Abstract
Background
Mutations involving isocitrate dehydrogenase 1 (IDH 1) occur in a high proportion of diffuse gliomas, with implications on diagnosis and prognosis. About 90% involve exon 4 at codon 132, replacing amino acid arginine with histidine (R132H). Rarer ones include R132C, R132S, R132G, R132L, R132V, and R132P. Most authors have used DNA-based methods to assess IDH1 status. Preliminary studies comparing imunohistochemistry (IHC) with IDH1-R132H mutation-specific antibodies have shown concordance with DNA sequencing and no cross-reactivity with wild-type IDH1 or other mutant proteins. The present study compares results of IHC with DNA sequencing in diffuse gliomas.
Materials and methods
Fifty diffuse gliomas with frozen tissue samples for DNA sequencing and adequate tissue in paraffin blocks for IHC using IDH1-R132H specific antibody were assessed for IDH1 mutations.
Results
Concordance of findings between IHC and DNA sequencing was noted in 88% (44/50) cases. All 6 cases with discrepancy were immunopositive with DIA-H09 antibody. While in 3 of these 6 cases, DNA sequencing failed to reveal any mutations, R132L (arginine replaced by leucine) mutation was found in the rest 3 cases. Interestingly, of the immunopositive cases, 46.6% (14/30) showed immunostaining in only a fraction of tumor cells.
Conclusions
IHC is an easy and quick method of detecting IDH1-R132H mutations, but there may be some discrepancies between IHC and DNA sequencing. Although there were no false-negative cases, cross-reactivity with IDH1-R132L was seen in 3, a finding not reported thus far. Because of more universal availability of IHC over genetic testing, cross-reactivity and staining heterogeneity may have bearing over its use in detecting IDH1-R132H mutation in gliomas.
Keywords: diffuse gliomas, DNA sequencing, IDH1-R132H, IDH1-R132L, immunohistochemistry
The IDH1 gene on chromosome 2q33.3 encodes for isocitrate dehydrogenase 1 (IDH1), located in the cytoplasm and the peroxisomes. This enzyme catalyzes NADPH production via oxidative decarboxylation of isocitrate to alpha-ketoglutarate in the Krebs citric acid cycle.1 In 2008, for the first time, Parsons et al introduced to the medicine world the role of IDH1 in the pathogenesis of glioblastoma multiforme (GBM). In their genome-wide sequencing analysis, recurrent somatic mutations specifically involving the amino acid arginine at position 132 were detected in 12% of the GBM specimens.2 Subsequent studies have shown that IDH1 mutation is an early step in gliomagenesis and has been reported to occur in grades II and III astrocytomas, oligodendrogliomas (OG), oligoastrocytomas (OA), and secondary GBM.3–12 Hartmann et al, in their analysis of 1010 diffuse glioma tumors, demonstrated that most cases of diffuse astrocytomas (DA; 72.7%, 165/227), anaplastic astrocytomas (AA; 64.0%, 146/228), OG (82.0%, 105/128), anaplastic oligodendrogliomas (AOG; 69.5%, 121/174), OA (81.6%, 62/76), and anaplastic oligoastrocytomas (AOA; 66.1%, 117/177) had IDH1 mutations.13 Of importance, these mutations appear to be specific for these tumors as primary GBM, pilocytic astrocytoma World Health Organization (WHO) grade I and other central nervous system (CNS) and non-CNS neoplasms, with the exception of acute myeloid leukemia and cartilaginous neoplasms, harbor this genetic alteration much less frequently.3–11,14–19
Different types of mutations have been described, and the most frequent is G to A transitions at position 395 of the IDH1 transcript. This results in substitution of the amino acid arginine with histidine (R132H). Rarer ones include R132C, R132S, R132G, R132L, R132V and R132P.2,4,5,8–11,13,17,20,21 Mutations involving IDH2, a homologous gene, have also been detected in gliomas but at a much lower frequency ranging from 2% to 5%.8,10,13,17,21,22
Although these mutations are rare in the pediatric age group, in patients aged ≥18 years, they seem to be associated with younger age at presentation and have a favorable impact on the overall and progression-free survival associated with grade II-IV gliomas.2,6,7,10,12,13,22–28
IDH1 testing is being used as a standard diagnostic tool in many neuropathology laboratories. It is useful in differentiating gliomas from nonneoplastic CNS lesions,21,29–31 diffuse astrocytoma WHO grade II from pilocytic astrocytoma grade I,15,32 anaplastic astrocytomas WHO grade III from GBM,32 primary from secondary GBM,6,32 and astrocytomas from ependymomas.32
Most studies of IDH mutations are based on DNA sequencing, which is labor intensive, requiring trained personnel and sophisticated equipment, not available at every center. Moreover, false-negative results may be obtained in cases of inadequate tumor DNA availability because of small biopsy samples, extensive necrosis, or admixture with normal tissue elements. Alternate rapid methods, some based on routinely processed tissue specimens, have been recently suggested.17,21,26,29–36 Of these, a significant development was the introduction of mAb H0933 and IMab-134 antibodies, which are specific for the most common IDH1 mutation: R132H. In the present study, which is a continuation of our previous study on IDH1 mutation as assessed by direct DNA sequencing,11 we compared the results of immunohistochemistry (IHC) using mAb H09 for IDH1-R132 mutations with those of DNA sequencing in different types and grades of gliomas.
