Abstract
UCP3 (uncoupling protein 3) and its homologues UCP2 and UCP1 are regulators of mitochondrial function. UCP2 is known to have a short half-life of approx. 1 h, owing to its rapid degradation by the cytosolic 26S proteasome, whereas UCP1 is turned over much more slowly by mitochondrial autophagy. In the present study we investigate whether UCP3 also has a short half-life, and whether the proteasome is involved inUCP3 degradation. UCP3 half-life was examined in the mouse C2C12 myoblast cell line by inhibiting protein synthesis with cycloheximide and monitoring UCP3 protein levels by immunoblot analysis. We show that UCP3 has a short half-life of 0.5–4 h. Rapid degradation was prevented by a cocktail of proteasome inhibitors, supporting a proteasomal mechanism for turnover. In addition, this phenotype is recapitulated in vitro: UCP3 was degraded in mitochondria isolated from rat skeletal muscle or brown adipose tissue with a half-life of 0.5–4 h, but only in the presence of a purified 26S proteasomal fraction. This in vitro proteolysis was also sensitive to proteasome inhibition. This phenotype is in direct contrast with the related proteins UCP1 and the adenine nucleotide translocase, which have long half-lives. Therefore UCP3 is turned over rapidly in multiple cell types in a proteasome-dependent manner.
Keywords: half-life, mitochondrion, proteasome, protein degradation, uncoupling protein 1 (UCP1), uncoupling protein 3 (UCP3)
INTRODUCTION
The UCPs (uncoupling proteins) are a subset of the mitochondrial solute carrier family: transporters of metabolites, protons and nucleotides across the mitochondrial inner membrane. UCP2 and UCP3 are homologues of the archetypal uncoupling protein UCP1, which acts as a protonophore to uncouple electron transport from ATP synthesis and generate heat in brown adipose tissue mitochondria [1,2]. UCP2 [3] and UCP3 [4] are expressed in multiple tissues at considerably lower levels than UCP1, and their roles in metabolic regulation are less clear [5].
UCP3 mRNA expression has been reported in many tissues [6,7], although it is most prevalent in skeletal muscle and brown adipose tissue. UCP3 mRNA expression is up-regulated by the thyroid hormone T3 [8–10], cold exposure [11,12], fasting [3,6,13–15], non-esterified fatty acids [16,17] and hypoxia [18]. UCP3 protein has been detected in skeletal muscle at concentrations several hundred-fold less than UCP1 in brown adipose tissue [4], and is also found in thymus [19,20], brown adipose tissue [7] and pancreatic islet cells [21].
The biochemical functions and physiological roles of UCP3 remain cryptic, although several models have been proposed [5,22]. By causing mild uncoupling, UCP3 attenuates the production of ROS (reactive oxygen species) and of oxidative damage, especially during fatty acid oxidation [23–25]. Other suggested roles include export from mitochondria of fatty acids [26,27] or lipid hydroperoxides [28,29], although fatty acid oxidation and export were recently shown to be independent of UCP3 [30].
UCP3 is closely related to UCP2, with which it shares 73% amino acid identity in humans [6]. For a mitochondrial inner membrane protein, UCP2 has a remarkably short half-life of ~1 h in cells [31–33]. The cytosolic proteasome appears to be responsible for UCP2 turnover in an INS-1E pancreatic β-cell line. There are two lines of evidence for this. First, degradation is prevented by a variety of proteasome inhibitors. Secondly, degradation with similar kinetics, sensitive to the proteasome inhibitors, can be reconstituted in isolated INS-1E mitochondria by adding back fractions containing the 26S cytosolic proteasome and ubiquitination machinery [34].
UCP3 is also related to UCP1, with which it shares 57% amino acid identity in humans [6]. However, UCP1 has a much longer half-life, of ~1–4 days [31,35,36]. Unlike UCP2, UCP1 is thought to be degraded by autophagy [36], since its turnover increases co-ordinately with the turnover of other mitochondrial proteins in brown adipose tissue from noradrenaline (norepinephrine)-treated mice.
Nothing is known about the deactivation or turnover of UCP3. In the present study, we test whether UCP3 turnover is similar in kinetics and mechanism to turnover of UCP2 or, alternatively, to turnover of UCP1. We provide evidence that UCP3 is rapidly turned over with an apparent half-life of 0.5–4 h, similar to UCP2. This turnover is sensitive to proteasomal inhibition and, in a cell-free system, is dependent on the presence of purified 26S cytosolic proteasome fractions. These results are consistent with UCP3 degradation mediated by the proteasome and suggest similarities between UCP2 and UCP3 that are not shared by UCP1 or other family members.
