Abstract
Development of natural competence in S. pneumoniae entails coordinated expression of two sets of genes. Early gene expression depends on ComE, a response regulator activated by the pheromone CSP (Competence-Stimulating-Peptide). Subsequently, an early gene product (the alternative sigma factor ComX) activates expression of late genes, establishing the competent state. Expression of both sets of genes is transient, rapidly shut off by a mechanism that depends on the late gene, dprA. It has been thought that the rapid shutoff of late gene expression is the combined result of auto-inhibition of ComE and the instability of ComX. However, this explanation seems incomplete, because of evidence for ComX-dependent repressor(s) that might also be important for shutting off the response to CSP and identifying dprA as such a gene. We screened individual late gene mutants to investigate further the roles of ComX-dependent genes in competence termination. A ΔdprA mutant displayed a prolonged late gene expression pattern, whereas mutants lacking cbpD, cibABC, cglEFG, coiA, ssbB, celAB, cclA, cglABCD, cflAB, or radA, exhibited a wild-type temporal expression pattern. Thus, no other gene than dprA was found to be involved in shutoff. DprA limits the amounts of ComX and another early gene product, ComW, by restriction of early gene expression rather than by promoting proteolysis. To ask if DprA also affects late gene expression, we decoupled late gene expression from early gene regulation. Because DprA did not limit ComX activity under these conditions, we also conclude that ComX activity is limited by another mechanism not involving DprA.
Introduction
Streptococcus pneumoniae, a gram positive bacterium inhabiting the human upper respiratory tract, is an opportunistic pathogen which causes many kinds of infection, such as pneumonia, meningitis and otitis. One of the most remarkable features of this microbe is its ability of natural genetic transformation, or competence, discovery of which led to the identification of DNA as the genetic material [1]. In laboratory cultures of S. pneumoniae, an outburst of competence occurs during the mid-log phase, emerging and shutting off rapidly, within about 30 minutes. Recent research has uncovered several aspects of the regulatory process by which virtually all cells of an unsynchronized pneumococcal culture shift, in concert, from an incompetent state to a fully competent state [2]. It starts with the synthesis of a polypeptide ComC, which is exported through a cell membrane transporter ComAB, and processed to a 17-amino-acid peptide, named CSP (competence stimulating peptide). Acting as a pheromone, CSP binds ComD and activates the two component system: histine kinase ComD and response regulator ComE. Phosphorylated ComE is thought to act as a transcriptional activator, recognizing an imperfect direct repeat in front of the promoters of several genes, termed early genes. Among these are comAB and comCDE, which act to produce more CSP, and comX and comW, whose products together turn on expression of the set of genes termed late genes. ComX acts as an alternative sigma factor recognizing a promoter sequence termed the "cinbox” or “combox". Finally, some late genes up-regulated by ComX participate in a variety of functions related to transformation, including DNA uptake and processing, recombination, fratricide, and immunity to fratricide, while others have roles that remain to be discovered, but are not required for transformation. While it is clear that the auto-catalytic peptide pheromone CSP serves to coordinate development of competence among the cells of a culture, the signals or stresses that trigger the developmental cycle are only beginning to be discovered [3]–[5].
During the period of maximal competence, also termed the X state [3], transcription is dominated by an excess of the alternative sigma factor, ComX, which is otherwise entirely absent from the cell. To escape from this state, key connections in the circuit must be interrupted decisively. A dramatic temporal pattern of mRNA accumulation and loss in response to an acute dose of the CSP signal attests to the coordination and strength of these effects [6]–[8]. A brief period of early gene expression is followed by a brief period of late gene expression and a somewhat longer period of competence reflecting the activities of the accumulated late gene products.
The lag in expression of the late genes is explained by the role of the early gene product ComX as an alternative sigma factor driving expression of late genes from specific non-canonical promoters. However, as rapidly as this chain of responses to CSP is established, its effects are nearly as rapidly reversed [6], [7], [9], [10], leading to the remarkably transient nature of competence. Early gene mRNA quickly accumulates, increasing at least 100-fold between 2 and 10 min after a sudden increase in CSP level, but these messages then disappear just as quickly, leaving less than 10% of maximal levels in the cell by 15 min. As no specifically targeted anti-mRNA mechanism is known, this suggests that ComE-directed transcription stops abruptly by 10 min. While the ComE protein itself is stable [11], a prime candidate for the cause of this change is a change in the amount of phosphorylated ComE (ComEP). Transcripts of late genes follow a delayed but similar temporal course, peaking at 13 min and largely disappearing by 17 min, implying as well that ComX activity is largely dissipated by 13 min. Although labile, ComX protein is present well past 20 min, suggesting that some inhibition of ComX activity occurs before it physically decays. A further indication of an antagonist of ComX activity was recently provided: while mutation of ClpP stabilized ComX, it failed to prolong its transcriptional activity, detected either as prolonged transformation or as transcription of late genes [12]. Thus, there appear to be at least two targets of competence shutoff mechanisms, the activity of ComEP (which might be inactivated by a phosphatase) and the activity of ComX.
Martin et al [13] reported that the ComE R120S mutant exhibits a much delayed shutoff of competence, with transformation continuing past 90 min, much longer than the 20-min period of competence in wild type cultures. This behavior indicates that ComE activity is an important determinant of the exit from competence and that cells in a competent culture can support an extended period of transformation. Indeed, Claverys and Håvarstein [2] proposed that ComE itself acts to shut off its own activity when accumulated to a high enough level. Thus, it has been thought that exit from competence might be a two-step process involving, first, dephosphorylation or other inactivation of the stable regulator, ComE, followed by degradation of the unstable proteins, ComX and ComW [2], [14]. More recently the case for the importance of the second, proteolytic, step was clouded when it was observed that competence still shuts off in a strain where ComX and ComW are stabilized by inactivation of cognate proteases [12]. Furthermore, competence induced by ectopic expression of comX and comW, without participation of other early genes, also follows a course leading to rapid shutoff [15]. These results together indicate that there is at least one factor shutting off competence by targeting ComX directly, independent of proteolysis of ComX and independent of any regulatory effects on expression of early genes.
Additional clues to the mechanism by which competence is terminated are provided by the phenotypes of comX and dprA mutants. While comX mutants do not transform, or become competent by the criterion of expression of late genes, they do respond to CSP, with over-expression of ComX [9], [16] and other early genes [7]. The temporal pattern of this response is remarkable: it is not rapidly reversed, as in wild type; instead the induced early gene expression continues for generations [16]. This simple result immediately suggests that one or more late gene products may be important for the reversal of the CSP response, although another logical possibility is that ComX itself acts as such a repressor. A remarkably similar regulatory phenotype was described for mutants defective in the late gene, dprA. Bergé reported that the dprA mutation blocked transformation (later explained by roles of DprA in stabilization or recombination of donor DNA fragments), but also that it causes an exacerbated (∼60 min) growth arrest upon CSP treatment, while simultaneously permitting continued expression of the comCDE early competence operon [17].
To search more broadly for late gene(s) implicated in reversal of the response to CSP, Peterson et al [7] examined the competence kinetics of many mutants defective in genes that were induced in competent cells but not required for transformation (green genes in Figure 1). None of the transformable mutant strains tested displayed an extended period of competence. Mirouze et al. subsequently demonstrated that it was possible to complement the regulatory defect of a comX mutant in escape from the CSP response, restoring transient expression of the comCDE operon, by ectopic comX-independent expression of dprA, under control of an early class promoter [18]. They proposed that the relevant activity of DprA might be either to promote de-phosphorylation of ComE or to block phospho-transfer to ComE by ComD, either of which would be expected to cause a broad effect on all early genes in addition to the effect they observed on the comCDE operon.
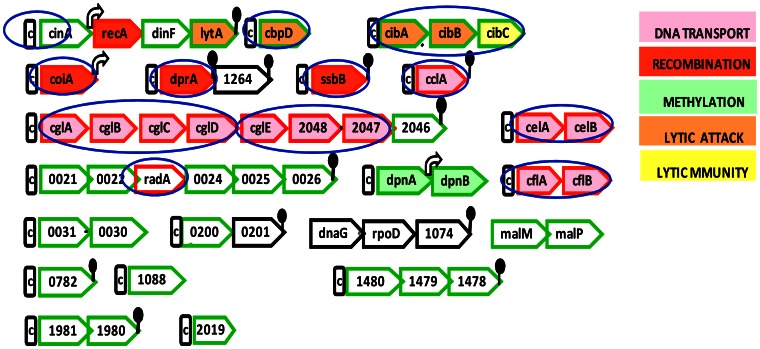
Figure 1. Organization of late genes of the S. pneumoniae competence regulon.
Internal deletion mutants studied in this paper are indicated by blue ovals. C represents the cinbox promoter, TACGAATA. Fill colors symbolize the functions of proteins, as indicated by the key on the right. Bent arrow and lollipop stand for constitutive promoter and terminator respectively. Red borders indicate genes required for transformation; green borders indicate genes not required for transformation. Black borders indicate genes whose importance for transformation has not been directly determined. Genes described by numbers (ORF numbers in TIGR4) are those whose functions in competence are unknown. The ORF numbers for named late genes are: cinA(sp1941), cbpD(sp2201), cibA(sp0125), coiA(sp0978), dprA(sp1266), ssbB(sp1908), cclA(sp1808), cglA(sp2053), celA(sp0054), cflA(sp2208). The late gene clusters and associated cinbox are as in Peterson et al [9], but without cases of apparent read-through transcription identified in [34].