Materials and Methods
Tumor Specimens
Tumor samples were obtained fresh at the time of surgery from the operation theatre of the Neurosurgery Department at the All India Institute of Medical Sciences, New Delhi, India. All experiments using the human samples were approved by the ethical committee of our institution. There were 8 DA (WHO grade II), 7 AA WHO grade III, 20 GBMs WHO grade IV, 5 grade II OG, 4 AOG, 3 grade II OA, and 3 AOA. Portions of resected tumors were snap-frozen in liquid nitrogen and stored at -80°C until use, and the rest of the tissue was formalin-fixed and paraffin-embedded for routine histopathology and IHC. The hematoxylin and eosin (H&E)–stained slides were reviewed by 4 independent neuropathologists (M.C.S., S.A., V.S., and C.S.), who were not aware of the results of the genetic analysis, and consensus diagnoses were made according to WHO classification (2007).37 Clinical parameters, including age, sex, duration of symptoms, and relevant clinical history, of all the patients were recorded. Cases of GBM were classified as primary if there was no history and histomorphological evidence of a diffuse or anaplastic glial tumor and when the duration of symptoms was <3 months. A case was categorized as secondary GBM only if there was a history of surgery for lower grade precursor tumors.38,39 Thus, there were 15 primary and 5 secondary GBM cases.
Tissue Procurement and DNA Preparation
Frozen tumor specimens were embedded in freezing medium, sectioned at 5 μm, and stained with H&E. Subsequent 15 serial sections of 40 μm were taken separately for DNA isolation and stored immediately in liquid nitrogen cooled vials. Flanking sections measuring 5 μm were analyzed histologically for presence of adequate tumor tissue, to look for areas of necrosis and normal cerebral cortex. Those sections where the flanking H&E sections showed tumor content >80%, with none or minimal necrosis and no normal tissue were used for DNA extraction. DNA from the tumor tissue was extracted using Genelute mammalian DNA isolation Kit (M/s. Sigma Aldrich, St. Louis, MO) according to the manufacturer's protocol.
IDH1 Mutational Analysis
Mutations in exon 4 of IDH1 were determined by direct sequencing in all the cases. Primer sequences used were forward 5′AATGAGCTCTATATGCCATCACTG3′and reverse 5′ TTCATACCTTGCTTAATGGGTGT3′. PCR amplification was performed in a total of 10 µL reaction mixture containing 50 ng of tumor DNA, 1 µL of 10× PCR buffer, 0.8 µL of 10 mM dNTPs, 0.25 µL of each forward and reverse primers, and 0.2 µL of AmpliTaq Gold PCR Master Mix (Applied Biosystems, Inc., Foster City, CA). Initial denaturation was performed at 95°C for 5 min. This was followed by 35 cycles of amplification consisting of denaturation at 95°C for 1 min, annealing at 57°C for 45 s, and extension at 72°C for 2 min. Bidirectional sequencing was performed using ABI 3730 sequencer (Applied Biosystems).
In cases in which the results of DNA sequencing and IHC were discrepant, repeat genetic testing was done on formalin-fixed paraffin embedded (FFPE) tissue. For this, paraffin blocks with adequate representative tumor tissue >80% and with a minimal necrotic zone were chosen. This was confirmed by assessing H&E-stained flanking sections. Forward and reverse primers used included 5′CGGTCTTCAGAGAAGCCATT3′ and 5′GCAAAATCACATTATTGCCAAC3′, respectively. For PCR, initial denaturation was performed at 95°C for 10 min. This was followed by 37 cycles of amplification consisting of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s.
Immunohistochemistry
Immunohistochemistry for IDH1-R132H was done on 5-microns–thick formalin-fixed, paraffin-embedded tumor sections with use of antibody specific for the mutant IDH1-R132H protein (H09, Dianova, dil 1:100). Labeled streptavidin biotin kit (Universal) was used as a detection system (Dako, Denmark). Antigen retrieval was performed in citrate buffer (pH, 6.0) in a microwave oven.
