Abstract
The post-translational modification of protein by acetylation has been emerging as a prevalent modification in enzymes that catalyze intermediary metabolism. However, the dynamics of protein acetylation during adipocyte differentiation that involves a major shift in cellular metabolism is not known. In this study, we investigated the temporal changes in acetylation during adipocyte differentiation. Almost all acetylated proteins identified showed a sequential change in acetylation during the differentiation process. While the majority of the acetylated proteins showed a sequential upregulation during adipocyte differentiation, in a few proteins a sequential downregulation of protein acetylation was also observed. Our findings suggest that a wide-ranging temporal change in protein acetylation occurs during adipocyte differentiation including differentially expressed proteins signifying an important role in adipocyte differentiation.
Keywords: adipogenesis, ubiquitylation, post-translational modification, 3T3-L1, 2-D electrophoresis, mass spectrometry
The acetylation of the ε-NH2 of lysine residues in proteins has emerged as a key post-translational modification in cellular regulation mainly through the modification of transcription regulators and histones.1 In addition, recently lysine acetylation has been found as a prevalent post-translational modification in proteins involved in intermediary metabolism and majority of the enzymes involved in glucose and fatty acid metabolism have been identified to be acetylated in human liver tissue.2 Furthermore, chemically distinct histone deacetylase (HDAC) inhibitors have been shown to prevent adipocyte differentiation.3 Similarly, overexpression of NAD-dependent deacetylase sirtuin-2 (SIRT2) has been found to suppress adipocyte differentiation.4,5 It has been shown that SIRT2 mediated suppression of adipocyte differentiation involves the deacetylation of forkhead box protein O1 (FOXO1) and the repression of peroxisome proliferator-activated receptor gamma (PPARγ) activity, a critical transcription factor in adipocyte differentiation.4,5 It has been suggested that SIRT2 that acts as an important regulator of adipocyte differentiation through modulation of FOXO1 acetylation/phosphorylation and activity may play a role in controlling adipose tissue mass and function.4 In addition to PPARγ, transcription factor CCAAT/enhancer binding protein β (C/EBPβ) has an important role in adipocyte differentiation.6 Acetylation and deacetylation regulate C/EBPβ in mediating gene transcription.7 Acetylation of C/EBPβ at a specific lysine residue is an important regulatory event that contributes to its ability to transactivate target genes, including those associated with adipocyte differentiation and function.7 Collectively, these studies point toward an important role of lysine acetylation in the regulation of proteins involved in adipocyte differentiation. Furthermore, this would suggest that protein acetylation is tightly regulated during the differentiation process as forced alteration in protein acetylation (either through HDAC or SIRT2) leads to impaired adipogenesis. However, the temporal changes in lysine acetylation during adipocyte differentiation that is marked by the upregulation of a number of transcription factors and involved a significant shift in cellular metabolism is not known.
Moreover, several studies have successfully exploited two-dimensional (2-D) gel electrophoresis in combination with mass spectrometry to identify differentially expressed proteins during adipocyte differentiation.8-12 There is no doubt that such studies have revealed the identity of a number of proteins involved in adipocyte differentiation including metabolic enzymes, mitochondrial proteins and transcription factors.6-12 However, no careful attention has been made to decipher that the altered protein spots identified on the 2-D gels represents only the differentially expressed proteins or also represents protein isoforms as a consequence of post-translational modification in proteins. For example, metabolic enzymes, mitochondrial proteins and transcription factors that are known to play a critical roles in the differentiation process are regulated through post-translational modifications such as phosphorylation and acetylation.10,13 Such modifications may alter protein stability and function, translocation to different cellular compartments and protein-protein interaction. Protein modification by phosphorylation is known to alter the pI of the substrate protein.1 In addition to phosphorylation, protein modification by acetylation has been recently reported to be a widespread modification among enzymes involved in intermediary metabolism.2 Since lysine side chains are cationic at physiological pH, ε-NH2 acetylation will quench the positive charges resulting in alterations in the pI of proteins.14 It is highly likely that differential protein spots on a 2-D gel also represents protein isoforms produced as a result of various post-translation modifications of proteins during the differentiation process including protein modification by lysine acetylation. Given the considerable increase in obesity and obesity-associated diseases worldwide, it is necessary to understand the molecular basis of adipocyte differentiation and its control. In this study we have explored the temporal change in protein acetylation during adipocyte differentiation using 3T3-L1 preadipocytes as a model cell line and a combination of proteomic approaches.
