Abstract
From their inception, Y chromosomes in plants and animals are subjected to the powerful effects of Müller’s ratchet, a process spurred by suppression of recombination that results in a rapid accumulation of mutations and repetitive elements. These mutations eventually lead to gene loss and degeneration of the Y chromosome. Y chromosomes in mammals are ancient, whereas most sex chromosomes in plants and many in insects and fish evolved recently. Sex type in papaya is controlled by a pair of nascent sex chromosomes that evolved around 7 million years ago. The papaya X and Yh were recently sequenced, providing valuable insight into the early stages of sex chromosome evolution. Here we discuss the fruits of this work with a focus on the repeat accumulation, gene trafficking and promiscuous DNA sequences found in the slowly degenerating Yh chromosome of papaya.
Keywords: chloroplast genomic DNA, gene loss and gain, heterochromatin, retrotransposons, Y chromosome degeneration
Papaya as a Model for Sex Chromosome Evolution
Sex chromosomes are widespread in plants and animals, and they have evolved independently numerous times.1,2 But despite their independent origins, sex chromosomes are all shaped by a common set of forces and share a number of features. Sex chromosomes evolve from autosomes and arise through suppression of recombination around sex determination loci. The lack of recombination fixes the genes that control sex, fostering separate male and female individuals. Once the recombination ceased, the sex chromosomes diverge, creating distinct chromosome types. In humans, males are heterogametics with XY chromosomes, whereas in birds, females are heterogametic with ZW chromosomes.
Without recombination, purifying selection is relaxed in Y and W chromosomes, causing a slow accumulation of deleterious mutations through a process known as Müller’s ratchet.3 Recombination removes harmful mutations, but in the case of heteromorphic sex chromosomes, there is no homologous chromosome to recombine with, and this process is eliminated. Furthermore, selection of favorable alleles can “hitchhike” along mildly deleterious mutations.4 The accumulation of deleterious mutations eventually causes the Y and W chromosomes to degenerate, rapidly losing genes and gaining repeat sequences. Y chromosomes from plants and animals vary in age and are at different stages of evolution. Mammalian Y chromosomes are the most ancient, evolving around 166 million years ago.5 The human Y chromosome is extensively degraded and three times smaller than the X, retaining only 78 genes in the male specific region.6 In contrast, the young Y chromosome of white campion, Silene latifolia, evolved about 10 million years ago and has since ballooned to 570 Mb in size; 150 Mb larger than its X counterpart.7,8 The sex chromosomes in papaya are young with limited gene loss and expansion, serving as a model for the early stages of sex chromosome evolution.
Sex determination in papaya (Carica papaya L.) is controlled by a pair of nascent sex chromosomes, differentiated by an 8.1 Mbp, recombinationally suppressed, hermaphrodite-specific region on the Y chromosome (HSY).9 There are two slightly different Y chromosomes in papaya: Y controlling male and Yh controlling hermaphrodite sex. Any combination of the Y and Yh chromosomes, YY, Y Yh and Yh Yh, are lethal resulting in genotypes of XY, X Yh and XX for males, hermaphrodites and females respectively. The X and Yh chromosome (here on referred to as HSY) of papaya were recently sequenced and annotated using a tedious BAC by BAC approach.10,11 Together, we sequenced a total of 68 overlapping BACs for the 8.1 Mb HSY and 43 BACs for the 3.5 Mb X. The papaya X and HSY represent the second pair of sex chromosomes to be completely sequenced, and have uncovered a number of findings in the early stages of sex chromosome evolution.
Retrotransposon Accumulation and Novel Sex Specific Repeats
Repeat accumulation is common in Y chromosomes from animals,12,13 and has recently been documented in several plants, including hemp (Cannabis stativa)14; liverwort (Marchantia polymorpha)15; white campion (Silene latifolia)16; and papaya.17 The overabundance of repeat sequences in sex chromosomes is due to suppressed recombination and the lack of a mechanism to remove them.
