Abstract
It was recently shown that the expansion of CD4+ T cells during a primary immune reaction to a peptide from cytochrome c decreases approximately 0.5 log for every log increase in the number of cognate precursor cells, and that this remains valid over more than four orders of magnitude [Quiel, et al., Antigen-stimulated CD4 T-cell expansion is inversely and log-linearly related to precursor number. PNAS, 2011, 108: 3312]. This observed “power law” was explained by a mechanism where non-dividing mature T cells inhibit the proliferation of less-differentiated cells of the same specificity. Here we interpret the same data by a mechanism where CD4+ T cells acquire cognate peptide-MHC (pMHC) complexes from the surface of antigen presenting cells (APCs), thereby increasing the loss rate of pMHC. We show that a mathematical model implementing this “T cell grazing" mechanism, and having a T cell proliferation rate that is determined by the concentration of pMHC, explains the data equally well. As a consequence, the data no longer unequivocally support the previous explanation, and the increased loss of pMHC complexes on APCs at high T cell densities is an equally valid interpretation of this striking data.
2 Introduction
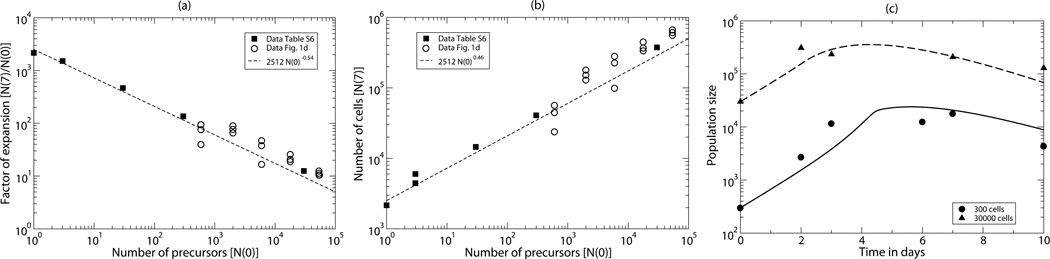
T cell responses to antigens are characterized by an expansion phase due to the antigen-specific activation and clonal expansion of cognate T cells, and a contraction phase during which cell densities approach a stable level that is called the memory phase [1]. Several studies have demonstrated that the level of expansion during the expansion phase depends on the number of precursor cells recruited into the immune response [2, 3, 4, 5, 6, 7, 8]. The most quantitative of these studies was recently performed by Quiel et al., [7] who varied the initial precursor density by more than four orders of magnitude, i.e., from one to more than 30000 cells per mouse, and measured the fold expansion at day seven of the response (i.e., somewhere in the contraction phase). Their most striking result was that in a log-log representation, the fold expansion declines linearly with the precursor density over this wide range (see Fig. 1a), i.e., the fold expansion drops approximately 0.5 log for every log increase in the number of precursors [7]. Since physiologically measured precursor frequencies cover a similar range, the tight relationship between the fold expansion and the precursor density was taken to reflect a fundamental property of antigen-mediated immune responses [7]. Phenomenologically the population density after one week was very well described by cells (see Fig. 1b), where N(0) is number of naive precursors in the mouse [8], reflecting the same 0.5 log decrease in the fold expansion per log increase in the precursor density. Time courses of responses starting with 300 or 30000 cells revealed that the peak of the response occurred earlier when the initial precursor density was higher (Fig. 1c).
Figure 1.
The data presented by Quiel et al., [7]. (a) The fold expansion at day 7, N(7)/N(0), as a function of the number of precursors, N(0). The short-dashed line is the regression line 2512×N(0)−0.54 fitted by Bocharov et al., [8]. (b) The population size at day 7 as a function of the number of precursors, N(0). The short-dashed line is the equivalent regression line 2512×N(0)0.46 fitted by Bocharov et al., [8]. The data was taken from Table S6 in Bocharov et al., [8] and by digitizing the data from Fig. 1d in Quiel et al., [7]. (c) The time course data of responses starting with N(0)=300 (bullets) or N(0)=30000 (triangles) 5C.C7 T cells, and the best fit to this data by the simplified model of Bocharov et al., [8] (solid and long-dashed lines). The data was taken from Table S3 in Bocharov et al., [8].