Although DprA thus seems necessary for shutoff of the (early) comCDE operon and sufficient for ComX-independent shutoff of comCDE transcription, its regulatory target remains uncertain, and it is unknown whether additional late genes might contribute as well to exit from the X state. To identify possible additional pieces of this puzzle, we looked further among the late genes for products that act to promote the exit from competence. Although the late genes outlined in red in Fig. 1 (cclA, celA, cflA, cglA, coiA, radA, recA, and ssbB) are required for transformation and have critical roles in DNA transport, processing, or recombination, they had not been examined for possible roles in competence termination, perhaps because of the defective transformation in such mutants. This impediment could be circumvented with an indirect method, simply by monitoring shutoff of competence gene expression, instead of decay of transformability per se as an indicator of exit from the competent state. Since the late gene expression pattern is itself a reliable indicator of competence kinetics [17], [19] and late gene products are the effectors of transformation, the expression pattern of a late gene revealed by a simple LacZ assay in a ComX+ background will best reflect competence development and persistence, even in non-transformable mutants. Using this strategy, with a lacZ transcriptional reporter at the ssbB late gene, we examined mutants defective in additional late genes that are required for transformation. The mutant lacking dprA displayed, as expected, a prolonged late gene expression pattern, whereas mutants lacking cbpD, cibABC, cglEFG, coiA, ssbB, celAB, cclA, cglABCD, cflAB, or radA, exhibited the wild-type pattern of rapid extinction of late gene expression. We further report that closer examination of the effects of DprA on competence regulation revealed that DprA interacts with ComE to control early gene expression but does not affect proteolysis of labile early gene products, while a second, dprA-independent and non-proteolytic, mechanism targets ComX activity during escape from the X-state.
Materials and Methods
Bacterial Strains, Oligonucleotides, Plasmids, and Culture Media
All pneumococcal strains used in this study are described in Table 1. The transformation-proficient strains are the derivatives of the Rx1 strain [20] which carries a mis-sense mutation in cps3D [21]. CP2000 carries a deletion of all capsular polysaccharide synthesis genes between aliA and dexB, respectively. CPM7 is a derivative of CP1250 with the lacZ reporter plasmid pEVP3 inserted in the ssbB locus, with a truncated ssbB upstream of the insert and an intact ssbB downstream. All the strains were grown in the casein hydrolysate/tryptone medium, CAT [16], [22]. Antibiotics were used at the following selective concentrations: erythromycin (Em), 0.05 µg/ml; streptomycin (Sm), 100 µg/ml; kanamycin (Kan), 200 µg/ml; and tetracycline (Tet), 0.25 µg/ml. CSP1 [23] was obtained as a custom peptide from Mimitopes LLC (Raleigh). Synthetic oligonucleotide primers, purchased from Eurofins MWG Operon (Huntsville), are listed in Table 2. E. coli strains were grown to late exponential phase in LB media [24], and stored with 20% sterile glycerol. For use, the frozen stocks were streaked onto LB agar plates with appropriate antibiotics, and a single colony was then inoculated into fresh medium. Ampicillin was used 0.1 µg/ml for selection of E. coli. Yeast strains s were grown in YPD or SD synthetic media [25], and stored with sterile glycerol at 20% final concentration. For use, frozen stocks were streaked onto YPD agar plate, and a single colony was then inoculated into fresh YPD or synthetic selective media. 1 mM or 3 mM 3AT (3-Amino-1,2,4-triazole) was used to select against histidine production. Plasmids used in this study are listed in Table 3.
Table 1. Bacterial strains used in this study.
| Strain | Description | Source (a) or reference |
| Streptococcus. pneumoniae | ||
| CPM7 | CP1250, but ssbB−::lacZ::ssbB+; SsbB+ SmR CmR | [22] |
| CP1250 | Rx, but cps3D hex cps3D malM511 str-1 bgl-1; Hex− Mal− SmR Bga− | [29] |
| CP1275 | CP1250, but ΔcbpD::PcKan; KanR | [7] |
| CP1279 | CP1250, but ΔcibABC::PcKan; KanR | [7] |
| CP1333 | CP1250, but ΔcglEFG::PcKan; KanR | [35] |
| CP1344 | CP1250, but ΔclpC::PCTet; TetR | [12] |
| CP1359 | CP1250, but ΔclpP::PcTet; TetR | [12] |
| CP1389 | CP1250, but ΔdprA::PcKan; KanR | [36] |
| CP1415 | CP1250, but ΔcomA::PcErm; EmR | [37] |
| CP1500 | Rx, hex nov-r1, byr-r, ery-r1,ery-r2, str-1;NovR EmR SmR, | [20] |
| CP1793 | CP1250, but ΔcoiA::PcKan; KanR | [36] |
| CP1851 | CP1250, but ΔclpE::PcErm; EmR | [12] |
| CP1862 | CP1250, but ΔcelAB::PcKan; KanR | This work |
| CP1863 | CP1250, but ΔcclA::PcKan; KanR | This work |
| CP1868 | CP1250, but ΔcglABCD::PcKan; KanR | This work |
| CP1869 | CP1250, but ΔcflAB::PcKan; KanR | This work |
| CP1890 | CP1250, but ssbB−::lacZ::ssbB+, ΔclpP::PcTet; CmR TetR | CPM7×CP1359 |
| CP1894 | CP1250, but ssbB−::lacZ::ssbB+, ΔdprA::PcKan; CmR KanR | CPM7×CP1389 |
| CP1895 | CP1250, but ssbB−::lacZ::ssbB+, ΔclpP, ΔdprA; CmR KanR TetR | CP1890×CP1389 |
| CP1896 | CP1250, but aga::comX::comW::PcKan; KanR | This work |
| CP1961 | CP2000, but aga::comX::comW, ssbB−::lacZ::ssbB+; CmR KanR | CP1896×CP2000×CPM7 |
| CP1962 | CP1961, but ΔclpE::PcErm; EmR | CP1961×CP1851 |
| CP1963 | CP1961, but ΔclpC::PCTet; TetR | CP1961×CP1344 |
| CP1902 | CP1961, but ΔclpC::PCTet, ΔclpE::PcErm; TetR EmR | CP1962×CP1344 |
| CP1932 | CP1902, but ΔdprA::PcKan; KanR | CP1902×CP1389 |
| CP2000 | CP1250, but Δcps; Hex− Mal− Cps− SmR Bga− | This work |
| CP2108 | CP2000, but ssbB−::lacZ::ssbB+, ΔcomA::PcErm; SmR CmR EmR | CP2000×CPM7×CP1415 |
| CP2109 | CP2108, but ΔcbpD::PcKan; KanR | CP2108×CP1275 |
| CP2110 | CP2108, but ΔcibABC::PcKan; KanR | CP2108×CP1279 |
| CP2111 | CP2108, but ΔcoiA::PcKan; KanR | CP2108×CP1793 |
| CP2112 | CP2108, but ΔcglEFG::PcKan; KanR | CP2108×CP1333 |
| CP2113 | CP2108, but ΔdprA::PcKan; KanR | CP2108×CP1389 |
| CP2114 | CP2108, but ΔcclA::PcKan; KanR | CP2108×CP1863 |
| CP2115 | CP2108, but ΔcflAB::PcKan; KanR | CP2108×CP1869 |
| CP2116 | CP2108, but ΔcelAB::PcKan; KanR | CP2108×CP1862 |
| CP2117 | CP2108, but ΔcglABCD::PcKan; KanR | CP2108×CP1868 |
| CP2118 | CP1250, but ΔradA::PcSpc; SpcR | This work |
| CP2119 | CP2108, but ΔradA::PcSpc; SpcR | CP2108×CP2118 |
| CP2125 | CP2108, but ΔclpP::PcTet; TetR | CP2108×CP1359 |
| CP2126 | CP2125, but ΔcbpD::PcKan; KanR | CP2125×CP1275 |
| CP2127 | CP2125, but ΔcibABC::PcKan; KanR | CP2125×CP1279 |
| CP2128 | CP2125, but ΔcglEFG ::PcKan; KanR | CP2125×CP1333 |
| CP2129 | CP2125, but ΔdprA::PcKan; KanR | CP2125×CP1389 |
| CP2130 | CP2125, but ΔcoiA::PcKan; KanR | CP2125×CP1793 |
| CP2131 | CP2125, but ΔcelAB::PcKan; KanR | CP2125×CP1862 |
| CP2132 | CP2125, but ΔcclA::PcKan; KanR | CP2125×CP1863 |
| CP2133 | CP2125, but ΔcglABCD::PcKan; KanR | CP2125×CP1868 |
| CP2134 | CP2125, but ΔcflAB::PcKan; KanR | CP2125×CP1869 |
| CP2135 | CP2125, but ΔradA::PcSpc; SpcR | CP2125×CP2119 |
| CP2139 | CP2108, but ΔssbB::PcKan; KanR | This work |
| CP2140 | CP2125, but ΔssbB::PcKan; KanR | This work |
| CP2143 | CP2108, but ΔPc-cinA::PcKan; KanR | This work |
| CP2144 | CP2125, but ΔPc-cinA::PcKan; KanR | This work |
| Escherichia coli | ||
| DH5α | F-recA1, endA1 hsdR17 phoA supE44 thi-1 gyrA96 | Invitrogen |
| Saccharomyces cerevisiae | ||
| NSY468 | MATa, trp1-901, leu2-3, l 12, ura3-52, his3-200, gal4Δ, gal80ΔGAL2-ADE2, LYS2::GAL1-HIS3, met2::GAL7-lacZ | [38] |
| NSY752 | MATα, trp1-901, leu2-3, l 12, ura3-52, his3-200, gal4Δ, gal80ΔGAL2-ADE2, LYS2::GAL1-HIS3, met2::GAL7-lacZ | [38] |
Crosses are indicated as recipient X donor genomic DNA.