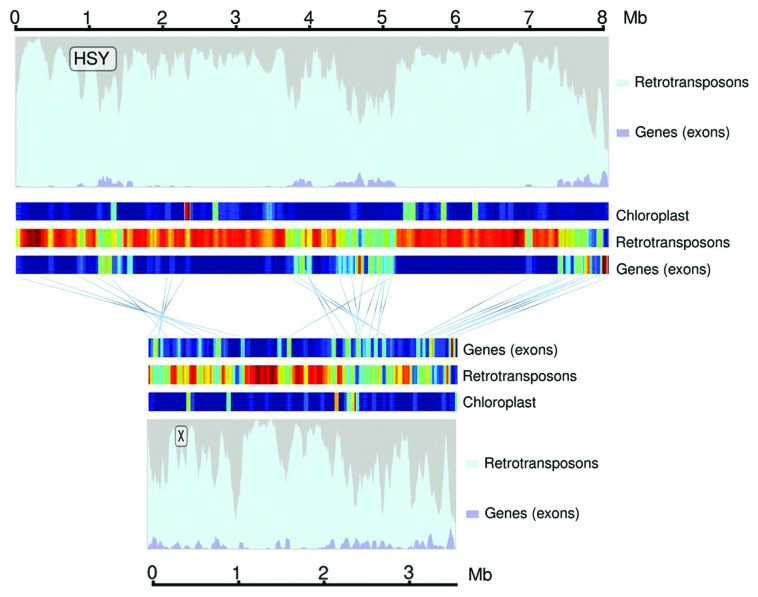
The papaya HSY is largely repetitive, and the accumulation of repeats accounts for much of its expansion in relation to the X. 79.3% of the HSY sequence is composed of repeat sequences, compared with 67.2% of the X.10 The papaya genome is about 51% repetitive,18 suggesting that repeat accumulation has occurred extensively in both the X and HSY. A closer look at the repeats indicates that most are retroelements with an abundance of Ty1-copia and Ty3-gypsy retrotransposons (Fig. 1). Ty3-gypsey elements are twice as abundant in the X and HSY as in the autosome, with Ty1-copia elements showing a similar but less remarkable expansion. DNA transposons are found in both the X and HSY, but at numbers comparable to the autosome. The origin and classification of many transposable elements in the papaya HSY is unknown, as repeated integration and shuffling has fragmented many of the repeats. In addition to the transposable elements, we identified 157 pairs of direct and inverted duplications that represent 6% of the HSY. This is remarkably higher than the X which contains 8 pairs of duplications which make up a mere 0.5% of the total sequence.10
Figure 1. Sequence comparison of the HSY and X chromosomes of papaya. Major DNA components were classified into exons (blue) and retrotransposons (cyan) and plotted using a sliding window of 50 kb and a shift of 10 kb. The gray portion represents unclassified DNA content. Heatmaps showing relative abundance of exons, retrotransposon and chloroplast fragments were plotted from low (blue), medium (yellow), high (red) ranges. Paired genes in the X and HSY are connected using blue lines and clearly show the two large scale inversions and the collinear region.
Two sequence blocks account for much of the expansion seen in the HSY. These blocks are mostly composed of repeats, with 84.6% and 87.2% of the sequences identified as repetitive. Ty3-gypsy elements are the most abundant repeat in these regions, followed by other retroelements, inverted duplications, microsatellites and unclassified sex specific repeats. Not surprisingly, the expansion blocks are extremely gene poor and correspond to the heterochromatic knob structures found on the HSY. The expanded regions have no homology to the X, suggesting they arose after divergence. Free from the selective pressures enforced by recombination, these regions gradually expanded, accumulating more repeats.
We identified 21 sex specific repeats (20 in HSY and 1 in X) which are absent from the autosome and have no homology to other plant repeats. Sex specific repeats in the HSY are mostly localized to the heterochromatic regions and together represent 10.7% of the total sequence. X specific repeats are more dispersed and constitute 3.5% of the sequence. The origin of the sex specific repeats in papaya is unknown, but this phenomenon is common in plant sex chromosomes. Hemp contains male specific LINE-like retrotransposons, and in liverwort, sex specific repeats harbor male specific genes.14,15
Gene Loss and Trafficking in the HSY
The Y chromosome in humans is an ancient relic of the ancestral autocome from which the X and Y have diverged; it is largely degenerated, having lost the vast majority of its original gene content throughout the course of its evolution. The papaya sex chromosomes are much younger, and most genes are conserved between the X and HSY. 106 protein coding genes were identified in the X and HSY, including 50 paired genes found in both chromosomes as well as 22 HSY-specific and 34 X specific genes. One quarter of the genes found in the HSY are pseudogenes, compared with 14% in the X. The high percentage of pseudogenes in the HSY are likely a result of Müllers ratchet, and the inability to select against slightly deleterious mutations. The HSY has lost at least one essential gene to the forces of Müllers ratchet, as the Yh Yh genotype is lethal, and embryos abort 25–50 d after pollination.19 Gene density in the HSY is seven times lower than the genome-wide-average with one gene per 112.5 kb (Fig. 1). The low gene density is the consequence of accumulation of repetitive sequences and degeneration of its gene content.