The experimental system involved the adoptive transfer of CD4+ 5C.C7 TCR transgenic T cells that are specific for a peptide from cytochrome c peptide presented on I-Ek. The magnitude of the response was measured by an extremely sensitive PCR analysis specifically amplifying the CDR3 region of the Vβ chain of the 5C.C7 TCR, allowing the detection the progeny of a single cell in a mouse [7]. Importantly, the effect shown in Fig. 1 was not due to a reduced recruitment of precursor cells at precursor high densities, because the difference in clonal expansion came about at a late stage in the response. Measuring the fold expansion at day two, i.e., before the release of proliferated cells from the lymphoid tissue, during responses starting with 300 or 30000 precursors, it was found that both populations expanded about 10-fold [7]. Recording cell division by killing mice at days 3.5, 4.5, 5.5, 6.5, and 7.5, after pulsing them with BrdU for their last 6 h, it was found that about 60% of the cells were BrdU+ at days 3.5 and 4.5 when the response started with 300 cells, and that these percentages decline to 40% and 23%, when the response started with 30000 cells (see Fig. 2d) [7]. Thus the fraction of dividing cells is smaller and declines earlier when responses start with more precursor cells, and such a difference in the expansion rates can account for the observed effect [7].
Figure 2.
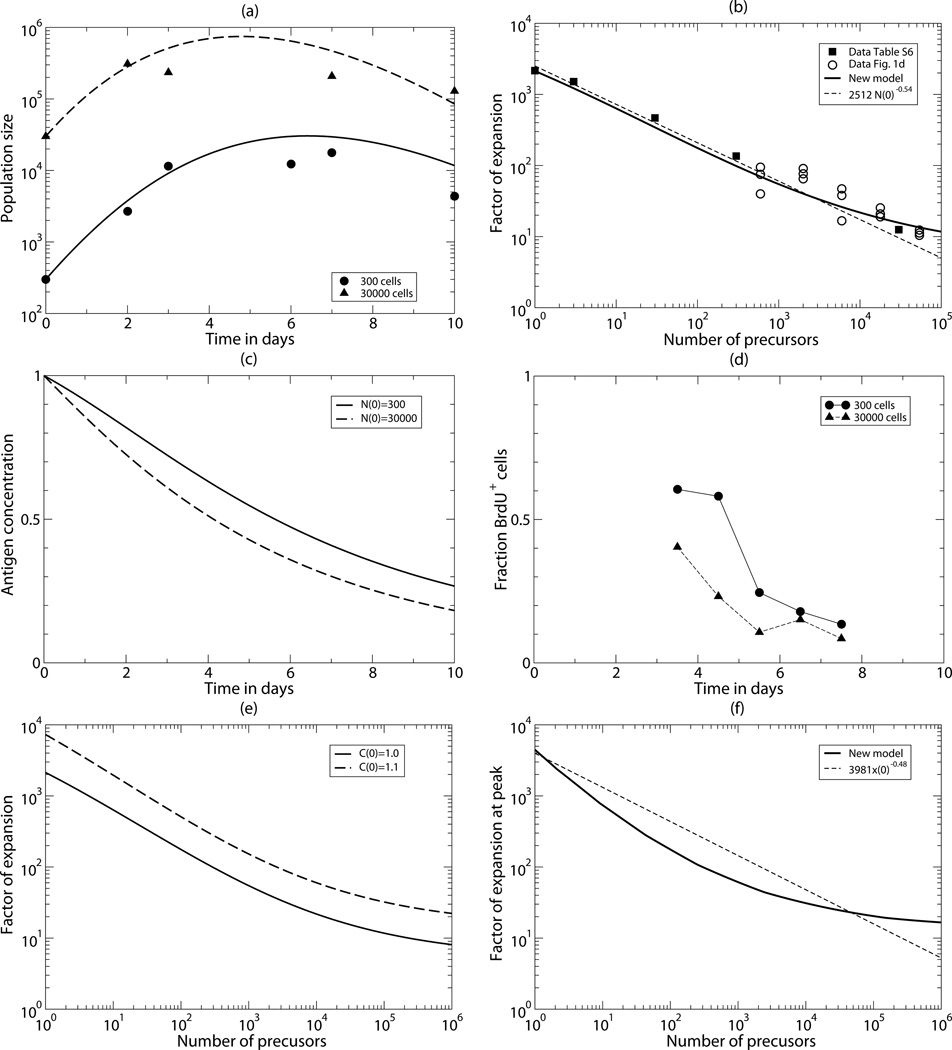
Comparing the data to the best fit of the new model. (a) The time course of the total number of T cells starting with N(0)=300 (bullets & solid line) or N(0)=30000 (triangles & long-dashed line) CD4+ 5C.C7 T cells. Because the time courses run fairly parallel until day 3, this is in good agreement with the data showing a similar fold expansion at day 3 [7]. (b) The fold expansion, N(7)/N(0)), as a function of the number of precursors, N(0). The heavy line is the prediction of the new model, and the short-dashed line is the regression line 2512×T(0)−0.54 fitted by Bocharov et al., [8]. (c) The pMHC concentration, C, as a measure of the fraction cells entering division. (d) The measured fraction of BrdU+ T cells after a short pulse of 6 hours, which should also be proportional to the fraction of cells entering division. The BrdU data was taken from Table S4 in Bocharov et al., [8]. (e) The effect of 10% increase in the initial antigen concentration. (f) The maximum clone size during the response predicted by the new model (solid line). The short-dashed line is the regression line 3981×N(0)−0.48 fitted to the prediction of the Bocharov et al., [8] model. Parameters were estimated by fitting Eq. (3) to the data in Panels (a) and (b). Their values and confidence intervals are given in Table 1.
