Abstract
Retinoblastoma (Rb1) has been described as an essential player in white adipocyte differentiation in mice. No studies have been reported thus far in human adipose tissue or human adipocytes. We aimed to investigate the possible role and regulation of RB1 in adipose tissue in obesity using human samples and animal and cell models. Adipose RB1 (mRNA, protein, and activity) was negatively associated with BMI and insulin resistance (HOMA-IR) while positively associated with the expression of adipogenic genes (PPARγ and IRS1) in both visceral and subcutaneous human adipose tissue. BMI increase was the main contributor to adipose RB1 downregulation. In rats, adipose Rb1 gene expression and activity decreased in parallel to dietary-induced weight gain and returned to baseline with weight loss. RB1 gene and protein expression and activity increased significantly during human adipocyte differentiation. In fully differentiated adipocytes, transient knockdown of Rb1 led to loss of the adipogenic phenotype. In conclusion, Rb1 seems to play a permissive role for human adipose tissue function, being downregulated in obesity and increased during differentiation of human adipocytes. Rb1 knockdown findings further implicate Rb1 as necessary for maintenance of adipogenic characteristics in fully differentiated adipocytes.
Adipose tissue plays a central role in the management of systemic energy storage as well as in many other processes. This is due to its capacity to store triglycerides and to secrete many proteins that have a major impact on energy homeostasis. Several studies suggest that one of the key factors that link excess caloric intake and a positive energy balance with metabolic disturbance is the inability to appropriately expand the adipose tissue, which occurs in parallel to the decrease of its adipogenic capacity (1,2).
The retinoblastoma gene (RB1) was the first tumor suppressor gene to be recognized and cloned (3,4). The gene encodes the retinoblastoma protein (pRb), a central regulator of the cell cycle (5), which is required for several cellular processes (6) including apoptosis, cell proliferation, and differentiation in skeletal myocytes, osteoblasts, and adipocytes (7–9).
The involvement of pRb in adipocyte differentiation was first suggested in 1994 (10) in connection with the ability of pRb to activate C/EBP-mediated transcription (10–13). Mouse embryonic fibroblasts and 3T3-L1 lacking the Rb1 gene are unable to undergo adipose conversion in response to treatment with standard adipogenic inducers (11). The defective differentiation of Rb1−/− mouse embryonic fibroblasts, however, can be bypassed by administration of a peroxisome proliferator–activated receptor (PPAR)γ ligand or by partial inhibition of extracellular signal–regulated kinase 1/2 activity (14,15).
In mice, pRb has been described as an essential player in white adipocyte differentiation and as a negative modulator of brown adipocyte differentiation (16,17). In fact, several studies in both mouse and rat models have shown that pRb inactivation leads to or associates with the acquisition of brown-like features (browning) in white fat depots (18–20).
To our knowledge, no previous studies have explored the possible role of Rb1 in fully differentiated 3T3-L1 adipocytes during human adipocyte differentiation or in whole human adipose tissue or the effects of diet-induced weight changes on adipose tissue Rb1 gene expression.
In the current study, we aimed 1) to explore RB1 expression and activity in human adipose tissue in relation to obesity and during human adipocyte differentiation, 2) to study the effects of diet-induced weight gain and loss on Rb1 expression and activity in rat adipose tissue, and 3) to investigate the possible role of Rb1 in the adipogenic maintenance of fully differentiated 3T3-L1 adipocytes.
RESEARCH DESIGN AND METHODS
Subject recruitment.
Samples from two independent cohorts were analyzed: 152 adipose tissue samples (76 visceral and 76 subcutaneous) from a group of Caucasian men (n = 19) and women (n = 57) with BMI between 20 and 58 kg/m2 and 64 adipose tissues (32 visceral and 32 subcutaneous) from a group of Caucasian morbidly obese men (n = 5) and women (n = 27) with different degrees of insulin sensitivity. Insulin sensitivity was measured using euglycemic-hyperinsulinemic clamp.
Both cohorts were recruited at the Endocrinology Service of the Hospital Universitari Dr. Josep Trueta (Girona, Spain). All subjects reviewed that their body weight had been stable for at least 3 months before the study and gave written informed consent after the purpose, nature, and potential risks of the study were explained to them.
Adipose tissue samples were obtained from subcutaneous and visceral depots during elective surgical procedures (cholecystectomy, surgery of abdominal hernia, and gastric bypass surgery), washed, fragmented, and immediately flash frozen in liquid nitrogen before being stored at −80°C.
Anthropometric measurements.
BMI was calculated as weight (in kilograms) divided by the square of height (in meters). According to this anthropometric parameter, subjects were classified as nonobese (BMI <30 kg/m2) and obese (BMI ≥30 kg/m2) following World Health Organization guidelines.
Protein preparation.
Proteins were extracted from adipose tissue and adipocytes by using a Polytron PT-1200C homogenizer (Kinematica, Lucerne, Switzerland) directly in radioimmunoprecipitation assay (RIPA) buffer (0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 150 mmol/L NaCl, and 50 mmol/L Tris-HCl, pH 8.0), supplemented with protease inhibitors (1 mmol/L phenylmethylsulfonyl fluoride). Cellular debris and lipids were eliminated by centrifugation of the solubilized samples at 13,000 rpm for 60 min at 4°C, recovering the soluble fraction below the fat supernatant and avoiding the nonhomogenized material at the bottom of the centrifuge tube. Protein concentration was determined using the RC/DC Protein Assay (Bio-Rad Laboratories, Hercules, CA).
Total and phosphorylated (Thr821) pRb protein determination.
Total and phosphorylated pRb protein levels in RIPA extracts of human adipose tissue and adipocyte samples were measured using a human Rb total ELISA kit (KHO0011; Invitrogen) and a human Rb (pT821) ELISA kit (KHO0021; Invitrogen) following the manufacturer instructions or by immunoblotting after denaturing electrophoresis (see below), using the primary antibodies included in the ELISA kits.
Effects of diet-induced weight gain and loss in rats.
