Abstract
Heterochromatin causes epigenetic repression that can be transmitted through multiple cell divisions. However, the mechanisms underlying silencing and stability of heterochromatin are not fully understood. We show that heterochromatin differs from euchromatin in histone turnover, and identify histone deacetylase (HDAC) Clr3 as a factor required for inhibiting histone turnover across heterochromatin domains in Schizosaccharomyces pombe. Loss of RNAi factors, Clr4 methyltransferase, or HP1 proteins involved in HDAC localization causes increased histone turnover across pericentromeric domains. Clr3 also affects histone turnover at the silent mating–type region where it can be recruited by alternative mechanisms acting in parallel to H3K9me–HP1. Importantly, the JmjC–domain protein Epe1 promotes histone exchange, and loss of Epe1 suppresses both histone turnover and defects in heterochromatic silencing. Our results suggest that heterochromatic silencing factors preclude histone turnover to promote silencing and inheritance of repressive chromatin.
In eukaryotic cells, genomic DNA is folded with histone and non–histone proteins to form discrete structural and functional chromatin domains1. The genome is organized into euchromatin and heterochromatin domains by mechanisms that involve posttranslational modifications of histones and remodeling of nucleosomes2-4. Whereas heterochromatin is typically condensed and generally inhibitory to transcriptional machinery, euchromatin is less condensed and more readily transcribed2. Hypoacetylation of histones is one hallmark that distinguishes heterochromatin from euchromatin, and is believed to be a critical feature required for the assembly of repressive chromatin2,4,5. Moreover, histone H3 lysine 9 (H3K9) is specifically methylated in heterochromatic regions of the genome2,3. H3K9me serves to recruit conserved HP1 protein family members6-8. Heterochromatin plays an important role in the regulation of gene expression, and also protects genome integrity by inhibiting unwanted recombination between repetitive DNA elements and facilitating proper segregation of chromosomes2.
Studies using the fission yeast Schizosaccharomyces pombe have contributed greatly to our understanding of heterochromatin assembly and its biological significance2. While H3K9me and associated factors can be detected at several sites across the S. pombe genome9, heterochromatin is preferentially enriched across large domains at centromeres, subtelomeres and the silent mating–type locus10. At centromeres, pericentromeric regions containing tandem and inverted arrays of the dg and dh repeats are assembled into heterochromatin10,11. Similarly, heterochromatin coats extended domains at subtelomeres, as well as a 20–kb domain at the mating–type region that includes the silent mating–type cassettes mat2 and mat3, and a cenH element that shares strong homology with dg and dh repeats10,12.
The dg and dh repeats and their homologous sequences are transcribed by RNA polymerase II10,13,14, producing transcripts that are processed into siRNAs by RNAi factors, including Argonaute (Ago1), Dicer (Dcr1) and RNA–dependent RNA polymerase (Rdp1)15. The processing of repeat–derived transcripts by RNAi is coupled to the loading of heterochromatin proteins. siRNAs are bound by Ago1, a subunit of the RNA–induced transcriptional silencing (RITS) complex that is composed of 2 additional proteins: Chp1 and Tas3 (ref. 16). siRNA–bound Ago1 assists in the localization of RITS to chromatin, which in turn is believed to target Clr4, a homolog of mammalian SUV39h that methylates H3K9 to nucleate heterochromatin17,18. Additional RNAi–independent mechanisms nucleate heterochromatin at the mat locus and at centromeres19,20. Once nucleated, heterochromatin spreads across domains defined by boundary DNA elements21-23.
H3K9me facilitates the localization of chromodomain proteins required for diverse heterochromatin functions. Binding of the RITS subunit Chp1 to H3K9me is critical for the processing of heterochromatic repeat transcripts22,24. H3K9me also mediates recruitment of HP1 family proteins Chp2 and Swi6, which in turn associate with factors involved in transcriptional gene silencing (TGS), such as Snf2–HDAC Repressor Complex (SHREC) containing Clr3 HDAC, Asf1–HIRA histone chaperone and Clr6 HDAC complexes25-30. The anti–silencing factor Epe1 also associates with Swi6 and facilitates transcription of heterochromatic repeats31-33. While heterochromatin precludes RNAPII accessibility at target loci, Epe1 counteracts these effects to allow RNAPII transcription of centromeric repeats that is required to generate siRNA precursors. The exact mechanisms by which TGS effectors and Epe1 modulate heterochromatin are not fully understood.
The accessibility of DNA in eukaryotic genomes is largely determined by nucleosome stability34,35. In euchromatin, histone exchange at regulatory elements provides access to factors involved in transcription and other chromosomal processes36-39. However, heterochromatic sequences are generally inaccessible to trans–acting factors. TGS effectors such as SHREC and Asf1–HIRA have been shown to influence nucleosome occupancy at discrete sites within heterochromatin domains including pericentromeric regions and the silent mating–type region27,30,40. However, these localized changes in nucleosome occupancy that are restricted to specific sites cannot fully explain changes across extended domains that impact global expression patterns. A previous study reported that Clr4 and Swi6 impact histone dynamics at heterochromatic loci41. However, the relationship between nucleosome turnover and the epigenetic stability of heterochromatin was not explored. Moreover, whether heterochromatin–associated TGS effectors such as SHREC, which localize throughout heterochromatin domains, can preclude turnover of nucleosomes to assemble repressive chromatin domains was not examined.
To gain insight into the mechanisms by which heterochromatin factors mediate the assembly and propagation of repressive chromatin, we investigated the relationship between epigenetic stability of heterochromatin and replication–independent nucleosome turnover. Our detailed analyses using chromatin immunoprecipitation combined with microarray (ChIP–chip) show that heterochromatic silencing machinery prevents histone exchange across large chromosomal domains both at centromeres and the silent mating-type region. Clr3 HDAC, a component of a TGS effector, which is targeted to heterochromatic loci by H3K9me–HP1 or alternative recruitment mechanisms, is required to inhibit histone turnover. We also demonstrate that Epe1 counteracts heterochromatic silencing by promoting nucleosome turnover. These results suggest that histone deacetylation, which is one of the most conserved features of silenced chromatin domains from yeast to humans, promotes heterochromatin stability by inhibiting turnover of histones.