The effect did not depend on regulatory T cells, because (1) there were <1% Foxp3+ among the 5C.C7 cells, and this percentage did not depend on the precursor density, (2) the percentage Foxp3+ cells among the endogenous CD4+ T cells was normal (i.e., about 10%), and did not depend on the precursor density, and (3) because similar fold expansions were observed in mice receiving OT-II cells from wild type donors, or from Scurfy donor mice that have no functional regulatory T cells [7]. Several cytokines, like IFN-γ, IL-2, IL-7, and IL-15, and various inhibitory molecules, like Fas, TNF-α receptor, and CTLA-4, had hardly any effect on the differences in the fold expansion. Importantly, the effect was antigen specific because adding large numbers of T cells of another specificity hardly affected the fold expansion [7]. Increasing the antigen dose, or increasing the density of CD11+ dendtritic cells, increased the fold expansion, but for all precursor densities in a similar manner, implying that the observed reduction of the fold expansion cannot be due to limiting antigen presentation at high precursor numbers only [7].
The data was interpreted by developing a mathematical model which allowed cognate T cells, after their priming by antigen, to differentiate from slowly dividing cells, to rapidly proliferating cells, to non-dividing cells, to non-divided mature cells [8]. The mature cells in this developmental cascade down-regulate the differentiation of slowly dividing cells into rapidly proliferating cells. Intuitively, one can understand that this allows for an initial expansion that remains fairly independent of the initial precursor frequency, because the regulatory mature cells only appear later. When they appear, they readily account for the observed earlier and increased down-regulation of the expansion at large population sizes (Fig. 1c & Fig. 2d). Combining the data and the modeling, it was suggested that the data are best explained by down-regulatory feedbacks from differentiated (memory) cells on the clonal expansion of less-differentiated cells [7, 8].
The mathematical model defined a cascade from proliferating to mature non-dividing cells, and consisted of two populations of dividing cells and two populations of mature cells, i.e., of four differential equations [8]. Due to the inclusion of several feedback loops, and proliferation functions allowing for competitive effects of cell density on the division rate, the model contained 15 parameters that were all estimated by fitting the model to the data. The model described the data well, and it was found that the fit remained of similar quality when one of the feedback loops was eliminated from the model. This simplified model had 10 parameters that were also estimated by fitting. The mere fact that the models describe the data well demonstrates that the interpretation in terms of down-regulatory effects from differentiated cells on less-differentiated dividing cells is a possible explanation. Because both models are complicated, and have a large number of parameters, the excellent agreement between model and data fails to prove however that this is the only interpretation of the data. The authors indeed acknowledge that they cannot exclude other explanations like competition between cognate cells for presented peptides [7].