Two observers (M.C.S. and S.A.) together evaluated the results of IHC on a multiheaded microscope and were blinded to the results of the genetic analysis. Combined cytoplasmic and nuclear staining was interpreted as immunopositive. A 3-tiered semiquantitative system used in the previous study by Preusser M et al40 was followed: negative, if no tumor cell was immunopositive; partly positive (focal positivity), if there was admixture of immunopositive and immunonegative tumor cells or areas of immunopositive and immunonegative tumor cells were adjacent to each other; complete positivity (diffuse positivity), if all the tumor cells were immunopositive.
Results (Table 1)
Table 1.
Details of the cases included in the study
| Case no. | Age/Sex | Diagnosis | IHC for IDH1 (H09) | Pattern of immunopositivity | IDH1 mutation on initial DNA sequencing | IDH1 mutation on repeat genetic testing | Type of mutation |
|---|---|---|---|---|---|---|---|
| 1 | 30/M | OG II | Neg | - | Absent | Absent | - |
| 2 | 32/F | OG II | Pos | Diffuse | Present | Present | R132H |
| 3 | 50/M | OG II | Pos | Diffuse | Present | Present | R132H |
| 4 | 48/M | OG II | Pos | Diffuse | Present | Present | R132H |
| 5 | 42/M | OG II | Neg | - | Absent | Absent | - |
| 6 | 30/M | OG III | Pos | Diffuse | Present | Present | R132H |
| 7 | 40/M | OG III | Pos | Diffuse | Present | Present | R132H |
| 8 | 28/M | OG III | Pos | Focal | Present | Present | R132L |
| 9 | 38/M | OG III | Pos | Diffuse | Present | Present | R132H |
| 10 | 43/M | OA II | Pos | Focal | Present | Present | R132H |
| 11 | 45/M | OA II | Pos | Focal (Oligodendroglial component) | Present | Present | R132H |
| 12 | 39/M | OA II | Neg | - | Absent | Absent | - |
| 13 | 32/M | OA III | Pos | Diffuse | Present | Present | R132H |
| 14 | 24/M | OA III | Pos | Focal | Absent | Present | R132L |
| 15 | 28/M | OA III | Pos | Focal (Oligodendroglial component) | Present | Present | R132H |
| 16 | 32/M | DA II | Pos | Focal | Present | Present | R132H |
| 17 | 35/M | DA II | Pos | Focal | Absent | Absent | - |
| 18 | 32/M | DA II | Pos | Focal | Present | Present | R132H |
| 19 | 26/M | DA II | Pos | Focal | Absent | Absent | - |
| 20 | 17/M | DA II | Pos | Diffuse | Present | Present | R132H |
| 21 | 26/M | DA II | Neg | - | Present | Present | R132L |
| 22 | 28/M | DA II | Pos | Focal | Present | Present | R132H |
| 23 | 34/F | DA II | Pos | Focal | Present | Present | R132H |
| 24 | 24/M | AA III | Pos | Focal | Present | Present | R132L |
| 25 | 42/M | AA III | Pos | Diffuse | Present | Present | R132H |
| 26 | 22/M | AA III | Pos | Diffuse | Present | Present | R132H |
| 27 | 30/M | AA III | Pos | Diffuse | Present | Present | R132H |
| 28 | 13/M | AA III | Neg | - | Absent | Absent | - |
| 29 | 34/M | AA III | Neg | - | Present | Present | R132C |
| 30 | 28/M | AA III | Pos | Diffuse | Present | Present | R132H |
| 31 | 55/M | Pri GBM | Neg | - | Absent | Absent | - |
| 32 | 40/M | Sec GBM | Pos | Focal | Present | Present | R132H |
| 33 | 44/M | Sec GBM | Pos | Diffuse | Present | Present | R132H |
| 34 | 65/M | Pri GBM | Neg | - | Absent | Absent | - |
| 35 | 48/M | Sec GBM | Pos | Diffuse | Present | Present | R132H |
| 36 | 70/M | Pri GBM | Neg | - | Absent | Absent | - |
| 37 | 27/M | Pri GBM | Neg | - | Absent | Absent | - |
| 38 | 40/M | Pri GBM | Pos | Focal | Present | Present | R132H |
| 39 | 48/F | Pri GBM | Neg | - | Absent | Absent | - |
| 40 | 39/M | Pri GBM | Neg | - | Absent | Absent | - |
| 41 | 36/M | Sec GBM | Neg | - | Absent | Absent | - |
| 42 | 50/M | Pri GBM | Neg | - | Absent | Absent | - |
| 43 | 50/M | Pri GBM | Neg | - | Absent | Absent | - |
| 44 | 67/M | Pri GBM | Neg | - | Absent | Absent | - |
| 45 | 65/M | Pri GBM | Neg | - | Absent | Absent | - |
| 46 | 52/M | Pri GBM | Neg | - | Absent | Absent | - |
| 47 | 10/M | Sec GBM | Neg | - | Absent | Absent | - |
| 48 | 60/F | Pri GBM | Neg | - | Absent | Absent | - |
| 49 | 40/M | Pri GBM | Pos | Diffuse | Present | Present | R132H |
| 50 | 60/F | Pri GBM | Pos | Diffuse | Absent | Absent | - |
Abbreviations: AA, Anaplastic astrocytoma; GBM, Glioblastoma multiforme; IHC, Immunohistochemistry; Neg, Negative; OA: Oligoastrocytoma; OG, Oligodendroglioma; Pos, Positive; Pri, Primary; Sec, Secondary.