EXPERIMENTAL
C2C12 cell culture and UCP3 turnover in C2C12 cells
C2C12 cells (passage < 10) were grown and maintained in DMEM (Dulbecco’s modified Eagle’s medium; Gibco) supplemented with 100 i.u./ml penicillin, 100 µg/ml streptomycin (Gibco) and 10% (v/v) FCS (foetal calf serum), and passaged every 3 days. For determination of the UCP3 half-life, cells were seeded at 1.5×104 cells/well in six-well plates and differentiated for 4 days using DMEM containing 1% FCS. At time zero, protein synthesis was inhibited with cycloheximide, then cells were harvested at different times by trypsinization and pelleting (800 g for 3 min). Pellets were resuspended in Laemmli gel-loading buffer [10% (w/v) SDS, 250 mM Tris/HCl (pH 6.8), 5 mM EDTA, 50% (v/v) glycerol, 5% (v/v) 2-mercaptoethanol and 0.05% Bromophenol Blue], boiled for 10 min and vortex-mixed vigorously. Where indicated, cells were treated with a PIC (proteasome inhibitor cocktail; Calbiochem, catalogue number 539 160) for 2 h prior to the addition of cycloheximide at time zero.
Mitochondrial isolation from skeletal muscle and brown adipose tissue
Female Wistar rats (Harlan Laboratories) were housed at 22±2°C, 45±15% humidity, 12/12 h light/dark cycle, with standard chow and water ad libitum, and tissue was collected at 4–6 weeks of age following killing by CO2 and cervical dislocation. All animal experiments were carried out following standards and guidelines of the American Veterinary Medical Association (AUMA) and the Association for the Assessment and Accreditation of Laboratory Animal Care (AALAC) and approved by the Buck Institute Institutional Animal Care and Use Committee (IACUC) (protocol number 10080). Skeletal muscle mitochondria were isolated as previously described [37]. Briefly, skeletal muscle from the hind limbs and dorsal region of one animal was placed into CP1 medium [100 mM KCl, 5 mM Tris/HCl (pH 7.4 at 4°C) and 2 mM EGTA]. All steps were performed on ice or at 4°C. Skeletal muscle was finely minced on a pre-cooled surface, rinsed five times in CP1 medium, suspended in CP2 medium [CP1 medium with 0.5% defatted BSA, 5 mM MgCl2, 1 mM ATP and protease type VIII (catalogue number P5380) at 245.7 units/100 ml] at approx. 40 ml of CP2 medium per 5 g of muscle, then subjected to slow polytron disaggregation in three 3 s pulses and incubated with stirring for 6 min. Homogenized tissue was centrifuged at 500 g for 10 min, the supernatant was filtered through gauze, and the mitochondria were pelleted at 10400 g for 10 min. After gentle resuspension in CP1 medium and centrifugation at 10400 g (10 min), the pellet was again resuspended in CP1 medium and centrifuged at 3800 g (10 min). The final pellet was resuspended in a small volume of CP1 medium.
Brown adipose tissue mitochondria were isolated as previously described [38]. Briefly, brown adipose tissue from one animal was collected into buffer A (250 mM sucrose, 10 mM Tes, 1 mM EDTA and 1% defatted BSA) on ice and tissue was chopped and then disaggregated using a Dounce homogenizer. The homogenate was transferred to a centrifuge tube through two layers of gauze and centrifuged at 8500 g for 10 min. The pellet was gently resuspended in buffer B (250 mM sucrose, 20 mM Tes, 1 mM EGTA and 0.4% BSA) and centrifuged at 700 g for 10 min. The supernatant was subsequently centrifuged at 8500 g for 10 min, and the final pellet was resuspended in buffer B. Protein concentrations were determined using the biuret assay.
Reconstitution of UCP3 degradation in vitro
Isolated mitochondria were incubated with highly purified 26S proteasome (Enzo Life Sciences, catalogue number PW9310) and ubiquitin fractions (Boston Biochem, catalogue number K-960, or Calbiochem, catalogue number 662096), as well as succinate and an ATP-regenerating system as previously described [34]. Experiments were carried out in STE buffer [250 mM sucrose, 5 mM Tris/HCl (pH 7.4 at 4°C) and 2 mM EGTA] at 37 °C. Where indicated, samples were pre-incubated with the PIC for 30 min.
Immunoblot analysis
Samples were diluted in Laemmli buffer, boiled for 10 min and vigorously vortex-mixed. Proteins were separated by SDS/PAGE (12.5% gel) [39] and transferred on to a 0.1 µM Protran nitrocellulose membrane (Whatman) using the semi-dry method (20 V for 30 min). Membranes were blocked for 1–2 h using 5% (w/v) non-fat dried skimmed milk in PBST (PBS supplemented with 0.1% Tween 20) or TBST [Tris-buffered saline (20 mM Tris/HCl and 137 mM NaCl, pH7.6) supplemented with 0.1% Tween 20] and probed with 0.4 µg/ml anti- UCP3 rabbit primary antibody (Affinity Bioreagents, catalogue number PA1-055), 0.2 µg/ml anti-β-actin rabbit polyclonal IgG antibody (Abcam, catalogue number ab8227), 0.1 µg/ml anti-UCP1 antibody (Sigma, catalogue number U6382) or 0.2 µg/ml anti-ANT (adenine nucleotide translocase) goat polyclonal IgG antibody (Santa Cruz Biotechnology, catalogue number sc- 9300). The secondary antibodies were peroxidase-conjugated, either 0.08 µg/ml goat anti-rabbit IgG (Pierce, catalogue number 31463) or 0.04 µg/ml rabbit anti-goat IgG (Pierce, catalogue number 31433). Where required, membranes were stripped using Restore PLUS Western blot stripping buffer (Pierce, catalogue number 46430), and reblotted according to the manufacturer’s protocol. Immunoblots were developed using a Lumigen ECL (enhanced chemiluminescence) Plus Western Blotting Detection system (Amersham Biosciences). Signals were quantified by analysing band intensities using ImageJ software (http://rsb.info.nih.gov/ij/); all data lay in the region where the signal was linearly related to protein loaded. All data were normalized to loading controls: β-actin (cells) or ANT (mitochondria).