Rb1 gene expression was analyzed by real time Q-PCR in retroperitoneal white adipose tissue (rWAT) of male Wistar rats (Charles River Laboratories España SA, Barcelona, Spain) fed for 4 months (from month 2 to month 6 of age) a normal-fat diet (10% of energy as fat), a high-fat diet (60% of energy as fat), or a cafeteria diet (19) and in a group of male Wistar rats fed the cafeteria diet followed by a normal-fat diet for 2 months (POST-CAF group). Six to seven animals were analyzed per group. Adiposity index was calculated by dividing the sum of the weight of all white adipose tissue depots by the body weight and multiplying by 100. Methodology used for analysis by immunoblotting of pRb in rat tissues is detailed as Supplementary Data.
RB1 gene expression during human preadipocyte differentiation.
Isolated human preadipocytes from lean (BMI <25 kg/m2) and obese (BMI >30 kg/m2) subjects (Zen-Bio, Research Triangle Park, NC) were plated on T-75 cell culture flasks and cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM)/Nutrient Mix F-12 medium (1:1, v/v) supplemented with 10 units/mL penicillin/streptomycin, 10% FBS, 1% HEPES, and 1% glutamine (all from GIBCO, Invitrogen, Barcelona, Spain). One week later, the isolated and expanded human subcutaneous preadipocytes were cultured (∼40,000 cells/cm2) in 12-well plates with preadipocyte medium (Zen-Bio) composed of DMEM/Nutrient Mix F-12 medium (1:1, v/v), HEPES, FBS, penicillin, and streptomycin in a humidified 37°C incubator with 5% CO2. Twenty-four hours after plating, cells were checked for complete confluence (day 0) and differentiation was induced using differentiation medium (Zen-Bio) composed of preadipocyte medium, human insulin, dexamethasone, isobutylmethylxanthine, and PPARγ agonists (rosiglitazone). After 7 days, differentiation medium was replaced with fresh adipocyte medium (Zen-Bio) composed of DMEM/Nutrient Mix F-12 medium (1:1, v/v), HEPES, FBS, biotin, pantothenate, human insulin, dexamethasone, penicillin, streptomycin, and amphotericin. Fourteen days after the initiation of differentiation, cells appeared rounded with large lipid droplets apparent in the cytoplasm. Cells were then considered mature adipocytes, harvested, and stored at −80°C for protein and RNA extraction for study of RB1 protein and gene expression levels after human adipocyte differentiation. The experiment was performed in triplicate for each sample. Adipogenic differentiation was verified by fatty acid synthase (FASN) (Hs00188012_m1; Applied Biosystems, Madrid, Spain) and adiponectin (Adipoq) (Hs00605917_m1; Applied Biosystems) gene expression.
3T3-L1 preadipocyte differentiation and short hairpin RNA– and small interfering RNA–mediated knockdown of Rb1.
The embryonic fibroblast mouse cell line 3T3-L1 (American Type Culture Collection) was maintained in DMEM containing 20 mmol/L glucose, 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Two days after confluence, insulin (5 μg/mL), dexamethasone (0.5 μmol/L), and isobutylmethylxanthine (0.5 mmol/L) mixture was added for 2 days, followed by 5 days with insulin (5 μg/mL) alone. Differentiation was verified by morphological assessment and adipogenic gene expression.
Permanent silencing was performed using Rb1-targeted and control short hairpin RNA (shRNA) lentiviral particles (sc-29468-V and sc-108080; Santa Cruz Biotechnology, Santa Cruz, CA) and following the manufacturer instructions. Positive 3T3-L1 preadipocytes harboring shRNA cassette for Rb1 were selected by puromycin (3 μg/mL) selection 60 h after infection.
Transient silencing was performed in fully differentiated adipocytes using mouse Rb1 (ON-TARGETplus SMARTpool mouse Rb1, L-047474–00; Dharmacon) or control (ON-TARGETplus nontargeting pool, D-001810–01–05; Dharmacon) small interfering RNAs (siRNAs) introduced by electroporation or lipid transfection. For electroporation, cells were suspended by mild trypsinization and electroporated using the BioRad kit according to the manufacturer’s recommendations (GP Electrop.Buffer; BioRad). Approximately 2 million cells in 200 μL GP Electrop.Buffer (BioRad) and 250 nmol/L siRNA were used, and the adipocytes were reseeded afterward into 12-well plates. In transfection using a lipid-based formulation (siPORT NeoFx; Life Technologies SA, Madrid, Spain), Rb1 siRNA (25 nmol/L) and the transfection reagent siPORT NeoFX (0.3%) were assayed, and a successful transfection was achieved using a reverse transfection protocol (21) with 8 × 104 cells during 36 h.
Oil Red O staining and analysis.
For Oil Red O staining, cells were washed twice with PBS, fixed in 4% formaldehyde for 1 h, and stained for 30 min with 0.2% Oil Red O solution in 60% isopropanol. Cells were then washed several times with water, and excess water was evaporated by placing the stained cultures at ∼32°C. For quantification of the extent of adipose conversion, 0.2 mL isopropanol was added to the stained culture dish. The extracted dye was immediately removed by gentle pipetting, and optical density was determined spectrophotometrically at 500 nm using a multiwell plate reader (Anthos Labtec 2010 1.7).
Gene expression.
RNA was prepared from human adipose tissues and cell culture samples using RNeasy Lipid Tissue Mini Kit (QIAgen; Izasa SA, Barcelona, Spain). The integrity of each RNA sample was checked by Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). Total RNA was quantified by means of spectrophotometer (GeneQuant, GE Health Care, Piscataway NJ) and reverse transcribed to cDNA using a High Capacity cDNA Archive kit (Applied Biosystems) according to the manufacturer’s protocol.
Real-time quantitative PCR was conducted with Light Cycler 480 Probes Master (Roche) using TaqMan technology suitable for relative genetic expression quantification. Commercially available and prevalidated TaqMan primer/probes sets were used, and they are described in Supplementary Data.