RESULTS
Hetero– and eu–chromatin domains differ in histone turnover
Nucleosome stability affects the accessibility of underlying DNA sequences34. We therefore wondered whether heterochromatin and euchromatin differ in the dynamic properties of nucleosomes within these domains. To measure histone turnover, we used S. pombe cells expressing carboxy–terminal FLAG–tagged histone H3 (H3–FLAG) under the control of the inv1 promoter, which can be rapidly induced by shifting the growth medium carbon source from glucose to sucrose42 (Fig. 1a). H3–FLAG expression was induced after blocking DNA replication by hydroxyurea (HU) (Fig. 1b,c). We found that the tagged histone H3 was incorporated into chromatin, as indicated by methylation of tagged H3 at lysine 4 (Supplementary Fig. 1b). Cross–linked chromatin was digested to mononucleosomes by micrococcal nuclease (MNase), followed by immunoprecipitation of FLAG–tagged H3. DNA isolated from immunoprecipitated chromatin was subjected to microarray analyses using a custom tiling array that covers heterochromatic regions at 10 base pair resolution and includes pericentromeric repeats, the silent mating–type region and ~225–kilobases of a euchromatic region from chromosome 2. Consistent with results from budding yeast and Drosophila36-39, we observed widespread exchange of histones in euchromatic regions (Fig. 1d,e). In particular, histone exchange at gene promoters was more prominent, whereas we measured relatively modest levels of H3 replacement across the body of genes (Supplementary Fig. 1a). Therefore, the low nucleosome occupancy at S. pombe promoters43 might reflect the intrinsic dynamic behavior of histones at these sites.
Figure 1. Differential patterns of histone turnover define heterochromatin and euchromatin domains.

(a) Schematic depiction of the experimental design used to investigate histone H3 replacement. (b) Western blot (WB) analysis of the induction of H3–FLAG fusion protein expression upon a switch in carbon source from glucose (glu) to sucrose (suc) for 2h. Ponceau S staining is shown as loading control. (c) FACS analysis of DNA replication arrest and synchronization of cells. N and 2N refer to DNA content before and after DNA replication, respectively. (d) Distribution of H3 replacement across a pericentromeric domain and the adjacent euchromatic region. H3 replacement values, as measured by MNase–ChIP–chip (ChIP vs Input), are plotted in alignment with the map of the right pericentromeric region of cen2 and the neighboring euchromatic region (top). Heterochromatin domain containing the inner centromeric repeat (imr2R) as well as the dg and dh repeat elements is indicated. Vertical lines indicate tRNAs. Nucleosome occupancy (measured by MNase hypersensitivity30) is shown for comparison (bottom). (e) Distribution of H3 replacement and nucleosome occupancy across the mating type region illustrated shown as in d. Arrows mark the inverted repeats IR–L and IR–R that act as boundary elements. The dotted lines and gap between mat2P and mat3M represent the probes that correspond to the cenH region, which are omitted due to cross–hybridization with centromeric repeats. Since non–switching mat1M–Smt0 cells were used to perform experiments, signal mapping to mat3M may represent cross hybridization to the mat1M locus.
In contrast to euchromatin, the pericentromeric and silent mating–type regions are enriched in heterochromatin10 and showed lower histone H3 replacement (Fig. 1d,e). These heterochromatic domains are largely occupied by nucleosomes, as indicated by relatively few MNase hypersensitive sites as compared with euchromatic regions (Fig. 1d,e). We also detected H3 turnover at tRNA clusters and inverted repeat (IR) heterochromatin boundaries at centromeres and the mat locus, respectively (Fig. 1d,e). The histone exchange at these sites might directly delimit the spread of heterochromatin, as suggested in other systems36,39, or help to expose binding sites for trans–acting factors. Indeed, tRNAs and IRs, which are refractory to heterochromatin30, are bound by the transcription factor TFIIIC23.
RNAi and Clr4 regulate histone exchange at centromeres
The marked differences in histone replacement across heterochromatin domains compared with euchromatic regions prompted us to ask whether heterochromatin machinery prevents nucleosome turnover. As a first step, we investigated the effects of loss of Dcr1 and Clr4, which causes severe defects in heterochromatin assembly at centromeres6,10,15. Deletion of either dcr1 or clr4 caused an increase in histone H3 turnover across pericentromeric regions (Fig. 2a). The increase in histone turnover in mutants as compared to wild type was not due to variation between microarrays, and could be reproduced in biological replicates. To quantitate these differences, we compared normalized H3 turnover values in each mutant to wild type by calculating the fold enrichment values across pericentromeric heterochromatin relative to a euchromatic region (Supplementary Fig. 2). These analyses revealed a more than 2–fold increase in histone turnover in dcr1 or clr4 mutants as compared to wild type. The observed changes extended beyond the dg and dh repeats and include the entire heterochromatin coated domain (Fig. 2a). Importantly, the elevated levels of histone exchange throughout pericentromeric regions in clr4Δ cells could be detected readily within 15 minutes after H3–FLAG induction (Supplementary Fig. 3b). Since H3–FLAG is barely detectable at this time point (Supplementary Fig. 3a), the increased H3 exchange in heterochromatin–defective mutants is not likely due to H3–FLAG overexpression. The observed differences in histone turnover in wild–type and mutant cells were not attributable to gross changes in histone occupancy in HU treated cells, as revealed by ChIP–chip analyses of endogenous H3 distributed across pericentromeric regions (Supplementary Fig. 3c). Moreover, the normalization of H3 turnover to total H3 measured under conditions used to induce H3–FLAG expression revealed that the increased histone exchange in dcr1Δ and clr4Δ is not due to an increased H3 occupancy (Supplementary Fig. 4a,b). As expected, neither dcr1Δ nor clr4Δ showed a major increase in H3 replacement at euchromatic regions (Supplementary Fig. 5).