Data analysis
Unless otherwise stated, values are means±S.E.M. for n independent cell cultures, animal tissue collections, mitochondrial isolations, experimental assays and immunoblots. Where appropriate, data were fitted with non-linear regression curves using Prism 5 software. Statistical analysis of the different treatments was performed by repeated-measures ANOVA of all matching non-zero time points with Dunnett’s post-hoc testing. P values therefore apply to the treatment course at all times except zero. Values of P < 0.05 were considered significant.
RESULTS
UCP3 degradation in C2C12 myoblasts is rapid and proteasome-dependent
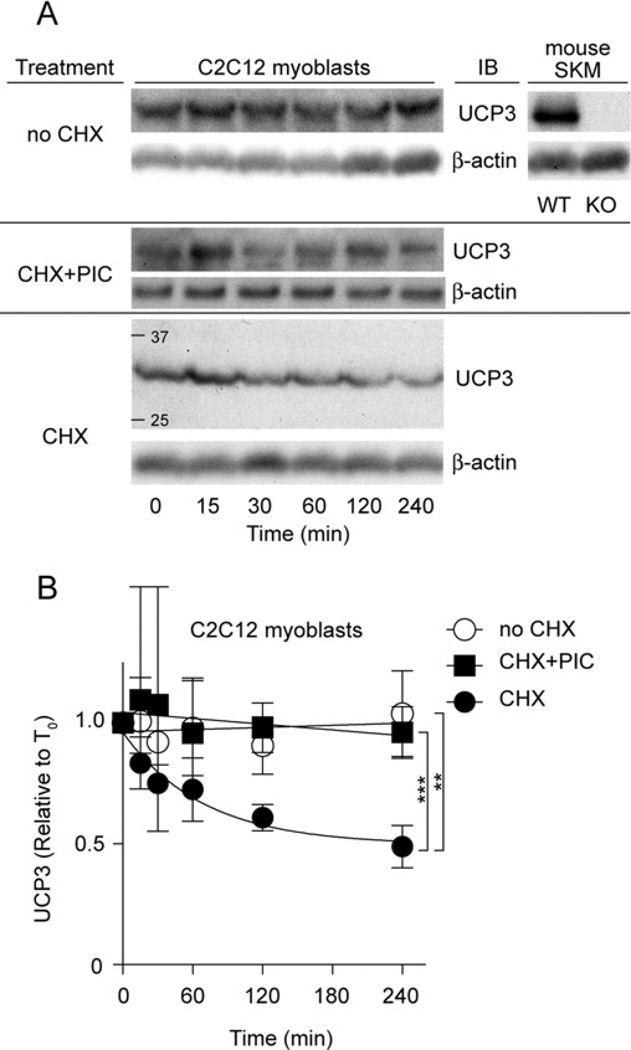
UCP2 has a short half-life in cells [31,32], and we have reported the role of the cytosolic ubiquitin–proteasome system in its degradation [34]. To test whether UCP3 is similarly degraded, we treated differentiated UCP3-containing C2C12myoblasts with cycloheximide to inhibit protein synthesis (Figure 1A). Specificity of the UCP3 antibody was verified by immunoblotting skeletal muscle mitochondria from WT (wild-type) and KO (knockout; Ucp3-ablated) mice [23]. Figure 1(A) shows that UCP3 content decreased with time after cycloheximide addition. Figure 1(B) quantifies the rapid kinetics of UCP3 degradation from four replicate experiments, and shows that UCP3 had a half-life of 0.5–4 h (depending on the end-point used in the calculation), similar to UCP2 (~1 h, [31,32]), but considerably slower than UCP1 (~1–4 days [31,35,36]). Similar to our findings for UCP2 [34], UCP3 turnover was blocked by pharmacological inhibition of the proteasome using a PIC (Figure 1B), suggesting a role for the 26S proteasome in UCP3 degradation.
Figure 1. Proteasome inhibitors block UCP3 degradation in C2C12 cells.