One alternative explanation for the observed reduction in the fold expansion is an increased loss of peptide-MHC (pMHC) complexes on antigen presenting cells (APCs) at high densities of precursor cells. During cognate interactions with APCs, CD4+ T cells tend to acquire a variety of cell surface molecules, including the specific pMHC complexes binding the T cell receptors in the immunological synapse [9, 10, 11, 12, 13, 14]. This leads to a form of antigen specific competition between T cells binding the same pMHC on the same APCs [15, 10]. This effect would be perfectly consistent with the observations of Quiel et al., [7] because T cells of another specificity hardly affected the fold expansion. Their observations that increasing the antigen concentration, or the density of CD11+ dendritic cells, increased the fold expansion at all precursor densities in a similar manner [7], indeed suggest that pMHCs on APCs become a limiting resource at all precursor densities tested. We therefore investigate whether this striking data is also consistent with competition for presented peptides in a system where the pMHC are being grazed by other cells of the same specificity. Our approach is to develop a caricature model that only implements the essentials of clonal expansion and contraction, and the loss of pMHC by cognate T cells binding APCs. Since this simple model consisting of just five parameters describes the data equally well, we conclude that the data equally support this alternative explanation.
3 Model
To investigate whether or not the data presented in Fig. 1 can also be explained in terms of a model where T cells divide in response to pMHC, and where the availability of pMHC declines as a function of the number of T cells that form conjugates with the APCs, we developed a simple model describing the number of 5C.C7 CD4+ T cells in a mouse, N, and the concentration of pMHC, C, on an average APC. The concentration of pMHC is scaled by its maximal value, i.e., upon immunization with cytochrome c, we set C(0)=1, and let it decline afterwards. pMHC decline by normal turnover, and are consumed by the 5C.C7 CD4+ T cells. Because APCs can only bind a limited number of T cells at one time, the rate at which T cells consume pMHC should saturate at large T cell densities, and we model this by a Hill function with saturation constant h and shape parameter n. We model proliferation by assuming that individual T cells divide at a rate proportional to their cognate pMHC concentration.
The pMHC concentration depends on the de novo generation of peptides from the depot of cytochrome c that was injected at day zero, and which we assume declines exponentially at rate kc. For the average concentration of pMHC on an APC we write
| (1) |
where P is rate at which pMHC are produced from cytochrome c, kd is the dissociation rate of peptides from MHC molecules, and kg is the maximum rate at which T cells graze pMHC. The dissociation rate, kd, typically ranges between several hours to more than 10 hours [16, 17, 18, 19]. Since we have no information on the concentration of cytochrome c over time, nor on the rate at which pMHC are produced from the full protein, we scale the pMHC concentration by its maximum concentration P/kd, so that C is at most 1. In terms of this scaled concentration, we obtain in the absence of T cells dC/dt=kde−kct−kdC, which for C(0)=1 obeys the solution
| (2) |
which defines a bi-phasic decline from the initial value C(0)=1 to zero. The addition of T cell grazing can only increase the rate at which C(t) declines, and we indeed obtain that 0<C(t)≤1. This has the immediate advantage that we can define the maximum proliferation rate of the 5C.C7 CD4+ T cells as a proportionality constant, p, reflecting the initial division rate of a 5C.C7 CD4+ T cell (in days). The full model can be written as the following two differential equations:
| (3) |
where N is the total number of cognate CD4+ T cells in a mouse, C is the scaled concentration of pMHC on an APC, p and d are proliferation and death rates, respectively, and kc, kd and are loss rates of pMHC (all rates are per day; see Table 1). We initialize the model with varying numbers of precursors cells by setting N(0) to various values between 1 and 106cells per mouse. A standard dose of cytochrome c corresponds to an initial pMHC concentration of C(0)=1, and an increased dose of antigen is modeled by setting C(0)=1.1.
Table 1.