The reaction was performed in a final volume of 7 μL. The cycle program consisted of an initial denaturing of 10 min at 95°C followed by 40 cycles of 15 s denaturizing phase at 95°C and 1 min annealing and extension phase at 60°C. A threshold cycle (Ct value) was obtained for each amplification curve, and a ΔCt value was first calculated by subtracting the Ct value for human PPIA RNA from the Ct value for each sample. Fold changes compared with the endogenous control were then determined by calculating 2−ΔCt, so gene expression results are expressed as expression ratio relative to PPIA gene expression according to the manufacturers’ guidelines.
For rat tissue samples, total RNA was extracted from frozen rWAT samples using TriPure reagent (Roche), purified using an E.Z.N.A. Total RNA Kit I (Omega Bio-Tek) and retrotranscribed to cDNA. Quantitative PCR was run using SybrGreen Master Mix on a StepOnePlus instrument essentially as previously described (22). Analysis of quantitative PCR data was done using the LinRegPCR (23) or the StepOnePlus software. Eif4e, Eif2s1, or both were used as reference genes (24). Primers for Rb1, Eif4e, and Eif2s1 are provided in Supplementary Data.
Immunoblotting analysis.
RIPA protein extracts of human adipose tissues and cell culture samples (25–50 μg) were separated by SDS-PAGE and transferred to nitrocellulose membranes by conventional procedures. Membranes were immunoblotted with primary antibodies against pThr821Rb1 and Rb1 (KHO0021 and KHO0011, respectively; Invitrogen); Rb1, FASN, PPARγ, Adipoq, Ucp1, and β-actin (sc-74562, sc-20140, sc-7273, sc-26497, sc-28766, and sc-47778, respectively; Santa Cruz Biotechnology); and Prdm16 (AP18024PU-N; Acris Antibodies). Anti-rabbit IgG and anti-mouse IgG coupled to horseradish peroxidase were used as secondary antibody. Horseradish peroxidase activity was detected by chemiluminescence, and quantification of protein expression was performed using scion image software.
Statistics.
Statistical analyses were performed using SPSS 12.0 software for Windows (SPSS, Chicago, IL). All assays were performed at least in duplicate and are reported as means ± SD. The relation between variables was analyzed by ANOVA (using minimum significant difference [DMS] and Bonferroni post hoc tests), simple correlation (Pearson or Spearman test), and multiple linear regression models (using stepwise method). One-way ANOVA was used to compare the effects of weight gain and loss and differences between groups.
RESULTS
RB1 gene expression in human adipose tissue
First cohort.
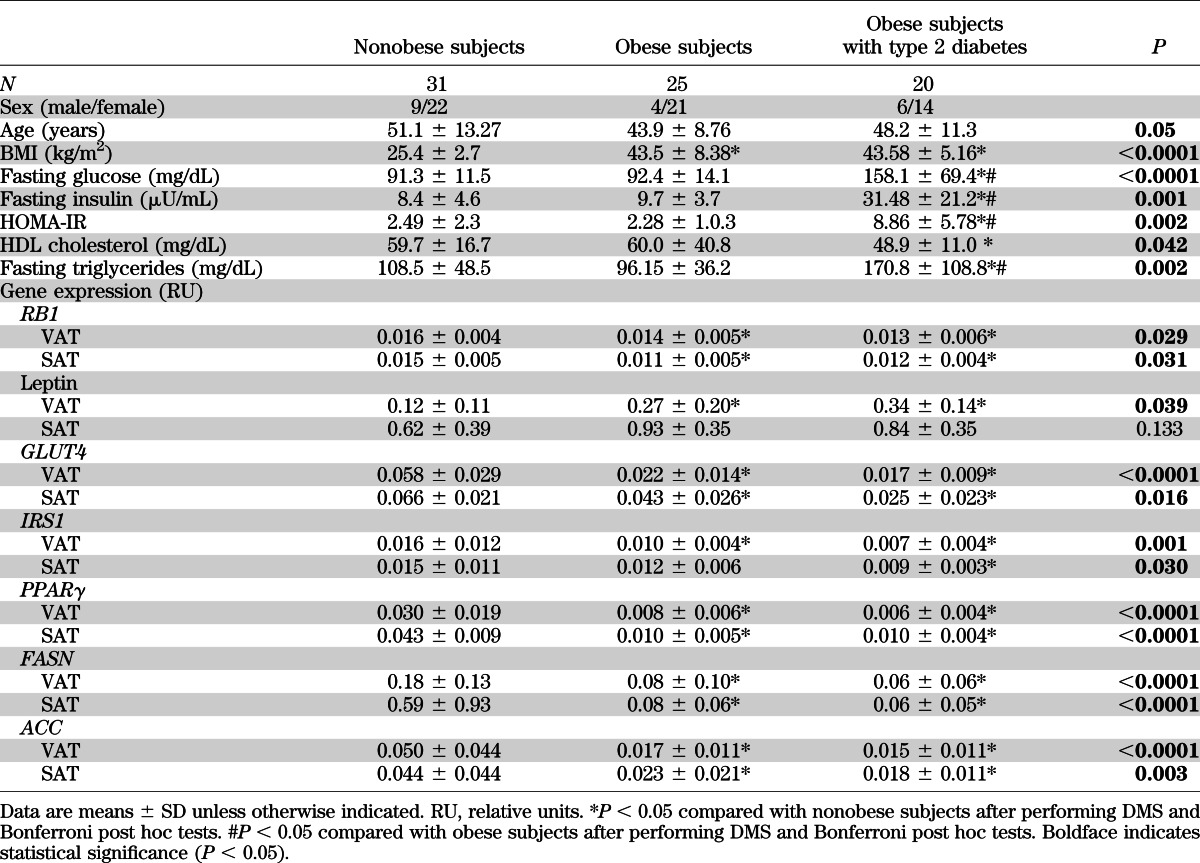
Mean anthropometric, clinical, and adipose gene expression data of participants from the first human cohort are described in Table 1. RB1 gene expression in adipose tissues decreased significantly in association with obesity, together with expression of adipogenic/lipogenic genes. No differences were found among obese subjects depending on the presence of type 2 diabetes.