We used the 50 paired genes to survey the syneny relationship between the X and HSY. Three distinct regions are evident, including two large scale inversions and a collinear region (Fig. 1). Genes in the two large inversions form distinct evolutionary strata which indicate that the first inversion occurred about five million years before the second, which occurred about 1.9 million years ago. The first inversion likely contains the sex determination loci, and the lack of recombination in this inverted region spurred the development of sex chromosomes. The second inversion expanded the size of the HSY. Genes are conserved and collinear in the third region and sequence differences gradually fade to zero as the sex chromosome transitions to the pseudoautosomal region.
Ten of the HSY specific genes we identified have copies elsewhere in the genome, including seven protein colding genes and three pseudogenes, and are likely the result of translocation from the autosome. Translocated genes are the product of two similar mechanisms: transposons capturing portions of neighboring genes through transduplication20 or intron-less mRNAs that are reverse transcribed and integrated into a new chromosomal location via retrotransposition.21 Most transposed genes are pseudogenes containing a few exons from the autosomal copy, but the rest are free to mutate and acquire novel functions. Newly acquired genes that are absent from the X may play a role in sex determination, or sex specific traits such as flower and fruit morphology. Most of the autosome derived genes in the HSY are less than 300 bp and display no homology to known proteins, with a few notable exceptions. CpYh19 contains a partial MADS box domain sequence and may be involved in flower development. PCpYh1 has homology to HB22 from Arabidopsis, a Zinc finger homeodomain protein involved in embryo development and seed dormancy.22 Translocated genes with known functions also include an ATP synthase subunit and photosystem II protein D1. In S. latifolia, a gene involved in floral development AP3Y, was translocated from the autosome to the Y chromosome, and may be involved in sex determination.23 A similar phenomenon may be occurring in papaya, several of these translocated genes on the HSY could play a role in sexual reproduction.
Chloroplast DNA Accumulation in the Absence of Recombination
Organelle derived sequences are abundant in plant nuclear genomes, and organelle to nucleus transfers are common. The Arabidopsis genome, for instance, contains a 620 kb fragment of mitochondria DNA, and the rice genome contains a 113 kb fragment of chloroplast DNA.24,25 The papaya genome contains a number of insertions totaling 785,000 bp of chloroplast and 858,190 bp of mitochondria DNA, representing 0.28% and 0.31% of the genome respectively.18 Most nuclear chloroplast DNAs are less than 1 kb in length, as insertions are quickly shuffled and eliminated, with an estimated 80% of insertions lost within a million years of integration.26
In the absence of recombination, the papaya HSY has accumulated a staggering amount of chloroplast DNA (Fig. 1). The HSY has 195 insertions totaling 93,824bp, representing 1.15% of the total sequence.27 Chloroplast insertions are four times higher than the genome average and 12 times higher than the X chromosome. Insertions range in size from 50–4,500 bp including 20 insertions larger than 1 kb. In the autosome, these large fragments would have been shuffled and eventually lost due to recombination, but the lack of recombination in the HSY has instead, fixed them.
Perhaps the most interesting feature of the accumulated chloroplast DNA is a fragment of the chloroplast genome containing the rsp15 gene scattered throughout the HSY 23 times. These rsp15 fragments are nearly identical to each other but divergent from the chloroplast with estimated insertion times ranging from 20–25 million years ago, well before the inception of sex chromosomes in papaya. It is unlikely that the rsp15 fragments arose from independent insertion events; they were probably transposed across the HSY tagging along with active retrotransposons. Indeed, intact retrotransposons are found in close proximity to most rsp15 fragments, supporting this hypothesis. No copies of the rsp15 fragment are found elsewhere in the papaya genome or in the X chromosome.