(A) C2C12 cells were treated with 10 µg/ml cycloheximide (CHX) at time zero. Cells were harvested at the times shown, separated by SDS/PAGE (1×105 cells/lane) and immunoblotted (IB) for UCP3 (and β-actin). Immunoblot signals were UCP3-specific, being present in skeletal muscle mitochondria from WT, but not KO, mice. One representative blot is shown. (B) UCP3 degradation kinetics based on immunoblot signals. Where indicated, cells were pre-incubated with PIC (containing 10 µM MG132, 10 µM lactacystin and 30 µM PI-1) for 2 h before addition of CHX. Values are means±S.E.M. (n =4), corrected for loading (β-actin). Statistical significance was determined by repeated-measures ANOVA (comparison of matching non-zero time points) with Dunnett’s post-hoc testing (**P < 0.01, ***P < 0.001).
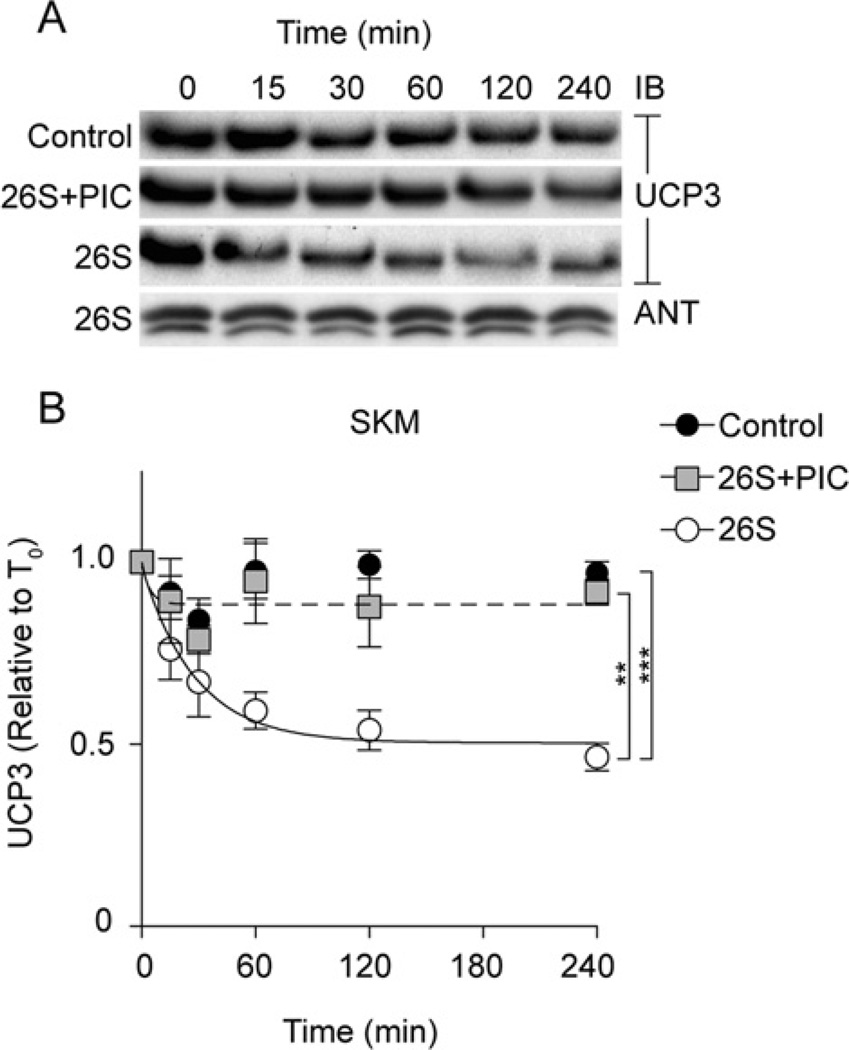
UCP3 in isolated skeletal muscle mitochondria degrades in vitro
To test whether the UCP3 degradation seen in cell culture could be observed in vitro, and whether it was dependent on proteasome activity, we used a recently developed assay in which commercial ubiquitin and proteasome fractions, succinate (for mitochondrial energization) and an ATP-regeneration system are added to isolated mitochondria [34]. In the absence of 26S proteasome, UCP3 was stable in isolated skeletal muscle mitochondria (Figure 2, control). However, when the 26S fraction was added, UCP3 degradation was reconstituted (Figure 2, 26S). Figure 2(B) quantifies the rapid kinetics of UCP3 degradation from three to five replicate experiments, and shows that UCP3 had a half-life of 0.5–4 h (depending on the end-point used in the calculation), similar to the half-life of UCP3 in C2C12 cells (Figure 1). Pre-incubation of the cell-free system with the PIC also blocked UCP3 proteolysis (Figure 2, 26S+PIC). In contrast, another mitochondrial inner membrane solute carrier, ANT, was not similarly degraded in this cell-free system (Figure 2A), suggesting that this proteolytic phenomenon is not a general feature of mitochondrial inner membrane proteins.
Figure 2. Reconstitution of UCP3 degradation in skeletal muscle mitochondria.