TABLE 1.
Anthropometric, clinical, and adipose gene expression features of the first human cohort
In both subcutaneous (SAT) and visceral (VAT) adipose tissues, RB1 gene expression was negatively associated with BMI (r = –0.269, P = 0.019, and r = –0.332, P = 0.003, respectively) and positively associated with PPARγ (r = 0.460, P < 0.0001, and r = 0.414, P = 0.001) and IRS1 (r = 0.413, P = 0.001, and r = 0.358, P = 0.004) gene expression (Table 2). In addition, adipose RB1 gene expression was negatively associated with LEP (r = –0.295, P = 0.01) and positively associated with FASN (r = 0.251, P = 0.036) and ACC (r = 0.240, P = 0.042) gene expression in SAT and positively associated with GLUT4 (r = 0.392, P = 0.002) gene expression in VAT (Table 2). In a multiple linear regression model, PPARγ (β=0.44, P = 0.02) gene expression contributed independently to RB1 gene expression variance after BMI and IRS1, FASN, ACC, and LEP gene expressions in SAT were controlled for. In VAT, IRS1 (β = 0.44, P = 0.02) gene expression contributed independently to RB1 gene expression variance after BMI and PPARγ and GLUT4 gene expressions were controlled for.
TABLE 2.
Correlations of RB1 gene expression with anthropometric and clinical characteristics of first human cohort
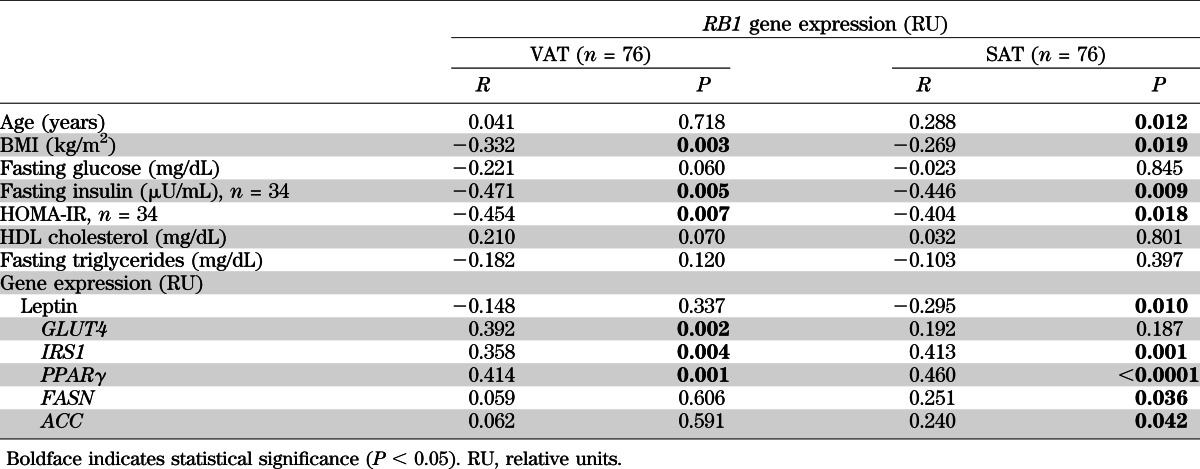
In a subgroup of 34 participants in whom fasting insulin was measured, RB1 gene expression in both SAT and VAT was negatively associated with fasting insulin (r = –0.446, P = 0.009, and r = –0.471, P = 0.005, respectively) and homeostasis model assessment of insulin resistance (HOMA-IR) (r = –0.404, P = 0.018, and r = –0.454, P = 0.007) (Table 2). In the multiple linear regression model, BMI (β = –0.42, P = 0.01, in SAT and β = –0.36, P = 0.03, in VAT) contributed independently to RB1 gene expression variance after fasting insulin or HOMA-IR was controlled for.
Second cohort.
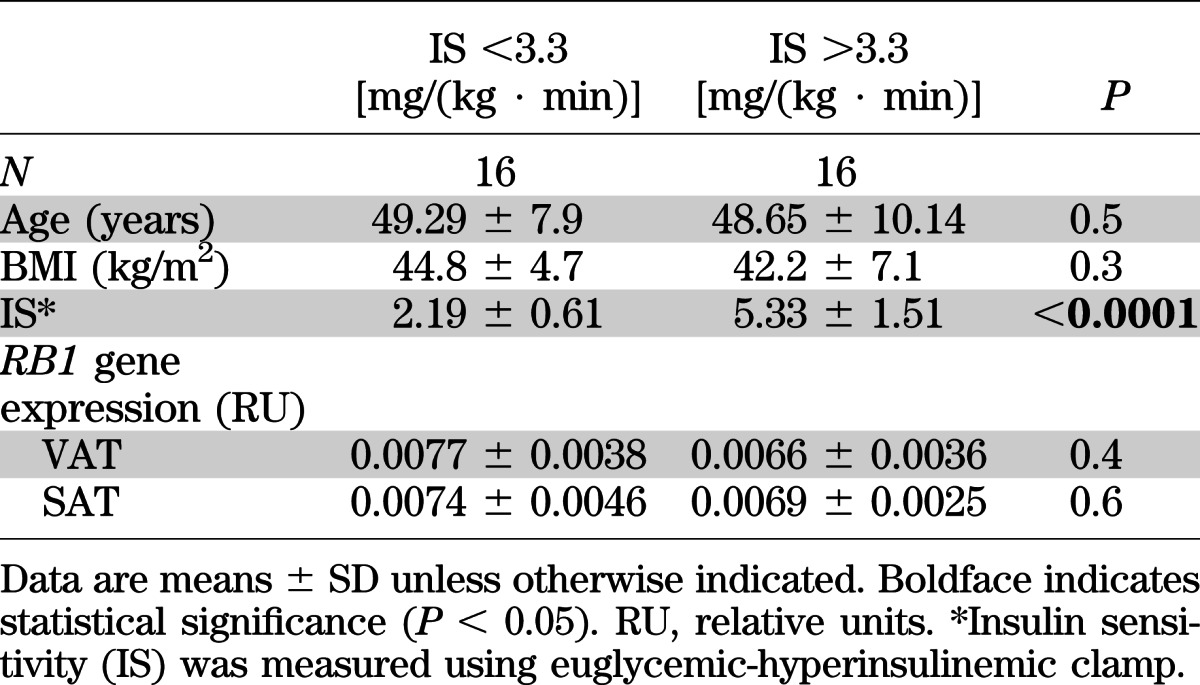
A second cohort made up of morbid obese subjects with different degrees of insulin sensitivity (measured using euglycemic-hyperinsulinemic clamp) was used to investigate the possible relationship between RB1 gene expression in adipose tissues and insulin sensitivity excluding the effects of obesity. Mean anthropometric and clinical characteristics as well as adipose RB1 gene expression levels in this second cohort are described in Table 3. No correlation was found between RB1 gene expression in SAT or VAT and insulin sensitivity (r = –0.205, P = 0.30, in SAT and r = 0.161, P = 0.42, in VAT).
