Abstract
Systems biology is now recognized as a needed approach to understand the dynamics of inter- and intra-cellular processes. Redox processes are at the foundation of nearly all aspects of biology. Free radicals, related oxidants, and antioxidants are central to the basic functioning of cells and tissues. They set the cellular redox environment and therefore are key to regulation of biochemical pathways and networks, thereby influencing organism health. To understand how short-lived, quasi-stable species, such as superoxide, hydrogen peroxide, and nitric oxide, connect to the metabolome, proteome, lipidome, and genome we need absolute quantitative information on all redox active compounds as well as thermodynamic and kinetic information on their reactions, i.e. knowledge of the complete redoxome. Central to the state of the redoxome are the interactive details of the superoxide/peroxide formation and removal systems. Quantitative information is essential to establish the dynamic mathematical models needed to reveal the temporal evolution of biochemical pathways and networks. This new field of Quantitative Redox Biology will allow researchers to identify new targets for intervention to advance our efforts to achieve optimal human health.
Keywords: Quantitative redox biology, free radicals, antioxidants, redoxome, Nernst equation
1. Introduction
One of the major goals in biology in this new century is to completely understand the chemistry, biochemistry, and all aspects of the regulatory pathways of cells, tissues and organisms. Systems biology, the focus of a special issue of FEBS Letters (1), is an approach to achieve this goal. The human genome project, formally begun in 1990, is the first step in this major undertaking and has resulted in a wealth of data that is being used to understand the nature and functioning of genes and their corresponding proteins. Defining the proteome is the second step in this big effort. Once all the genes and proteins are characterized the third level will be the understanding of the pathways and networks that orchestrate cell, tissue, and organism function. To reach this third level we must have knowledge of an additional, fundamental aspect of biology, the redoxome. The redoxome is a composite of quantitative information on the redox enzymes and proteins as well as the unstable, quasi stable, and redox active species that determine the redox environment of cells and tissues. These unstable, quasi stable, and redox active species are fundamental regulators of genes, proteins, and connecting pathways and networks.
It is now recognized by the greater research community that redox active molecules in conjunction with reactive oxygen and nitrogen species (ROS and RNS) are at the base of the regulation of biological processes (2, 3, 4, 5). These very reactive species set the redox state of the many redox couples and pathways in cells and tissues thereby leading to health or disease (6). Missing in all of the above is quantitative information on these transient, unstable, and quasi-stable redox active species that provide the connection between metabolism, protein function, lipid use and function, and gene expression, Figure 1. This starts with the use of dioxygen by cells (7) and subsequent formation of reactive oxygen and nitrogen species, the concentration and redox state of the many thiols and disulfides (cysteine, glutathione, thioredoxin, glutaredoxin, the peroxiredoxins, mixed protein thiols etc.) as well as the actual concentrations of antioxidant enzymes (the superoxide dismutases, glutathione peroxidases, peroxiredoxins, catalase, etc.). These enzymes have traditionally been considered solely as antioxidant enzymes; however, their cellular role reaches much further. Because they control the steady-state levels of ROS and RNS, they are involved in the regulation of signaling processes (8, 9, 10, 11, 12, 13, 14, 15, 16, 17). More appropriate would be to refer to these substances as redox proteins and enzymes.
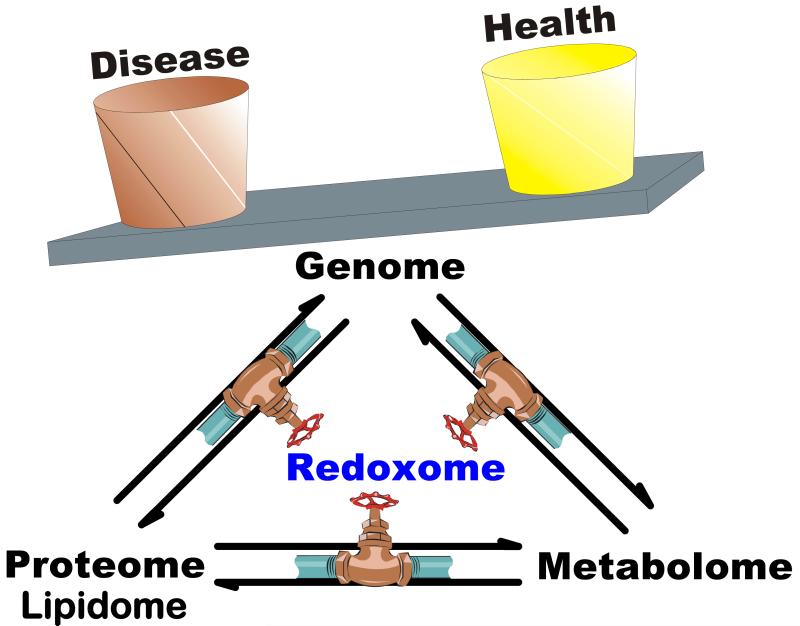
Figure 1. The redoxome as a determinant of health and disease.