Table 2. Primers used for strain construction.
| Primer | Sequence (5′-3′) | Size (kb) | Construct | |
| Kan | ||||
| DAM303 | AAGGGCCCGTTTGATTTTTAATG | 0.8 | KanR marker | |
| DAM304 | AGGATCCATCGATACAAATTCCTC | |||
| Δcps3 | ||||
| TTM01 | ATCATGACCTCCCTCGTATTGT | 0.8 | Upstreamfragment | |
| TTM02 | CGCGGATCCTTAATAGTGGGAATTTG | |||
| DAM823 | CGCGGATCCTTGGAGTTAGAATAGGGCA | 1.5 | Downstreamfragment | |
| DAM827 | GCCTCATCACCAGCCTCAGTAAC | |||
| ΔssbB | ||||
| DAM934 | GCATGGGCCCTGAAGAAAGCAGACAAGTAAGC | 0.7 | Upstreamfragment | |
| DAM935 | GGCCTATCTGACAATTCCTG | |||
| BVD 104 | ATGGATCCTGCCATTTTAAGAATTAAAAAGTC | 1.0 | Downstreamfragment | |
| BVD 105 | GACTCTTCGATGGTGATGACACCGTCTTTG | |||
| ΔcoiA | ||||
| BVD26 | AAACGGGAGTCTATCAAACGTCGTGAGCAA | 0.8 | Upstreamfragment | |
| BVD27 | ATGGATCCTGAATTCCCTCCTTTTCTATATCAT | |||
| BVD28 | ATGGGCCCGAATAGAAAGGATGGAGGAATCTAA | 1.5 | Downstreamfragment | |
| BVD29 | GTAGACATCGTACATCTTGAGATCTGAAAT | |||
| ΔcibABC | ||||
| DAM305 | CAAGGACTGACTAGGTAAACAGC | 0.7 | Upstreamfragment | |
| DAM306 | GCTAGGATCCGAGGGCACTCTTGTCTGG | |||
| DAM307 | ACGAGGGCCCGATAGCAAAAGCAAATAA | 0.9 | Downstreamfragment | |
| DAM308 | CAAGAGGCCGTGTTCTTCGAG | |||
| ΔcbpD | ||||
| DAM313 | AGCTTTCTCGTGGTGTAGAACAAC | 1.6 | Upstreamfragment | |
| DAM314 | ACGAGGATCCGATCCATTTCCTCTGGAATA | |||
| DAM315 | AGCAGGGCCCAGGTCTCTGGTAAGTGGTAT | 0.8 | Downstreamfragment | |
| DAM316 | CTCTCAAGGTCGCCCAGCTATG | |||
| ΔcglEFG | ||||
| DAM419 | CTGTAATTGAGCCTCCGTTACCAATATG | 1.2 | Upstream fragment | |
| DAM420 | ATGGATCCGAGTCTGGTTGCTATGATTAGTCT | |||
| DAM421 | ATGGGCCCTTAGCTACCCTCAAGACTTCTTC | 1.5 | Downstream fragment | |
| DAM422 | TTGTGCAGACCTACTTGACAGCCTATTATG | |||
| ΔdprA | ||||
| DAM563 | GATAGAGGCGATAAGCATGGCACATAGTAA | 1.0 | Upstream fragment | |
| DAM564 | ATGGGCCC-TGCCATCATTTGATTCAAGAAG | |||
| DAM565 | GGATCC-ATAACGGCTGGATTACGGCAACCT | 2.0 | Downstream fragment | |
| DAM566 | GATTGGGAACTCGCTTGCGTCCTATGACTGA | |||
| ΔcelA | ||||
| DAM659 | CTAATTCTGGAGCAGGCGGCCATGTG | 1.1 | Upstream fragment | |
| DAM660 | CGCGGATCC-TTTCAACTGCTTATTTATTTGC | |||
| DAM661 | ACGTGGGCCC-GGAAGGATAAATGTTGTAGATTAG | 0.8 | Downstream fragment | |
| DAM662 | TGAGCCAGCATTTGGCCTGACTGAG | |||
| ΔcclA | ||||
| DAM663 | TGTTGAGTGGCGACGATAAATAAGG | 1.1 | Upstream fragment | |
| DAM664 | CGCGGATCC-TAGTATAATGGAGAAACATAGATAAG | |||
| DAM665 | ACGTGGGCCC-TTGTTTGATAAAGTCCAATTTC | 1.2 | Downstream fragment | |
| DAM666 | AACAAGCCATTTGGCAGTTTGAGTC | |||
| ΔcglABCD | ||||
| DAM680 | TGCAGCGTAGCCATTATTGGTTCAG | 1.2 | Upstreamfragment | |
| DAM679 | CGCGGATCC-TCCTCACCTATACTATTCGCAAAG | |||
| DAM682 | ACGTGGGCCC-TGATTTTACTGGAAGCAGTAGTC | 1.0 | Downstreamfragment | |
| DAM681 | ATCCGTACGAACCCTCGTCACTAAG | |||
| ΔcflAB | ||||
| DAM684 | TTCAATCATGCTAAGGGCAATACGG | 1.2 | Upstreamfragment | |
| DAM683 | CGCGGATCC-AATCATGGAATTTAGGACAATTAAAG | |||
| DAM686 | ACGTGGGCCC-TCATAAAAACAAAAATGTTTAG | 1.0 | Downstreamfragment | |
| DAM685 | ACGTGGGCCCTCATAAAAACAAAAATGTTTAG | |||
| ΔPcom-cinA | ||||
| DAM936 | GCATGGGCCCCGCAGGAATTTTCCTACGATTG | 1.0 | Upstreamfragment | |
| DAM937 | CAAGGGACAGAAACCTTAGC | |||
| DAM938 | GTCAGGATCCGAGTGGCAGGACCAGATAG | 1.2 | Downstreamfragment | |
| DAM939 | GGTGCTCTGCCAAGTATTTC | |||
| aga::comXcomW::PcKan | ||||
| DAM786 | AAACTGGGTGGAAGTCTAGAAAGTC | 1.4 | aga::comX | |
| PL82 | CGCGGATCCTGACTTACTAATGGGTACG | |||
| DAM791 | ATCGAATTCGGATCCGTTTGATTTTTAATGG | 0.4 | PcKan:rafE | |
| DAM793 | AACATCGGTATAGCCAGCACCTTCC | |||
| DAM794 | ATCGGATCCAAAAAAGAAAAGGAGTATTTGA | 2.0 | comW | |
| DAM790 | CTAGAATTCCTCAACAAGAAATAAACCCCC | |||
| pACT2 or pGBDUC2 inserts | ||||
| DAM967 | gtaggatccAGTTATTTATGAAAATCACAAACTATGAAATCT | 0.9 | pGBDUC2::dprA | |
| DAM968 | gcatgtcgacTTAAAATTCAAATTCCGCAAGAACATC | |||
| DAM969 | cagtggatccAAAGAGTAATGGATTTATTTGGATTTG | 1.3 | pACT2::comD | |
| DAM970 | gcatctcgagCTTTCATTCAAATTCCCTCTTAAATCTA | |||
| DAM971 | gtacggatccGAATGAAAGTTTTAATTTTAGAAGATGTTATTG | 0.8 | pACT2::comE | |
| DAM972 | gcatctcgagTCAATCACTTTTGAGATTTTTTCTCTAA | |||
| DAM973 | gtacggatccAGGGGAAAATTATGATTAAAGAATTGTAT | 0.5 | pACT2::comX | |
| DAM974 | gcatctcgagCTAATGGGTACGGATAGTAAACTC | |||
| DAM975 | gtcaggatccTTATGTTACAAAAAATTTATGAGCAGATG | 0.3 | pACT2::comW | |
| DAM976 | gcatctcgagTACTAAAATTACCTCAACAAGAAATAAAC | |||
| DAM989 | gactggatccGAATGGCGAAAAAACCAAAAAAATTA | 1.2 | pACT2::recA | |
| DAM990 | gtcactcgagCAGCTTATTCTTCAATTTCGATTTCA | |||
Table 3. Plasmids used in this studies.
| Plasmid | Description | Source |
| pACT2 | shuttle vector for yeast 2-hybrid, carrying Gal4 activating domain (AD) | Clontech Labs, Inc. |
| pGBDUC2 | shuttle vector for yeast 2-hybrid, carrying Gal4 DNA binding domain (BD) | [38] |
| pACT2-comD | pACT2 derivative, carrying AD-comD fusion | This work |
| pACT2-comE | pACT2 derivative, carrying AD-comE fusion | This work |
| pACT2-comX | pACT2 derivative, carrying AD-comX fusion | This work |
| pACT2-comW | pACT2 derivative, carrying AD-comW fusion | This work |
| pACT2-dprA | pACT2 derivative, carrying AD-dprA fusion | This work |
| pACT2-recA | pACT2 derivative, carrying AD-recA fusion | This work |
| pGBDUC2-dprA | pGBDUC2 derivative, carrying BD-dprA fusion | This work |
Transformation
For S. pneumoniae, cultures grown in CAT to OD550 = 0.03 were exposed to DNA (0.1 µg/ml) for 45 min after the treatment with CSP (0.25 µg/ml), CaCl2 (0.5 mM), and bovine serum album (0.04%). Transformants were selected in CAT solidified with 1.5% agar containing appropriate antibiotics. For E. coli, 60 µl desalted, electro-transformable cells in 10% glycerol were mixed with 3 µl plasmid DNA (∼100 ng) and electroporated with voltage 2.0 kV, resistance 200 Ω and capacitance 25 µF. After recovering in 1 ml LB broth medium at 37°C for one hour, the cell culture was spun down and plated. For yeast, 1 ml OD600>5 haploid cells were spun down and resuspended in 0.5 ml PEGLET buffer (40% PEG, 0.1 M LiAC, 10 mM EDTA, 0.1 M Tris-HCl, pH 7.5). About 100 µg ssDNA (salmon sperm DNA, Sigma) plus 0.5∼1 µg plasmid DNA were added to the resuspended cells. The cell culture was seated on the bench at room temperature overnight. Next day, the cells were spun down again and plated.
