Abstract
Insertion sequences (IS) are abundant in the bacterial fish pathogen Aeromonas salmonicida genome. IS are involved in rearrangement events that lead to the loss of virulence. In previous work, we studied a plasmid rearrangement that causes the deletion of the type three secretion system in A. salmonicida, resulting in a loss of virulence. We showed that the rearrangement is caused by the recombination of two IS (ISAS11) on an unstable plasmid (pAsa5). However, many rearrangements cannot be explained by our experimental approach and are thought to be the result of more complex or incomplete rearrangement events, as suggested by other plasmid loss profiles observed in various A. salmonicida strains. In this commentary, we examine the genetic instability of A. salmonicida indicating that its genome is rapidly evolving.
Keywords: furunculosis, Aeromonas salmonicida, type three secretion system, plasmid, DNA rearrangement, insertion sequence, IS256
Aeromonas salmonicida is a bacterial fish pathogen that causes furunculosis, a disease affecting salmonids (salmon, trout, etc.). This psychrophilic bacterium is known to lose its virulence at 22°C or higher.1-3 The absence of virulence was initially explained by the loss of the A-layer (also called the S-layer), a protective, hydrophobic, protein array located at the outer boundary of the bacterial cell wall.3 The A-layer contributes to virulence by promoting bacterial adherence to host cells and by protecting bacteria from host proteolytic activities.4 Temperatures at or above 22°C were subsequently shown to cause the loss of the type three secretion system (TTSS), another virulence factor.2 The TTSS is a well-characterized virulence factor encountered in many Gram-negative pathogens. The components of the TTSS are assembled into a needle-like apparatus that injects various pathway-disrupting toxins into host cells.5 In A. salmonicida, the TTSS is a prerequisite for establishing an infection.6,7
In the case of both the A-layer and the TTSS, the loss of virulence is caused by a genetic rearrangement that results in a permanent avirulent phenotype. The gene encoding the A protein (vapA) can be disrupted in many ways. Belland and Trust first observed a 5′ deletion in vapA. However, vapA can also be disrupted by the transposition of the endogenous insertion sequences (IS) ISAS1 and ISAS2.8,9 ISAS1 can also be inserted in abcA, a gene located downstream from vapA that is required for O-polysaccharide synthesis and transport.10 Since the A-layer binds to O-polysaccharides, this insertion also disrupts A-layer formation.11
For the TTSS, the genetic alteration occurs on pAsa5 (or pASvirA), a 155 kb plasmid bearing most TTSS-encoding genes. In 2003, Stuber et al. first reported the loss of the TTSS due to the loss of the plasmid.2 However, more recent results by Daher et al. have shown that the loss of the TTSS is caused by a rearrangement of pAsa5, which leads to the deletion of the TTSS locus alone or together with other contiguous regions of the plasmid.1 Interestingly, a loss of virulence associated with deletions in a large plasmid was observed as early as 1989.12 In retrospect, the plasmid considered by the authors was probably pAsa5. The frequency of these rearrangements appears to be strain-dependent. Stuber et al. reported that strain JF2267 completely lost the TTSS after only six hours at 25°C.2 Interestingly, strain A449 did not lose the TTSS when grown in liquid medium.13 However, a sub-population bearing TTSS rearrangements appears when A449 is grown as colonies on agar medium.1
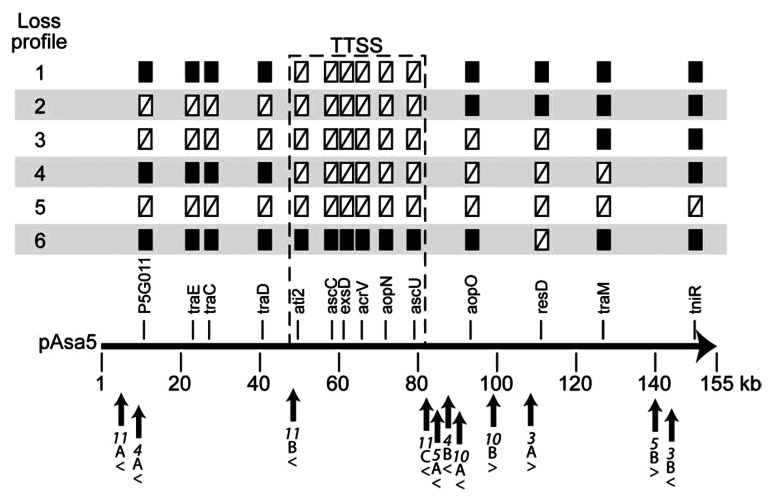
In their work, Daher et al. first assessed the integrity of pAsa5 and the TTSS in various A. salmonicida isolates.1 Based on an analysis of 14 genes on pAsa5, they discovered that 42% of the isolates had undergone pAsa5 rearrangements, with all but one losing the entire TTSS locus (Fig. 1). Daher et al. then assessed the rearrangement of pAsa5 under controlled conditions. They grew three unaltered pAsa5-bearing strains (A449, 01-B526 and 01-B516) at 25°C for two weeks and then assayed the 14 pAsa5 genes.1 Under these conditions, pAsa5 rearranged at a low frequency, but the TTSS locus was always lost in the process. With one exception, they observed two loss profiles, which we later called loss profile 1 and loss profile 2 (Fig. 1).14 Loss profile 1 corresponds to the loss of the TTSS locus alone, while loss profile 2 corresponds to the loss of both the TTSS locus and a fragment of about 40 kb upstream from the TTSS locus. A frequency comparison showed that loss profile 1 is dominant while loss profile 2 is infrequent. Sub-strains in which pAsa5 rearrangements occur under controlled conditions also lose their virulence.1 Daher et al. concluded that growth at 25°C can trigger rearrangements in pAsa5 and, as a result, cause a loss of virulence, and that the integrity of pAsa5 can be threatened when A. salmonicida strains are routinely grown under these conditions.1