Figure 2. Clr4 and RNAi are required to suppress H3 replacement at centromeres but not at the silent mating–type locus.

(a) H3 replacement across the right pericentromeric region of cen2 in clr4Δ, dcr1Δ or wild–type (WT) cells was measured by MNase–ChIP–chip method as described in Fig 1. (b) H3 replacement across the silent mating–type region in clr4Δ or dcr1Δ, measured and shown as in a.
Since S. pombe cells spend the majority of their vegetative life cycle in the G2 phase of the cell cycle, we wondered if the impact of clr4Δ on H3 exchange could be recapitulated in G2–arrested cells. Loss of Clr4 also resulted in a marked increase in H3 turnover when H3–FLAG was expressed in cells blocked at the G2–M boundary (Supplementary Fig. 6). Together, these results suggest involvement of heterochromatin assembly factors in suppression of histone replacement across pericentromeric domains.
Heterochromatin and histone exchange at the mat locus
Heterochromatin at the mat region is nucleated by redundant mechanisms that, in addition to RNAi, involve sequence–specific DNA binding factors19,22. We asked whether RNAi plays a role in the control of histone replacement across the silent mating–type region. Consistent with previous results showing that RNAi is dispensable for the maintenance of heterochromatin at the mat locus21, the loss of Dcr1 had little or no effect on nucleosome replacement at this region (Fig. 2b).
We next investigated the effect of loss of Clr4, which is essential for H3K9me across the silent mating–type region. In contrast to a previous observation that Clr4 affects histone replacement across the entire silent mating–type region41, our analyses did not detect domain–wide increase in nucleosome turnover across this region in clr4Δ cells (Fig. 2b). This may be due to differences in the resolution of the techniques used in the 2 studies. While previous work compared the levels of histone deposition at heterochromatic regions to a reference euchromatic region using conventional ChIP, we measured H3 turnover using high–resolution tiling microarrays. Notably, loss of Clr4 also did not cause a major increase in H3 exchange in G2–arrested cells at the mat region (Supplementary Fig. 6c). These observations are consistent with the finding that loss of Clr4 alone has only a minor effect on the silencing of endogenous mat2 and mat3 loci44, but is required for spreading of repressive chromatin and silencing of reporter genes artificially inserted in this region17,22.
HP1 proteins cooperate to control histone exchange
HP1 family proteins Chp2 and Swi6, which bind methylated H3K9, are enriched across heterochromatin domains and perform overlapping functions in transcriptional gene silencing29. Whereas the chp2Δ or swi6Δ single mutants modestly affect the expression level of target loci, the chp2Δswi6Δ double mutant causes a severe loss of heterochromatic silencing at centromeres29. To explore whether HP1 proteins play a role in preventing histone turnover, we analyzed histone H3 exchange in the single and double HP1 mutant cells (Fig. 3). The loss of Chp2 or Swi6 alone slightly increased histone replacement at pericentromeric regions (Fig. 3a). However, the double mutant that lacks both of these factors showed more than 2–fold increase in H3 replacement, comparable with that observed in clr4Δ or dcr1Δ cells (Supplementary Fig. 2 and Fig. 3a). In contrast, loss of either or both of the HP1 proteins only modestly altered H3 replacement at the silent mating–type region (Fig. 3b). Together, these observations suggest that HP1 proteins mediate the downstream effects of the Clr4 methyltransferase to suppress histone exchange at pericentromeric regions. However, like Clr4, loss of these proteins has little effect at the mat locus, raising the possibility that factor(s) working independently of the H3K9me–HP1 pathway compensate to preclude histone turnover at this domain.
Figure 3. HP1 proteins Chp2 and Swi6 cooperate to prevent nucleosome turnover across a pericentromeric loci.

(a) Histone H3 replacement in swi6Δ chp2Δ, chp2Δ, swi6Δ, or wild–type (WT) cells was measured by MNase–ChIP–chip method as described in Fig.1 and plotted in alignment with the map of the right pericentromeric region of cen2. (b) Histone H3 turnover across the silent mating–type region. H3 replacement was measured and shown as in a.
Clr3 HDAC is required for suppression of histone turnover
HP1 proteins might directly suppress histone exchange at heterochromatic loci, but H3K9me–HP1 also serves as a platform for the recruitment as well as spreading of effectors involved in transcriptional silencing. In light of the observations that Clr4 and HP1 proteins have modest effects on histone turnover at the silent mating–type region (Figs. 2b and 3b), we wondered whether TGS effectors targeted by HP1–dependent or –independent mechanisms are critical for preventing histone turnover. Indeed, while Clr4 and HP1 are essential for localization of SHREC across pericentromeric regions27,29, the components of SHREC, including Clr3 HDAC, are targeted to the silent mat region by additional recruitment mechanisms26,45. We therefore tested whether Clr3 is involved in precluding histone turnover, which may also explain the differential effects of Clr4 and HP1 on nucleosome replacement at centromeres and the mat locus. Interestingly, clr3Δ cells showed a domain–wide increase in histone turnover both at the silent–mating type region and the pericentromeric domains (Fig. 4a,b). The increase in H3 turnover correlated with defective silencing (Supplementary Fig. 7)26,27, and supports a functional connection between histone replacement and heterochromatic silencing.
Figure 4. Clr3 HDAC is required for suppression of histone H3 exchange across heterochromatin domains.