Interestingly, only 10% of the chloroplast fragments are shared between the X and HSY. Most fragments either integrated after sex chromosome inception, or were eliminated from one of the sexes. The lack of recombination and prevalence of large fragments in the HSY suggests elimination isn’t responsible. Unlike chloroplast insertions, mitochondria genome insertions are less abundant in the HSY than the rest of the papaya genome, encompassing only 16,458 bp or 0.1% of the total sequence. Mitochondria insertions are short with an average size of 310 bp and only two are larger than 1 kb. Chloroplast and mitochondria integrations are equally prevalent in the papaya autosome, raising the question of why chloroplast integrations are more frequent in the HSY.
Organelle DNA accumulation isn’t limited to the papaya HSY; the Y chromosome of humans has been shown to preferentially accumulate mitochondrial DNA28 and the Y chromosome of S. latifolia is accumulating chloroplast DNA.29 In contrast to the papaya HSY, the Y chromosome in liverwort contains hundreds of mitochondria derived sequences, but only 3 plastid insertions.30 The accumulation of organelle DNA is a result of suppressed recombination, but a mechanism explaining the preference of mitochondrial or chloroplast sequences is currently unknown. Sequencing additional plant sex chromosomes may shed light on this phenomenon.
Future Prospects of Sex Chromosome Evolution
The study of sex chromosome evolution is often hindered by a lack of complete sequence information. Until recently, only the Y chromosomes from humans, rhesus monkey, chimpanzee and liverwort, and the chicken Z were available for comparative analysis. Sequencing sex chromosomes is difficult, as the heteromorphic chromosomes are highly repetitive, contain duplicated regions and have areas of homology between the chromosome types, making whole genome shotgun sequencing not suitable for assembling sex chromosomes. This is further complicated by the lack of informative genetic markers for sequence anchoring and orientation due to the suppressed recombination in the Y or Z chromosome. Even with current next generation sequencing technologies, the best approach for sequencing sex chromosomes is a laborious and tedious BAC by BAC approach.
The absence of recombination wreaks havoc on the gene content and structure in heteromorphic sex chromosomes. Ancient Y chromosomes like those found in humans and other mammals are highly degenerate, gene poor and are drastically shrunken in size compared with their corresponding X chromosomes. It’s difficult to trace the events that shaped these ancient Y chromosomes or the early stages of their evolution. The papaya X and Yh are only 7 million years old and provide detailed information of what occurs early on in sex chromosome evolution in this sex chromosome system.
Despite its young age, the papaya HSY is already showing signs of degeneration with changes in gene content and repeat expansion. Retrotransposons have expanded the HSY to almost three times the size of the X and have translocated 10 genes from the autosome in the process. In addition to the Ty3-gyspy retrotransposons and Ty1-copia elements, we identified over 20 HSY specific repeats which together, represent 10% of the total sequence. Furthermore, a fragment of the chloroplast genome has translocated 23 times in the HSY, evidence of highly active retrotransposons. Chloroplast integrations are far more common in the HSY than the rest of the genome, reflecting the function of recombination to shuffle and eliminate promiscuous DNA.