Creation of strain CP2000
To create the capsule-less strain CP2000 by use of the Janus replacement cassette [26], CP1250 was transformed with genomic DNA from strain R6J (R6S but cps::kan-rpsL +) [27], using kanamycin for selection. One transformant with the correct structure was retained as strain CP1999. CP1999 was transformed with a donor DNA created by the ligation of two BamHI digested PCR fragments. The first fragment was created by amplifying CP1500 genomic DNA with primers TTM01 and TTM02; the second fragment was created by amplifying the same genomic DNA with primers DAM823 and DAM827. A SmR transformant with the correct structure, removing genes between dexB and aliA, was retained as CP2000, after verification by PCR and sequencing of the new junction/deletion.
Construction of parental strains CP2108 and CP2125
The parent strain CP2108 was made by the crosses among three strains: CP2000, CPM7, and CP1415. CP2000 (Δcps), was transformed with CPM7 genomic DNA; a CmR transformant with pEVP3 inserted at ssbB and exhibiting CSP-dependent β-galactosidase activity was named CP2107. This new strain was further transformed with CP1415 genomic DNA, to incorporate a disrupted comA gene. The disrupted comA gene in an EmR transformant was confirmed by PCR and the strain was named CP2108. Then CP2108 was transformed with CP1359 (ΔclpP::PcTet) genomic DNA to make CP2125.
Creation of ectopic comXcomW strain, CP1896
To create strain CP1896 (aga::comX::comW::PcKan), two fragments were amplified from CP1372 DNA: one contained part of aga and a complete comX (primers DAM786 and PL82); the second contained a KnR cassette and part of rafE (primers DAM791 and DAM792). A third fragment containing comW was amplified from CP1500 using DAM794 and DAM790. After digestion by BamHI (Fermentas), and/or by EcoRI (Fermentas), the three fragments were purified, ligated, and used directly as donor for transforming strain CP1250 as described above. One KnR transformant was retained as strain CP1896 after sequencing the insert, which exactly matched the predicted sequence.
Preparation of late gene mutants
Late gene mutants used in this paper were obtained by transforming the parent strains (CP2108 and CP2125) with either the genomic DNA of strains containing the corresponding late gene disruption (ΔcbpD, ΔcibABC, ΔcglEFG, ΔcoiA, ΔdprA, and ΔradA), or with donor DNA synthesized by molecular cloning (ΔPccinA, ΔcelAB, ΔcclA, ΔcglABCD, ΔcflAB, and ΔssbB). For synthesizing donor DNA, two pairs of primers were used to amplify the upstream and downstream sequences individually. Then, the two sequences were ligated with a KanR cassette to make up the donor DNA for transformation.
Induction of competence using CSP
Cultures of pneumococcus were started by inoculating 200 ml complete CAT medium plus 10 mM HCl with 1/100 volume of a frozen stock of cells (OD550 0.1). At the first visible turbidity during growth at 37°C, 10 ml was transferred to a 18 mm by 150 mm tube for monitoring optical density. When the culture reached an OD550 of about 0.06, it was induced to competence with CaCl2 (to 0.5 mM), BSA (to 0.002%), and CSP (to 250 ng/ml). Samples were taken periodically from the culture for various analyses as described in subsequent sections.
Induction of competence using raffinose
Cultures of pneumococcus were started by inoculating 200 ml complete CAT medium plus 10 mM HCl with 1/100 volume of a frozen stock of cells (OD550 0.1). At the first visible turbidity during growth at 37°C, 10 ml was transferred to a 18 mm by 150 mm tube for monitoring optical density. When the culture reached an OD550 of about 0.05, the culture was transferred to 30°C. When the culture reached an OD550 of about 0.1, it was induced to competence with CaCl2 (to 0.5 mM), BSA (to 0.002%), and raffinose (to 0.1% w/v). Samples were taken periodically from the culture for various analyses as described in subsequent sections.
Sampling for western analysis
For Western blot analyses 1.8-ml samples were withdrawn from the culture, chilled rapidly on dry ice, without freezing, and then kept at 4°C until harvesting by centrifugation (10,000×g, 2 min, 4°C). After each cell pellet was resuspended with 35 µl loading buffer (50 mM Tris-HCl (pH 6.8), 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 100 mM dithiothreitol) and heated at 95°C for 10 min, 15 µl of the lysate was loaded into one lane of an SDS-PAGE gel.
SDS-PAGE
SDS-PAGE was done as described previously [24] using the Bio-Rad Mini-Protean II gel apparatus. Each gel was a 15-well, 1.5-mm-thick discontinuous gel composed of a 5% stacking gel and a 15% resolving gel prepared according to the manufacturer’s recommendations. Protein samples were prepared by mixing with one volume of loading buffer (100 mM Tris-HCl (pH 6.8), 4% SDS, 0.2% bromophenol blue, 20% glycerol, and 200 mM dithiothreitol) and heating at 95°C for 10 minutes. The gels were run in 25 mM Tris, 250 mM glycine, 0.1% SDS, pH 8.0 at 65 V until the dye reached the resolving gel, then at 120 V until the dye reached the bottom of the gel.
Western analysis
For Western analysis, SDS-PAGE gels were run as described above, washed 3 times for 5 min with 100 ml deionized water, and equilibrated for 15 min in transfer buffer (25 mM Tris, 192 mM glycine, 10% (v/v) methanol, pH 8.0). The proteins were trans-blotted from the gel to a PVDF membrane (Immobilon Psq, Millipore) in transfer buffer for 2 hr at 36 V at 4°C. The membrane was then blocked overnight at 4°C in TBS-T (20 mM Tris-HCl (pH 7.6), 137 mM NaCl, 1% (v/v) Tween-20) with 5% nonfat dry milk. The membrane was incubated for 90 min at room temperature with primary antibody, specific for ComW or ComX, diluted by 1∶3000 in 5 ml TBS-T with 1% nonfat dry milk in a small plastic pouch. After washing 3 times with 100 ml TBS-T, then incubating for 1 hr at room temperature with secondary antibody (Anti-Rabbit IgG conjugated to HRP, Amersham) diluted 1∶20 000 in TBS-T with 1% nonfat dry milk and then washing 3 times with 100 ml TBS-T, the position of secondary antibody on the membrane was detected using an ECL substrate (ECL Plus, Amersham) and either Hyblot CL film (Denville Scientific) or the Alpha Imager CCD Camera (Alpha Innotech). Typical exposure times were 1 to 5 min for the film and 5 to 15 min for the CCD camera. Quantification was done by spot densitometry using AlphaEaseFC (Alpha Innotech).
β-galactosidase assay
For measurement of β-galactosidase activity, a 0.4-ml sample of liquid culture was chilled on ice. After adding 100 µl 5×Z buffer (300 mM Na2HPO4, 200 mM NaH2PO4, 50 mM KCl, 5 mM MgSO4, 250 mM β-mercaptoethanol, 0.5% TritonX-100) and incubation at 37 C for 10 min to lyse the cells, 150 µl of the resulting lysate was combined with 50 µl of o-nitrophenyl-β-D-galactopyranoside solution (4 mg/ml o-nitrophenyl-β-D-galactopyranoside, 60 mM Na2HPO4, 40 mM NaH2PO4) in a 96-well microplate. The plate was incubated at 37 C, and absorbance at 420 nm was read every 10 min for 90 min in Spectra Max M2 of Molecular Devices. The initial slope of the absorbance curve was used to calculate LacZ activity, reported in Miller units.
α-galactosidase assay
For α-galactosidase activity measurement, a 0.4-ml culture sample was added to 0.1 ml of 5×lysis buffer (405 mM Na2HPO4, 95 mM NaH2PO4, 6 mM MgCl2, 27 mM β-mercaptoethanol, 0.5% Triton X-100) and incubated at 37°C for 10 min. After 150 µL of the resulting lysate was added to 50 µL of p-nitrophenyl-β-D-galactopyranoside (PNPG) solution (0.9 mg/ml PNPG, 81 mM Na2HPO4, 19 mM NaH2PO4) in a 96 well microplate, absorbance was measured at 405 nm every 5 min for 30 min at room temperature by a microplate reader (Molecular Devices’ Spectramax M2).
Yeast two-hybrid assay
A yeast two-hybrid assay was established by inserting dprA (prey) into yeast plasmid pGBDUC2 (containing DNA binding domain of GAL4 transcription factor), and inserting comD, comE, comX, comW, and recA (baits) into yeast plasmid pACT2 (containing transcription-activating domain of GAL4), using PCR followed by restriction enzyme digestion and ligation at BamHI and XhoI, or SalI sites. The two plasmids with corresponding inserts were first transformed into E. coli (DH5α) to replicate and purified with QIAprep Miniprep kit. The purified plasmid DNA was then transformed into yeast haploids PNS468 (MATE-a) and PNS752 (MATE-α) respectively. The two transformed haploid cells with different mating types were selected in SD medium without leucine (SD-Leu) or SD medium without uracil (SD-Ura) and mated to make diploids in SD agar plate without both (SD-Leu-Ura). The diploids were collected and stored at −80°C in 20% glycerol stocks. Diploids were grown again on YPD agar plates and frog-replicated onto four test agar plates: SD-Leu-Ura; SD-Leu-Ura-His; SD-Leu-Ura-His+1 mM 3AT; SD-Leu-Ura-His+3 mM 3AT [28]. The four test plates were photographed daily during incubation at 26°C for 7 days.