Figure 1. Various pAsa5 rearrangements observed in A. salmonicida isolates. This figure reports the rearrangement patterns observed by Daher et al. as well as the numerous insertion sequences (IS) and their locations in the pAsa5 plasmid.1 We studied loss profiles 1 and 2 and showed that IS11 is involved in pAsa5 rearrangements. Loss profiles 1 and 2 have been observed in isolates as well as clones following controlled exposure to stressful conditions.1 Loss profiles 3 to 6 have been observed only in a few isolates and may result from more complex rearrangements involving other IS carried by pAsa5. The 14 genes targeted by Daher et al. in the PCR assay are shown below the loss profiles.1 The locations of IS in pAsa5 are indicated by arrows. Arrowheads below the name of the ISs show their orientation. The variety of genes lost is greater downstream from the TTSS locus than upstream. This may be due to the IS clusters in this region inducing more complex rearrangements.
In our work, we investigated the causes underlying the rearrangement observed by Daher et al.14 We focused on the loss profiles 1 and 2 observed under controlled conditions. We were particularly interested in the multiple IS flanking the lost region (Fig. 1). Since these IS were copies of the same IS (ISAS11, or IS11, from the IS256 family),15 the rearrangement, and thus the loss of virulence, may have resulted from the homologous recombination of two copies of these mobile genetic elements (Fig. 2).16 We attempted to assess the mechanism involved using a disjoint-joint primer PCR assay that generates amplicons only if the sequences flanking the loss profiles are brought sufficiently close together after the recombination. Loss profile 2, which was readily explained using this approach, was the result of the recombination of IS11A and IS11C. Loss profile 1 was shown to be caused by the recombination of IS11B and IS11C only in a fraction of clones. Most loss profile 1 rearrangements could not be explained using this approach. This might be due to other recombination events involving one or two other copies of IS11 in the A. salmonicida genome (one in the chromosome and one in pAsal1, a plasmid bore by 01-B516 and 01-B526, but not by A449). We then unsuccessfully attempted to assess further IS11 recombinations in TTSS-deleted sub-strains in which the initial PCR approach failed to link pAsa5 rearrangements with an IS11-dependent recombination process. Repeat regions at primers targeting sites in many cases led to non-specific PCR amplifications and, as such, prevented the assessment by PCR of more complex rearrangements. However, we showed that pAsal1 could be also affected by growth at 25°C. pAsal1, which is not present in the A449 reference strain, can be cured in some TTSS-deleted sub-strains originating from the 01-B516 and 01-B526 parental strains more likely because it includes an IS11.14
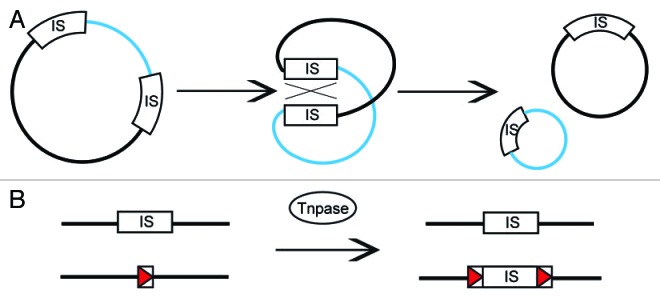
Figure 2. Homologous recombination and transposition: two IS-mediated gene inactivation patterns identified in A. salmonicida. (A) Homologous recombination events involving two IS in a plasmid can lead to deletions. Such events are transposase-independent and can occur because the two IS are identical. In such cases, the DNA fragment bearing the replication origin is replicated and propagated, while the other fragment is deleted. This deletion pattern occurs in pAsa5 plasmid and is mediated by IS11 leading to the deletion of the TTSS locus.14(B) A transposition event involving a single IS can lead to gene inactivation. An IS can use its transposase (Tnpase) to insert at an insertion site (red arrow). The transposition causes the duplication of the insertion site. In A. salmonicida, ISAS1 and ISAS2 can transpose in the vapA and abcA genes, which are essential for the proper assembly and display of the A-layer.10,11 In the case of ISAS1 and ISAS2, the transposition is coupled with duplication, leaving a copy of the IS on the donor strand.9