Sex chromosomes in plants display a range of evolutionary stages including incipient, early stage and late stage. To understand the underlying processes controlling sex chromosome evolution, it is necessary to study organisms with sex chromosomes at each of these stages. The sequenced sex chromosomes in papaya shed light on these processes, but sequencing sex chromosomes at additional stages is needed for a more complete picture of how sex chromosomes evolve.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/23462
References
- 1.Marshall Graves JA, Shetty S. Sex from W to Z: evolution of vertebrate sex chromosomes and sex determining genes. J Exp Zool. 2001;290:449–62. doi: 10.1002/jez.1088. [DOI] [PubMed] [Google Scholar]
- 2.Ming R, Bendahmane A, Renner SS. Sex chromosomes in land plants. Annu Rev Plant Biol. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- 3.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 4.Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987;116:161–7. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–73. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 7.Delph LF, Arntz AM, Scotti-Saintagne C, Scotti I. The genomic architecture of sexual dimorphism in the dioecious plant Silene latifolia. Evolution. 2010;64:2873–86. doi: 10.1111/j.1558-5646.2010.01048.x. [DOI] [PubMed] [Google Scholar]
- 8.Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–54. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Na JK, Wang J, Murray JE, Gschwend AR, Zhang W, Yu Q, et al. Construction of physical maps for the sex-specific regions of papaya sex chromosomes. BMC Genomics. 2012;13:176. doi: 10.1186/1471-2164-13-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Na JK, Yu Q, Gschwend AR, Han J, Zeng F, et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci U S A. 2012;109:13710–5. doi: 10.1073/pnas.1207833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gschwend AR, Yu Q, Tong EJ, Zeng F, Han J, VanBuren R, et al. Rapid divergence and expansion of the X chromosome in papaya. Proc Natl Acad Sci U S A. 2012;109:13716–21. doi: 10.1073/pnas.1121096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinemann S, Steinemann M. Y chromosomes: born to be destroyed. Bioessays. 2005;27:1076–83. doi: 10.1002/bies.20288. [DOI] [PubMed] [Google Scholar]
- 13.Erlandsson R, Wilson JF, Pääbo S. Sex chromosomal transposable element accumulation and male-driven substitutional evolution in humans. Mol Biol Evol. 2000;17:804–12. doi: 10.1093/oxfordjournals.molbev.a026359. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto K, Ohmido N, Fukui K, Kamada H, Satoh S. Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol Biol. 2000;44:723–32. doi: 10.1023/A:1026574405717. [DOI] [PubMed] [Google Scholar]
- 15.Okada S, Sone T, Fujisawa M, Nakayama S, Takenaka M, Ishizaki K, et al. The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc Natl Acad Sci U S A. 2001;98:9454–9. doi: 10.1073/pnas.171304798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant S, Houben A, Vyskot B, Siroky J, Pan WH, Macas J, et al. Genetics of sex determination in flowering plants. Dev Genet. 1994;15:214–30. doi: 10.1002/dvg.1020150304. [DOI] [Google Scholar]
- 17.Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–52. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- 18.Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–6. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu CT. Study on sex inheritance and horticultural characteristics of hermaphrodite papaya MS thesis, National Pingtung University of Science and Technology, Pingtung, Republic of China, 2000. [Google Scholar]
- 20.Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 2008;18:1924–37. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neufeld TP, Carthew RW, Rubin GM. Evolution of gene position: chromosomal arrangement and sequence comparison of the Drosophila melanogaster and Drosophila virilis sina and Rh4 genes. Proc Natl Acad Sci U S A. 1991;88:10203–7. doi: 10.1073/pnas.88.22.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, et al. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–14. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- 23.Matsunaga S, Isono E, Kejnovsky E, Vyskot B, Dolezel J, Kawano S, et al. Duplicative transfer of a MADS box gene to a plant Y chromosome. Mol Biol Evol. 2003;20:1062–9. doi: 10.1093/molbev/msg114. [DOI] [PubMed] [Google Scholar]
- 24.Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–8. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 25.Rice Chromosome 10 Sequencing Consortium In-depth view of structure, activity, and evolution of rice chromosome 10. Science. 2003;300:1566–9. doi: 10.1126/science.1083523. [DOI] [PubMed] [Google Scholar]
- 26.Matsuo M, Ito Y, Yamauchi R, Obokata J. The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast-nuclear DNA flux. Plant Cell. 2005;17:665–75. doi: 10.1105/tpc.104.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanBuren R, Ming R. Organelle DNA accumulation in the recently evolved papaya sex chromosomes. doi: 10.1007/s00438-013-0747-7. submitted. [DOI] [PubMed] [Google Scholar]
- 28.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 29.Kejnovsky E, Kubat Z, Hobza R, Lengerova M, Sato S, Tabata S, et al. Accumulation of chloroplast DNA sequences on the Y chromosome of Silene latifolia. Genetica. 2006;128:167–75. doi: 10.1007/s10709-005-5701-0. [DOI] [PubMed] [Google Scholar]
- 30.Yamato KT, Ishizaki K, Fujisawa M, Okada S, Nakayama S, Fujishita M, et al. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc Natl Acad Sci U S A. 2007;104:6472–7. doi: 10.1073/pnas.0609054104. [DOI] [PMC free article] [PubMed] [Google Scholar]