Results
Among Candidate Late Competence Genes Examined, Only dprA is Required for Normal Shutoff of Late Gene Expression in a Wild-type Background
To investigate the roles of additional late genes in competence termination, we monitored the expression pattern of a late gene transcriptional reporter, which is established as a reliable indicator of competence development [17]. For this purpose, we constructed a parent strain that contains a lacZ reporter at the late gene, ssbB and is also comA deficient, so as to avoid any potential growth deficiency that might be caused by prolonged competence in a mutant deficient in exit from competence. The late gene expression pattern in response to CSP in this reporter parent strain was verified as identical to that reported previously [7], [9], [16], [29]. Mutations in genes with known roles in transformation were crossed into the parent strain. Included were cibABC and cbpD, which affect fratricide, and cinA, linked to the cinbox promoter required for induction of recA by CSP. The internal deletion of each gene or operon was replaced by a KnR marker. Altogether, we examined 12 mutants, defective in a total of about 20 late genes (circled with in blue ovals in Fig. 1): ΔcbpD, ΔcibABC, ΔssbB, ΔcglEFG, ΔcoiA, ΔdprA, ΔcelAB, ΔcclA, ΔcglABCD, ΔcflAB, ΔradA and ΔPc-cinA. Each mutation’s structure was confirmed by PCR, and the border of each deletion is shown in Fig. 1. Since the transformation efficiency of these mutants was already known to be very low, we verified that transformation rates for the new strains were comparable to the values expected from the literature (Table 4).
Table 4. Comparison of transformation efficiency of new strains with literature.
| New allelecombination | Relative transformation rate | |||
| Strain(s) | Mutation | Literatureb | Experimentala | |
| ClpP+ | ClpP− | |||
| CP1250, CP2000 | Δcps | 1 | ||
| CPM7 | ssbB−::lacZ::ssbB+ | 1 | ||
| CP1359 | ΔclpP::PcTet | 1 | ||
| CP2108, CP2125 | ΔcomA::PcErm | 1 | 1 | 1 |
| CP2109, CP2126 | ΔcbpD::PcKan | 1 | 1 | 1 |
| CP2110, CP2127 | ΔcibABC::PcKan | 1 | 1 | .4 |
| CP2111, CP2130 | ΔcoiA::PcKan | .01 | .001 | .001 |
| CP2112, CP2128 | ΔcglEFG::PcKan | 0 | .001 | .001 |
| CP2113, CP2129 | ΔdprA::PcKan | 0 | .0001 | .001 |
| CP2116, CP2131 | ΔcelAB::PcKan | 0 | .001 | .001 |
| CP2114, CP2132 | ΔcclA::PcKan | 0 | .001 | .0001 |
| CP2117, CP2133 | ΔcglABCD::PcKan | 0 | .0001 | .0001 |
| CP2115, CP2134 | ΔcflAB::PcKan | 0 | .0001 | .0001 |
| CP2119, CP2135 | ΔradA::PcSpc | 0 | .001 | .001 |
| CP2139, CP2140 | ΔssbB::PcKan | .3 | .3 | 0.5 |
| CP2143, CP2144 | Äcinbox-ÄcinA::PcKan | 1 | 1 | 1 |
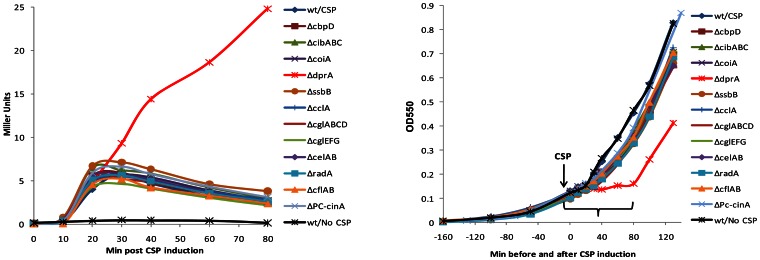
To determine the effect of each of the 12 mutations on exit from competence, each mutant reporter strain was treated with CSP under standard conditions and sampled 0, 10, 20, 30, 40, 60, or 80 minutes later for LacZ assay. The enzyme levels, which indicate late gene expression patterns, are shown as a function of the time after CSP induction in Fig. 2. Nearly all of these mutants displayed normal (brief) patterns of late gene expression, with a burst of LacZ synthesis restricted to the period between 10 and 30 min after addition of CSP. The single exception was the dprA mutant, in which late gene expression continued beyond 60 min The pattern shown in Fig. 2 reproduces the growth defect reported by Bergé [17] for the R6 strain, and further supports his interpretation by showing directly that expression of a late gene continues for an unusually long time after exposure of a dprA mutant to CSP; that is, the cells appear to remain in an active X-state for a greatly extended period when DprA is missing.
Figure 2. Survey of effect of late gene mutations on exit from the competent state.
A. Late gene expression patterns were monitored in wild type and late gene mutants after CSP induction using a lacZ reporter inserted at the late gene ssbB. β-galactosidase activity (Miller units) was measured in culture samples harvested at indicated times after CSP induction. Each strain is indicated by its mutated competence gene (see Table 1). B. Growth patterns monitored as culture optical density at 550 nm before and after CSP treatment. Bracket indicates period of LacZ assay shown in panel A.
The greatly prolonged expression of a late gene suggests a prolonged presence of ComX and thus that DprA affects not only expression of the comCDE operon, but also expression of the duplicate comX genes. Combined with the report of Mirouze [18] that premature expression of dprA repairs the transience defect of a comX mutant, these results suggest that DprA is both necessary and sufficient for the shutoff of early gene expression. From the absence of any similar effect of the other late gene mutations on the temporal pattern of ssbB expression we conclude further that none of the other late gene products examined has a strong role in shutoff of early gene expression. Because of the uniquely strong effect of DprA on shutoff, it is of interest to determine its effects on gene expression in more detail, both to know if it is the only shutoff agent, and to identify its regulatory targets.
The Kinetics of Exit from Competence is not Altered in a ClpP Protease-deficient Background
We were concerned to be able to detect regulation of comX at multiple levels. While the effect of DprA shown above is dramatic, as expected for a factor controlling early gene expression, the change in late gene expression kinetics might be more subtle for mutation of a gene reducing ComX activity directly. Thus, for all late gene mutants tested above, except the ΔdprA mutant, levels of ComX and ComW would be expected to start to decline by ∼30 minutes after CSP induction, simply due to the rapid dprA-dependent halt to early gene expression and subsequent decay of ComX and ComW that had accumulated during the brief window of ComE activity. Therefore, any extension of late gene expression occasioned by mutation of a gene acting specifically to suppress ComX activity during this window might be modest in length and difficult to detect. To extend the window of ComX availability and thus improve the chance of detecting such a ComX-dependent shutoff gene targeting ComX itself, we decided to study the temporal pattern of late gene expression in late gene mutants in a protease-deficient background (ΔclpP), in which both ComX and ComW would be stabilized. ClpP and ClpE are largely responsible for the proteolysis of ComX and in strains deficient for either of the two proteins, ComX becomes stable [30]. Similarly, ClpP and ClpC are largely responsible for the proteolysis of ComW and in strains deficient for one or the other protease subunit, ComW is stable [12].
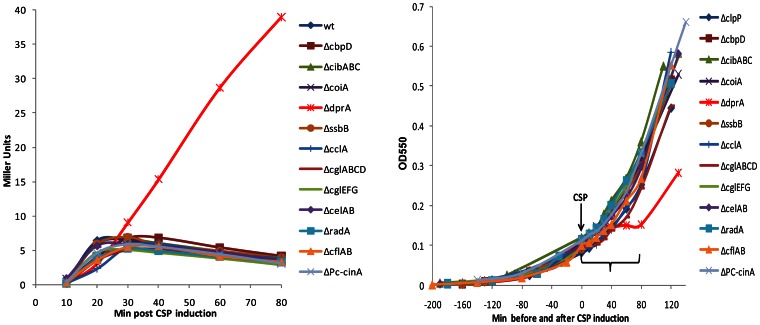
Adopting the same strategy as in the previous section, a new parent strain, CP2125, was made from the original one by disruption of the clpP gene, to increase stability of both ComW and ComX proteins. From this new parent strain, we again obtained 12 late gene mutants: ΔcbpD, ΔcibABC, ΔssbB, ΔcglEFG, ΔcoiA, ΔdprA, ΔcelAB, ΔcclA, ΔcglABCD, ΔcflAB, ΔradA and ΔPc-cinA, and verified their structures and competence phenotypes as above. These mutants were analyzed for their late gene expression pattern after induction by CSP. In the ΔclpP background, we observed the same result as in the protease-proficient wild type: only the dprA mutant displayed prolonged late gene expression (with an elevated β-galactosidase activity, which may be due to the deficiency in host Clp proteases (Fig. 3). Since our survey of ∼19 late genes other than dprA did not reveal any whose loss extends the X-state when the half-lives of ComX and ComW are prolonged by interruption of their proteolysis, we conclude that none of these gene products is individually responsible for suppression of ComX activity during exit from competence.
Figure 3. Survey of effect of late gene mutations on exit from the competent state in the protease deficient ΔclpP background.
A. After CSP induction, late gene expression patterns were monitored in wild type and late gene mutants in ΔclpP background. The lacZ reporter and the measurement of β-galactosidase activity were done as described for Figure 2. Each strain is indicated by its mutant competence gene as listed in Table 1. B. Growth patterns monitored as culture optical density at 550 nm before and after CSP treatment. Bracket indicates period of LacZ assay shown in panel A.