The reconstituted system (see the Experimental section) consisted of 1 mg/ml isolated rat skeletal muscle mitochondria in STE buffer (pH 7.4) incubated at 37°C with an ATP-regeneration system (0.5 mM ATP, 10 mM phosphocreatine and 20 units/ml creatine kinase), ubiquitination fractions (50 µg of ubiquitin, 1.6 µg of fraction 1 and 1.6 µg of fraction 2), 2 µg of 26S proteasome fraction, 20 mM succinate and 50 µM PIC as indicated. Aliquots were removed at the time points shown. (A) Proteins (25 µg of protein/lane) were separated by SDS/PAGE and immunoblotted (IB) for UCP3 and ANT. One representative blot is shown. (B) UCP3 degradation kinetics based on immunoblot signals. Values are means±S.E.M. (n =3–5), normalized to ANT. Statistical significance was determined by repeated-measures ANOVA (comparison of matching non-zero time points) with Dunnett’s post-hoc testing (*P < 0.05, ***P < 0.001).
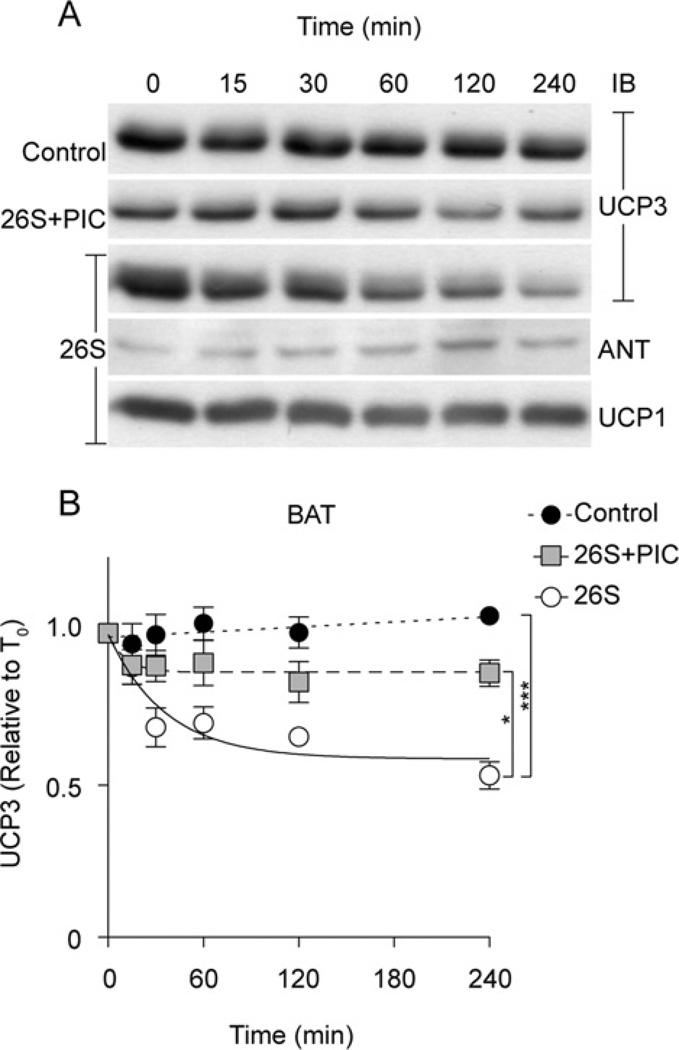
UCP3 degradation occurs in multiple tissue types
We also tested whether rapid UCP3 degradation could be reconstituted in vitro with mitochondria isolated from brown adipose tissue. As with skeletal muscle (Figure 2), UCP3 was rapidly degraded by added 26S proteasome fractions in brown adipose tissue mitochondria, with a half-life of 0.5–4 h, and this degradation was inhibited by the PIC (Figure 3). Since brown adipose tissue mitochondria also contain UCP1 and ANT, we were able to directly compare UCP3 half-life to that of UCP1 or ANT. Consistent with previous studies [31,35,36], UCP1 degradation was not apparent during the 4 h time course, unlike UCP3 (Figure 3) or UCP2 [34]. Similarly, ANT was not degraded in the reconstituted system.
Figure 3. Reconstitution of UCP3 degradation in brown adipose tissue mitochondria.
The reconstituted ubiquitin–proteasome/mitochondria system consisted of 1 mg/ml isolated rat brown adipose tissue mitochondria in STE buffer (pH 7.4) incubated at 37°C with an ATP-regeneration system (0.5 mM ATP, 10 mM phosphocreatine and 20 units/ml creatine kinase), ubiquitination fractions (50 µg of ubiquitin, 1.6 µg of fraction 1, 1.6 µg of fraction 2), 2 µg of 26S proteasome fraction, 20 mM succinate and 50 µM PIC-1 as indicated. Aliquots were removed at the time points shown. (A) Proteins (25 µg of protein/lane) were separated by SDS/PAGE and immunoblotted (IB) for UCP3, UCP1 and ANT. (B) UCP3 degradation kinetics based on immunoblot signals. Values are means±S.E.M. (n =3–5), normalized to ANT. Statistical significance was determined by repeated-measures ANOVA (comparison of matching non-zero time points) with Dunnett’s post-hoc testing (**P < 0.01, ***P < 0.001).