TABLE 3.
Anthropometric and clinical features and adipose RB1 gene expression of second cohort
pRb activity in human adipose tissue.
The activity of pRb was analyzed in 18 SAT and 28 VAT human samples measuring by ELISA the levels of pRb protein phosphorylated at Thr 821 and of total pRb protein and calculating the ratio between them. Typically, more phosphorylation indicates less Rb1 protein activity (i.e., less capacity of pRb to interact with transcription factors and other interacting proteins [25]). In concordance with RB1 mRNA levels, in both SAT and VAT total pRb protein levels decreased (r = –0.46, P = 0.05, and r = –0.41, P = 0.03, respectively) and levels of phosphorylated pRb increased (r = 0.55, P = 0.03, and r = 0.39, P = 0.04) in association with obesity, resulting in an increased phosphorylated pRb–to–total pRb ratio indicative of reduced pRb activity (Fig. 1A). The ELISA measurements were validated by immunoblotting (Fig. 1B).
FIG. 1.
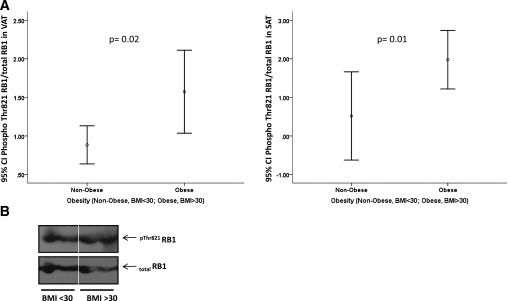
A: pThr821RB1–to–total RB1 protein ratio in SAT and VAT of nonobese and obese subjects. pThr821RB1 and total RB1 were measured using specific ELISA kits. B: Representative immunoblots of pThr821RB1 and total RB1 protein in VAT of nonobese and obese subjects measured by immunoblotting using the ELISA antibodies. Phospho, phosphorylated.
Effects of diet-induced weight gain and loss in rats.
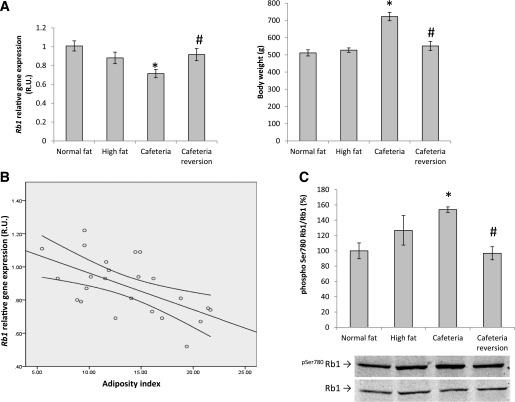
To gain further insight into the effects of obesity on Rb1 gene expression in adipose tissue, we explored the impact of diet-induced weight changes on Rb1 mRNA levels in rats fed with a cafeteria-style high-fat diet. Confirming the association found in human obesity, adipose Rb1 gene expression decreased significantly in response to cafeteria diet–induced weight gain in rats. Furthermore, when the cafeteria diet was replaced by a normal diet and body weight returned toward normality, adipose Rb1 gene expression was reestablished (Fig. 2A). In fact, adipose Rb1 gene expression was negatively correlated with the adiposity index of the animals (r = –0.587, P = 0.003) (Fig. 2B). In keeping with the mRNA data, adipose pRb activity decreased significantly with cafeteria diet–induced weight gain, as indicated by increased pSer780pRb–to–pRb ratio, and returned to basal levels after weight loss (Fig. 2C).
FIG. 2.
A: Effect of high-fat and cafeteria diet on Rb1 gene expression in rWAT and body weight. B: Correlation between Rb1 gene expression in rWAT and adiposity index. C: Effect of high-fat and cafeteria diet on pSer780Rb1/Rb1 protein in rWAT. *P < 0.05 in comparison with normal-fat diet; #P < 0.05 in comparison with cafeteria diet. Phospho, phosphorylated.
Changes in RB1 during human adipocyte differentiation.
RB1 gene and protein expression and activity were monitored during differentiation of primary preadipocytes derived from human VAT and SAT. In both cases, RB1 gene expression decreased transiently at day 2 and then increased gradually in parallel to adipogenic gene expression (Fig. 3A) and lipid droplet formation. Similar to gene expression, pRb total protein quantity increased significantly during human adipocyte differentiation (Fig. 3B), as did pRb activity (Fig. 3B [reduced Thr821 phosphorylation indicative of increased activity]). ELISA measurements were validated by immunoblotting analyses (Fig. 3C and D).
FIG. 3.