Parameter values and their 95% confidence intervals.
| Name | Interpretation | Dimension | Value | Range |
|---|---|---|---|---|
| p | maximum proliferation rate | per day | 2.71 | 2.12—11.2 |
| d | death rate | per day | 1.21 | 0.50—9.92 |
| h | saturation constant | cells | 1307 | 309—4.5×105 |
| kg | maximum grazing rate | per day | 0.20 | 0.022—0.604 |
| n | slope (Hill coeffcient) | — | 0.31 | 0.15—0.62 |
In the Results section we will show that Eq. (3) describes the data well even if we set the parameter, i.e., when the loss of pMHC concentration from its initial value C(0)=1 is completely due to the grazing by the T cells. This means that at high T cell densities (i.e., whenever N>>h) we obtain that dC/dt~−kgC. This yields an exponential decay of pMHC, C(t)=C(0)e−kgt. Since the peak of the T cell response occurs when pC=d, we can solve for the time of the peak from d/p=e−kgt (when C(0)=1). Thus, large T cell populations are predicted to start with an expansion phase and peak at day t=ln[p/d]/kg. The number of T cells at the peak, Nmax, can then be obtained by substituting the time of the peak into the full solution of dN/dt, i.e.,
| (4) |
The model was simulated using the variable time step Runge-Kutta integrator provided by Press et al., [20], and was fitted to the data using the Levenberg-Marquardt algorithm [21]. Confidence intervals were obtained by bootstrapping the residuals 50 times [22].
4 Results
The model, Eq. (3), has seven parameters, and was fitted to the data from the two published time courses, starting with either 300 or 30000 T cells (Fig. 2a), and to the two published data sets on the measured fold-expansion at day 7 for various different precursor densities (Fig. 2b). The new model describes the data well. One salient feature of the data was that for both initial conditions the population size expanded 10-fold during the first two days of the response [7]. Comparing the corresponding initial expansions in Fig. 2a reveals that the new model capture this very well Fitting the data, it turned out that most of the net loss of pMHC was estimated to be due to the uptake by T cells, and that the quality of the fit was hardly reduced when the normal production and turnover of pMHC was ignored by setting (see Eq. (3)). Conversely, the model fails to describe the data if we do not allow for consumption of pMHC by T cells, by setting kg =0. Even the five remaining parameters have large 95% confidence limits because a high maximum rate, p, of T cell proliferation can be compensated by a high saturation constant, h, and/or a high death rate d (see Table 1). The best estimate of the proliferation rate of p=2.7 per day is a reasonable value, however, as similar values are obtained when CD4+ T cells proliferate vigorously during an acute LCMV infection [23]. The fact that Eq. (3) with only five parameters describes the data well (see Fig. 2a & b), demonstrates that the loss of pMHC by the uptake of cognate T cells is a sufficient explanation for the striking observations made by Quiel et al., [7].
To test whether our alternative explanation is also consistent with the other data in the original publication [7] we compare the activation rate of the T cells in our model with the BrdU data taken at various time points in the mice (Fig. 2d). Since the fraction of cells entering division is proportional to the concentration of pMHC, we depict in Fig. 2c the predicted time course of the pMHC decay after starting with 300 or 30000 precursor cells. Although the actual values of the pMHC concentration in the model (Fig. 2c) cannot be compared directly with the fraction of cells becoming labeled during a 6 h pulse with BrdU (Fig. 2d), the model and the data are in general agreement as the predicted division rate pC is somewhat lower and declines earlier when the immune response starts with 30000 cells, as is seen in the data (Fig. 2d).
The model also correctly predicts that the fold expansion increases similarly for all precursor densities if the antigen concentration is increased. Increasing the initial pMHC concentration by 10% we find the same increase in the fold expansion for a wide range of precursor densities (Fig. 2e), such that the dependence of the fold expansion on the initial density is not affected. Like Bocharov et al., [8], we can use the model to predict the fold expansion at the peak of the response, rather than that at day 7 (which is somewhere in the contraction phase); see Fig. 2f. However, since there is very little data on the maximum size of the response, these two predictions cannot be compared with data. An interesting difference between the two models is that at very large precursor densities the regression model of Bocharov et al., [8] predicts that the maximum clone size will be smaller than the number of precursor cells, whereas Eq. (3) predicts a 13-fold expansion at the peak of the response when the precursor density is very large (Fig. 2f). The fold expansion for large precursor densities can be solved analytically (see Eq. (4)), and for the estimated parameters, the peak corresponds to a 13-fold expansion occurring around day 4. Close inspection of the fit of the regression line of Bocharov et al., [8] to the predicted data (their Fig. 2D) indeed reveals a deviation at large precursor numbers, and solving their simplified mechanistic model numerically confirmed that their mechanism also predicts that there should always be an initial expansion phase.