DISCUSSION
The results of the present study show that UCP3 has a very short half-life of 0.5–4 h in both C2C12 cells and mitochondria from skeletal muscle or brown adipose tissue incubated with the 26S cytosolic proteasome. This degradation is proteasome-dependent, since it is inhibited by a PIC in C2C12 cells, and the proteasome is required in the reconstituted system in vitro.
The only other mitochondrial inner membrane protein known to display similarly rapid proteasome-dependent turnover is the closely related UCP2 [34]. The proteolytic mechanism is remarkable because it involves the degradation of these mitochondrial inner membrane proteins by cytosolic machinery and because it differs from the proteolysis of UCP1. The latter is degraded via autophagy [36] and has a half-life that is one to two orders of magnitude larger than those of UCP2 and UCP3.
The half-life of UCP1 is consistent with the reported half-lives of other mitochondrial inner membrane proteins, which typically fall within a range of 20–100+ h [40,41]. In contrast, the half-lives of both UCP2 and UCP3 stand out as exceptionally short, and we therefore suggestive of a separate degradation mechanism. The requirement of the proteasome for UCP3 degradation in vitro supports this model.
The rapid degradation of UCP2 and UCP3 led us to search for possible motifs associated with ubiquitination and proteasomal degradation. Such motifs include the D-box, KEN box and PEST motifs. Analysis (http://bioinfo2.weizmann.ac.il/~danag/d-box/form.html) of 86 UCP sequences in 45 species [42] revealed a D-box-like motif (RGVLGTILTM; motif in bold) present in 100% of UCP3 and ~70% of UCP2 sequences from higher vertebrates, but was absent from UCP1 and ANT. Whether it is involved in degradation of UCP2 and UCP3 remains to be determined.
In summary, in the present paper we report the rapid turnover of the uncoupling protein UCP3, and provide strong support for its degradation by the cytosolic 26S proteasome in both cellular and reconstituted cell-free systems. We hypothesize that rapid UCP3 turnover may be a means of dynamically regulating UCP3 levels in various physiological contexts, including adaptation to fasting and protection from oxidative damage.
ACKNOWLEDGEMENTS
We thank Dr Martin Jastroch (Buck Institute for Age Research, Novato, CA, U.S.A.) for providing collated UCP sequences.
FUNDING
This work was supported by the Medical Research Council (UK); the School of Clinical Medicine, University of Cambridge (UK); and the National Institutes of Health [grant numbers P01 AG025901, PL1 AG032118, P30 AG025708].
Abbreviations used
- ANT
adenine nucleotide translocase
- DMEM
Dulbecco’s modified Eagle’s medium
- FCS
foetal calf serum
- KO
knockout
- PIC
proteasome inhibitor cocktail
- UCP
uncoupling protein
- WT
wild-type.
Footnotes
AUTHOR CONTRIBUTION
Vian Azzu, Shona Mookerjee and Martin Brand designed the study and wrote the manuscript. Vian Azzu and Shona Mookerjee collected the data and analysed the results.
REFERENCES
- 1.Nicholls DG, Locke RM. Thermogenic mechanisms in brown fat. Physiol. Rev. 1984;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 3.Pecqueur C, Alves-Guerra MC, Gelly C, Lévi-Meyrueis C, Couplan E, Collins S, Ricquier D, Bouillaud F, Miroux B. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress, and evidence for translational regulation. J. Biol. Chem. 2001;276:8705–8712. doi: 10.1074/jbc.M006938200. [DOI] [PubMed] [Google Scholar]
- 4.Harper JA, Stuart JA, Jekabsons MB, Roussel D, Brindle KM, Dickinson K, Jones RB, Brand MD. Artifactual uncoupling by uncoupling protein 3 in yeast mitochondria at the concentrations found in mouse and rat skeletal-muscle mitochondria. Biochem. J. 2002;361:49–56. doi: 10.1042/0264-6021:3610049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2005;2:85–93. doi: 10.1016/j.cmet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, Seydoux J, Muzzin P, Giacobino JP. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 7.Vidal-Puig AJ, Solanes G, Grujic D, Flier JS, Lowell BB. UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem. Biophys. Res. Commun. 1997;235:79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- 8.Larkin S, Mull E, Miao W, Pittner R, Albrandt K, Moore C, Young A, Denaro M, Beaumont K. Regulation of the third member of the uncoupling protein family, UCP3, by cold and thyroid hormone. Biochem. Biophys. Res. Commun. 1997;240:222–227. doi: 10.1006/bbrc.1997.7636. [DOI] [PubMed] [Google Scholar]