The redoxome is a composite of all the information on the unstable, quasi stable, and redox active species that determine the redox environment of cells and tissues. The redoxome has control over many aspects of the metabolome, lipidome, proteome, and genome. However, each provides feedback into the redoxome, making it a central player in establishing the balance between health and disease.
2. The Antioxidant Network
Oxidation is essential for life. However, unwanted and uncontrolled oxidations can lead to cell damage, tissue and organ failure, disease, and early death. These detrimental oxidations are the result of both normal and aberrant metabolism that produce oxygen-centered and nitrogen-centered free radicals and related oxidants. An antioxidant/redox network consisting of coupled enzyme systems and small molecule-antioxidants (reductants) exists that remove these oxidants (Figure 2) guarding against the potential damage these species can inflict (18, 19). Although these redox active species can be harmful, they are central to the growth, development, and functioning of healthy living systems (12, 20, 21, 22).
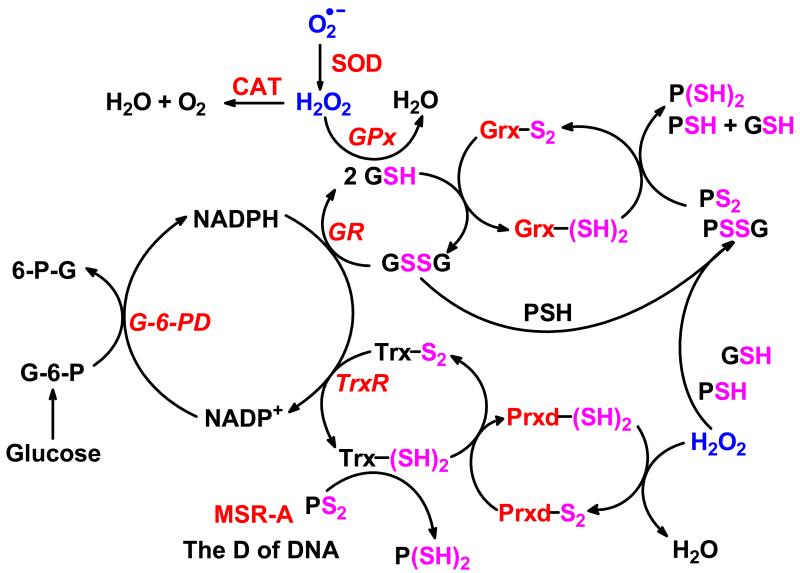
Figure 2. Cellular redox couples and peroxide-associated enzymes are tightly connected.
Three redox couples (glutathione disulfide/glutathione; thioredoxin-(S-S)/thioredoxin-(SH)2; and NADP+/NADPH) are involved in the peroxide-removal system. The NADP+/NADPH redox couple provides the reducing equivalents for some of the enzymes involved in generating and removing peroxide. The peroxide removal system has three nodes. Node-1: catalase (CAT) when acting in the catalytic mode requires no reducing equivalents to remove hydrogen peroxide. Node 2: the GR/GSH/GPx node (i.e., glutathione disulfide reductase/glutathione/glutathione peroxidase node) requires electrons from NADPH for peroxide-removal (55). Node-3: the TrxR/Trx/Prxd (i.e. thioredoxin reductase/thioredoxin/peroxiredoxin) node also requires NADPH as a source of reducing equivalents to reduce hydrogen peroxide to water (56, 57, 58).