5 Discussion
A simple model where the loss of pMHC complexes increases with the density of cognate CD4+ T cells is capable of explaining the observed 0.5 log reduction in the fold expansion of T cells with every log increase in the precursor density over more than 4 orders of magnitude [7, 8]. Since T cells are known to specifically pick up cognate pMHC from APC [9, 11, 12, 13, 14], we provide a simple and realistic interpretation of the data. Hence, our main result is that consumption of pMHC by cognate T cells is a natural alternative interpretation of the data.
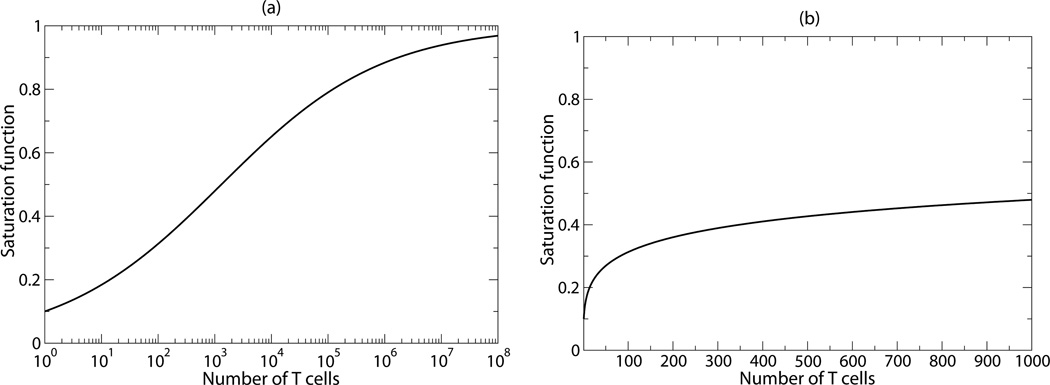
The saturation function that describes the net loss rate of pMHC as a function of the number of grazing T cells estimated by the fitting procedure is depicted in Fig. 3. Unfortunately, this function remains phenomenological, and the exponent of the Hill function, n=0.31, has no mechanistic underpinning. Since we have been unable to estimate the production rate of pMHC and the normal dissociation rate of peptides from the data, the quantitative interpretation of the rate at which T cells graze pMHC also remains uncertain. Additionally, one could make the model more realistic by including an initial time lag between the onset of CD4+ T cell proliferation, and/or explicitly account for the uptake of cytochrome c by antigen processing cells, its fragmentation into short peptides, the transportation of peptide loaded MHC molecules to the cell surface, and the full activation of the precursor cells. Finally, it is likely that these CD4+ T cells complete several divisions following their initial activation, and only need to see pMHC again later in their clonal expansion phase [24, 25, 26, 27, 28]. Such a partially programmed response would make it easier to explain that the initial expansion hardly depends on the precursor density. Given that estimating just five parameters from the current data already gives rise to large confidence intervals (see Table 1) illustrates that the data do not allow us to reliably fit more complicated models to the data. It would be interesting to extend our model with a more mechanistic description of the increased loss of pMHC generated by cognate T cells, if data on the loss rate of pMHC in the system becomes available.
Figure 3.
The saturation function fitted by the model on a logarithmic (a) and a linear scale (b). The panels depict with parameters h=1307 cells and n=0.31
Since the two very different models behave so similarly, we need new experimental data to establish which of the interpretations of the data is correct. Previously, several authors have suggested that T cells compete for access to pMHC on APCs [29, 30, 31, 17, 32, 33, 34, 4] and that T cells pick up pMHC from APCs [9, 11, 12, 13, 6, 14]. However, to test which mechanism is responsible for the reduction in T cell expansion over more than 4 orders of magnitude in precursor densities, it would be best to directly test the proposed mechanisms. This seems possible for the new interpretation involving the increased loss of cognate pMHC on APC at high densities of T cells, since pMHC densities can in principle be measured directly ex vivo by activating cognate T cells with APC taken out at various time points [35, 36]. Directly testing the maturation and feedback model is more difficult because which T cell subsets are responsible for the negative feedback is not known [8], nor are the molecular interactions underlying the proposed feedback.