- 9.Jekabsons MB, Gregoire FM, Schonfeld-Warden NA, Warden CH, Horwitz BA. T3 stimulates resting metabolism and UCP-2 and UCP-3 mRNA but not nonphosphorylating mitochondrial respiration in mice. Am. J. Physiol. 1999;277:E380–E389. doi: 10.1152/ajpendo.1999.277.2.E380. [DOI] [PubMed] [Google Scholar]
- 10.Gong DW, Monemdjou S, Gavrilova O, Leon LR, Marcus-Samuels B, Chou CJ, Everett C, Kozak LP, Li C, Deng C, et al. Lack of obesity and normal response to fasting and thyroid hormone in mice lacking uncoupling protein-3. J. Biol. Chem. 2000;275:16251–16257. doi: 10.1074/jbc.M910177199. [DOI] [PubMed] [Google Scholar]
- 11.Lin B, Coughlin S, Pilch PF. Bidirectional regulation of uncoupling protein-3 and GLUT-4 mRNA in skeletal muscle by cold. Am. J. Physiol. 1998;275:E386–E391. doi: 10.1152/ajpendo.1998.275.3.E386. [DOI] [PubMed] [Google Scholar]
- 12.Schrauwen P, Westerterp-Plantenga MS, Kornips E, Schaart G, van Marken Lichtenbelt WD. The effect of mild cold exposure on UCP3 mRNA expression and UCP3 protein content in humans. Int. J. Obes. Relat. Metab. Disord. 2002;26:450–457. doi: 10.1038/sj.ijo.0801943. [DOI] [PubMed] [Google Scholar]
- 13.Gong DW, He Y, Karas M, Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, β3-adrenergic agonists, and leptin. J. Biol. Chem. 1997;272:24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- 14.Boss O, Samec S, Kühne F, Bijlenga P, Assimacopoulos-Jeannet F, Seydoux J, Giacobino JP, Muzzin P. Uncoupling protein-3 expression in rodent skeletal muscle is modulated by food intake but not by changes in environmental temperature. J. Biol. Chem. 1998;273:5–8. doi: 10.1074/jbc.273.1.5. [DOI] [PubMed] [Google Scholar]
- 15.Cadenas S, Buckingham JA, Samec S, Seydoux J, Din N, Dulloo AG, Brand MD. UCP2 and UCP3 rise in starved rat skeletal muscle but mitochondrial proton conductance is unchanged. FEBS Lett. 1999;462:257–260. doi: 10.1016/s0014-5793(99)01540-9. [DOI] [PubMed] [Google Scholar]
- 16.Weigle DS, Selfridge LE, Schwartz MW, Seeley RJ, Cummings DE, Havel PJ, Kuijper JL, BeltrandelRio H. Elevated free fatty acids induce uncoupling protein 3 expression in muscle: a potential explanation for the effect of fasting. Diabetes. 1998;47:298–302. doi: 10.2337/diab.47.2.298. [DOI] [PubMed] [Google Scholar]
- 17.Hoeks J, Hesselink MK, Schrauwen P. Involvement of UCP3 in mild uncoupling and lipotoxicity. Exp. Gerontol. 2006;41:658–662. doi: 10.1016/j.exger.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Flandin P, Donati Y, Barazzone-Argiroffo C, Muzzin P. Hyperoxia-mediated oxidative stress increases expression of UCP3 mRNA and protein in skeletal muscle. FEBS Lett. 2005;579:3411–3415. doi: 10.1016/j.febslet.2005.04.084. [DOI] [PubMed] [Google Scholar]
- 19.Carroll AM, Haines LR, Pearson TW, Fallon PG, Walsh CM, Brennan CM, Breen EP, Porter RK. Identification of a functioning mitochondrial uncoupling protein 1 in thymus. J. Biol. Chem. 2005;280:15534–15543. doi: 10.1074/jbc.M413315200. [DOI] [PubMed] [Google Scholar]
- 20.Adams AE, Hanrahan O, Nolan DN, Voorheis HP, Fallon P, Porter RK. Images of mitochondrial UCP1 in mouse thymocytes using confocal microscopy. Biochim. Biophys. Acta. 2008;1777:115–117. doi: 10.1016/j.bbabio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Maedler K, Shu L, Haataja L. UCP-2 and UCP-3 proteins are differentially regulated in pancreatic β-cells. PLoS ONE. 2008;3:e1397. doi: 10.1371/journal.pone.0001397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bézaire V, Seifert EL, Harper ME. Uncoupling protein-3: clues in an ongoing mitochondrial mystery. FASEB. J. 2007;21:312–324. doi: 10.1096/fj.06-6966rev. [DOI] [PubMed] [Google Scholar]
- 23.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 24.Echtay KS, Pakay JL, Esteves TC, Brand MD. Hydroxynonenal and uncoupling proteins: a model for protection against oxidative damage. Biofactors. 2005;24:119–130. doi: 10.1002/biof.5520240114. [DOI] [PubMed] [Google Scholar]