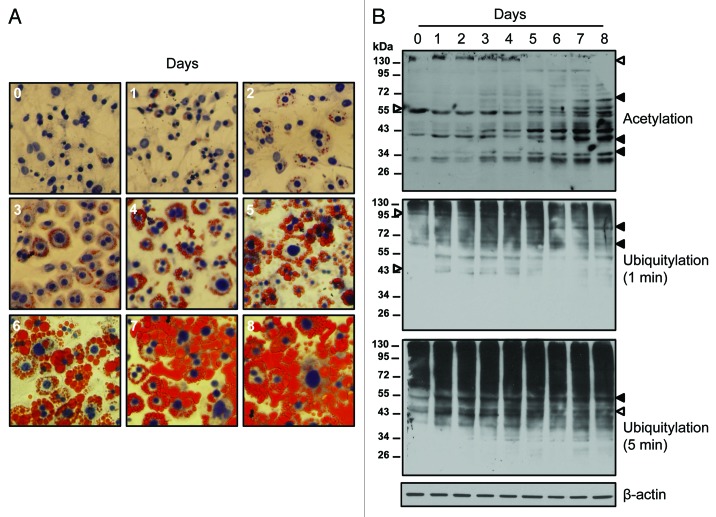
To determine the dynamics of protein acetylation during adipocyte differentiation, 3T3-L1 cells were first grown in a preadipocyte growth medium and subsequently in the differentiation medium (PromoCell GmBH) containing insulin (0.5 μg/ml), dexamethasone (400 ng/ml) and isobutylmethylxanthine (44 μg/ml) for 72 h. Subsequently adipocyte differentiation was followed in the nutrition medium (PromoCell GmBH) as per the manufacturer’s protocol. The adipocyte differentiation was followed for eight days. Each day during the differentiation process, cells were processed for oil-red-O staining and photomicrography to monitor the differentiation process (Fig. 1A).15 Cell lysates prepared at each time point were analyzed by SDS-PAGE followed by immunoblotting using anti-lysine acetylation specific antibody (Cell Signaling Technology). Almost all acetylated protein bands identified showed sequential changes in acetylation during the differentiation process (Fig. 1B). While majority of the acetylated proteins showed a sequential upregulation during adipocyte differentiation, in a few cases a sequential downregulation in protein acetylation was also observed (Fig. 1B). In general, the downregulation in protein acetylation was observed during the early phase (days 1–4) of the differentiation process whereas the upregulation was found during the advanced phase (days 4–8) of the differentiation process (Fig. 1B). In this context it should be noted that in an earlier report by Catalioto et al. chemically distinct HDAC inhibitors were found to prevent the conversion of preadipocytes to adipocytes at an early stage of the differentiation process.3 This would mean that downregulation of protein acetylation is a critical step at the early stage of the differentiation program. However, in a separate study overexpression of SIRT2 that possesses NAD-dependent deacetylase activity has been shown to suppress adipocyte differentiation mediated through the deacetylation of FOXO1 and subsequent repression of PPARγ activity.5 An overexpression of SIRT2 inhibits differentiation, whereas reducing SIRT2 expression promotes adipogenesis suggesting an important role of protein acetylation in adipocyte differentiation.4,5 Both effects are accompanied by their corresponding changes in the expression of PPARγ, C/EBPα, and genes marking terminal differentiation of adipocyte, including glucose transporter-4, adipocyte protein-2 and fatty acid synthase.4 Moreover, the sterol regulatory element-binding proteins (SREBPs) family of transcription factors that control cholesterol and lipid homeostasis and play important roles in adipocyte differentiation has been shown to be stabilized by coactivator-dependent acetylation.16 These studies suggest that changes in protein acetylation has an important role in both the early and the advance stages of the differentiation program. Whether inverse and dynamic changes in protein acetylation during the early and the advance stages of adipocyte differentiation that we have found are associated with each other remains to be determined. Taken together our data suggest that a wide ranging and sequential change in protein lysine acetylation occurs during adipocyte differentiation signifying an important role in the differentiation process.
Figure 1. Temporal changes in the lysine acetylation in proteins during adipocyte differentiation. (A) Photomicrographs showing differentiation of 3T3-L1 preadipocytes into adipocytes in response to differentiation medium (PromoCell, Germany) as determined by Oil-red-O staining. Cells were counterstained with hematoxylin. Representative photomicrographs of three different experiments are shown. (B) Immunoblots showing sequential changes protein acetylation and ubiquitylation (open arrow head, downregulation; closed arrow head, upregulation) during adipocyte differentiation from day 0 through day 8. Cell lysates were prepared from 3T3-L1 cells at each time points [as shown in (A)] and equal amount of proteins (30 μg/lane) were subjected to immunoblot analysis using modification specific antibodies obtained from Cell Signaling Technology. Two different exposures of the ubiquitylation blot are shown to better visualize the ubiquitylated bands in the upper and the lower part of the immunoblot. Anti-β-actin immunoblot is shown as a loading control. Experiments were repeated for three times. Representative immunoblots of three different experiments are shown.