Accumulation of the Early Gene Products, ComX and ComW, is Increased and Prolonged in a dprA Mutant
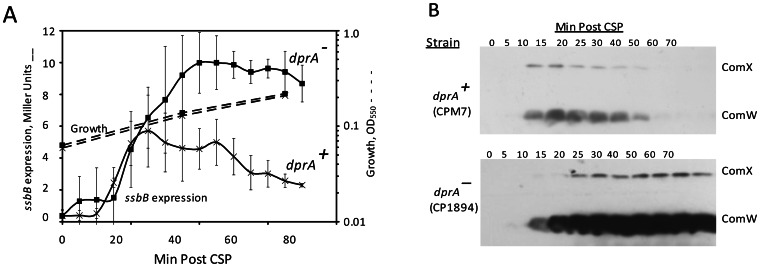
The prolonged late gene expression in a ΔdprA mutant might in principle reflect abrogation of a direct effect of DprA on late gene expression or interruption of an effect of DprA at the level of ComE, affecting early gene expression. These possibilities could be distinguished by comparing the accumulation of the early gene products, ComX and ComW, in a dprA mutant to that in wild type. If they displayed comparable levels of ComX and ComW, it would suggest action of DprA on late gene expression; whereas higher levels of ComX and ComW in the dprA strain would indicate that DprA shuts off expression of these additional early genes. To test the possibility that DprA negatively regulates the amounts of ComX and ComW produced during competence development, we used Western blotting to compare the levels of ComX and ComW in dprA + and dprA − strains upon treatment with CSP. As shown in Figure 4, both ComX and ComW accumulated too much higher levels in the dprA mutant and remained at a high level even after 70 min of exposure to CSP. In contrast, in the dprA+ strain ComX and ComW reached a lower maximum at 15–20 min, and then declined below detectable levels by 50 min. We conclude that dprA mutation prolonged competence by enhancing the accumulation of the early gene products, ComX and ComW.
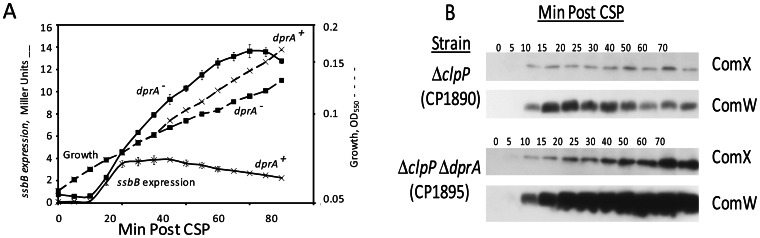
Figure 4. Prolonged appearance of ComX and ComW in a dprA mutant.
A. Late gene expresion in strains CPM7 (x, dprA +) and CP1894 (□, ΔdprA) was monitored by measuring β-galactosidase activity (Miller units) (−) in a culture treated with CSP, using a lacZ transcriptional fusion to the ssbB promoter. Points are averages of three samples and error bars represent their standard deviation. –, growth of parallel non-CSP treated cultures. B. Western analysis of samples taken in parallel to samples for late gene expression run in a separate gel, transferred to a separate membrane for each strain and probed with antisera specific to both ComX and ComW using the ECL substrate and imaged on a separate film for each strain. Each lane represents a cell lysate from 0.6 ml of culture.
The Effect of DprA on the Accumulation of ComX and ComW does not Depend on ClpP
In light of the lability of ComX and ComW proteins, the difference in the CSP-induced levels of ComX and ComW between dprA − and dprA + strains might have two quite different explanations: 1) DprA may promote the proteolysis of ComX and ComW, acting as chaperon or adaptor that greatly stimulates the targeting of these proteins to the proteases responsible for their lability, ClpEP and ClpCP, respectively; or 2) DprA may inhibit the production of ComX and ComW, at either translational or transcriptional stages. To distinguish experimentally whether the increase in amounts of ComX and ComW caused by dprA mutation reflects a negative effect on proteolysis of the labile ComX and ComW proteins or, in parallel to the effect of DprA on expression of the comCDE operon, an enhancement of the rate of synthesis of these proteins, we compared the amounts of ComX and ComW in a ΔclpPdprA+ mutant to the amounts in a ΔclpP dprA − mutant. As ClpP is necessary for the proteolysis of both ComX and ComW, both proteins are stable in a ΔclpP background and an anti-proteolytic mechanism would have no effect in that context.
Late gene expression differed radically between the two strains was (Figure 5), consistent with the result in Figure 4. The amounts of ComX and ComW, as determined by Western blot, continued to increase for more than 50 min in the dprA clpP mutant, whereas the levels of the two proteins became constant after 15 min in the dprA+ clpP mutant strain. The continued accumulation of both proteins well after the time that transcription of early genes would stop in a wild type strain [7] shows that DprA acts to block transcription (or translation) of both genes, not to promote proteolysis of these labile early proteins. Since ComX and ComW belong to distinct operons in the early class, and DprA suppresses transcription of the early operon comCDE [17], this now shows that three early operons respond similarly to deprivation of DprA. We conclude that DprA suppresses the transcription of at least three, and likely all, early genes.
Figure 5. Prolonged accumulation of ComX and ComW in aΔclpPdprA − mutant but not in a ΔclpP mutant.
A. Late gene expression in strains CP1890 (x, dprA+ ΔclpP) and CP1895 (□, ΔdprA ΔclpP) was monitored by measuring β-galactosidase activity (Miller units) (–) in a culture treated with CSP. The lacZ reporter was in a transcriptional fusion to the ssbB promoter. Points are averages of three samples and error bars represent their standard deviation. –, growth of the cultures. B. Western blot analysis of samples taken in parallel to samples for late gene expression run in a separate gel, transferred to a separate membrane for each strain and probed with antisera specific to both ComX and ComW using the ECL substrate and imaged on a separate film for each strain. Each lane represents a cell lysate from 0.6 ml of culture.
DprA Interacts with ComE
In view of the effect of DprA on early gene expression, we made a limited search to distinguish among possible molecular targets, through a yeast two-hybrid screen for protein interactions. ComD or ComE, the major players in turning on early gene expression, were the major candidates in our screen. Because DprA interacts with RecA [31], a recA insert was adopted as a positive control. Furthermore, because of results indicating that DprA does not affect late gene expression (described below), comX and comW inserts were included as negative controls. After confirmation of the constructions, pGBDUC2 and pGBDUC2-dprA were transformed into yeast haploid NSY752(α). On the other hand, pACT2, pACT2-comD, pACT2-comE, pACT2-comX, pACT2-comW and pACT2-recA were transformed into yeast haploid NSY468(a). Six diploids were obtained by mating between NSY752(α) and NSY468(a) cell lines, as indicated in Figure 6.
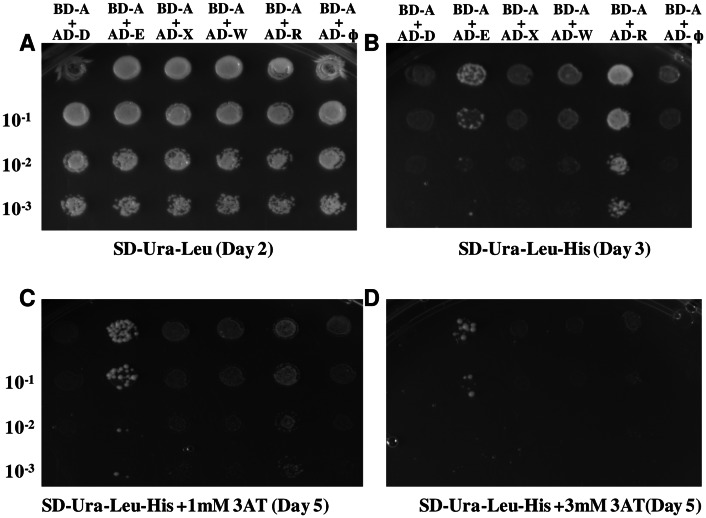
Figure 6. Screening of candidate DprA targets with yeast two-hybrid assay.
Images were taken at indicated time during the incubation of the diploids (BD-A+AD-D, BD-A+AD-E, BD-A+AD-X, BD-A+AD-W, BD-A+AD-R, BD-A+AD-<$>\raster(70%)="rg1"<$>) in four kinds of agar plates for five days: SD-Ura-Leu (A), SD-Ura-Leu-His (B), SD-Ura-Leu-His+1 mM 3AT (C) and SD-Ura-Leu-His+3 mM 3AT (D). AD, activating domain of pACT2; BD, DNA binding domain of pGBDCU2; A, dprA; D, comD; E, comE; X, comX; W, comW; R, recA; SD, synthetic defined medium; Ura, uracil; Leu, leucine; His, histidine; 3AT, 3-Amino-1,2,4-triazole.
After incubation of the diploids in four kinds of media (SD-Ura-Leu, SD-Ura-Leu-His, SD-Ura-Leu-His+1 mM 3AT and SD-Ura-Leu-His+3 mM 3AT), all diploids grew on SD-Ura-Leu (Figure 6A), confirming that plasmids pGBDUC2 and pACT2 (or their derivatives) were harbored respectively in the diploids. But in the plates without histidine (SD-Ura-Leu-His), only the diploids containing inserts dprA+comE and dprA+recA produced colonies. This indicates that DprA can interact with ComE as strongly as it interacts with RecA (Figure 6B). The growth in plates without histidine but with the competitive inhibitor of the production of histidine, 3AT, further confirmed the interaction between DprA and ComE is positive and strong, perhaps even stronger than the interaction between DprA and RecA because the diploid with dprA and recA inserts was unable to grow in the media SD-Ura-Leu-His+1 mM 3AT and SD-Ura-Leu-His+3 mM 3AT after a week incubation (Figure 6C, 6D). Further verification of the interaction between DprA and ComE was achieved with yeast two-hybrid by incorporating more negative controls, including double empty vectors and one empty vector with one vector plus insert (data not shown). Taken together, these results suggest a direct interaction between DprA and ComE, an interaction that might mediate its strong negative effect on expression of early genes.