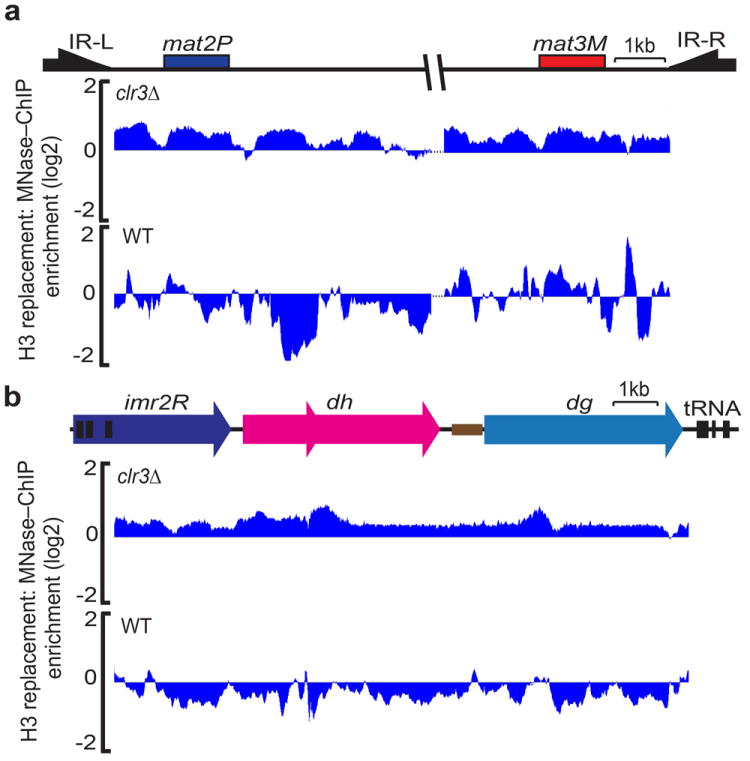
Histone H3 replacement was measured across the silent mating type region (a) and pericentromeric heterochromatin (b) in clr3Δ or wild–type (WT) cells as described in Fig.1.
Heterochromatin and co–transcriptional histone exchange
Given the potential impact of RNAPII transcription on histone replacement46-48, we considered that the increase in H3 turnover in the clr3Δ mutant might be linked to elevated levels of RNAPII transcription of heterochromatic sequences. However, we found that high levels of histone H3 turnover within the silent mating–type region were observed in clr3Δ even at sites that show no detectable increase in RNAPII transcription (Fig. 5a,b). This result suggests that changes in H3 replacement across the heterochromatin domain cannot solely be explained by transcription–coupled turnover of nucleosomes.
Figure 5. Increased H3 turnover in heterochromatin mutants is not solely due to changes in RNAPII transcription.
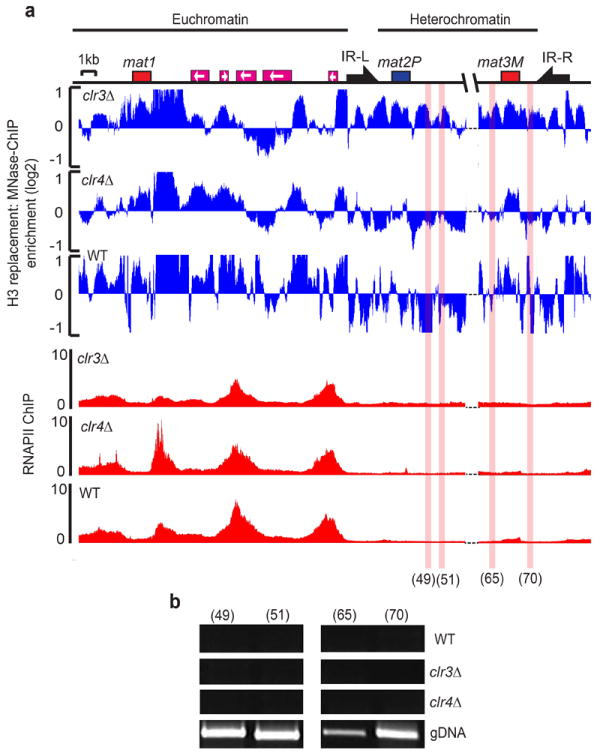
(a) Histone H3 replacement (blue) was measured across the mating type locus using MNase–ChIP–on–Chip analysis as described in Fig 1. RNAPII occupancy (ChIP vs Input) was measured by ChIP–on–Chip in clr3Δ, clr4Δ or wild–type (WT) cells and plotted in alignment with the map (red). (b) RT–PCR analysis performed using total RNA samples isolated from clr3Δ, clr4Δ or wild–type (WT) cells. Genomic DNA (gDNA) was used as a control. The locations amplified by primer pairs 49, 51, 65 and 70 are highlighted with red shading (see online methods for primer references). Heterochromatin and euchromatin portions of the mating–type region are indicated at the top.
Epe1 stimulates histone turnover across heterochromatin
As mentioned previously, the JmjC domain containing anti–silencing factor Epe1 binds Swi6 and counteracts heterochromatic silencing by Clr3 (refs. 31-33,49). Because JmjC proteins have been shown to demethylate histones49, Epe1 could function as a histone demethylase. However, no such activity has been detected49. Considering that the Clr3 HDAC is required for inhibiting turnover of nucleosomes at heterochromatic loci (this study) and that Epe1 genetically interacts with Clr331, we wondered if Epe1 promotes nucleosome turnover. To test this hypothesis, we first investigated the effects of epe1Δ on histone replacement in ago1Δ cells. Similar to dcr1Δ, ago1Δ cells showed about a 2–fold increase in H3 replacement across the pericentromeric heterochromatin domain (Fig. 6a and Supplementary Fig. 2). However, H3 turnover was suppressed in ago1Δ epe1Δ cells, to levels comparable with wild–type cells (Fig. 6a and Supplementary Fig. 2). Importantly, the suppression of H3 replacement caused by epe1Δ correlated with the restoration of centromeric silencing (Fig. 6b), as well as with the suppression of sensitivity to the microtubule–destabilizing drug thiabendazole (TBZ) observed in ago1Δ cells (Fig. 6c).
Figure 6. The JmjC domain–containing protein Epe1 promotes histone turnover across the pericentromeric regions.