In addition to acetylation, post-translational modification of protein by ubiquitylation that also occur at the ε-NH2 of lysine residue, has been emerging as an important regulatory mechanism for protein function in addition to its important role in protein degradation.17 For example, ubiquitylation has been found to be involved in signal transduction, transcription and metabolism.18 Furthermore, a large scale mapping of site specific acetylation and ubiquitylation in proteins have revealed a number of common sites in a wide range of proteins.2,17 This would mean that in a number of sites in a variety of proteins acetylation and ubiquitylation occurs in a mutually exclusive manner. For example, the acetylated residue in SREBP1a is also regulated by ubiquitylation, and acetylation inhibits this process.16 To explore whether dynamic changes in protein acetylation that we have found during adipocyte differentiation also involve concurrent changes in ubiquitylation, membranes were stripped and reprobed with anti-ubiquitin antibody (Cell Signaling Technology). Similar to changes in protein acetylation, a number of proteins were detected showing both the upregulation and the downregulation of ubiquitylation during adipocyte differentiation (Fig. 1B). A comparison of the acetylation and ubiquitylation immunoblots indicated a wide-ranging putative association between these two modifications (Fig. 1B). It is possible that acetylation/ubiquitylation balance can control the lifetime of proteins and their functional consequences during the differentiation process.
Next, we examined the functional consequence of SIRT2 specific inhibitor (AGK2, Tocris Bioscience) on protein acetylation during adipocyte differentiation (Fig. 2). An apparent increase in acetylation was observed only in a few proteins suggesting involvement of multiple acetate cycling enzymes during adipocyte differentiation. Interestingly acetylation was also found to be downregulated in some proteins (corresponding to molecular masses of prohibitin and aconitase 2 as identified by mass spectrometry) most likely due to downregulation of protein level itself. To confirm this, membranes were reprobed with prohibitin and aconitase 2 specific antibodies. Both proteins were found to be downregulated during the advance stage of the differentiation process suggesting that sequential changes in protein acetylation during adipocyte differentiation has a role in the regulation of differentially expressed proteins.
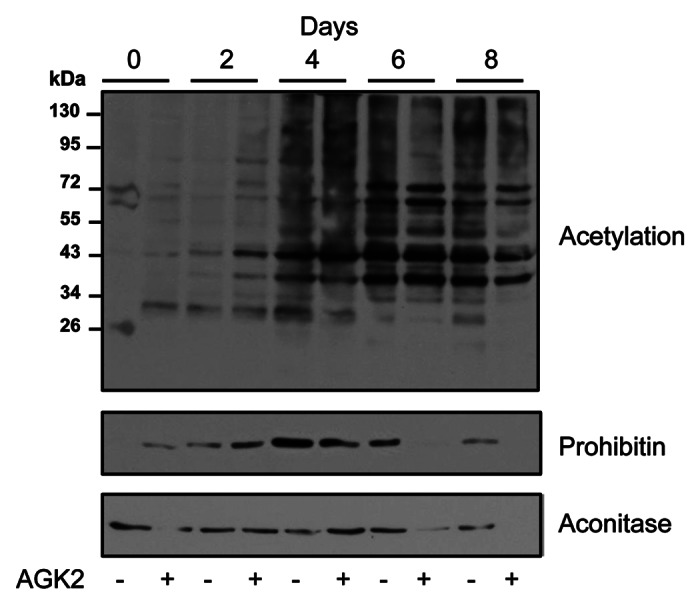
Figure 2. Effect of SIRT2 inhibitor on protein acetylation during adipocyte differentiation. Differentiation of 3T3-L1 preadipocytes into adipocytes was followed (as described in Fig. 1) in the presence and absence of SIRT2 specific inhibitor AGK2 (20 nM). Cell lysates were prepared at different time points and equal amount of total proteins (30 μg/lane) were analyzed by immunoblotting using anti-lysine acetylation specific antibody. Membranes were reprobed with anti-prohibitin and anti-aconitase 2 specific antibodies. Representative immunoblots of three different experiments are shown.