DNA Sequencing (Fig. 1)
Fig. 1.

Representative electropherograms of the various IDH1 mutations as obtained by DNA sequencing in the current series of diffuse gliomas.
Of the 50 gliomas included in the study, in initial DNA sequencing, 28 cases were found to bear heterozygous IDH1 mutations, all localized to codon 132 of IDH1. There was substitution of the amino acid arginine by histidine (R132H) in 48% (24/50), by leucine (R132L) in 6% (3/50), and by cysteine (R132C) in 2% (1/50).
Immunohistochemistry (Fig. 2)
Fig. 2.
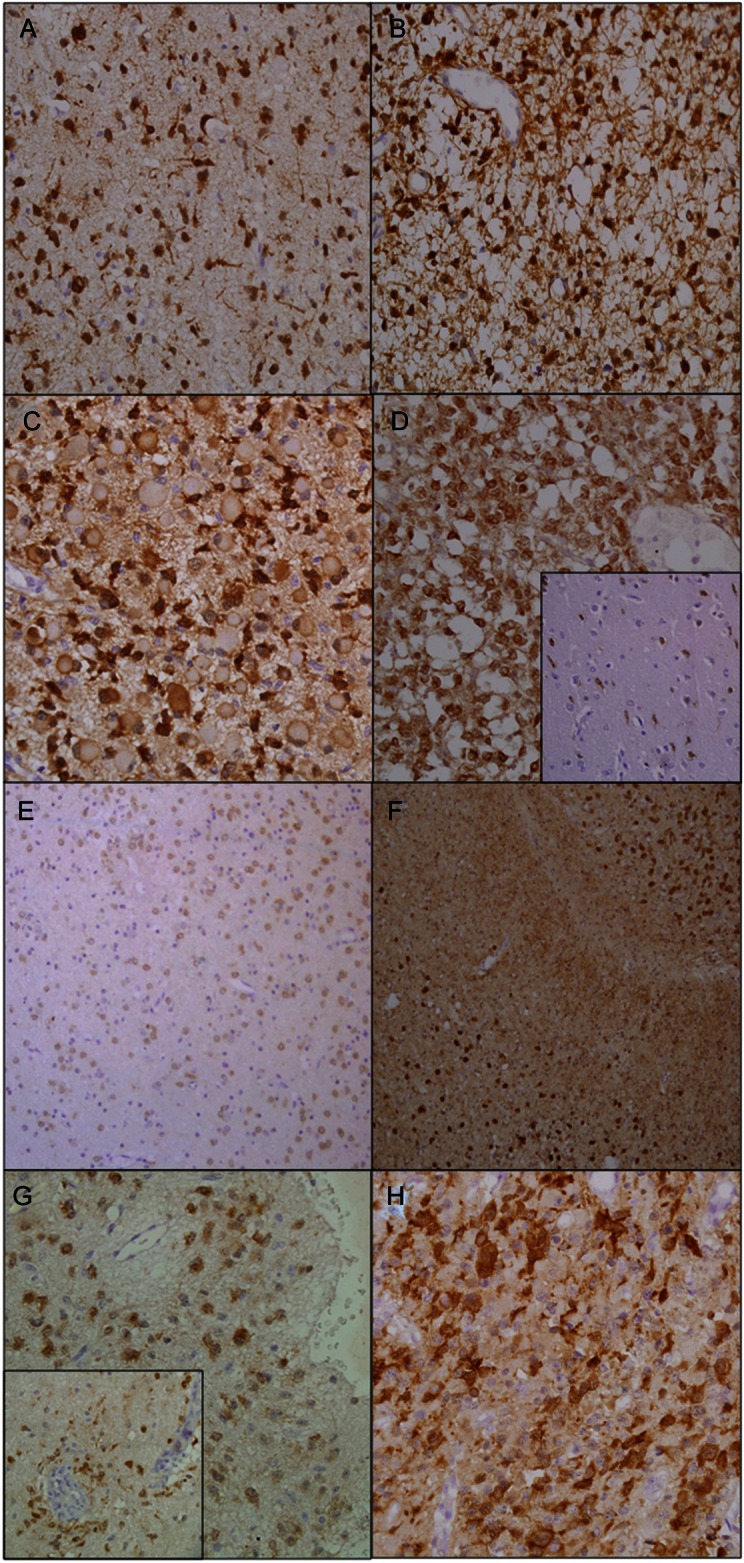
Showing different patterns of immunopositivity in different types of gliomas. (A) Cytoplasmic and weak nuclear positivity with uni- and bipolar cytoplasmic processes (Diffuse atrocytoma, grade II) (×400). (B) Anaplastic astrocytoma (grade III) showing multipolar stellate cytoplasmic processes of the tumor cells (×400). (C) Gemistocytic astrocytoma (grade III) showing peripheral accentuation of staining and central paler unstained area (×400). (D) Anaplastic oligoastrocytoma (grade III) showing immunopositivity in all tumor cells. Inset showing immunopositive tumor cells at the infiltrating edge of the same case (×400). (E) Mixed oligoastrocytoma (grade II) in which oligodendroglial component is immunopositive but the astrocytic cells negative (×200). (F) Biphasic anaplastic oligoastrocytoma (grade III) showing immunopositivity of oligodendroglial component but the astrocytic component is negative (×400). (G) Subpial and perivascular (inset) infiltration of tumor cells in cases of oligoastrocytoma grade II and anaplastic oligoastrocytoma grade III, respectively (×400). (H) GBM IV showing strong immunoreactivity in few of the tumor cells with short cytoplasmic processes (×400).