(a) Histone H3 replacement in Epe1 overexpressing (nmt1–epe1), ago1Δ epe1Δ, ago1Δ, epe1Δ or wild–type (WT) cells was measured by the MNase–ChIP–chip method as described in Fig. 1 and plotted in alignment with the map of the right pericentromeric region of cen2. (b) RT–PCR analysis was used to measure expression at the cen–dh locus in epe1Δ, ago1Δ, ago1Δepe1Δ or wild–type (WT) cells. RNA isolated from epe1Δ, ago1Δ, ago1Δepe1Δ or wild–type (WT) cells was used to perform RT–PCR analysis with primer sets specific to centromeric dh repeats (cen–dh) or act1 loading control. (c) TBZ sensitivity of epe1Δ, ago1Δ, ago1Δepe1Δ or wild–type (WT) cells. Ten–fold serial dilutions of the indicated cultures were grown on rich medium (YEA) in the presence (10μg/ml) or absence of TBZ.
Epe1 levels are tightly regulated50, and its overproduction impairs heterochromatic silencing31,33,51. Strikingly, overexpression of Epe1 caused more than a 2–fold increase in the replacement of H3 across the pericentromeric domain (Fig. 6a and Supplementary Fig. 2). Taken together, these results suggest a novel role for a JmjC family protein in the dynamic turnover of nucleosomes, and establish an important functional link between histone replacement and heterochromatic silencing.
Histone turnover and epigenetic stability of heterochromatin
A remarkable feature of heterochromatin is that the silenced chromatin state can be propagated through multiple cell divisions21. Our previous analyses have shown that replacement of a portion of the region between the silent mating–type cassettes (referred to as the K–region), which includes the cenH heterochromatin nucleation center, with ura4+ (KΔ∷ura4+) results in a metastable locus52. Cells containing KΔ∷ura4+ display alternate silenced (ura4–off) and expressed (ura4–on) epigenetic states. This variegation is due to defects in de novo heterochromatin assembly in KΔ∷ura4+ (ref. 21). Once established, however, the heterochromatic state is stably inherited in cis21. Importantly, the cis–inheritance of heterochromatin requires binding of Clr4 to H3K9me via its chromodomain17.
These studies suggest that pre–existing methylated H3K9 provides the initial binding site for Clr4 to establish a feedback loop for the clonal propagation of heterochromatin. This model predicts that factors that prevent the loss of methylated histones would be essential for heterochromatin maintenance. We therefore investigated whether cells carrying ura4–off and ura4–on epigenetic states differ in levels of Clr3 association and turnover of histones at this locus (Fig. 7). ChIP experiments revealed that ura4–off cells show considerable enrichment of Clr3 at the KΔ∷ura4+ locus (Fig. 7a). More importantly, nucleosome turnover was suppressed at the silent mating–type region of ura4–off cells (Fig. 7b). In contrast, the levels of Clr3 associated with this region were lower in ura4–on cells, and these cells showed higher levels of histone replacement (Fig. 7a,b). The increase in histone replacement in ura4–on cells, as compared with ura4–off cells, correlated with the reduction in H3K9me at KΔ∷ura4+ (Fig. 7c). Together with the observations that loss of Clr3 or the transient treatment of cells with an HDAC inhibitor affects the maintenance of preassembled heterochromatin at the mat locus26,53, these results suggest that the intrinsic nature of heterochromatin to prevent nucleosome turnover via activities such as Clr3 might be linked to the stable propagation of these structures.
Figure 7. Clr3–dependent suppression of histone turnover correlates with epigenetic stability of heterochromatin.

(a) ChIP analysis of Clr3 localization of at the silent mat region. Strains expressing Myc tagged Clr3 in KΔ∷ura4+ ura4–on or ura4–off state were used to perform ChIP. ChIP DNA was analyzed by semi–quantitative competitive PCR using primers that amplify both full–length KΔ∷ura4+ and endogenous mini–ura4 (ura4DSE) as internal control. The relative enrichments were determined by calculating the ratio of the band intensities of [ChIP KΔ∷ura4+ ÷ ChIP ura4DSE] ÷ [Input KΔ∷ura4+ ÷ Input ura4DSE]. Results were confirmed by quantitative real–time PCR (qPCR). Relative enrichment of KΔ∷ura4+ was normalized against untagged negative control and the mean enrichment is presented. Error bars represent standard error of the mean calculated from 3 independent biological replicates (n=3) (b) H3 replacement was measured in KΔ∷ura4+ ura4–on or ura4–off cells. The endogenous ura4+ was deleted in the strains used. (c) ChIP analysis of H3K9me2 levels at KΔ∷ura4+ ura4–on cells. Experiments were performed with the same strains used in a. H3K9me levels were confirmed by qPCR and the mean enrichment is presented. Error bars represent standard error of the mean calculated from 4 independent biological replicates (n=4).
DISCUSSION
The heterochromatin assembly pathway is remarkably conserved from fission yeast to humans. The repressive heterochromatin defined by methylation of H3K9 and associated HP1 proteins possesses the ability to spread across chromosomes and to be epigenetically transmitted through mitotic and meiotic cell divisions6,8,21. HP1 proteins recruit various effectors including chromatin–modifying activities that modulate chromatin structure2. In S. pombe, the loss of HP1 proteins and their associated activities causes defects in nucleosome occupancy and defective heterochromatic silencing27,30,40. However, changes in steady–state nucleosome patterns are restricted to a few discrete sites30,40, and the mechanism for global repression across heterochromatin domains has remained unclear.
Our results define a critical feature that differentiates heterochromatin from euchromatin and provide important insights into repression and propagation of heterochromatin domains (Fig. 8). We found that the Clr3 HDAC, which can be targeted by multiple mechanisms depending upon the chromosomal context26,27,45, is required to suppress histone turnover across heterochromatin. Loss of Clr3 or factors involved in its localization, such as Clr4, affect acetylation of histones at heterochromatic loci54 (Supplementary Fig. 8). Therefore, deacetylation of a combination of histone residues may prevent histone turnover by nucleosome remodeling factors that require acetylated histones55. In addition, Clr3 affects the subnuclear localization of certain target loci and this spatial reorganization correlates with changes in chromatin structure56. Therefore, it is possible that Clr3 may also impact nucleosome dynamics by modulating genome organization.
