Abstract
Microbes have several mechanisms that promote evolutionary adaptation in stressful environments. The corresponding molecular pathways promote diversity through modulating rates of recombination, mutation or influence the activity of transposable genetic elements. Recent experimental studies suggest an evolutionary conflict between these mechanisms. Specifically, presence of mismatch repair mutator alleles in a bacterial population dramatically reduced fixation of bacterial insertion sequence elements. When rare, these elements had only a limited impact on adaptive evolution compared with other mutation-generating pathways. IS elements may initially spread like molecular parasites, but once present in many copies in a given genome, they might become generators of novelty during bacterial evolution.
Keywords: insertion sequence elements, evolution of mutation rate
Insertion sequence (IS) elements are small transposable genetic elements widely distributed in bacterial genomes.1 They are generally very short and contain only the genetic information essential for their transposition.2 By inserting to new genomic locations, they frequently inactivate or upregulate flanking genes. By inducing recombination events, they also cause deletions and inversions of large genomic segments.1 Several lines of observations point to the direction that the net effects of transposon insertions are harmful for the host.3 First, direct experimental evidences indicate that enhanced mobilization of transposable elements are generally deleterious.4-6 Second, most IS families are found only in a limited number of species.6,7 However, within any one genome, they are typically present in many copies which are very similar to each other.7 This is reminiscent to the evolutionary dynamics of other genomic parasites,8 such as retroviruses. Both retroviruses and IS elements have entered bacterial genomes only very recently and spread through horizontal gene transfer across species. Third, to minimize damage they may cause during insertions, these elements have become suppressed by host regulatory factors,9 or reside in genomic regions where they cause less harm.6
If harmful, why are they present, even if transiently, in bacterial genomes? One answer may be that in sexual populations, IS elements spread as selfish entities10 even if they deliver no beneficial effects.11 Indeed, these elements are nearly always autonomous, i.e., the genes necessary for transpositions are encoded by the elements and not by the host genome. One prediction of the theory is that bacterial species with numerous IS elements should also have more genetic exchange.12
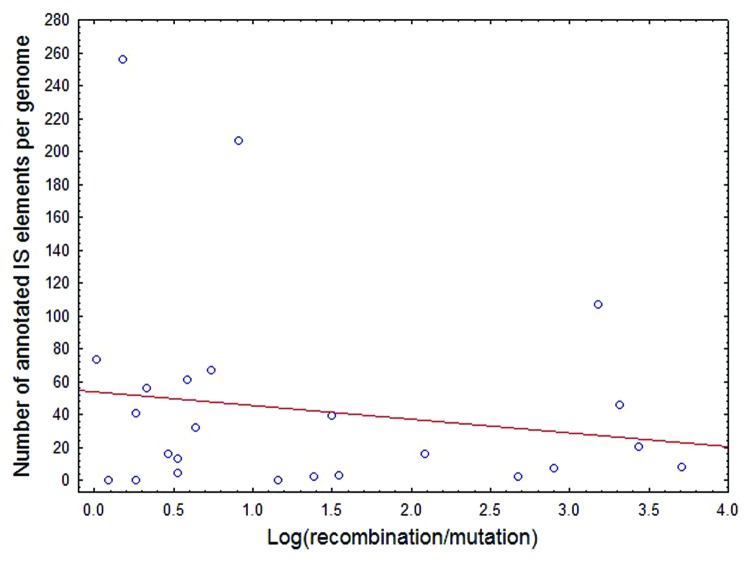
A preliminary analysis failed to find strong support for this idea. Recently, Multi Locus Sequence Typing (MLST) data sets of bacterial and archaeal species were analyzed to explore the ecological and phylogenetic determinants of recombination frequency differences across species.13 The authors compiled a data set on the estimated ratio of nucleotide changes as the result of recombination relative to point mutations13 for 48 species. In agreement with previous studies, the data suggest that homologous recombination rates vary widely between species. Another work6 reconstructed the distribution of IS elements in a wide range of bacterial genomes with the aid of the ISfinder database.14 We merged the two data sets and found that there was no general association between recombination rate and the number of IS elements across genomes (Fig. 1). Needless to say, this is only a preliminary analysis, and more sophisticated approaches are required to settle this issue. It may also be the case than even rare genetic exchange can promote spread of insertion sequence elements. With all these limitations in mind, this result at the very least invites speculations on complementary explanations which are consistent with the phylogenetic patterns observed.
Figure 1. No positive relationship between relative recombination rate and the number of IS elements across bacterial genomes. The recombination relative to point mutation values (r/m) are from ref. 13, while the number of IS elements per genome was estimated by Touchon and Rocha.6 Spearman correlation, r = -0.16, p = 0.43, n = 25.
The mutator theory is one such alternative.15 It claims that as transposable elements initiate mutational events, they can efficiently boost host genetic adaptation. Tight genetic linkage between IS insertion and the chance advantageous mutation created may drive their joint fixation in the population.