With use of H09, mAb specific for R132H, immunoreactivity was noted in 60% (30/50) cases, of which 53.3% (16/30) showed diffuse positivity and 46.6% (14/30) showed immunopositivity in a fraction of tumor cells. The antibody stained the cytoplasm and weakly the nucleus of the tumor cells. Endothelial cells and residual normal and reactive glial cells did not show immunopositivity. Background staining was absent in most but, wherever present, did not hamper interpretability of results.
Table 2.
Comparison of results of DNA sequencing with immunohistochemistry
| Cases | Initial DNA sequencing | Repeat DNA sequencing | Percentage of cases immunopositive with H09 |
|---|---|---|---|
| With R132H mutation | 48% (24/50) | 48% (24/50) | 100% (24/24) |
| With R132L mutation | 6% (3/50) | 8% (4/50) | 75% (3/4) |
| With R132C mutation | 2% (1/50) | 2% (1/50) | None |
| With no IDH1 mutation | 44% (22/50) | 42% (21/50) | 14.3% (3/21) |
In astrocytic tumors, H09 highlighted uni- and bipolar processes emanating from the neoplastic cells (Fig. 2A). AA (WHO grade III), in addition, showed thin multipolar stellate processes (Fig. 2B). Gemistocytic astrocytoma (WHO grade III) showed peripheral accentuation of staining with central paler unstained zone (Fig. 2C). Strong cytoplasmic staining, with very few cytoplasmic processes, was observed in oligodendrogliomas in both WHO grades II and III tumors (Fig. 2D). However, in the infiltrating zones, these cells appeared to acquire uni- to bipolar processes similar to those of astrocytomas (Fig. 2D inset).
In all but 2 cases of OA, both the astrocytic and the oligodendroglial components were immunoreactive. In the remaining 2 cases, both of mixed OA, one being WHO grade II (Fig. 2E) and the other grade III (Fig. 2F), however, immunopositivity was observed in only the oligodendroglial component. Tumor cells showing condensation in the subpial (Fig. 2G) and perivascular (Fig. 2G inset) zones in 2 of the oligoastrocytic tumors were also positive. GBM (WHO grade IV) showed cytoplasmic and weak nuclear positivity with absent or short cytoplasmic processes (Fig. 2H).
Cases With Discrepant Results of Genetic Testing and IHC (Fig. 3)
Fig. 3.

Photomicrographs of cases with mismatched results of IHC and genetic testing. (A and B) Diffuse astrocytoma grade II (H&E ×400) showing immunopositivity of cytoplasm and cytoplasmic processes (IHC ×400). (B-inset) Another area of the same tumor showing only a fraction of the tumor cells to be immunoreactive (IHC ×400). (C and D) Case of DA II (H&E ×400) immunopositive with DIA-H09 (IHC ×400). (D-inset) Photomicrograph from a different area demonstrating a proportion of the tumor cells to be immunonegative (IHC ×400). On genetic analysis, both these cases failed to show IDH1mutation. (E and F) Anaplastic astrocytoma grade III (H&E ×400) with R132L mutations showing immunopositivity for IDH1-R132H (IHC ×400). (G and H) Anaplastic oligodendroglioma grade III (H&E ×400), showing (H) strong immunopositivity in a subset of tumor cells (IHC ×400). Genetic testing revealed R132L mutations.