Late Gene Expression is Independent of DprA after Ectopic Induction of comX and comW
The prolonged transcription of the late gene reporter seen in dprA mutants contrasts with the rapid extinction of late gene expression in clpP mutants despite the continued presence of high levels of the otherwise labile ComX and ComW [12], suggesting that DprA or some other late com gene product might also play an additional regulatory role by affecting late gene transcription directly, supplementing any effects on early gene expression and the amount of ComX and ComW produced.
Since ComX and ComW are the only early gene products required for high levels of late gene transcription and cells can become fully competent when comX and comW are ectopically expressed under CSP-independent regulation [15], we sought to decouple late gene expression from early gene expression by introducing both comX and comW at the aga locus, to allow their expression under regulation by the raffinose-inducible aga promoter. By inducing competence development with raffinose while by-passing expression of the other early genes, DprA’s possible effect on late gene expression could thus be separated from its effect on early gene expression. The gene that codes for α-galactosidase, aga, was retained in the construct, so that expression from the Aga promoter could be monitored by measuring α-galactosidase activity [32], while the ssbB::lacZ reporter allowed monitoring of late gene expression by measuring β-galactosidase activity. To stabilize ComX and ComW, the new strain was also deficient in the ClpE and ClpC ATPases. The new strain, with ectopically regulated comX and comW, CP1902 (aga::comX::comW, ΔclpC, ΔclpE, ssbB::lacZ::ssbB+), was transformed with the dprA mutation of strain CP1389, to create the isogenic dprA derivative, strain CP1932.
Since the entire CSP sensing circuit is intact in both the ectopic comX comW strain CP1902 and the isogenic dprA derivative (CP1932), the two strains could be treated with CSP to induce competence development through the activity of early genes, or treated instead with raffinose, to induce competence without expression of any early genes other than comX and comW. When treated with CSP, the two different strains responded with different expression patterns, as expected, confirming the effect of the dprA mutation on the ‘early’ regulatory pathway (Fig. 7). When the two strains were treated instead with raffinose, the patterns of late gene expression in the dprA + and dprA − strains were identical (Fig. 7), except for a somewhat broader curve compared to CSP-induced late gene expression. The broader pattern reflects a slower rise in raffinose-induced ComX and ComW expression. Similar results were obtained with the aga::comX nis::comW strain constructed by Luo (data not shown). As dprA thus had no effect on late gene expression driven by ectopically expressed ComX and ComW, we conclude that DprA does not affect late gene expression by any direct effect on the activity of the alternative sigma factor, ComX. Instead, expression of the early genes via the CSP sensing circuit is necessary to reveal an effect of DprA on late gene expression. Logically, this also suggests that the difference in expression in the CSP treated dprA − vs dprA + cells is due solely to DprA’s effect on the regulation of early gene expression.
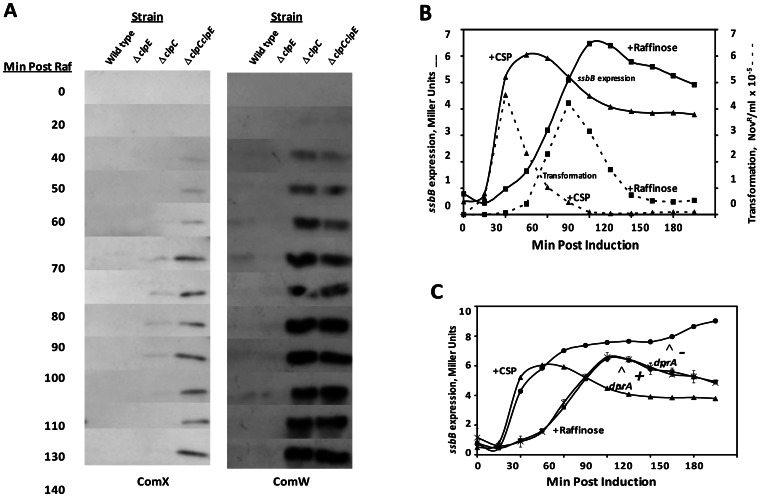
Figure 7. Lack of effect of DprA on late gene expression in an ectopic comXcomW strain.
A. Western blot analysis of samples taken at different times after induction with raffinose. Wild type, CP1896 (aga::comX::comW); ΔclpE, CP1962 (CP1896, but ΔclpE); ΔclpC, CP1963 (CP1896, but ΔclpC); ΔclpE ΔclpC, CP1964 (CP1896, but ΔclpE ΔclpC). B. Kinetics of competence induction and late gene expression induced with either CSP (Δ) or raffinose (□). During treatment of CP1902 (aga::comX::comW, ssbB::lacZ, ΔclpC,ΔclpE,) with either CSP (250 ng/ml) or raffinose (0.1%) at 30°C, samples were taken in parallel, to monitor transformation (–) and α-gal activity (–). β-gal activity is expressed in Miller Units with respect to the OD of the culture at each time point. C. Late gene expression of CP1902 treated with either CSP (Δ) or raffinose (□) or CP1932 (CP1902, ΔdprA) with either CSP (•) or raffinose (x) at as 30°C for B.
CSP Induces Higher Levels of ComX and ComW in a dprA− Background than does Raffinose in the Ectopic ComX/ComW Expression Regime
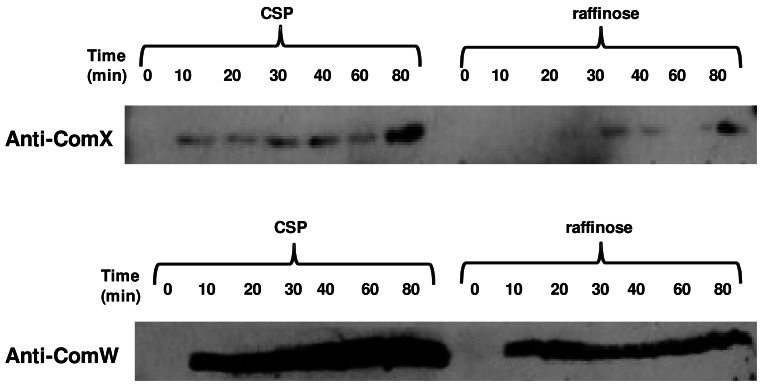
Since late gene expression induced by raffinose was independent of a functional dprA gene, we infer that there may be another regulator, which reverses late gene expression soon after it begins. We further hypothesize that the apparent failure of this inhibition in the dprA mutant induced by CSP (Fig. 2 and Fig. 5) could be explained if ComX and ComW were produced in amounts far above those accumulated in the ectopically induced cells of Fig. 7. When ComX and ComW were produced in much higher amount, they might overwhelm the hypothetical regulator, without being affected by it.. To test this hypothesis, we compared levels of both ComX and ComW in CP1932 induced by CSP to the levels in the same strain induced by raffinose, sampling during 80 minutes after addition of the respective inducers. Two parallel SDS-gels were run, one probed with anti-ComX antibody and another probed with anti-ComW antibody. CSP-induced levels of both ComX and ComW were indeed much higher than those induced by raffinose (Figure 8), even while late gene expression, as represented by a ssbB reporter, though prolonged, was not increased in rate (Fig. 1, Fig. 7). Thus, the greatly elevated ratio of ComX to a hypothetical late gene product acting as an inhibitor could explain why late gene expression patterns in dprA mutants shut off after raffinose induction but not after induction by the native CSP pheromone (in Figure 7), and indicates the existence of another competence repressor.
Figure 8. Reduced levels of ComX and ComW under ectopic regulation of competence.
Late gene expression was induced in strain CP1932 (aga::comX::comW, ssbB::lacZ, ΔclpC, ΔclpE ) by 250 ng/ml CSP or 0.1% raffinose respectively at OD 0.1. The cultures were sampled at 0, 10, 20, 30, 40, 60, and 80 min after induction for western blotting assay. Two SDS-PAGE gels were run in parallel and probed with different antibodies: Top, PVDF membrane probed with anti-ComX antibody; Bottom, PVDF membrane probed with anti-ComW antibody.
Discussion
The activity of alternative sigma factors is often controlled quite strictly at multiple levels, perhaps because they can cause a global shift in gene expression, which could be especially harmful if carried out under inappropriate circumstances. In the case of the alternative sigma factor central to competence development in S. pneumoniae, at least five distinct mechanisms of regulation are already established or glimpsed. (I) Transcription of comX depends on a TCSTS that coordinates within local populations and responds to unknown elicitors. (II) Production of ComX protein depends on an early competence gene, comW. (III) Separately, activity of ComX depends on ComW. (IV) ComX is labile, targeted by the ClpEClpP ATP-dependent protease. (V) ComW is labile, targeted by the ClpCClpP ATP-dependent protease. With the observations reported here on escape from the X state, two additional mechanisms that regulate the activity of ComX or ComW can be considered, both apparently forming negative feedback loops that ensure that induction of late gene expression is self-limiting and transient. One mechanism requires DprA and targets ComE, perhaps through direct interaction between these two proteins themselves; the other is less well defined, but is independent of dprA and appears to target ComX or other determinant of transcription of late genes.