Acknowledgments
Most of this paper was written at the Santa Fe Institute and it was finished at the KITP at UCSB.
Footnotes
Portions of this work were done under the auspices of the U. S. Department of Energy under contract DE-AC52-06NA25396 and supported by NIH grants AI028433, OD011095, P01-AI071195, and P20-GM103452, and contract HHSN272201000055C (ASP). This research was supported in part by the National Science Foundation under Grant No. NSF PHY11-25915.
References
- 1.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 2.Kemp RA, Powell TJ, Dwyer DW, Dutton RW. Cutting edge: regulation of CD8+ T cell effector population size. J. Immunol. 2004 Sep;173:2923–2927. doi: 10.4049/jimmunol.173.5.2923. [DOI] [PubMed] [Google Scholar]
- 3.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006 Apr;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 4.Foulds KE, Shen H. Clonal competition inhibits the proliferation and differentiation of adoptively transferred TCR transgenic CD4 T cells in response to infection. J. Immunol. 2006 Mar;176:3037–3043. doi: 10.4049/jimmunol.176.5.3037. [DOI] [PubMed] [Google Scholar]
- 5.Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8+ T cell response to infection. Immunity. 2007 Jun;26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helft J, Jacquet A, Joncker NT, Grandjean I, Dorothee G, Kissenpfennig A, Malissen B, Matzinger P, Lantz O. Antigen-specific T-T interactions regulate CD4 T-cell expansion. Blood. 2008 Aug;112:1249–1258. doi: 10.1182/blood-2007-09-114389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quiel J, Caucheteux S, Laurence A, Singh NJ, Bocharov G, Ben-Sasson SZ, Grossman Z, Paul WE. Antigen-stimulated CD4 T-cell expansion is inversely and log-linearly related to precursor number. Proc. Natl. Acad. Sci. U.S.A. 2011 Feb;108:3312–3317. doi: 10.1073/pnas.1018525108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bocharov G, Quiel J, Luzyanina T, Alon H, Chiglintsev E, Chereshnev V, Meier-Schellersheim M, Paul WE, Grossman Z. Feedback regulation of proliferation vs. differentiation rates explains the dependence of CD4 T-cell expansion on precursor number. Proc. Natl. Acad. Sci. U.S.A. 2011 Feb;108:3318–3323. doi: 10.1073/pnas.1019706108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang JF, Yang Y, Sepulveda H, Shi W, Hwang I, Peterson PA, Jackson MR, Sprent J, Cai Z. TCR-mediated internalization of peptide-MHC complexes acquired by T cells. Science. 1999 Oct;286:952–954. doi: 10.1126/science.286.5441.952. [DOI] [PubMed] [Google Scholar]
- 10.Kedl RM, Schaefer BC, Kappler JW, Marrack P. T cells down-modulate peptide-MHC complexes on APCs in vivo. Nat. Immunol. 2002 Jan;3:27–32. doi: 10.1038/ni742. [DOI] [PubMed] [Google Scholar]
- 11.Wetzel SA, McKeithan TW, Parker DC. Peptide-specific intercellular transfer of MHC class II to CD4+ T cells directly from the immunological synapse upon cellular dissociation. J. Immunol. 2005 Jan;174:80–89. doi: 10.4049/jimmunol.174.1.80. [DOI] [PubMed] [Google Scholar]
- 12.Wetzel SA, Parker DC. MHC transfer from APC to T cells following antigen recognition. Crit. Rev. Immunol. 2006;26:1–21. doi: 10.1615/critrevimmunol.v26.i1.10. [DOI] [PubMed] [Google Scholar]
- 13.Sprent J. Swapping molecules during cell-cell interactions. Sci. STKE. 2005 Mar;2005:pe8. doi: 10.1126/stke.2732005pe8. [DOI] [PubMed] [Google Scholar]