Using a combination of proteomic approaches, a number of proteins have been identified that are differentially expressed during adipocyte differentiation.8-12 However, it is not clear whether differentially expressed protein spots identified using 2-D gel analyses represent only differentially expressed proteins or also include protein isoforms that are produced during the differentiation process as a result of post-translational modifications. A careful examination of sequential changes in protein acetylation during adipocyte differentiation appears to indicate formation of protein isoform most likely as a result of changes in post-translational modification in them (Fig. 1B). Such changes may involve acetylation alone or simultaneous changes in protein phosphorylation, as acetylation and phosphorylation in proteins are known to influence each other.1,14 For example, phosphorylation is known to stimulate acetylation in histones, p53 and nuclear factor κB emphasizing a temporal hierarchy to waves of such post-translational modifications.19-21 Similarly, recruitment of p300/CBP is enabled by phosphorylation, and once recruited, p300/CBP can then acetylate the substrate protein.22
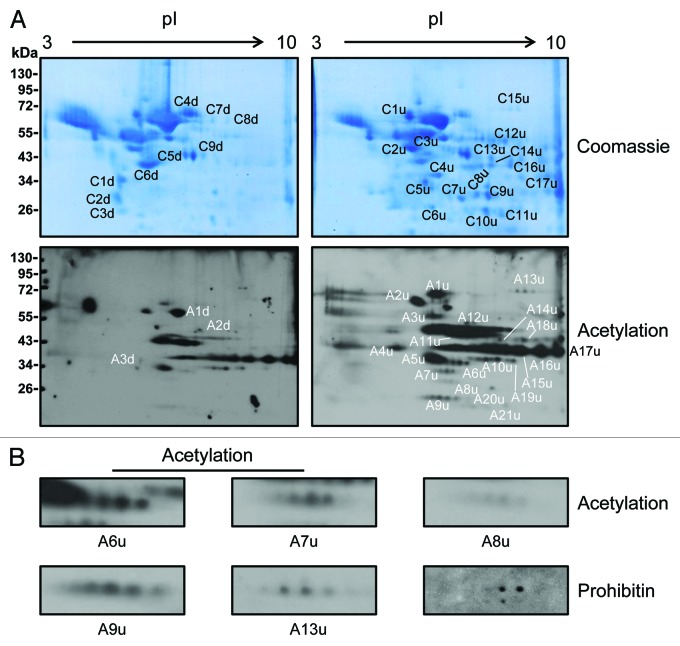
To explore whether changes in protein acetylation during adipocyte differentiation generates protein isoforms, cell lysates prepared from preadipocytes (day 0) and adipocytes (day 8) were analyzed by 2-D gel electrophoresis and immunoblotting using anti-lysine acetylation antibody.23 Multiple series of protein spots were identified on the immunoblot of proteins prepared from adipocyte indicating the formation of protein isoforms (Fig. 3A). To further confirm this, membranes were reprobed with anti-prohibitin antibody as prohibitin is known to undergo post-translational modification by acetylation and phosphorylation as well as upregulated during adipocyte differentiation.15,24,25 Indeed a series of prohibitin spots were identified matching acetylation spots suggesting formation of prohibitin isoforms during adipocyte differentiation (Fig. 3B). Acetylated protein isoforms observed during adipocyte differentiation may be produced due to a series of acetylation or subsequent phosphorylation as a result of acetylation in proteins. In this way acetylation alone or in combination with phosphorylation may play an important role in the regulation of protein function and in protein-protein interaction during adipocyte differentiation. For example, acetylated lysine side chains can be specifically recognized by bromodomains in partner proteins.26
Figure 3. Proteomic changes during adipocyte differentiation. (A) Two-dimensional gel electrophoretic analysis of 3T3-L1 cell lysates prepared from preadipocytes (day 0) and adipocytes (day 8). The upper panel showing Coomassie blue stained gels and the lower panel showing anti-lysine acetylation immunoblots. Representative photomicrographs of three different experiments are shown. (B) Magnified view of putative acetylation isoforms of five different proteins from adipocytes (day 8) anti-lysine immunoblot as shown in (A). Membrane was reprobed with anti-prohibitin antibody to confirm the formation acetylation isoform of prohibitin in adipocytes. Notations used to indicate various protein spots: C, Coomassie blue stained protein spot; d, downregulation; u, upregulation; A, acetylated protein spot.
To determine the identity of proteins having altered acetylation during the adipocyte differentiation, identical spots were excised in an overlay from Coomassie stained gels and processed for protein identification by mass spectrometry.23 In addition, differentially expressed proteins in preadipocytes (day 0) and adipocytes (day 8) were also excised from Coomassie stained 2-D gels and processed for identification by mass spectrometry.23 A number of mitochondrial proteins and metabolic enzymes were identified in both cases (i.e., differentially expressed and differentially acetylated; Tables 1 and 2). However, one of the limitations of our strategy used in this study is relatively low number of acetylated proteins identified as no enrichment methods have been applied for the acetylated proteins. This may be the reason why only abundantly expressed metabolic enzymes/proteins are identified and not the transcription factors known to be upregulated during adipocyte differentiation. Interestingly, some of the proteins (heat shock protein 60, glycerol-3-phosphate dehydrogenase 1, chaperonin containing Tcp1, transaldolase 1, aldo-keto reductase family 1 member B3, prohibitin, enolase 1, aconitase 2, fructose biphosphate aldolase A and voltage-dependent anion channel 2) were identified in both cases (Tables 1 and 2) confirming that post-translational modifications during adipocyte differentiation contributes to the repertoire of differentially expressed proteins detected using 2-D gel electrophoresis. In addition, this would indicate that some of the differentially expressed proteins are regulated by acetylation during the differentiation process. As metabolic enzymes and mitochondrial proteins play an important role in adipocyte differentiation,15,27 this would indicate that the net result of the upregulation of protein acetylation during adipocyte differentiation is to enhance the function and the stability of protein components important for metabolic shift and mitochondrial activity. Interestingly among metabolic enzymes, a number of enzymes involved in amino acid metabolism are also identified in addition to enzymes involved in carbohydrate and lipid metabolism (Tables 1 and 2). As cellular metabolism in adipocyte is concerted toward the biosynthesis of lipid and intermediary metabolites can be utilized through anaplerotic process this would imply that enhanced lipogenesis during the differentiation process involves a substantial shift in both glucose and amino acid metabolism.