- 25.Talbot DA, Brand MD. Uncoupling protein 3 protects aconitase against inactivation in isolated skeletal muscle mitochondria. Biochim. Biophys. Acta. 2005;1709:150–156. doi: 10.1016/j.bbabio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Himms-Hagen J, Harper ME. Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: an hypothesis. Exp. Biol. Med. 2001;226:78–84. doi: 10.1177/153537020122600204. [DOI] [PubMed] [Google Scholar]
- 27.Schrauwen P, Saris WH, Hesselink MK. An alternative function for human uncoupling protein 3: protection of mitochondria against accumulation of nonesterified fatty acids inside the mitochondrial matrix. FASEB J. 2001;15:2497–2502. doi: 10.1096/fj.01-0400hyp. [DOI] [PubMed] [Google Scholar]
- 28.Goglia F, Skulachev VP. A function for novel uncoupling proteins: antioxidant defense of mitochondrial matrix by translocating fatty acid peroxides from the inner to the outer membrane leaflet. FASEB J. 2003;17:1585–1591. doi: 10.1096/fj.03-0159hyp. [DOI] [PubMed] [Google Scholar]
- 29.Jaburek M, Miyamoto S, Di Mascio P, Garlid KD, Jezek P. Hydroperoxy fatty acid cycling mediated by mitochondrial uncoupling protein UCP2. J. Biol. Chem. 2004;279:53097–53102. doi: 10.1074/jbc.M405339200. [DOI] [PubMed] [Google Scholar]
- 30.Seifert EL, Bézaire V, Estey C, Harper ME. Essential role for uncoupling protein-3 in mitochondrial adaptation to fasting but not in fatty acid oxidation or fatty acid anion export. J. Biol. Chem. 2008;283:25124–25131. doi: 10.1074/jbc.M803871200. [DOI] [PubMed] [Google Scholar]
- 31.Rousset S, Mozo J, Dujardin G, Emre Y, Masscheleyn S, Ricquier D, Cassard-Doulcier AM. UCP2 is a mitochondrial transporter with an unusual very short half-life. FEBS Lett. 2007;581:479–482. doi: 10.1016/j.febslet.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Azzu V, Affourtit C, Breen EP, Parker N, Brand MD. Dynamic regulation of uncoupling protein 2 content in INS-1E insulinoma cells. Biochim. Biophys. Acta. 2008;1777:1378–1383. doi: 10.1016/j.bbabio.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giardina TM, Steer JH, Lo SZ, Joyce DA. Uncoupling protein-2 accumulates rapidly in the inner mitochondrial membrane during mitochondrial reactive oxygen stress in macrophages. Biochim. Biophys. Acta. 2008;1777:118–129. doi: 10.1016/j.bbabio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Azzu V, Brand MD. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 2009 doi: 10.1242/jcs.060004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puigserver P, Herron D, Gianotti M, Palou A, Cannon B, Nedergaard J. Induction and degradation of the uncoupling protein thermogenin in brown adipocytes in vitro and in vivo: evidence for a rapidly degradable pool. Biochem. J. 1992;284:393–398. doi: 10.1042/bj2840393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moazed B, Desautels M. Differentiation-dependent expression of cathepsin D and importance of lysosomal proteolysis in the degradation of UCP1 in brown adipocytes. Can. J. Physiol. Pharmacol. 2002;80:515–525. doi: 10.1139/y02-067. [DOI] [PubMed] [Google Scholar]
- 37.Rolfe DFS, Hulbert AJ, Brand MD. Characteristics of mitochondrial proton leak and control of oxidative phosphorylation in the major oxygen-consuming tissues of the rat. Biochim. Biophys. Acta. 1994;1188:405–416. doi: 10.1016/0005-2728(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 38.Echtay KS, Esteves TC, Pakay JL, Jekabsons MB, Lambert AJ, Portero-Otin M, Pamplona R, Vidal-Puig AJ, Wang S, Roebuck SJ, Brand MD. A signalling role for 4-hydroxy-2-nonenal in regulation of mitochondrial uncoupling. EMBO J. 2003;22:4103–4110. doi: 10.1093/emboj/cdg412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Lipsky NG, Pedersen PL. Mitochondrial turnover in animal cells. Half-lives of mitochondria and mitochondrial subfractions of rat liver based on [14C]bicarbonate incorporation. J. Biol. Chem. 1981;256:8652–8657. [PubMed] [Google Scholar]
- 41.Hare JF, Hodges R. Turnover of mitochondrial inner membrane proteins in hepatoma monolayer cultures. J. Biol. Chem. 1982;257:3575–3580. [PubMed] [Google Scholar]
- 42.Jastroch M, Withers KW, Taudien S, Frappell PB, Helwig M, Fromme T, Hirschberg V, Heldmaier G, McAllan BM, Firth BT, et al. Marsupial uncoupling protein 1 sheds light on the evolution of mammalian nonshivering thermogenesis. Physiol. Genomics. 2008;32:161–169. doi: 10.1152/physiolgenomics.00183.2007. [DOI] [PubMed] [Google Scholar]