As mentioned earlier, some loss profile 1 rearrangements could not be explained using our PCR approach and may result from more complex or imperfect recombinations.14 Daher et al. also reported the existence of more complex recombination patterns than those studied in our work (Fig. 1).1 These recombinations resulted from routinely growing bacterial isolates for long periods at inappropriate temperatures.1 This may also provide indirect proof that rearrangements are not limited to perfect homologous recombinations between copies of the same IS, that is, ISAS11. There is evidence indicating that the genome of A. salmonicida is prone to rearrangements and thus to rapid genomic evolution, as first postulated by Reith et al., who resolved the first A. salmonicida genome (A449).15 In addition to pAsa5, the most striking example of rearrangements came from the sequencing of the pAsa6 plasmid in an A. salmonicida strain isolated from a diseased turbot.17 pAsa6 is an 18 kb plasmid that bears open reading frames sharing strong homology with various pAsa5 genes, but in a discontinuous manner. pAsa6 contains many putative transposases and transposase fragments from the A. salmonicida genome as well as a TTSS-effector AopH homolog. The researchers who first identified pAsa6 suggested that it may have originated from pAsa5, or that pAsa5 may be a combination of pAsa6 and another plasmid, a hypothesis they favored.17 The A. salmonicida genome, in addition to its IS, also contains many regions that share high levels of identity that may have arisen from genome duplication. Our work highlights several of these regions, including sequences in two A. salmonicida plasmids (pAsa3 and pAsal1) that share a high level of identity.14 Two regions in pAsa5 also exhibit a high level of identity, with a 5 kb fragment directly upstream from the TTSS locus being almost identical to another sequence elsewhere in pAsa5.14 A-layer inactivation, either by deletion or by IS insertions in vapA and abcA, is another example of rearrangement in A. salmonicida. However, in this case, the IS undergoes transposition rather than homologous recombination as with pAsa5 (Fig. 2). This transposase-mediated event allows ISAS1 and ISAS2 to integrate at an insertion site, creating direct repeats and disrupting the gene. In this case, the IS is duplicated during transposition, leaving a copy at the donor site.9
The A. salmonicida genome also teems with IS that provide many other rearrangement possibilities that have been observed in other microorganisms. For instance, Desulfitobacterium sp strain Y51 displays a rearrangement pattern similar to that of A. salmonicida, with two IS256 families framed by an occasionally deleted region that codes for PceA reductive dehydrogenase.18 Desulfitobacterium sp strain Y51 can also be affected by a defective IS256 rearrangement, as it is the case for Enterococcus faecalis gentamicin resistance cassette.18,19 IS256 is also an important component of biofilm regulation and genome plasticity in Staphylococcus epidermis because of its multiple rearrangements, including homologous recombination.20 In S. aureus, the IS256 transposition rate is modified by sub-lethal concentrations of antibiotics such as ciprofloxacin and vancomycin.21 Other substances can also modify the mobility rate of transposons that harbour IS. For instance, caffeine, ethionine, acriflavine, procaine and cinnamaldehyde can modulate Tn10 activity.22
Since the A. salmonicida genome was shown to comprises several copies of IS, it might be possible to modulate IS mobility to better control A. salmonicida outbreaks. This is particularly relevant given that at least two distinct recombination events (one for vapA and one for the TTSS) lead to a significant loss of virulence. However, this application may be limited by the occasionally low recombination frequencies of IS, at least in the case of the TTSS. Nevertheless, it would be worthwhile to further characterize A. salmonicida genomic mobility, at least as a model of contemporary bacterial evolution. Additional genome sequences such as the recently published 01-B526 sequence23 might provide new examples of IS-related bacterial rearrangements and ultimately improve our understanding of the driving forces behind bacterial evolution that are involved in host-pathogen relationships.