Table 1. List of differentially expressed proteins identified by mass spectrometry.
| Spot ID | UniprotKB Ref No. | Probability log(e) score* | Expected pI | Mr | Protein | Known modification status in mouse/human |
||
|---|---|---|---|---|---|---|---|---|
| Ac | Ub | MS | ||||||
| C1d |
Q7TQI3 |
-22.8 |
4.85 |
31.2 |
Ubiquitin thioesterase (Otub1) |
Yes |
Yes |
Yes |
| C2d |
P17918 |
-39.7 |
4.66 |
28.8 |
Proliferating cell nuclear antigen (PCNA) |
Yes |
Yes |
Yes |
| C3d |
P63101 |
-43.4 |
4.73 |
27 |
14–3-3 zeta (tyrosine 3 monooxygenase) |
Yes |
Yes |
Yes |
| C4d |
Q3THK7 |
-44.8 |
6.29 |
76.7 |
Glutamine amidotransfrase |
Yes |
Yes |
Yes |
| C5d |
P17182 |
-43.1 |
6.37 |
47.1 |
Enolase 1 |
Yes |
Yes |
Yes |
| C6d |
P63260 |
-756.5 |
5.31 |
41.8 |
Actin gamma cytoplasmic 1 (Actg1) |
Yes |
Yes |
Yes |
| C7d |
Q99PU7 |
|
6.33 |
80.4 |
Ubiquitin carboxyl-terminal hydrolase |
No |
Yes |
No |
| C8d |
P56480 |
-961.9 |
5.19 |
56.3 |
ATP synthase subunit β (Atp5b) |
Yes |
Yes |
Yes |
| C9d |
Q9Z2I8 |
-113.1 |
6.58 |
46.8 |
Succinate- Coenzyme A ligase (Suclg2) |
Yes |
Yes |
No |
| C1u |
Q61696 |
-883.1 |
5.53 |
70.0 |
Heat shock protein 70 (HSP70) |
Yes |
Yes |
Yes |
| C2u |
P42932 |
-199.2 |
5.44 |
59.5 |
Chaperonin containing Tcp1, subunit 8 (Cct8) |
Yes |
Yes |
Yes |
| C3u |
P19226 |
-43.1 |
5.91 |
60.9 |
Heat shock protein 60 (Hspd1) |
Yes |
Yes |
Yes |
| C4u |
O88844 |
-95.5 |
6.73 |
46.6 |
Isocitrate dehydrogenase (Idh1) |
Yes |
Yes |
Yes |
| C5u |
Q93092 |
-53.4 |
6.57 |
37.4 |
Transaldolase (Taldo1) |
Yes |
Yes |
Yes |
| C6u |
P67778 |
-220.6 |
5.57 |
29.8 |
Prohibitin (Phb) |
Yes |
Yes |
Yes |
| C7u |
Q8K354 |
-245.7 |
6.15 |
30.9 |
Carbonyl reductase 3 (Cbr3) |
Yes |
Yes |
No |
| C8u |
P13707 |
-413.1 |
6.75 |
37.5 |
Glycerol-3-phosphate dehydrogenase 1 (Gpd1) |
No |
No |
No |
| C9u |
P45376 |
-275 |
6.71 |
35.7 |
Aldo-keto reductase family 1, member B3 (Akr1b3) |
Yes |
Yes |
Yes |
| C10u |
Q9WTP6 |
-97.5 |
6.96 |
26.4 |
Adenylate kinase 2 (Ak2) |
Yes |
Yes |
Yes |
| C11u |
Q60930 |
-64.4 |
7.44 |
31.7 |
Voltage-dependent anion channel 2 (Vdac2) |
Yes |
Yes |
Yes |
| C12u |
Q8BMF4 |
-19.2 |
8.81 |
67.9 |
Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (DLAT) |
Yes |
Yes |
Yes |
| C13u |
Q9DBL1 |
|
8.0 |
47.8 |
2-methylbutyryl-CoA dehydrogenase |
Yes |
Yes |
Yes |
| C14u |
P55302 |
-133.8 |
7.35 |
42.2 |
Low density lipoprotein receptor-related protein associated protein 1 (Lrpap1) |
Yes |
Yes |
No |
| C15u |
Q99KI0 |
-409.5 |
8.08 |
85.4 |
Aconitase 2 |
Yes |
Yes |
Yes |
| C16u |
P09411 |
-290.5 |
8.02 |
44.5 |
Phosphoglycerate kinase 1 (Pgk1) |
Yes |
Yes |
Yes |
| C17u | P05064 | -504.3 | 8.3 | 39.3 | Fructose biphosphate aldolase A (Aldoa) | Yes | Yes | Yes |
Base -10 log of expectation that this assignment is stochastic. Low expect score (< -3.0) corresponds to a confident identification. Ac, acetylation; C, differentially expressed Coomassie blue stained protein spot; d, downregulated; MS, mutual site(s); u, upregulated; Ub, ubiquitylation.