Immunopositivity was observed in 30 (60%) cases, of which 24 (80%) showed concordance with DNA sequencing. There were 6 cases that were immunopositive for R132H protein but showed either an absence of the corresponding mutation (CGT–CAT; in 4 tumors) or the presence of a variant mutation (CGT–CTT; in 2 cases) on primary DNA sequencing. In 5 of these 6 cases, the tumor cells showed moderate to strong immunoreactivity in only a fraction of the neoplastic cells (Fig. 3). Repeat genetic testing of these 6 cases from paraffin-embedded tissue revealed results similar to those of the prior DNA sequencing done using frozen tissue, except for one case of AOA (WHO grade III), which initially had shown lack of IDH1 mutation, but later was found to harbor CGT–CTT transversion, leading to substitution of arginine with leucine (R132L). Thus, there were 3 cases that were IDH1 mutation negative by genetic analysis (cases 17, 19, and 50) and 3 that had R132L (cases 8, 14, and 24) type of mutation but were still immunoreactive with H09.
Discussion
IDH1 mutation testing is being used as a diagnostic tool in many neuropathology centers and for prognostication of these patients in many clinical trials. For this purpose, IHC using mutant protein–specific mAb has been shown to be a reliable method with high sensitivity and specificity, combined with the advantages of the technique (i.e., ease of performance along with time and cost-effectiveness).32,40–42 Capper et al, in a comparative study between IHC and genetic testing, found that IHC was 100% sensitive and specific in detecting IDH1-R132H mutations.32 In their study, there were 9 cases with discrepant results obtained using the 2 methods. However, repeat genetic testing yielded results similar to those of IHC. Both antibodies, H09 and IMab-1, used for detection of the most common IDH1- R132H mutation are shown to be equally effective, and no cross-reactivity with either wild-type IDH1 or other variant mutant proteins has been reported.32,33,42 However, a recent study has documented improved results with H09, compared with with IMab-1, because of a higher signal-to-background ratio with the former.40
In the present study, we compared immunostaining results using H09 with direct DNA sequencing in 50 cases of diffuse glioma. The pattern of immunoreactivity was similar to that described by Capper et al.32 The antibody stained both the cytoplasm and the nuclei of the tumor cells, as also has been previously reported.32,40 None of the tumors included showed only nuclear labeling. Although IDH1 is located in the cytoplasm, the reason behind nuclear staining remains ambiguous. Possible explanations for this phenomenon include in vivo localization of the mutant protein to nucleus or by antigen diffusion (i.e., the penetration of the soluble protein into the nucleus during tissue processing).32,40
Heterogeneity of staining with only a fraction of tumor cells taking up the stain was noted in 46.6% (14/30) cases. In a recent study on IHC that used both antibodies H09 and IMab-1 in 95 diffuse glioma cases, focal immunostaining of only a proportion of tumor cells was noted in as many as 15% of the cases. This heterogeneity was not restricted to higher grade tumors but was also noted in grade II lesions,40 as was also the case in our study, thus, raising the possibility that IDH alterations may not be as early a genetic lesion as has been previously hypothesized. Preusser et al reported preferential staining of the oligodendroglial component in biphasic OA.40 In the current series, 2 oligoastrocytic tumors (one mixed OA -WHO grade II and another case of biphasic AOA-WHO grade III) showed immunolabeling of the oligodendroglial component only. This could be explained by the higher frequency of IDH1-R132H mutations in oligodendroglial tumors than in astrocytomas.10,13 Pusch et al reported a case of DA diffusely immunopositive for IDH1R132H, but on progression to secondary GBM, showed focal loss of the mutated IDH1 protein.43 These findings are in contrast to those by Capper et al.30,32 They reported diffuse homogenous staining in all the morphologically recognizable tumor cells in cases with IDH1-R132H mutations, thus suggesting use of IHC in picking even few tumor cells in the infiltrating edge and in small biopsies and in differentiating them from nonneoplastic glial cells.30,32 In view of the focal immunostaining for IDH-R132H IHC, it has been proposed that other markers, such as p53 and vIII mutant of epidermal growth factor receptor, may be used as adjuncts for diagnosis.29
Genetic testing revealed a relatively high rate of R132L mutations (4/50; 8%). On review of the available English literature, the incidence of this mutation has varied from 0.08% to 2%.4,10,11,13,20,21 The high frequency in our samples may be attributable to a selection bias, because only those cases with adequate tissue in paraffin blocks and available frozen tissue were included in the study.