Figure 8. Model showing effects of factors that impact epigenetic stability of heterochromatin.

HDAC recruited by HP1 or other mechanisms are required to suppress nucleosome turnover and promote epigenetic stability of heterochromatin. In contrast, Epe1, which also associates with Swi6(HP1), stimulates histone exchange. The balance between these opposing activities that affect nucleosome turnover may underlie the epigenetic switch between ‘OFF’ and ‘ON’ states.
RNAi components and Clr4, which recruit HP1 proteins, also affected histone turnover (Fig. 2). Oligomerization of HP1 bound to H3K9me may help bridge nucleosomes57 to prevent histone exchange. Alternatively, the effects of H3K9me–HP1 might be mediated through associated effectors such as SHREC26-29. Notably, the loss of Clr4 or HP1 has little effect on histone turnover at the mat region, where HDACs can be recruited by alternative mechanisms26,45. Other factors are also likely to impact histone turnover, such as Spt6, a chromatin assembly factor that affects heterochromatic silencing58. Asf1–HIRA facilitates histone deacetylation by HDAC Clr6, which together with Clr3 is essential for hypoacetylation of histones at heterochromatic loci30. Since Asf1–HIRA also associates with HP1, the nucleosome turnover observed upon loss of Clr4 or RNAi most likely reflects the cumulative effects of defective localization of SHREC and other effectors. Indeed, histone exchange levels observed at the centromeres of clr4Δ cells are higher than that of clr3Δ cells (Fig. 2a, 4b and Supplementary Fig. 2), suggesting additional functions for Clr4 and/or its associated factors in this process.
How does histone turnover affect epigenetic stability of heterochromatin and silencing? The retention of histones decorated with H3K9me is predicted to be critical for recruitment of Clr4 through its chromodomain17 to modify newly assembled nucleosomes, thus promoting the parental histone–modification pattern and clonal propagation of heterochromatin in cis. We note that Clr3 is essential for suppression of nucleosome turnover as well as transmission of epigenetic information and propagation of heterochromatin. Nucleosome stability also impacts the accessibility of the underlying DNA sequences to transcription machinery34,36,38,39,46 and the increased histone exchange in heterochromatin mutants may provide access to trans–acting factors, including transcriptional machinery that manifests in loss of silencing. In this respect, heterochromatin partially resembles the chromatin at open reading frames of genes in which suppression of histone exchange prevents activation of transcription from cryptic promoters48. Indeed, factors such as Asf1–HIRA and Clr6 HDAC affect both heterochromatic silencing and suppress cryptic promoters within genes30,59.
Our analyses also revealed that the JmjC protein Epe1, which counteracts the silencing effects of HDACs31,51, promotes histone turnover. The loss of Epe1 not only suppressed the elevated histone H3 exchange present in RNAi mutants, but also suppressed pericentromeric silencing (Fig. 6). In contrast, overexpression of Epe1 increased histone replacement across heterochromatin domains and impaired silencing at these loci. These observations provide an important functional link between histone turnover and heterochromatin silencing, and argue that JmjC proteins, in addition to their roles in demethylation of histones, may remodel chromatin by affecting turnover of histones49. The increased turnover of histones promoted by Epe1 may facilitate low–level transcription of heterochromatic repeats31, which is necessary to produce precursors for siRNAs required for heterochromatin assembly. Epe1 has also been shown to remodel heterochromatin at specific loci in response to signals that induce sexual differentiation9, indicating that programmed histone turnover may serve to modify epigenomic profiles during development.
Our observations indicate that the opposing activities of HDAC and Epe1 that associate with HP1 proteins regulate heterochromatic silencing by modulating histone turnover levels across these domains (Fig. 8). Interestingly, pericentromeric heterochromatin is required for proper localization of CENP–A nucleosomes involved in kinetochore assembly60. It is conceivable that heterochromatin enriched in HDACs indirectly stabilizes CENP–A nucleosomes by suppressing nucleosome turnover across the pericentromeres, thereby preventing promiscuous incorporation of CENP–A. Moreover, hypoacetylation of histones is the most conserved feature of silenced chromatin among diverse species2,4,5. Components of different silencing mechanisms, such as Polycomb, have been found to associate with HDACs and interestingly, these activities map to regions exhibiting low levels of histone exchange39. Therefore, it is possible that the inhibition of histone turnover by distinct silencing effectors will emerge as a unifying theme for maintaining epigenetic memory in all systems. Future investigations into the mechanisms that regulate histone turnover are expected to shed light on the dynamic control of heterochromatin and the organization of the genome into distinct chromatin domains.
ONLINE METHODS
Strains and plasmids
To express carboxy–terminal FLAG–tagged histone H3 (H3–FLAG), the hht2 gene open reading frame was cloned between NdeI and NotI sites of the pINV1 plasmid42. The FLAG epitope sequence was added to the C–terminus of hht2 by inclusion on the reverse primer during the PCR amplification. The resulting pINV1–H3.2–FLAG plasmid was transformed into strains with the indicated genetic backgrounds. The strain overexpressing epe1 contains the full strength nmt1 promoter integrated immediately upstream of the endogenous Epe1 promoter. All strains are MatM–Smt0 mating type except for KΔ∷ura4+, which are h90 mating type. The endogenous 1.8kb ura4+ locus is deleted in the strains containing KΔ∷ura4+. KΔ∷ura4+ (OFF) cells were recovered in medium containing 5–FOA.