Table 2. List of acetylated proteins identified by mass spectrometry.
| Spot ID | UniprotKB Ref No. | Probability log(e) score* | Expected pI | Mr | Protein | Known modification status in mouse/human |
||
|---|---|---|---|---|---|---|---|---|
| Ac | Ub | MS | ||||||
| A1d |
P19226 |
-860.7 |
5.91 |
60.9 |
Heat shock protein 60 (Hspd1) |
Yes |
Yes |
Yes |
| A2d |
Q9JLJ2 |
-87.6 |
6.63 |
53.5 |
Aldehyde dehydrogenase 9, subfamily A1 (Aldh9a1) |
Yes |
Yes |
No |
| A3d |
P13707 |
-23.0 |
6.75 |
37.5 |
Glycerol-3-phosphate dehydrogenase 1 (Gpd1) |
No |
No |
No |
| A1u |
P38647 |
-22.2 |
5.91 |
73.5 |
Heat shock protein 9a (Hspa9) |
Yes |
Yes |
Yes |
| A2u |
P42932 |
-66.6 |
5.44 |
59.5 |
Chaperonin containing Tcp1, subunit 8 (Cct8) |
Yes |
Yes |
Yes |
| A3u |
P27773 |
-177.9 |
5.88 |
56.6 |
Disulfide isomerase (Grp58) |
Yes |
Yes |
Yes |
| A4u |
P63260 |
-232.6 |
5.31 |
41.8 |
Actin gamma cytoplasmic 1 (Actg1) |
Yes |
No |
No |
| A5u |
P16125 |
-33.8 |
5.7 |
36.5 |
Lactate dehydrogenase (Ldhb) |
Yes |
Yes |
Yes |
| A6u |
Q93092 |
-101.6 |
6.57 |
37.4 |
Transaldolase (Taldo1) |
Yes |
Yes |
Yes |
| A7u |
P45376 |
-22.1 |
6.71 |
35.7 |
Aldo-keto reductase family 1, member B3 (Akr1b3) |
Yes |
Yes |
Yes |
| A8u |
P67778 |
-119.4 |
5.57 |
29.8 |
Prohibitin (Phb) |
Yes |
Yes |
Yes |
| A9u |
P70349 |
|
6.36 |
13.7 |
Histidine triad nucleotide-binding protein 1 (Hint1) |
Yes |
Yes |
Yes |
| A10u |
Q9D051 |
-112.3 |
6.41 |
38.9 |
Pyruvate dehydrogenase E1 component subunit β (mito) |
Yes |
Yes |
No |
| A11u |
P17182 |
-323.7 |
6.37 |
47.1 |
Enolase 1 |
Yes |
Yes |
Yes |
| A12u |
P47738 |
-267.1 |
7.53 |
56.5 |
Aldehyde dehydrogenase 2 (Aldh2) |
Yes |
Yes |
Yes |
| A13u |
Q99KI0 |
-88.2 |
8.08 |
85.4 |
Aconitase 2 |
Yes |
Yes |
Yes |
| A14u |
Q07417 |
-49.2 |
8.68 |
44.9 |
Acyl-Coenzyme A dehydrogenase (Acads) |
Yes |
No |
No |
| A15u |
Q921H8 |
-190.2 |
8.74 |
43.9 |
Acetyl-Coenzyme A acyltransferase 1 (Acaa1a) |
Yes |
Yes |
No |
| A16u |
Q8QZT1 |
-117.0 |
8.71 |
44.8 |
Acetyl Coenzyme A acetyltransferase 1 (Acat1) |
Yes |
Yes |
Yes |
| A17u |
P45952 |
-145.8 |
8.60 |
46.5 |
Acyl-Coenzyme A (Acadm) dehydrogenase, medium chain |
Yes |
Yes |
No |
| A18u |
Q9JHI5 |
-48.3 |
8.53 |
46.3 |
Isovaleryl coenzyme A dehydrogenase (Ivd) |
Yes |
Yes |
Yes |
| A19u |
P05064 |
-280.1 |
8.3 |
39.3 |
Fructose biphosphate aldolase A (Aldoa) |
Yes |
Yes |
Yes |
| A20u |
Q60930 |
-139.8 |
7.44 |
31.7 |
Voltage-dependent anion channel 2 (Vdac2) |
Yes |
Yes |
Yes |
| A21u | P04117 | -21.3 | 8.53 | 14.6 | Fatty acid binding protein 4 (Fabp4) | Yes | Yes | Yes |
Base -10 log of expectation that this assignment is stochastic. Low expect score (< -3.0) corresponds to a confident identification. A, differentially acetylated protein spot; d, downregulated; MS, mutual site(s); u, upregulated.