Proponents of the theory generally refer to data demonstrating that IS elements cause wide range of genomic rearrangements, both in nature16,17 and in laboratory settings.18,19 Some of these changes are clearly associated with favorable effects. However, given their ubiquity in bacterial genomes, it is hardly surprising that they occasionally generate such mutations. One should not confuse the difference between cause and effect in evolution. Let us clarify this difference by Michael Lynch’s analogy.20 Although free oxygen radicals arising from oxidative metabolism is a major source of mutations, no one would seriously argue that oxidative metabolism arose to promote evolutionary change. Similarly, the fact that IS elements generate occasionally beneficial mutations does not imply that they have been selected and maintained to enhance genetic adaptation. Skeptics may also add that IS elements do not provide a completely new source of variation otherwise not accessible.20,21 Indeed, IS insertions are unlikely to deliver slight variations in protein structure or fine tune enzymatic activities.
Clearly, more rigorous experimental underpinnings of the theory are required. There are at least two crucial issues which have been neglected by the community so far. First, we need to understand the evolutionary forces driving gradual accumulation of these elements in nascent bacterial genomes. Second, given the wealth of other molecular pathways that boost bacterial mutation rate under times of stress, the interplay between them must be considered. To what extent does the presence of one such mechanism in the population influence the evolutionary fate of IS elements? Here we briefly summarize our current, but still rather limited state of knowledge on these issues.
When a given family of IS elements invades an initially transposon-free bacterial genome, the process is expected to start with a single or very few copies. Do they produce a sufficient number of mutations to be favored by selection? Answering this question is not straightforward, as it requires comparison of evolvability of genotypes with differences in the number of residing IS elements only.
Our lab took advantage of the availability of a Escherichia coli MDS42, a strain with a reduced genome devoid of all mobile genetic elements and cryptic virulence factors.22 This strain has several beneficial properties for biotechnological applications.22 We reinserted a single, highly active IS element into the MDS42 genome.23 Under certain, but not all stressful conditions, a single IS element generated mutations with especially large beneficial effects. Thus, in agreement with expectations of the mutator theory, a single IS element could accelerate host adaptation in the lab.23
However, as evolution is opportunistic, this result must be considered in a broader context. Bacteria have a wide range of toolkits to create genetic diversity.24 The toolkit includes changed activity of DNA safeguarding mechanisms, the SOS response and other recombination pathways.24 Most notably, many hypermutator bacteria in nature have deficient methyl-directed mismatch repair (MMR) system. These genotypes are frequent in bacterial populations,25 and are associated with clinical infections.26,27 A single, loss of function mutation in MMR causes a hundredfold increase in genomic mutation rate. Given their ubiquity, it’s important to address how the presence of such MMR mutants interferes with the spread of IS elements in bacterial populations. Experiments revealed that the rate of extra mutations and hence the advantage delivered by a single IS is relatively low compared with such mutator alleles.23 As expected, IS-carrying strains were inferior to MMR mutators in competition.23
This is a potentially problematic result for the mutator theory of transposable elements. IS elements may have to reach a critical number in the host genome to have a substantial impact on genome evolution. Prior to that point, bacteria could adapt more efficiently by other means, including defective MMR alleles. At the moment, we can only speculate how transposition rate depends on IS copy number and how the transition from a selfish element to a mutator beneficial for the host can take place in natural populations (Fig. 2).
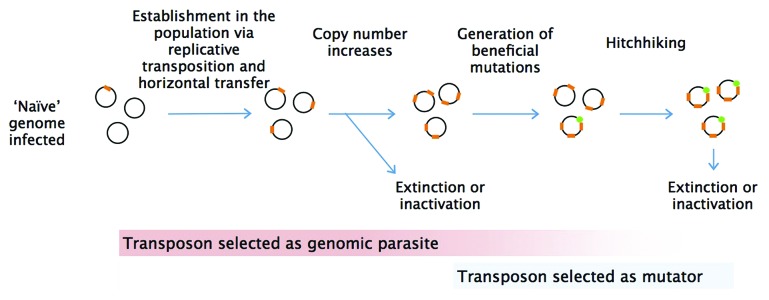
Figure 2. Initial establishment and maintenance of transposable elements in the population can be shaped by distinct selective forces. When rare, transposons likely spread and increase copy number as genomic parasites. Once present in sufficiently high copy numbers, they might act as mutators. By virtue of the frequent transposition events, they occasionally generate beneficial mutations which are closely linked to the transposons, hence allowing them to hitchhike and further spread in bacterial genomes.
Another, largely neglected issue is the long-term consequences of IS elements and other mutator alleles on survival. Once a mutator allele has spread in a bacterial population and stressful conditions are over, they will generate largely harmful mutations. Due to this long-term disadvantage, MMR deficient mutator alleles arise frequently, but low mutation rate can be restored through gain of functionial variants through horizontal transfer.28 IS elements may have a serious advantage over constitutive mutator alleles. They are activated only under stressful conditions,2,29 and hence they may not enhance mutation load substantially.