- 14.Hwang I, Ki D. Receptor-mediated T cell absorption of antigen presenting cell-derived molecules. Front. Biosci. 2011;16:411–421. doi: 10.2741/3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willis RA, Kappler JW, Marrack PC. CD8 T cell competition for dendritic cells in vivo is an early event in activation. Proc. Natl. Acad. Sci. U.S.A. 2006 Aug;103:12063–12068. doi: 10.1073/pnas.0605130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 17.Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999 Apr;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 18.Lazarski CA, Chaves FA, Jenks SA, Wu S, Richards KA, Weaver JM, Sant AJ. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005 Jul;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Harndahl M, Rasmussen M, Roder G, Dalgaard Pedersen I, Sorensen M, Nielsen M, Buus S. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur. J. Immunol. 2012 Jun;42:1405–1416. doi: 10.1002/eji.201141774. [DOI] [PubMed] [Google Scholar]
- 20.Press WH, Plannery BP, Teukolsky SA, Vetterling WT. Numerical Recipes in C. The Art of Scientific Computing. Cambridge: Cambridge U.P; 1988. [Google Scholar]
- 21.Marquardt DW. Finite difference algorithm for curve fitting. J. Soc. Ind. Appl. Math. 1963;11:431–441. [Google Scholar]
- 22.Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1986;1:54–77. [Google Scholar]
- 23.De Boer RJ, Homann D, Perelson AS. Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J. Immunol. 2003;171:3928–3935. doi: 10.4049/jimmunol.171.8.3928. [DOI] [PubMed] [Google Scholar]
- 24.Bajenoff M, Wurtz O, Guerder S. Repeated antigen exposure is necessary for the differentiation, but not the initial proliferation, of naive CD4+ T cells. J. Immunol. 2002;168:1723–1729. doi: 10.4049/jimmunol.168.4.1723. [DOI] [PubMed] [Google Scholar]
- 25.Lee WT, Pasos G, Cecchini L, Mittler JN. Continued antigen stimulation is not required during CD4+ T cell clonal expansion. J. Immunol. 2002;168:1682–1689. doi: 10.4049/jimmunol.168.4.1682. [DOI] [PubMed] [Google Scholar]
- 26.Foulds KE, Zenewicz LA, Shedlock DJ, Jiang J, Troy AE, Shen H. Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J. Immunol. 2002;168:1528–1532. doi: 10.4049/jimmunol.168.4.1528. [DOI] [PubMed] [Google Scholar]
- 27.Yarke CA, Dalheimer SL, Zhang N, Catron DM, Jenkins MK, Mueller DL. Proliferating CD4+ T cells undergo immediate growth arrest upon cessation of TCR signaling in vivo. J. Immunol. 2008 Jan;180:156–162. doi: 10.4049/jimmunol.180.1.156. [DOI] [PubMed] [Google Scholar]
- 28.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009 Mar;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- 30.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J. Exp. Med. 1999;189:701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borghans JAM, Taams LS, Wauben MH, De Boer RJ. Competition for antigenic sites during T cell proliferation: a mathematical interpretation of in vitro data. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10782–10787. doi: 10.1073/pnas.96.19.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith AL, Wikstrom ME, Fazekas de St Groth B. Visualizing T cell competition for peptide/MHC complexes: a specific mechanism to minimize the effect of precursor frequency. Immunity. 2000 Dec;13:783–794. doi: 10.1016/s1074-7613(00)00076-5. [DOI] [PubMed] [Google Scholar]
- 34.Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, Marrack P. T cells compete for access to antigen-bearing antigen-presenting cells. J. Exp. Med. 2000 Oct;192:1105–1113. doi: 10.1084/jem.192.8.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8+ T cells after infection. Nat. Immunol. 2002;3:619–626. doi: 10.1038/ni804. [DOI] [PubMed] [Google Scholar]
- 36.Badovinac VP, Porter BB, Harty JT. CD8+ T cell contraction is controlled by early inflammation. Nat. Immunol. 2004 Aug;5:809–817. doi: 10.1038/ni1098. [DOI] [PubMed] [Google Scholar]