To further validate the temporal upregulation in protein acetylation during adipocyte differentiation, prohibitin was immunoprecipitated at different time point during the differentiation process and analyzed by immunoblotting using anti-acetylation antibody. A sequential upregulation in prohibitin acetylation along with protein upregulation was found during adipocyte differentiation (Fig. 4). Next we search the known modification status (especially acetylation and ubiquitylation) of each protein identified by mass spectrometry using PhosphoSitePlus®—a curated database of commonly occurring known modifications in proteins.28 Interestingly, almost all proteins identified in this study are known to undergo acetylation and ubiquitylation and majority of them contain site(s) that have been reported to undergo both acetylation and ubiquitylation (Tables 1 and 2). This would suggest that in a number of proteins these two modifications occur in a mutually exclusive manner on certain sites which may explain the inverse association between acetylation and ubiquitylation during adipocyte differentiation. However, the inverse association as a result of acetylation and ubiquitylation at two distinct residues may not be ruled out. Similarly direct correlation between increased acetylation and ubiquitylation would indicate involvement of modifications at two different residues. Collectively this would indicate that interplay between acetylation and ubiquitylation may occur during adipocyte differentiation.
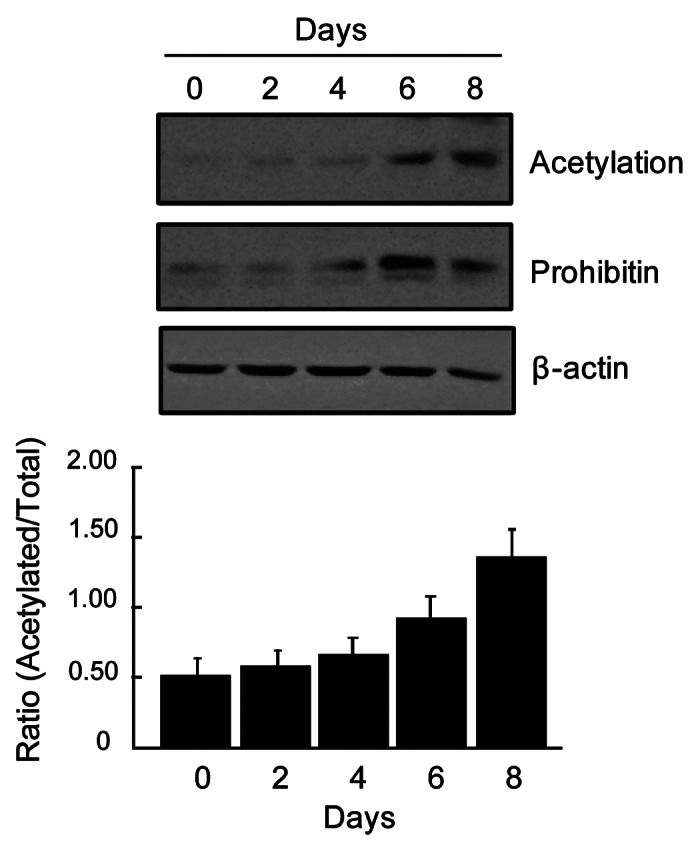
Figure 4. Temporal increase in prohibitin expression and acetylation during adipocyte differentiation. Upper panel: Immunoblots showing sequential upregulation of prohibitin acetylation and its protein level during adipocyte differentiation. Cell lysates (500 μg proteins) were processed for immunoprecipitation using anti-prohibitin antibody. Immunoprecipitates were analyzed by immunoblotting using anti-acetylation and anti-prohibitin antibodies as described in Figures 1and2. Anti-β-actin immunoblot is shown as a control (for pre-IP cell lysates). Lower panel: Histogram showing relative changes in prohibitin acetylation as shown in the upper panel.
In summary, our findings suggest that a sequential modification in protein lysine acetylation occurs in a number of proteins during adipocyte differentiation including differentially expressed proteins indicating an important role in the regulation of the differentiation program. Moreover, our data suggest that a sequential deacetylation in a few proteins occurs mainly at the early stage of the differentiation program whereas in majority of them acetylation is upregulated during the advance stage of the differentiation program. Whether chronological changes in protein deacetylation and acetylation during different stages of adipocyte differentiation are associated with each other remains to be determined and warrants further investigation.