By balancing redox reactions this network of cellular antioxidant/redox enzymes and proteins is essential for keeping cells functional and healthy. The members of the network are connected through substrates and products. One of the most important components of the antioxidant network is the superoxide-peroxide removal system (SPR). Peroxides are needed for cellular signaling but can also be very harmful to the cell when concentrations are out of balance. The SPR-system, as we currently know it, consists of several enzymes (SOD, CAT, GPx, Grx, TrxR and Prxd) that are intertwined with small molecule and protein redox couples, Figure 2. Many components of the SPR-system have isoenzymes or isoforms that are located in different organelles, Figure 3. The SPR-system forms a network that encompasses all organelles of the cell as well as the extracellular matrix.
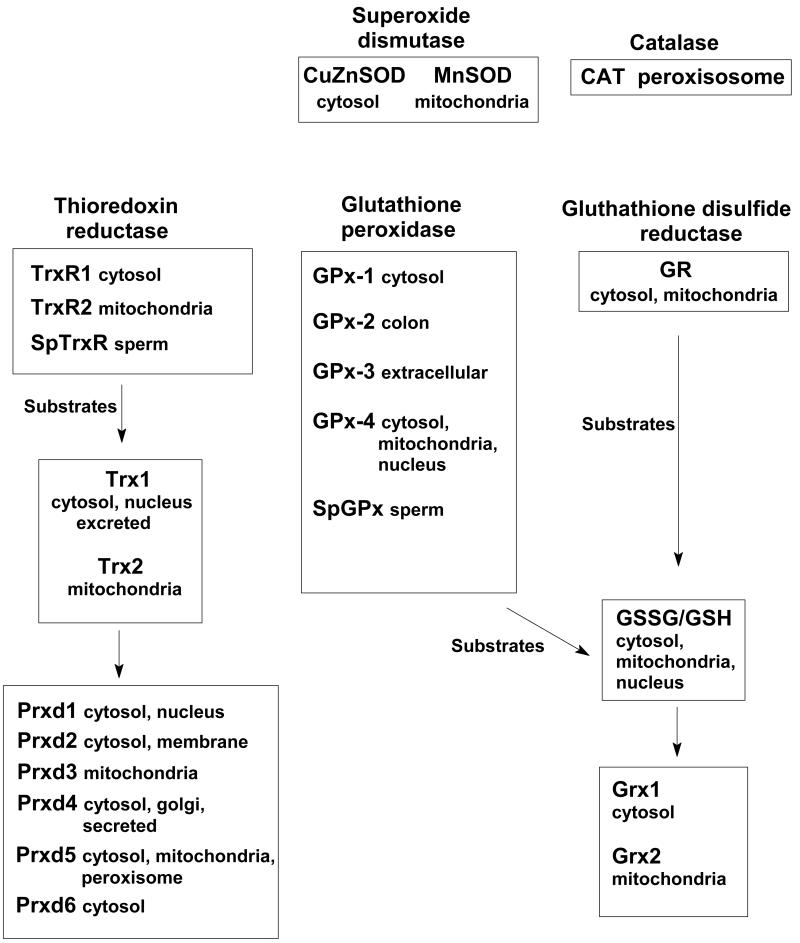
Figure 3. Enzymes of the SPR-system.
Superoxide- and peroxide-removal systems can be found in different locations in the cell. Shown is a subset of the many systems and isozymes and isoforms within the SPR-network that can remove superoxide and peroxides in cells. (It should be noted that Prxd6 is quite different than the other peroxiredoxins (59).)
It is widely recognized that ROS and RNS are key species that trigger cascades of signaling pathways (2). These signaling cascades can activate networks of coordinated gene expression; they can also effect post-translational changes in proteins. Systems biology, an approach to understand the dynamics and consequences of inter- and intra-cellular processes, will need to have at its base an understanding of the formation, reactions and interactions of reactive, transient species within the protein network. These species determine the redox state of small peptides and proteins thereby initiating signaling cascades that alter the biological state of the cell.