MNase–ChIP
Each experiment presented in Fig. 1-7 was reproduced with at least 2 biological replicates. For each MNase–ChIP experiment, fresh cells were grown on EMM–LEU agar plates for 3 days at 30°C. Cells were inoculated into 330ml EMM–LEU + 8% glucose at a density of OD600 ~0.02 and grown overnight at 30°C, with shaking at 250rpm. Cell synchronization by DNA replication arrest was started when cells reached OD600 ~0.1–0.15 (mid–log phase), by adding sterile Hydroxyurea (HU) at a final concentration of 15mM. Cells were synchronized by growing for 4 hours at 30°C, with shaking at 250rpm (except for KΔ∷ura4+, clr3Δ, chp2Δ and swi6Δchp2Δ cells, which were synchronized by adding 20mM HU and growing for 5 hours at 30°C). For nmt–epe1 cells, synchronization was optimized by incubating cells in 15mM HU for 4.5 hours. Cells were pelleted at 24°C and washed twice with 25ml of EMM–LEU–glucose + 4% sucrose containing 15mM HU (except for KΔ∷ura4+, clr3Δ, chp2Δ and swi6Δchp2Δ cells, where the concentration of HU was kept at 20mM throughout the experiment). After the last wash, cells were inoculated into 315ml EMM–LEU–glucose + 4% sucrose containing 15mM HU and grown for 2 hours at 30 °C, with shaking at 250rpm to induce the expression of H3–FLAG. Finally, cells were cross–linked for 20 minutes at room temperature by adding 1% formaldehyde.
For MNase–ChIP experiments, cells were resuspended in 400μl of ChIP cell lysis buffer (50 mM HEPES KOH pH 7.5, 140 mM NaCl, 1mM EDTA, 1% Triton, 0.1% Deoxycholate) supplemented with EDTA–free protease inhibitor cocktail (Roche). Cell lysis was performed using a Mini–BeadBeater–8 (BioSpec) with 1.25ml zirconia–silica beads (0.5mm diameter, BioSpec) by beating at full power twice for 3 minutes at 4°C, separated by a 5–minute interval. Cell lysate was filtered into fresh tubes by washing the disruption beads with an additional 400μl of ChIP cell lysis buffer, and cleared by centrifugation at 16060×g for 10 minutes at 4°C. The supernatant was removed and the pellet was washed with 500μl of MNase reaction buffer (50 mM Tris–HCl, 50mM NaCl, 5 mM CaCl2) supplemented by EDTA–free protease inhibitor cocktail (Roche), and centrifuged again at 16060×g for 10 minutes at 4°C. The supernatant was removed and the resulting pellet was resuspended in 500μl of MNase reaction buffer. Chromatin was fragmented with 0.5μl of MNase (NEB # M0247S, 2 × 106 gel units/ml), added along with 5.1μl of BSA (10mg/ml, NEB). Digestion was performed at 37°C for 30 minutes and stopped by adding 60μl of 0.5M EDTA. The reaction time and amount of enzyme was optimized to yield at least 80% mononucleosomal chromatin sample according to the amount of cells, using the enzyme batch lot number 0071107. Finally, the sample was cleared by centrifugation at 16060×g for 10 minutes at 4°C. 10% of the supernatant was kept as input sample, whereas the rest was diluted 1:1 with ChIP cell lysis buffer. For each ChIP reaction, 20μl packed M2–agarose beads (Sigma A2220) was pre–equilibrated with 1:1 MNase Buffer:ChIP lysis buffer and used for immunoprecipitation for 4 hours at 4°C, with slow rotation. After the immunoprecipitation step, beads were washed with 1:1 MNase Buffer:ChIP lysis buffer, then once with ChIP lysis buffer, twice with ChIP high–salt wash buffer (50 mM HEPES KOH pH 7.5, 500mM NaCl, 1mM EDTA, 1% Triton, 0.1% Deoxycholate), twice with ChIP wash buffer III (10 mM Tris pH 8, 0.25M LiCl, 0.5% NP–40, 0.5% Deoxycholate, 1mM EDTA), and once with TE buffer (50 mM Tris–HCl pH 8.0, 10mM EDTA) at 4°C. ChIP–DNA was eluted in 200μl elution buffer (50mM Tris pH 8.0, 10 mM EDTA, 100mM NaHCO3, 1% SDS) by incubating at 65 °C for 30 minutes with constant agitation. 40μg RNase A (Invitrogen) was added to each eluate and cross–linking was reversed in input samples and DNA by incubating for 12 hours at 65 °C. Finally, samples were treated with 20μg Proteinase K (Invitrogen) for 1 hour at 37 °C, and DNA was purified using QIAGEN PCR purification kits and spin columns.
Microarray analysis of MNase–ChIP and ChIP experiments
MNase–ChIP and ChIP samples were competitively hybridized with their respective input samples. Amplification, labeling, hybridization and analysis of the microarray experiments were performed as described in10, using the high–resolution tiling microarray platform as described in30. Microarray data analysis was performed using standard median normalization as described in10.
Nucleosome occupancy
Nucleosome occupancy was measured as described in30 and the data from the indicated study was plotted across centromere 2 and the mating–type region. For histone H3 occupancy after HU synchronization, a standard ChIP procedure was performed using anti–histone H3 antibody (Active Motif, cat. no: 39163 and Abcam, cat. no: ab1791). 3μl of each antibody was added per ChIP experiment.
RNAP II occupancy
RNAP II occupancy was measured using ChIP–on–chip analysis as described in29 with 8WG16 antibody (Covance, cat. no: MMS–126R). 5μl antibody was used per ChIP experiment.
FACS analysis
Cells were fixed with 70% ethanol at the indicated time points of the experiment (Fig. 1a and data not shown). Cells were washed twice with 50mM sodium citrate (dibasic) solution and incubated with 0.1μg/μl RNase A in 50 mM sodium citrate solution for 8 hours at 37°C. Cells were stained with SytoxGreen (2mM final concentration in 50 mM sodium citrate) and DNA content was analyzed by using BD FACSCalibur™ flow cytometer. BD Cell Quest Pro software was used for raw data acquisition and the FlowJo program was used for final data analysis. FACS analysis was performed for confirmation of all G1 arrested strains (data not shown).