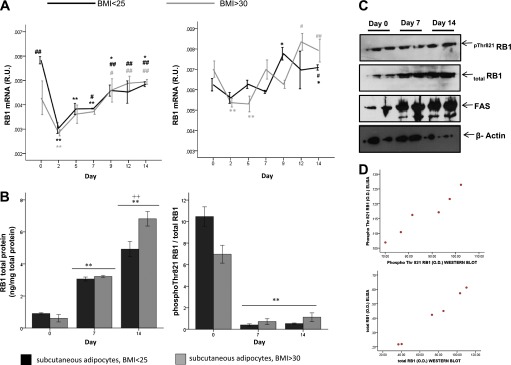
A and B: RB1 mRNA (A), RB1 total protein (B) (left panel), and RB1 activity (B) (right panel) during time course of differentiation of human preadipocytes isolated from the denoted fat depots of lean (BMI <25 kg/m2) or obese (BMI >30 kg/m2) subjects. Phosphorylated (Phospho) Thr821 RB1 and total RB1 were measured by ELISA. *P < 0.05 in comparison with day 0; **P < 0.005 in comparison with day 0; #P < 0.05 in comparison with day 2; ##P < 0.005 in comparison with day 2; ++P < 0.005 in comparison with day 7. C: Representative immunoblots of pThr821RB1 and total RB1 protein during time course of differentiation of human subcutaneous preadipocytes. Immunoblots for FASN as an adipocyte marker and β-actin as an internal control in the same cultures are also shown. D: Bivariate correlation between ELISA and Western blotting measurements of pThr821RB1 and total RB1 protein (r > 0.8, P < 0.01). O.D., optical density. (A high-quality color representation of this figure is available in the online issue.)
Effects of Rb1 knockdown on adipocyte gene expression.
Rb1 permanent knockdown in preadipocytes using shRNA lentiviral particles decreasing Rb1 expression by 50% led to decreased expression of adipogenic genes (Pparγ, Adipoq, and Fasn) and increased expression of brown adipocyte–related genes (Prdm16 and Ucp1) during differentiation, measured both at the mRNA and the protein levels (Fig. 4A and B). In line with the gene expression results, decreased accumulation of intracellular lipid droplets was apparent in the Rb1-silenced cells after 7 days of adipogenic conditions (Fig. 4C). In fully differentiated adipocytes, transient siRNA-induced Rb1 knockdown (36 h) (through electroporation or lipid-based formulation, decreasing Rb1 mRNA by 68 and 41%, respectively), led to loss of the adipocyte phenotype, as indicated by decreased adipogenic/lipogenic gene and protein expression (Fig. 5 and Supplementary Fig. 1). Although no effects were observed on the mRNA expression levels of the brown adipocyte–related genes Ucp1 and Prdm16, UCP1 protein was weakly detected after transient siRNA-induced Rb1 knockdown (Fig. 5).
FIG. 4.
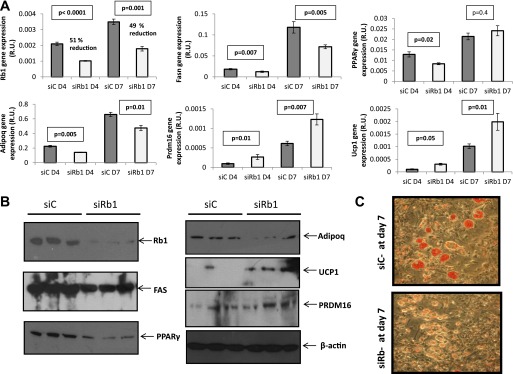
A: 3T3-L1 preadipocytes were treated with a nontargeted shRNA (siC) (control cells) or with a shRNA targeted to silence Rb1 (siRb1). Control cells and cells with permanent (∼50%) Rb1 knockdown were induced to differentiate using a standard adipogenic cocktail, and the mRNA expression levels of the denoted genes were measured at day 4 (D4) and day 7 (D7) of adipogenic differentiation. B: Same genes as in A were measured at the protein level in control and Rb1-silenced cells after 7 days of adipogenic differentiation; representative immunoblots are shown. C: Oil Red O staining of control and Rb1-silenced cells after 7 days of differentiation to test the accumulation of lipid droplets. R.U., relative units.
FIG. 5.
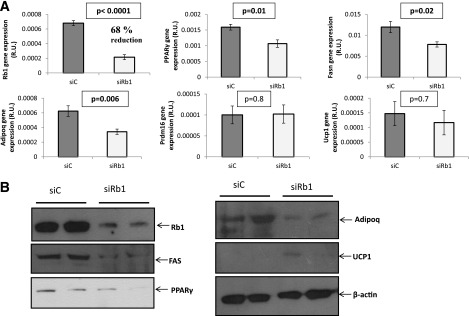
Effects of transient Rb1 knockdown (~70% reduction for 36 h) in fully differentiated 3T3-L1 adipocytes on the expression of the denoted genes at the mRNA (A) and the protein (B) level. Treatments: SiC, control siRNA; siRb1, Rb1 siRNA. siRNAs were introduced in the mature adipocytes by electroporation.
DISCUSSION
Rb1 has been described as a player in white adipocyte differentiation mainly on the basis of studies using immortalized murine cell lines, such as 3T3-L1. Studies dealing with pRb in human adipogenesis and with the regulation of adipose pRb in connection with human or rodent obesity are lacking. Moreover, most studies thus far have evaluated the role of Rb1 in adipogenesis but not in the mature adipocyte. To the best of our knowledge, this is the first study showing that adipose RB1 gene and protein expression and activity are significantly associated with human adipose tissue function and modulated in obesity and during human adipocyte differentiation. It is also the first to evidence a previously unappreciated role for the Rb1 protein in the maintenance of the mature adipocyte phenotype.