These issues represent only the tip of the iceberg. Evolutionists suggested several key candidate molecular systems driving bacterial evolution. Pioneering works claim that mutators and sex are conflicting adaptive strategies,30 stress-induced mutagenesis hinders evolution of constitutive mutators,31 and sex promotes mutational robustness.32 Future studies should investigate the interplay between these systems.
Acknowledgments
This work and related studies were supported by grants from European Research Council, Wellcome Trust and the “Lendület Program” of the Hungarian Academy of Sciences.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/23617
References
- 1.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–74. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siguier P, Filée J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol. 2006;9:526–31. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Sawyer SA, Dykhuizen DE, DuBose RF, Green L, Mutangadura-Mhlanga T, Wolczyk DF, et al. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics. 1987;115:51–63. doi: 10.1093/genetics/115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilke CM, Adams J. Fitness effects of Ty transposition in Saccharomyces cerevisiae. Genetics. 1992;131:31–42. doi: 10.1093/genetics/131.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elena SF, Ekunwe L, Hajela N, Oden SA, Lenski RE. Distribution of fitness effects caused by random insertion mutations in Escherichia coli. Genetica. 1998;102-103:349–58. doi: 10.1023/A:1017031008316. [DOI] [PubMed] [Google Scholar]
- 6.Touchon M, Rocha EP. Causes of insertion sequences abundance in prokaryotic genomes. Mol Biol Evol. 2007;24:969–81. doi: 10.1093/molbev/msm014. [DOI] [PubMed] [Google Scholar]
- 7.Wagner A. Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol Biol Evol. 2006;23:723–33. doi: 10.1093/molbev/msj085. [DOI] [PubMed] [Google Scholar]
- 8.Wagner A. Transposable elements as genomic diseases. Mol Biosyst. 2009;5:32–5. doi: 10.1039/b814624c. [DOI] [PubMed] [Google Scholar]
- 9.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–7. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 11.Hickey DA. Evolutionary dynamics of transposable elements in prokaryotes and eukaryotes. Genetica. 1992;86:269–74. doi: 10.1007/BF00133725. [DOI] [PubMed] [Google Scholar]
- 12.Zeyl C, Bell G, Green DM. Sex and the spread of retrotransposon Ty3 in experimental populations of Saccharomyces cerevisiae. Genetics. 1996;143:1567–77. doi: 10.1093/genetics/143.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 14.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(Database issue):D32–6. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao L, Vargas C, Spear BB, Cox EC. Transposable elements as mutator genes in evolution. Nature. 1983;303:633–5. doi: 10.1038/303633a0. [DOI] [PubMed] [Google Scholar]
- 16.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–32. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 17.Ooka T, Ogura Y, Asadulghani M, Ohnishi M, Nakayama K, Terajima J, et al. Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res. 2009;19:1809–16. doi: 10.1101/gr.089615.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider D, Lenski RE. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol. 2004;155:319–27. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–7. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 20.Lynch M. The Origins of Genome Architecture (Sinauer Press, 2007). [Google Scholar]
- 21.Stoebel DM, Dorman CJ. The effect of mobile element IS10 on experimental regulatory evolution in Escherichia coli. Mol Biol Evol. 2010;27:2105–12. doi: 10.1093/molbev/msq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pósfai G, Plunkett G, 3rd, Fehér T, Frisch D, Keil GM, Umenhoffer K, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–6. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 23.Fehér T, Bogos B, Méhi O, Fekete G, Csörgo B, Kovács K, et al. Competition between transposable elements and mutator genes in bacteria. Mol Biol Evol. 2012;29:3153–9. doi: 10.1093/molbev/mss122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aertsen A, Michiels CW. Diversify or die: generation of diversity in response to stress. Crit Rev Microbiol. 2005;31:69–78. doi: 10.1080/10408410590921718. [DOI] [PubMed] [Google Scholar]
- 25.Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, et al. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–4. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 26.Denamur E, Bonacorsi S, Giraud A, Duriez P, Hilali F, Amorin C, et al. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J Bacteriol. 2002;184:605–9. doi: 10.1128/JB.184.2.605-609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–4. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 28.Denamur E, Lecointre G, Darlu P, Tenaillon O, Acquaviva C, Sayada C, et al. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–21. doi: 10.1016/S0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 29.Levy MS, Balbinder E, Nagel R. Effect of mutations in SOS genes on UV-induced precise excision of Tn10 in Escherichia coli. Mutat Res. 1993;293:241–7. doi: 10.1016/0921-8777(93)90075-R. [DOI] [PubMed] [Google Scholar]
- 30.Tenaillon O, Le Nagard H, Godelle B, Taddei F. Mutators and sex in bacteria: conflict between adaptive strategies. Proc Natl Acad Sci U S A. 2000;97:10465–70. doi: 10.1073/pnas.180063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjedov I, Tenaillon O, Gérard B, Souza V, Denamur E, Radman M, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–9. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 32.Azevedo RB, Lohaus R, Srinivasan S, Dang KK, Burch CL. Sexual reproduction selects for robustness and negative epistasis in artificial gene networks. Nature. 2006;440:87–90. doi: 10.1038/nature04488. [DOI] [PubMed] [Google Scholar]