When comparing results of IHC with those of DNA sequencing (Table 2), 6 cases that were immunopositive for IDH1-R132H protein were found to lack the CGT-CAT transition. Strong staining reaction limited to the tumor cells along with minimal background ruled out misinterpretation. Of interest, initial genetic testing failed to reveal any mutation in 4 cases, and 2 tumors were found to carry R132L mutation (Table 1). Replicate results were obtained in 5 of the 6 cases on repeat DNA sequencing performed on paraffin-embedded tissue sections. Of note, the sixth case revealed no mutation on DNA sequencing using frozen tissue but showed R132L mutation on repeat DNA sequencing using paraffin-embedded tissue. Of interest, case 21 also had the R132L mutation but was negative by IHC. Thus, 3 of the 4 tumors with R132L mutation showed cross-reactivity with the antibody. Capper et al used Western blot on cell lysates, which overexpressed all variants of IDH1 mutation, including R132L, but found no cross-reactivity of the IDH1-R132H antibody.32 Similar results were obtained by Kato et al.34 Recently, guidelines for performing diagnostic IDH testing using IHC and DNA sequencing have been established.44 Although the present study was undertaken before these were published, the methodology used is similar and is well standardized, being used for routine neuropathology practice. Cell culture study demonstrating H09 binding the R132L mutant protein is required to establish this cross-reactivity. However, because of unavailability of this facility, it could not be performed. The reason for this cross-reactivity is not known and is a matter of discussion to be further elucidated in larger studies.
In addition, no IDH1 mutation could be detected, even on genetic retesting, in the rest of the 3 tumor specimens with discrepant results. In 2 of these 3 cases, IHC demonstrated presence of mutated protein in only a proportion of tumor cells. Although corresponding paraffin sections, on which IHC was performed, were used for repeat DNA sequencing, inadequacy of representative tumor tissue could be attributed to failure to demonstrate the mutations. Laser microdissection of tumor cells in cases with heterogenous immunopositivity and with discrepant results is required for genetic analysis but could not be performed in the present study.
Conclusion
Immunohistochemical testing for IDH1 mutation using antibody for IDH1-R132H in diagnostic neuropathology is being performed in more centers worldwide. Although it is more sensitive than DNA sequencing in detecting IDH1-R132H mutations, awareness to the possibilities of heterogenous staining pattern and cross-reactivity with variant mutant proteins is essential for correct interpretation of results. To the best of our knowledge, the present study for the first time reports cross-reactivity of IDH1-R132H monoclonal antibody with R132L protein in formalin-fixed paraffin-embedded sections. In view of the rarity of mutations other than R132H in gliomas, multicentric pooling and evaluation of larger number of such cases is needed.
Funding
This work was supported by the Indian Council of Medical Research, Neuro Sciences Centre, and the Department of Pathology, All India Institute of Medical Sciences, New Delhi, India.
Acknowledgments
We thank the Indian Council of Medical Research, Neuro Sciences Centre and Department of Pathology, All India Institute of Medical Sciences, New Delhi, India for funding; M/s Sandor Proteomics Pvt. Ltd, Hyderabad, and Mr. M. Kiran Kumar, for help in IDH1 mutational analysis; all consultants from Department of Neurosurgery, AIIMS; and all technical staff from neuropathology laboratory, AIIMS.
Conflict of interest statement. None declared.
References
- 1.Narahara K, Kimura S, Kikkawa K, et al. Probable assignment of soluble isocitrate dehydrogenase (IDH1) to 2q33.3. Hum Genet. 1985;71:37–40. doi: 10.1007/BF00295665. [DOI] [PubMed] [Google Scholar]
- 2.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 4.Bleeker FE, Lamba S, Leenstra S, et al. IDH1 mutations at residue p.R132(IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 5.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 7.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 8.Sonoda Y, Kumabe T, Nakamura T, et al. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 2009;100:1996–1998. doi: 10.1111/j.1349-7006.2009.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jha P, Suri V, Sharma V, et al. IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Exp Mol Pathol. 2011;91:385–393. doi: 10.1016/j.yexmp.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Mellai M, Piazzi A, Caldera V, et al. IDH1 and IDH2 mutations, immunohistochemistry and associations in a series of brain tumors. J Neurooncol. 2011;105:345–357. doi: 10.1007/s11060-011-0596-3. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 14.Kang MR, Kim MS, Oh JE, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 15.Korshunov A, Meyer J, Capper D, et al. Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol. 2009;118:401–405. doi: 10.1007/s00401-009-0550-z. [DOI] [PubMed] [Google Scholar]