We observed an inverse association between adipose RB1 levels/activity and human obesity in cross-sectional analysis, with this finding confirmed longitudinally in the rat diet interventional study, which indicated reciprocal changes in Rb1 gene expression/activity with weight gain and loss. Importantly, in both subcutaneous and visceral human adipose tissue, RB1 gene expression was negatively associated with BMI and positively associated with adipogenic gene expression (mainly, PPARγ and IRS1). Obesity is well-known to be characterized by a hypertrophied, dysfunctional adipose tissue, with an increased expression of proinflammatory cytokines (26) in parallel to a decreased expression of adipogenic/lipogenic genes (27–31). Changes in obesity are to some extent reminiscent of those occurring during dedifferentiation of isolated human adipocytes and adipose tissue explants—a process known to be promoted by cytokines (such as tumor necrosis factor-α) and prevented by adipogenic inductors (such as isobutylmethylxanthine) (32–34). Results in this study strongly suggest that RB1 is required for an optimal adipose tissue function, since Rb1 knockdown favored adipocyte dedifferentiation and, additionally, decreased RB1 levels correlated with decreased expression of adipogenic genes in the adipose tissue of obese subjects. Considering the well-known antiproliferative role of RB1 as a cell cycle break, reduced RB1 and adipogenic gene expression in human obesity may also be linked to and favor the hyperplasic component of obesity.
Dual effects of Rb1 on murine adipogenesis have been described. Thus, whereas a proadipogenic role of Rb1 is well established (10–12,35), some studies have shown that the activation of Rb1 during mitotic clonal expansion (at the first stage of adipocyte differentiation) leads to blunted adipogenesis (36–38). In fact, pRb is known to be hyperphosphorylated (inactive form) during clonal expansion of 3T3-L1 cells, corresponding to previously quiescent cells reentering the cell cycle. Later in adipogenesis, pRb becomes hypophosphorylated (active form) at the same time that its expression is induced (12,39,40). Similar changes in RB1 expression and activity have been demonstrated here during differentiation of human preadipocytes, including a decrease in RB1 gene expression at an early stage (day 2) followed by a gradual increase as adipogenesis further progresses.
Permanent knockdown of Rb1 in preadipocytes using shRNA lentiviral particles led to impaired white adipogenesis and promoted a brown-like adipocyte differentiation pattern with increased Prdm16 and Ucp1 gene expression—in agreement with previous studies using Rb1−/− knockout models (16–18). Transient Rb1 knockdown in fully differentiated adipocytes, in its turn, resulted in markedly reduced expression of adipogenic/lipogenic genes, suggesting that Rb1 is necessary, and not just permissive, for maintenance of the adipogenic/lipogenic capacity of mature adipocytes, and in good concordance with our findings in human adipose tissue samples. Browning due to Rb1 reduction in this model cannot be discarded, since a weak UCP1 protein band could be detected only after transient siRNA-induced Rb1 knockdown. However, Ucp1 and Prdm16 gene expression in the cells was not affected by Rb1 silencing. Additionally, no correlation was found between RB1 and UCP1 gene expression in human adipose tissue (data not shown).
In summary, RB1 expression and activity seem to be essential for maintenance of the adipogenic/lipogenic capacity of adipose tissue in the human, as they are reduced in parallel with adipogenic gene expression in the dysfunctional adipose tissue of obese subjects and increased during primary human preadipocyte differentiation. Rb1 knockdown findings further implicate Rb1 as necessary for maintenance of adipogenic characteristics in fully differentiated fat cells.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by research grants from the Ministerio de Educación y Ciencia (SAF2008-02073) and the European Union FP7 project DIABAT (HEALTH-F2-2011-278373). P.P. is supported by a PhD fellowship by Conselleria d'Educació, Cultura i Universitats, Govern de les Illes Balears, cofinanced by the European Social Fund. CIBERobn Fisiopatología de la Obesidad y Nutrición is an initiative from the Instituto de Salud Carlos III from Spain.
No potential conflicts of interest relevant to this article were reported.
J.M.M.-N., P.P., and M.S. researched data and wrote the manuscript. F.O., E.G.-R., and P.O. researched data. J.R. and W.R. contributed to discussion and reviewed the manuscript. A.P., M.L.B., and J.M.F.-R. researched data and wrote, reviewed, and edited the manuscript. M.L.B. and J.M.F.-R. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Parts of this study were presented in oral form at the 48th Annual Meeting of the European Association for the Study of Diabetes, Berlin, Germany, 1–5 October 2012.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-0977/-/DC1.
REFERENCES
- 1.Kim JY, van de Wall E, Laplante M, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest 2007;117:2621–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danforth E., Jr Failure of adipocyte differentiation causes type II diabetes mellitus? Nat Genet 2000;26:13. [DOI] [PubMed] [Google Scholar]
- 3.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995;81:323–330 [DOI] [PubMed] [Google Scholar]
- 4.Gordon GM, Du W. Conserved RB functions in development and tumor suppression. Protein Cell 2011;2:864–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobrinik D. Pocket proteins and cell cycle control. Oncogene 2005;24:2796–2809 [DOI] [PubMed] [Google Scholar]
- 6.Viatour P, Sage J. Newly identified aspects of tumor suppression by RB. Dis Model Mech 2011;4:581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipinski MM, Jacks J. The retinoblastoma gene family in differentiation and development. Oncogen 1999;18:873–883 [DOI] [PubMed] [Google Scholar]
- 8.Thomas DM, Yang HS, Alexander K, Hinds PW. Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol Ther 2003;2:124–130 [PubMed] [Google Scholar]
- 9.Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature 2010;466:1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puigserver P, Nadal-Ginard B, Palou A. Expression and interaction of C/EBPalpha adipogenic transcription factor and retinoblastoma protein in adipocytes during differentiation. Int J Obes Relat Metab Disord 1994;18:113 [Abstract] [Google Scholar]