RNA isolation and RT–PCR analysis
Total RNA was isolated from exponentially growing cultures of the indicated strains by using Epicentre MasterPure Yeast RNA purification kit. RT–PCR experiments were performed with 100ng of total RNA and 28 amplification cycles using QIAGEN one–step RT–PCR kit and following the manufacturer’s instructions. The primer set PROA13: GAAAACACATCGTTGTCTTCAGAG, PROA14: CGTCTTGTAGCTGCATGTGAA was used for the amplification of centromeric repeats (dh–cen), and the primer set PROA7: GAAGTACCCCATTGAGCACGG and PROA8: CAATTTCACGTTCGGCGGTAG was used for the amplification of the act1 locus. The primer sets 49, 51, 53, 55, 65 and 70 used for the silent mating–type region (Fig. 7b and Supplementary Fig. 7c) are described in12.
Western blotting
A total of 15ml of cells (O.D600 ~0.2–0.28) grown in media containing either glucose or sucrose were collected and frozen in liquid nitrogen. Proteins were extracted using the trichloroacetic acid (TCA) precipitation method. Briefly, cells were resuspended in 200μl of 20% TCA and mixed with 400μl acid–washed glass disruption beads. After bead beating for 3 minutes using the Mini–BeadBeater–8 (BioSpec), lysates were collected by washing the beads with 400μl of 5% TCA. The resulting 600μl lysate was centrifuged at 16060×g for 10 minutes at 4°C and supernatant was discarded. Precipitated proteins were dissolved in 2X NuPAGE sample buffer (Invitrogen) and analyzed by NuPAGE 12% protein gels, followed by Western blotting using anti–FLAG antibody (Sigma F7425) at 1:1000 dilution.
In order to investigate H3K4 methylation of ectopically expressed H3–FLAG protein, cells were resuspended in 500μl of ChIP cell lysis buffer (50 mM HEPES KOH pH 7.5, 140 mM NaCl, 1mM EDTA, 1% Triton, 0.1% deoxycholate) supplemented by protease inhibitor cocktail (Roche). Cell lysis was performed using the Mini–BeadBeater–8 (BioSpec) using 1.25ml of zirconia–silica beads (0.5mm diameter, BioSpec) and beating at full power once for 3 minutes at 4°C. Cell lysate was filtered into fresh tubes by washing the disruption beads with an additional 500μl of ChIP cell lysis buffer. After a brief sonication at 4°C, the lysate was cleared by centrifugation at 16060×g for 10 minutes at 4°C. The supernatant was mixed with 20μl packed M2–agarose beads and immunoprecipitation was performed for 4 hours by slowly rotating the samples at 4°C. Beads were washed three times in ChIP cell lysis buffer, and proteins were eluted in NuPAGE sample buffer (Invitrogen) by incubating at 96 °C for 10 minutes. Immunoprecipitates were analyzed on NuPAGE 12% protein gels, followed by Western blotting using anti–H3–K4me2 antibody (Upstate/EMD Millipore, cat. no: 07–030) in 1:1000 dilution.
Supplementary Material
Acknowledgments
We are thankful to J. Barrowman for editing the manuscript, K. Zhang, K. Yamane and E. Luk for discussions, members of the Grewal laboratory for their help, and P. Russell (The Scripps Research Institute, La Jolla, CA, USA) for the gift of pINV1 plasmid. This work is supported by the Intramural Research Program of the U.S. National Institutes of Health, National Cancer Institute. OA is supported by a EMBO long–term post–doctoral fellowship.
Footnotes
Author contributions O.A. and S.I.S.G. designed research, O.A. performed all experiments, S.M. helped with microarray probe design, O.A. and S.I.S.G. analyzed the data and wrote the paper.
References
- 1.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 5.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 7.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 8.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 9.Zofall M, et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cam HP, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 11.Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 12.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 13.Djupedal I, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato H, et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 15.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 16.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 18.Bayne EH, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 20.Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol. 2011;18:1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 22.Noma K, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 23.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 24.Schalch T, et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Sugiyama T, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 28.Motamedi MR, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–90. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer T, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamane K, et al. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Isaac S, et al. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics. 2007;175:1549–1560. doi: 10.1534/genetics.106.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 35.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 36.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 37.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia JF, Dumesic PA, Hartley PD, El-Samad H, Madhani HD. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 2010;24:1758–1771. doi: 10.1101/gad.1946410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi ES, Shin JA, Kim HS, Jang YK. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucl Acid Res. 2005;33:7102–7110. doi: 10.1093/nar/gki1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iacovoni JS, Russell P, Gaits F. A new inducible protein expression system in fission yeast based on the glucose-repressed inv1 promoter. Gene. 1999;232:53–58. doi: 10.1016/s0378-1119(99)00116-x. [DOI] [PubMed] [Google Scholar]
- 43.Lantermann AB, et al. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 44.Thon G, Cohen A, Klar AJ. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics. 1994;138:29–38. doi: 10.1093/genetics/138.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 46.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 47.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatesh S, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 49.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 50.Braun S, et al. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayoub N, et al. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol. 2003;23:4356–4370. doi: 10.1128/MCB.23.12.4356-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grewal SI, Klar AJ. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics. 1997;146:1221–1238. doi: 10.1093/genetics/146.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiren M, et al. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 2005;24:2906–2918. doi: 10.1038/sj.emboj.7600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasten M, et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alfredsson-Timmins J, Kristell C, Henningson F, Lyckman S, Bjerling P. Reorganization of chromatin is an early response to nitrogen starvation in Schizosaccharomyces pombe. Chromosoma. 2009;118:99–112. doi: 10.1007/s00412-008-0180-6. [DOI] [PubMed] [Google Scholar]
- 57.Canzio D, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiely CM, et al. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 2011;31:4193–4204. doi: 10.1128/MCB.05568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicolas E, et al. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 60.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