3. Changes in the Antioxidant Network can Translate into Changes in the Biological State
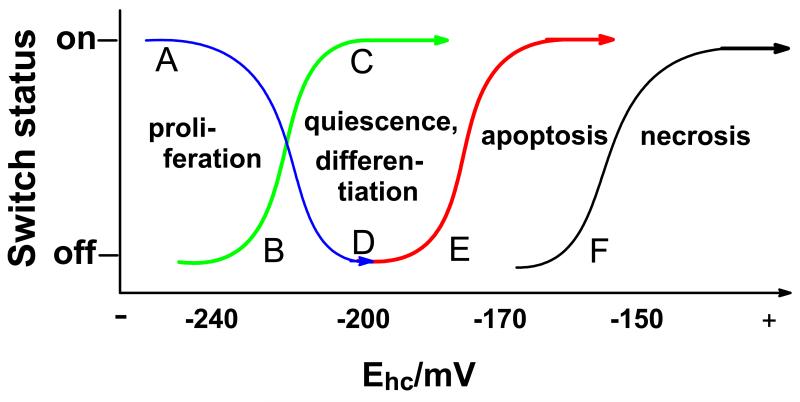
The status of the antioxidant/redox enzyme network and the biological state of a cell or organism are connected, Figure 4 (6, 23). The antioxidant/redox network together with the flux of ROS and RNS set the redox environment of cells and tissues. The overall redox environment of the cell is central in determining its biological state and function, e.g. proliferation, quiescence, differentiation, or cell death (6). Proliferating cells have a more reducing environment than differentiated cells. However, within the cell cycle there appears to be a redox cycle, where superoxide is associated with proliferation and hydrogen peroxide with quiescence (20, 24, 25, 26, 27). Cell death occurs when the cellular redox buffer becomes too oxidized, Figure 5. This can manifest itself as apoptosis or if severely oxidized necrosis (28, 29, 30). To understand redox biology, we must gather quantitative data on the redox environment of cells to gain the understanding needed to improve health.
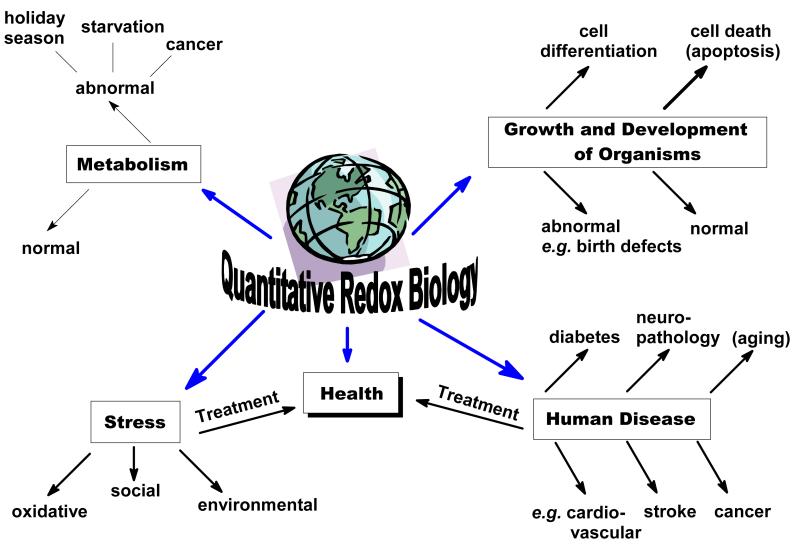
Figure 4. Redox Biology is central to events at the cellular and organism level.
Redox Biology centers on the mechanisms and biological consequences of the movement of electrons in systems that have reversible redox processes. Changes in the cellular redox environment result in signals that influence the biological status of cells and tissues. The signals can transform a cell from a normal status to an abnormal status, e.g. from differentiation to proliferation. Such changes on a cellular level can have consequences for the whole organism.
Figure 5. Biological states are tightly connected to the redox state of the principal cellular buffer, that is the redox state of the GSSG,2H+/2GSH redox pair.