- 11.Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev 1996;10:2794–2804 [DOI] [PubMed] [Google Scholar]
- 12.Puigserver P, Ribot J, Serra F, et al. Involvement of the retinoblastoma protein in brown and white adipocyte cell differentiation: functional and physical association with the adipogenic transcription factor C/EBPalpha. Eur J Cell Biol 1998;77:117–123 [DOI] [PubMed] [Google Scholar]
- 13.Classon M, Kennedy BK, Mulloy R, Harlow E. Opposing roles of pRB and p107 in adipocyte differentiation. Proc Natl Acad Sci USA 2000;97:10826–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen JB, Petersen RK, Larsen BM, Bartkova J, Alsner J, Kristiansen K. Activation of peroxisome proliferator-activated receptor gamma bypasses the function of the retinoblastoma protein in adipocyte differentiation. J Biol Chem 1999;274:2386–2393 [DOI] [PubMed] [Google Scholar]
- 15.Hansen JB, Petersen RK, Jørgensen C, Kristiansen K. Deregulated MAPK activity prevents adipocyte differentiation of fibroblasts lacking the retinoblastoma protein. J Biol Chem 2002;277:26335–26339 [DOI] [PubMed] [Google Scholar]
- 16.Hansen JB, Jørgensen C, Petersen RK, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA 2004;101:4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scimè A, Grenier G, Huh MS, et al. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab 2005;2:283–295 [DOI] [PubMed] [Google Scholar]
- 18.Dali-Youcef N, Mataki C, Coste A, et al. Adipose tissue-specific inactivation of the retinoblastoma protein protects against diabesity because of increased energy expenditure. Proc Natl Acad Sci USA 2007;104:10703–10708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercader J, Ribot J, Murano I, et al. Haploinsufficiency of the retinoblastoma protein gene reduces diet-induced obesity, insulin resistance, and hepatosteatosis in mice. Am J Physiol Endocrinol Metab 2009;297:E184–E193 [DOI] [PubMed] [Google Scholar]
- 20.Ström K, Hansson O, Lucas S, et al. Attainment of brown adipocyte features in white adipocytes of hormone-sensitive lipase null mice. PLoS One 2008;3:e1793 [DOI] [PMC free article] [PubMed]
- 21.Kilroy G, Burk DH, Floyd ZE. High efficiency lipid-based siRNA transfection of adipocytes in suspension. PLoS ONE 2009;4:e6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caimari A, Oliver P, Keijer J, Palou A. Peripheral blood mononuclear cells as a model to study the response of energy homeostasis-related genes to acute changes in feeding conditions. OMICS 2010;14:129–141 [DOI] [PubMed] [Google Scholar]
- 23.Ruijter JM, Ramakers C, Hoogaars WM, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009;37:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hruz T, Wyss M, Docquier M, et al. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics 2011;12:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickard A, Cichon AC, Barry A, et al. Inactivation of Rb in stromal fibroblasts promotes epithelial cell invasion. EMBO J 2012;31:3092–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taube A, Schlich R, Sell H, Eckardt K, Eckel J. Inflammation and metabolic dysfunction: links to cardiovascular diseases. Am J Physiol Heart Circ Physiol 2012;302:H2148–H2165 [DOI] [PubMed] [Google Scholar]
- 27.Dolinková M, Dostálová I, Lacinová Z, et al. The endocrine profile of subcutaneous and visceral adipose tissue of obese patients. Mol Cell Endocrinol 2008;291:63–70 [DOI] [PubMed] [Google Scholar]
- 28.Ranganathan G, Unal R, Pokrovskaya I, et al. The lipogenic enzymes DGAT1, FAS, and LPL in adipose tissue: effects of obesity, insulin resistance, and TZD treatment. J Lipid Res 2006;47:2444–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega FJ, Mayas D, Moreno-Navarrete JM, et al. The gene expression of the main lipogenic enzymes is downregulated in visceral adipose tissue of obese subjects. Obesity (Silver Spring) 2010;18:13–20 [DOI] [PubMed] [Google Scholar]
- 30.Kouidhi S, Berrhouma R, Rouissi K, et al. Human subcutaneous adipose tissue Glut 4 mRNA expression in obesity and type 2 diabetes. Acta Diabetol. 22 May 2011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 31.Veilleux A, Blouin K, Rhéaume C, Daris M, Marette A, Tchernof A. Glucose transporter 4 and insulin receptor substrate-1 messenger RNA expression in omental and subcutaneous adipose tissue in women. Metabolism 2009;58:624–631 [DOI] [PubMed] [Google Scholar]
- 32.Poloni A, Maurizi G, Leoni P, et al. Human dedifferentiated adipocytes show similar properties to bone marrow-derived mesenchymal stem cells. Stem Cells 2012;30:965–974 [DOI] [PubMed] [Google Scholar]
- 33.Gesta S, Lolmède K, Daviaud D, et al. Culture of human adipose tissue explants leads to profound alteration of adipocyte gene expression. Horm Metab Res 2003;35:158–163 [DOI] [PubMed] [Google Scholar]
- 34.Petruschke T, Hauner H. Tumor necrosis factor-alpha prevents the differentiation of human adipocyte precursor cells and causes delipidation of newly developed fat cells. J Clin Endocrinol Metab 1993;76:742–747 [DOI] [PubMed] [Google Scholar]
- 35.Hakim-Weber R, Krogsdam AM, Jørgensen C, et al. Transcriptional regulatory program in wild-type and retinoblastoma gene-deficient mouse embryonic fibroblasts during adipocyte differentiation. BMC Res Notes 2011;4:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang QQ, Otto TC, Lane MD. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc Natl Acad Sci USA 2003;100:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fajas L, Egler V, Reiter R, et al. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell 2002;3:903–910 [DOI] [PubMed] [Google Scholar]
- 38.Abella A, Dubus P, Malumbres M, et al. Cdk4 promotes adipogenesis through PPARgamma activation. Cell Metab 2005;2:239–249 [DOI] [PubMed] [Google Scholar]
- 39.Shao D, Lazar MA. Peroxisome proliferator activated receptor gamma, CCAAT/enhancer-binding protein alpha, and cell cycle status regulate the commitment to adipocyte differentiation. J Biol Chem 1997;272:21473–21478 [DOI] [PubMed] [Google Scholar]
- 40.Ribot J, Oliver P, Serra F, Palou A. Retinoic acid modulates the retinoblastoma protein during adipocyte terminal differentiation. Biochim Biophys Acta 2005;1740:249–257 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.