Since ComX and ComW are central players during competence development and the disappearance of ComX and ComW occurs at about the same time as the loss of transformation [7], [9], [12], [14], it is attractive to suggest that the disappearance of ComX and ComW itself accounts for the shutoff of late gene expression [2], [3]. However, the relative timing of events during response to CSP is difficult to reconcile with this simple mechanism. Specifically, late gene mRNA largely disappears even before the levels of ComX and ComW proteins begin to drop, suggesting that the activity of ComX is itself subject to some additional form of control [6], [8], [12], [14]. As some non-proteolytic factor thus seems to play a major additional role in the shut off of ComX activity, we began to look for possible candidates among ComX-induced late genes. Peterson et al [7] screened mutations of many of the late genes for effects on the rate of exit from competence, but found none that caused a pronounced extension of the period of transformation. Transformation defective mutants were not examined for this phenotype, however, prompting us to investigate the latter class of late genes directly. dprA mutants treated with CSP displayed a prolonged period of expression of the late gene ssbB, in addition to the prolonged expression of the early gene operon comCDE previously described as a mutant phenotype for dprA by Bergé [17] and by Mirouze et al [18]. Comparison of levels of ComX and ComW in dprA mutants vs wild type in protease defective backgrounds revealed that DprA not only turns off expression of the early gene operon comCDE, but also has a parallel effect on the early genes comX and comW, thus strongly suggesting an effect on expression of all early genes. Our present results suggest that the target of DprA action within the regulators of early gene expression may be ComE itself. This would provide a direct path to permitting exit from the competent state, although less direct effects of DprA or of its complexes with RecA or with ssDNA cannot be ruled out. As a response regulator of a two component system, phosphorylated ComE accounts for turning on early gene expression. It is our speculation that DprA, via interacting with ComE, might cause de-phosphorylation of ComE, or hinder its recruitment of RNAP to promoter regions, to control early gene expression. Further studies, including distinguishing the two mechanisms and identifying interacting surfaces in both proteins, are warranted.
Recently published results of a parallel study establishing a key role for DprA in exit from competence are based on comX-independent exit from competence when dprA is converted essentially into an early gene [33]. The present study, based on the complementary approach of removal of individual late genes to reveal which is needed for comX-dependent exit from competence, strengthens this conclusion.The apparent role of DprA in terminating late gene transcription could thus in principle be either a secondary effect of its inhibition of early gene expression or could reflect an additional direct effect on late gene expression. To see if DprA also restricted late gene expression directly by an effect on ComX, a new strain was created in this study for the ectopic expression of comX and comW. This new strain can develop competence upon induction of comX and comW by raffinose treatment. In this strain, the ability to transform was transient despite continued presence of ComX and ComW (Fig. 7), providing a good background to evaluate the possibility of a direct effect of DprA on late gene expression. In a dprA mutant derivative of this strain, the pattern of late gene expression following comX and comW induction perfectly matched the pattern in the dprA+ parent. This strongly suggests that DprA does not suppress ComX activity in late gene transcription, but that another gene may be responsible for limiting ComX activity to a short time window. This hypothetic regulator is unlikely to be CSP-induced because it could be induced by raffinose, but instead appears to be dependent, directly or indirectly, on induction of comX (or comW),
It remains a challenge to reconcile the different patterns of late gene expression in dprA mutants under CSP and raffinose inductions. Specifically, why is late gene expression prolonged in the dprA mutant if there is a separate (late) inhibitor directly targeting ComX? We propose that DprA is not the only factor that shuts off competence, but that a second independent inhibitor accounts for the termination of late gene expression in dprA mutants in the ectopic expression system as well as for the prompt termination of late gene transcription in the WT while ComX is still present [12], [14]. As this second repressor was apparently ineffective in CSP-induced cultures in the dprA mutant background, we hypothesize that ComX and ComW are accumulated to different levels with CSP and raffinose. In the ΔdprA mutant induced by CSP, where DprA, the factor normally curbing early gene expression, is removed, there would be a continuous supply of ComX and ComW at elevated levels, so that even if the second repressor could inactivate a normal amount of ComX or ComW, it could be overwhelmed by the unusually high amounts of these regulators. Direct comparison verified this inferred difference in the levels of ComX and ComW achieved under the two expression regimes, but the hypothetical factor responsible for the shut off of late gene expression remains unknown. While here we have ruled out ∼20 late gene products, several other late genes as well as the entire class of ‘delayed’ genes remain untested. Since competence in S. pneumoniae both imposes a stress on the competent cell itself and creates a potential hazard to nearby cells, it should not be surprising that its initiation and termination are both controlled at multiple levels.
Acknowledgments
We thank N. Mirouze, B. Martin, and J. P. Claverys for sharing unpublished results, J. P. Claverys for insightful critique of the manuscript, and Mercy Maccharia and Wael Abdel-Fattah for assistance with strain construction.
Funding Statement
This material is based upon work supported in part by the National Science Foundation Division of Molecular and Cellular Biosciences Genetic Mechanisms Program under Grant No. MCB1020863. No additional external funding received for this study. The NSF web site is at: https://www.fastlane.nsf.gov/fastlane.jsp. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Avery OT, Macleod CM, McCarty M (1944) Studies on the chemical nature of the substance inducing transformation of pneumococcal types : induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79: 137–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Claverys JP, Havarstein LS (2002) Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front Biosci 7: d1798–1814. [DOI] [PubMed] [Google Scholar]
- 3. Claverys JP, Prudhomme M, Martin B (2006) Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60: 451–475. [DOI] [PubMed] [Google Scholar]
- 4. Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP (2006) Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313: 89–92. [DOI] [PubMed] [Google Scholar]
- 5. Martin B, Granadel C, Campo N, Henard V, Prudhomme M, et al. (2010) Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol Microbiol 75: 1513–1528. [DOI] [PubMed] [Google Scholar]
- 6. Alloing G, Martin B, Granadel C, Claverys JP (1998) Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol Microbiol 29: 75–83. [DOI] [PubMed] [Google Scholar]
- 7. Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, et al. (2004) Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51: 1051–1070. [DOI] [PubMed] [Google Scholar]
- 8. Dagkessamanskaia A, Moscoso M, Henard V, Guiral S, Overweg K, et al. (2004) Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol 51: 1071–1086. [DOI] [PubMed] [Google Scholar]
- 9. Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA (2000) Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol 182: 6192–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo P, Li H, Morrison DA (2003) ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol Microbiol 50: 623–633. [DOI] [PubMed] [Google Scholar]
- 11. Ween O, Gaustad P, Havarstein LS (1999) Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol 33: 817–827. [DOI] [PubMed] [Google Scholar]
- 12. Piotrowski A, Luo P, Morrison DA (2009) Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW. J Bacteriol 191: 3359–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP (2000) Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol 38: 867–878. [DOI] [PubMed] [Google Scholar]
- 14. Luo P, Morrison DA (2003) Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J Bacteriol 185: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luo P, Li H, Morrison DA (2004) Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol Microbiol 54: 172–183. [DOI] [PubMed] [Google Scholar]
- 16. Lee MS, Morrison DA (1999) Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181: 5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergé M (2002) Compétence pour la transformation génétique chex la bactérie à Gram positif Streptococcus pneumoniae: étude du gène tardif dprA: Université Paul Sabatier de Toulouse (Sciences), Toulouse, France.
- 18.Mirouze N, Granadel C, Bergé M, Noirot P, Martin B, et al.. (2007) Identification of a late gene product required to shut off expression of early com genes in Streptococcus pneumoniae. 8th European Meeting on the Molecular Biology of the Pneumococcus. abstr F-08, p90.
- 19. Berge M, Moscoso M, Prudhomme M, Martin B, Claverys JP (2002) Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae. Mol Microbiol 45: 411–421. [DOI] [PubMed] [Google Scholar]
- 20. Cato A Jr, Guild WR (1968) Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double cross-over model. J Mol Biol 37: 157–178. [DOI] [PubMed] [Google Scholar]
- 21. Dillard JP, Vandersea MW, Yother J (1995) Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med 181: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee MS, Dougherty BA, Madeo AC, Morrison DA (1999) Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl Environ Microbiol 65: 1883–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, et al. (1996) Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol 178: 6087–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Russel DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- 25.Bai C, Elledge S (1997) Searching for interacting proteins with the two-hybrid system I. In: Bartel P, Fields S, editors. The Yeast Two-Hybrid System New York: Oxford University Press. pp. 1–28.
- 26. Sung CK, Li H, Claverys JP, Morrison DA (2001) An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67: 5190–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trzcinski K, Thompson CM, Lipsitch M (2003) Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl Environ Microbiol 69: 7364–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tokarev AA, Taussig D, Sundaram G, Lipatova Z, Liang Y, et al. (2009) TRAPP II complex assembly requires Trs33 or Trs65. Traffic 10: 1831–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pestova EV, Havarstein LS, Morrison DA (1996) Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21: 853–862. [DOI] [PubMed] [Google Scholar]
- 30.Luo P (2003) Genetic Transformation in Streptococcus Pneumoniae: Regulation by ComX an Alternative Sigma Factor [Ph.D. Thesis]. Chicago, IL: University of Illinois-Chicago. 207 p.
- 31. Mortier-Barriere I, Velten M, Dupaigne P, Mirouze N, Pietrement O, et al. (2007) A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130: 824–836. [DOI] [PubMed] [Google Scholar]
- 32. Rosenow C, Maniar M, Trias J (1999) Regulation of the α-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res 9: 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mirouze N, Berge MA, Soulet AL, Mortier-Barriere I, Quentin Y, et al. (2013) Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A 110: E1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oggioni M, Morrison D (2008) Cooperative regulation of competence development in Streptococcus pneumoniae: Cell-to-cell signaling via a peptide pheromone and an alternative sigma factor. In: Winans S, Bassler B, editors. Chemical Communication among Bacteria. Washington, DC: ASM Press. 345–362.
- 35. Sung CK, Morrison DA (2005) Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J Bacteriol 187: 3052–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Desai BV, Morrison DA (2006) An unstable competence-induced protein, CoiA, promotes processing of donor DNA after uptake during genetic transformation in Streptococcus pneumoniae. J Bacteriol 188: 5177–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morrison DA, Trombe MC, Hayden MK, Waszak GA, Chen JD (1984) Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAMβ1. J Bacteriol 159: 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]