molecular biomarkers in the clinical care of adults with glioblastomas. J Neurooncol. 2012;110(2):279–285. doi: 10.1007/s11060-012-0968-3. [DOI] [PubMed] [Google Scholar]