- 16.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsberg J, Wolter M, Seul H, et al. Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol. 2010;119:501–507. doi: 10.1007/s00401-010-0647-4. [DOI] [PubMed] [Google Scholar]
- 18.Sahm F, Capper D, Meyer J, et al. Immunohistochemical analysis of 1844 human epithelial and haematopoietic tumours and sarcomas for IDH1R132H mutation. Histopathology. 2011;58:1167–1172. doi: 10.1111/j.1365-2559.2011.03823.x. [DOI] [PubMed] [Google Scholar]
- 19.Amary MF, Bacsi K, Maggiani F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 20.Gravendeel LA, Kloosterhof NK, Bralten LB, et al. Segregation of non-p.R132H mutations in IDH1 in distinct molecular subtypes of glioma. Hum Mutat. 2010;31:E1186–99. doi: 10.1002/humu.21201. [DOI] [PubMed] [Google Scholar]
- 21.Horbinski C, Kofler J, Kelly LM, Murdoch GH, Nikiforova MN. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol. 2009;68:1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 22.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 23.Dubbink HJ, Taal W, van Marion R, et al. IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology. 2009;73:1792–1795. doi: 10.1212/WNL.0b013e3181c34ace. [DOI] [PubMed] [Google Scholar]
- 24.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 25.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27:5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 26.Bujko M, Kober P, Matyja E, et al. Prognostic value of IDH1 mutations identified with PCR-RFLP assay in glioblastoma patients. Mol Diagn Ther. 2010;14:163–169. doi: 10.1007/BF03256369. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 28.Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75:1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 29.Camelo-Piragua S, Jansen M, Ganguly A, Kim JC, Louis DN, Nutt CL. Mutant IDH1-specific immunohistochemistry distinguishes diffuse astrocytoma from astrocytosis. Acta Neuropathol. 2010;119:509–511. doi: 10.1007/s00401-009-0632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capper D, Sahm F, Hartmann C, Meyermann R, von Deimling A, Schittenhelm J. Application of mutant IDH1 antibody to differentiate diffuse glioma from nonneoplastic central nervous system lesions and therapy-induced changes. Am J Surg Pathol. 2010;34:1199–1204. doi: 10.1097/PAS.0b013e3181e7740d. [DOI] [PubMed] [Google Scholar]
- 31.Horbinski C, Kelly L, Nikiforov YE, Durso MB, Nikiforova MN. Detection of IDH1 and IDH2 mutations by fluorescence melting curve analysis as a diagnostic tool for brain biopsies. J Mol Diagn. 2010;12:487–492. doi: 10.2353/jmoldx.2010.090228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capper D, Weissert S, Balss J, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118:599–601. doi: 10.1007/s00401-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 34.Kato Y, Jin G, Kuan CT, McLendon RE, Yan H, Bigner DD. A monoclonal antibody IMab-1 specifically recognizes IDH1R132H, the most common glioma-derived mutation. Biochem Biophys Res Commun. 2009;390:547–551. doi: 10.1016/j.bbrc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer J, Pusch S, Balss J, et al. PCR- and restriction endonuclease-based detection of IDH1 mutations. Brain Pathol. 2010;20:298–300. doi: 10.1111/j.1750-3639.2009.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahm F, Capper D, Pusch S, et al. Detection of 2-hydroxyglutarate in formalin-fixed paraffin-embedded glioma specimens by gas chromatography/mass spectrometry. Brain Pathol. 2012;22:26–31. doi: 10.1111/j.1750-3639.2011.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleihues P, Louis DN, Wiestler OD, Burger PC, Scheithauer BW. WHO grading of tumours of the central nervous system. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO classification of tumours of the central nervous system. 4th ed. Lyon: IARC; 2007. pp. 10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 39.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 40.Preusser M, Wöhrer A, Stary S, Höftberger R, Streubel B, Hainfellner JA. Value and limitations of immunohistochemistry and gene sequencing for detection of the IDH1-R132H mutation in diffuse glioma biopsy specimens. J Neuropathol Exp Neurol. 2011;70:715–723. doi: 10.1097/NEN.0b013e31822713f0. [DOI] [PubMed] [Google Scholar]
- 41.Kaneko MK, Tian W, Takano S, et al. Establishment of a novel monoclonal antibody SMab-1 specific for IDH1-R132S mutation. Biochem Biophys Res Commun. 2011;406:608–613. doi: 10.1016/j.bbrc.2011.02.102. [DOI] [PubMed] [Google Scholar]
- 42.Takano S, Tian W, Matsuda M, et al. Detection of IDH1 mutation in human gliomas: comparison of immunohistochemistry and sequencing. Brain Tumor Pathol. 2011;28:115–123. doi: 10.1007/s10014-011-0023-7. [DOI] [PubMed] [Google Scholar]
- 43.Pusch S, Sahm F, Meyer J, Mittelbronn M, Hartmann C, von Deimling A. Glioma IDH1 mutation patterns off the beaten track. Neuropathol Appl Neurobiol. 2011;37:428–430. doi: 10.1111/j.1365-2990.2010.01127.x. [DOI] [PubMed] [Google Scholar]
- 44.Preusser M, Capper D, Hartmann C Euro-CNS Research Committee. IDH testing in diagnostic neuropathology: review and practical guideline article invited by the Euro-CNS research committee. Clin Neuropathol. 2011;30:217–230. doi: 10.5414/np300422. [DOI] [PubMed] [Google Scholar]