Acknowledgments
K.H.T. holds a scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). S.J.C. is a research scholar of the Fonds de la Recherche en Santé du Québec (FRSQ).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/23498
References
- 1.Daher RK, Filion G, Tan SG, Dallaire-Dufresne S, Paquet VE, Charette SJ. Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet Microbiol. 2011;152:353–60. doi: 10.1016/j.vetmic.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 2.Stuber K, Burr SE, Braun M, Wahli T, Frey J. Type III secretion genes in Aeromonas salmonicida subsp salmonicida are located on a large thermolabile virulence plasmid. J Clin Microbiol. 2003;41:3854–6. doi: 10.1128/JCM.41.8.3854-3856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishiguro EE, Kay WW, Ainsworth T, Chamberlain JB, Austen RA, Buckley JT, et al. Loss of virulence during culture of Aeromonas salmonicida at high temperature. J Bacteriol. 1981;148:333–40. doi: 10.1128/jb.148.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noonan B, Trust TJ. The synthesis, secretion and role in virulence of the paracrystalline surface protein layers of Aeromonas salmonicida and A. hydrophila. FEMS Microbiol Lett. 1997;154:1–7. doi: 10.1111/j.1574-6968.1997.tb12616.x. [DOI] [PubMed] [Google Scholar]
- 5.Büttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burr SE, Stuber K, Wahli T, Frey J. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J Bacteriol. 2002;184:5966–70. doi: 10.1128/JB.184.21.5966-5970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burr SE, Pugovkin D, Wahli T, Segner H, Frey J. Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology. 2005;151:2111–8. doi: 10.1099/mic.0.27926-0. [DOI] [PubMed] [Google Scholar]
- 8.Belland RJ, Trust TJ. Cloning of the gene for the surface array protein of Aeromonas salmonicida and evidence linking loss of expression with genetic deletion. J Bacteriol. 1987;169:4086–91. doi: 10.1128/jb.169.9.4086-4091.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustafson CE, Chu SJ, Trust TJ. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994;237:452–63. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- 10.Chu SJ, Noonan B, Cavaignac S, Trust TJ. Endogenous mutagenesis by an insertion sequence element identifies Aeromonas salmonicida AbcA as an ATP-binding cassette transport protein required for biogenesis of smooth lipopolysaccharide. Proc Natl Acad Sci U S A. 1995;92:5754–8. doi: 10.1073/pnas.92.12.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belland RJ, Trust TJ. Synthesis, export, and assembly of Aeromonas salmonicida A-layer analyzed by transposon mutagenesis. J Bacteriol. 1985;163:877–81. doi: 10.1128/jb.163.3.877-881.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belland RJ, Trust TJ. Aeromonas salmonicida plasmids: plasmid-directed synthesis of proteins in vitro and in Escherichia coli minicells. J Gen Microbiol. 1989;135:513–24. [Google Scholar]
- 13.Ebanks RO, Knickle LC, Goguen M, Boyd JM, Pinto DM, Reith M, et al. Expression of and secretion through the Aeromonas salmonicida type III secretion system. Microbiology. 2006;152:1275–86. doi: 10.1099/mic.0.28485-0. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka KH, Dallaire-Dufresne S, Daher RK, Frenette M, Charette SJ. An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS One. 2012;7:e33725. doi: 10.1371/journal.pone.0033725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics. 2008;9:427. doi: 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–74. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Najimi M, Balado M, Lemos ML, Osorio CR. Genetic characterization of pAsa6, a new plasmid from Aeromonas salmonicida subsp. salmonicida that encodes a type III effector protein AopH homolog. Plasmid. 2009;61:176–81. doi: 10.1016/j.plasmid.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Futagami T, Tsuboi Y, Suyama A, Goto M, Furukawa K. Emergence of two types of nondechlorinating variants in the tetrachloroethene-halorespiring Desulfitobacterium sp. strain Y51. Appl Microbiol Biotechnol. 2006;70:720–8. doi: 10.1007/s00253-005-0112-9. [DOI] [PubMed] [Google Scholar]
- 19.Casetta A, Hoï AB, de Cespédès G, Horaud T. Diversity of structures carrying the high-level gentamicin resistance gene (aac6-aph2) in Enterococcus faecalis strains isolated in France. Antimicrob Agents Chemother. 1998;42:2889–92. doi: 10.1128/aac.42.11.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenfelder SMK, Lange C, Eckart M, Hennig S, Kozytska S, Ziebuhr W. Success through diversity - how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int J Med Microbiol. 2010;300:380–6. doi: 10.1016/j.ijmm.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Nagel M, Reuter T, Jansen A, Szekat C, Bierbaum G. Influence of ciprofloxacin and vancomycin on mutation rate and transposition of IS256 in Staphylococcus aureus. Int J Med Microbiol. 2011;301:229–36. doi: 10.1016/j.ijmm.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 22.MacPhee DG, Hafner LM. Antimutagenic effects of chemicals on mutagenesis resulting from excision of a transposon in Salmonella typhimurium. Mutat Res. 1988;207:99–105. doi: 10.1016/0165-7992(88)90071-1. [DOI] [PubMed] [Google Scholar]
- 23.Charette SJ, Brochu F, Boyle B, Filion G, Tanaka KH, Derome N. Draft genome sequence of the virulent strain 01-B526 of the fish pathogen Aeromonas salmonicida. J Bacteriol. 2012;194:722–3. doi: 10.1128/JB.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]