Acknowledgments
This study is supported by funds from Natural Sciences and Engineering Council of Canada and Canada Foundation for Innovation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/adipocyte/article/21916
References
- 1.Walsh CT. Protein N-acetylation. In: Posttranslational modification of proteins: Expanding nature’s inventory. Colorado: Robert and Company Publishers; 2006; 151-170. [Google Scholar]
- 2.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catalioto RM, Maggi CA, Giuliani S. Chemically distinct HDAC inhibitors prevent adipose conversion of subcutaneous human white preadipocytes at an early stage of the differentiation program. Exp Cell Res. 2009;315:3267–80. doi: 10.1016/j.yexcr.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–14. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. 2009;20:801–8. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, Xie Y, Bucher NLR, Farmer SR. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995;9:2350–63. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 7.Ceseña TI, Cui TX, Subramanian L, Fulton CT, Iñiguez-Lluhí JA, Kwok RP, et al. Acetylation and deacetylation regulate CCAAT/enhancer binding protein beta at K39 in mediating gene transcription. Mol Cell Endocrinol. 2008;289:94–101. doi: 10.1016/j.mce.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Ye F, Zhang H, Yang Y-X, Hu H-D, Sze SK, Meng W, et al. Comparative proteome analysis of 3T3-L1 adipocyte differentiation using iTRAQ-coupled 2D LC-MS/MS. J Cell Biochem. 2011;112:3002–14. doi: 10.1002/jcb.23223. [DOI] [PubMed] [Google Scholar]
- 9.Molina H, Yang Y, Ruch T, Kim J-W, Mortensen P, Otto T, et al. Temporal profiling of the adipocyte proteome during differentiation using a five-plex SILAC based strategy. J Proteome Res. 2009;8:48–58. doi: 10.1021/pr800650r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed M, Neville MJ, Edelmann MJ, Kessler BM, Karpe F. Proteomic analysis of human adipose tissue after rosiglitazone treatment shows coordinated changes to promote glucose uptake. Obesity (Silver Spring) 2010;18:27–34. doi: 10.1038/oby.2009.208. [DOI] [PubMed] [Google Scholar]
- 11.Lee H-K, Lee B-H, Park S-A, Kim C-W. The proteomic analysis of an adipocyte differentiated from human mesenchymal stem cells using two-dimensional gel electrophoresis. Proteomics. 2006;6:1223–9. doi: 10.1002/pmic.200500385. [DOI] [PubMed] [Google Scholar]
- 12.Welsh GI, Griffiths MR, Webster KJ, Page MJ, Tavaré JM. Proteome analysis of adipogenesis. Proteomics. 2004;4:1042–51. doi: 10.1002/pmic.200300675. [DOI] [PubMed] [Google Scholar]
- 13.Kim WK, Jung H, Kim DH, Kim EY, Chung JW, Cho YS, et al. Regulation of adipogenic differentiation by LAR tyrosine phosphatase in human mesenchymal stem cells and 3T3-L1 preadipocytes. J Cell Sci. 2009;122:4160–7. doi: 10.1242/jcs.053009. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ande SR, Xu Z, Gu Y, Mishra S. Prohibitin has an important role in adipocyte differentiation. Int J Obes. 2012;36:1236–44. doi: 10.1038/ijo.2011.227. [DOI] [PubMed] [Google Scholar]
- 16.Giandomenico V, Simonsson M, Grönroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol Cell Biol. 2003;23:2587–99. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danielsen JMR, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics. 2011;10:M110–, 003590. doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra S, Saleh A, Espino PS, Davie JR, Murphy LJ. Phosphorylation of histones by tissue transglutaminase. J Biol Chem. 2006;281:5532–8. doi: 10.1074/jbc.M506864200. [DOI] [PubMed] [Google Scholar]
- 20.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–63. doi: 10.1016/S1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 21.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 22.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–15. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 23.Vessal M, Mishra S, Moulik S, Murphy LJ. Prohibitin attenuates insulin-stimulated glucose and fatty acid oxidation in adipose tissue by inhibition of pyruvate carboxylase. FEBS J. 2006;273:568–76. doi: 10.1111/j.1742-4658.2005.05090.x. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Lin Y, Kang T, Huang B, Xu W, Garcia-Barrio M, et al. Mitochondrial dysfunction and adipogenic reduction by prohibitin silencing in 3T3-L1 cells. PLoS One. 2012;7:e34315. doi: 10.1371/journal.pone.0034315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–6. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–91. doi: 10.1016/S0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 27.Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23:1085–94. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–61. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]