The half-cell reduction potential of the GSSG,2H+/2GSH couple correlates, or more likely determines, the biological state of the cell (6). The state of the GSH redox buffer can turn molecular switches on and off, leading to different biological states. The redox state of this buffer as well as related redox couples, such as the cystine,2H+/2cysteine couple, can inform approaches to interventions to improve health. Note that quiescence is a reversible state; cells can re-enter the cell cycle and proliferate. Differentiation is not reversible.
4. Quantitative Redox Biology
Although there is a wealth of data on antioxidants and redox active species in the literature, most of these data are not directly comparable because they are relative measurements that are dependent on the different assays and conditions used, and thus of use only in the context of a specific set of experiments. For example, typical assays for antioxidant enzymes determine the relative level of expression. Because these data are only qualitative it is difficult to compare results across laboratories. Information can also be lost when comparing antioxidant enzymes that reside in different locations. For example when comparing cytosolic SOD (CuZnSOD) with mitochondrial SOD (MnSOD) the units of activity will not show the bigger picture. In cells that show the same units of activity of each enzyme the actual concentration of MnSOD in the mitochondrial matrix will probably be 10-fold higher than the concentration of CuZnSOD in the cytosol and nucleus. This information could greatly alter interpretation of data by researchers.
Another example for the importance of quantitative measurements is in the determination of the redox environment of cells and tissues. First research in Quantitative Redox Biology suggests that redox states of important couples such as the GSSG,2H+/2GSH couple are comparable in different cell systems at specific biological states, e.g. proliferation, differentiation, Figure 5 (23, 31, 32, 33, 34). Quantitative data on [GSH] and [GSSG] were required for this insight. Quantitative data would enhance the overall efficiency of the research enterprise and provide much more insight into correlations and connections of biological processes and events. The redox state of specific redox couples can be used to quantitatively assess the cellular redox environment. The GSSG,2H+/2GSH couple is considered to be the major redox buffer of the cell; the [GSH] to [GSSG] ratio has often been used as an indicator of cellular redox environment. However, this relative measurement can be misleading. A better assessment is made by determining the half-cell reduction potential of this couple using the Nernst equation:
For example, [GSH] in cells ranges from approximately 1 to 10 mM. If a cell has 10 mM GSH, then a ratio of [GSH]/[GSSG] of 16.6 translates into a Ehc of −228 mV. However, in a cell with 1 mM GSH a ratio of 166 is required to achieve the same Ehc. These ratios would indicate that a cell with 10 mM GSH has a far more oxidizing environment and thus would be expected to be in a different biological state than cells with 1 mM GSH. However, the reduction potentials in both settings are the same, suggesting that the biological states would be similar.
The GSSG,2H+/2GSH is not the only redox couple that has a role in the redox biology of cells and organisms. The cystine,2H+/2cysteine couple also has a role, even though not necessarily as a major component of the cellar redox buffer (35, 36, 37).
To fully understand this and the consequences of differing roles and sizes of the cysteine, GSH, and NADPH pools in a wide variety of settings, a systems biology approach is needed (38, 39, 40, 41). Modeling can be used to determine the major pathways within the SPR-system that are used to establish an appropriate balance of oxidants and the redox environment; but modeling can also which elements of the network taste the redox environment and then react to changes (42). For example, changes in steady-state levels of superoxide due to changes in MnSOD can result in gene expression through the hypoxia inducible factor (HIF) system (43, 44). But changes in MnSOD will also change the flux of hydrogen peroxide in cells and tissues (45, 46), which could lead to different circuits being activated for the expression of an array of genes.
However, to apply systems biology detailed quantitative information on concentrations, locations, kinetic and thermodynamic properties of these redox active species are required. Most of this information is lacking. To address this major deficiency the scientific community must increase the effort to gather this type of information. The long-term goal would be to create a set of databases of the chemical reactions that describe all aspects of the redox biology of the cell and that determine the redox state of the many redox couples that define the biological state of cells and tissues. Publicly available tools such as CellDesigner™, which uses Systems Biology markup Language and is Systems Biology Workbench compliant, are being developed to make the mathematical modeling of networks and systems with cells and tissues accessible to a wide range of researchers (47, 48). These tools need quantitative information to bring the power of systems biology to the typical biological research laboratory.
5. The Redoxome in Systems Biology
For success, systems biology must bridge engineering, mathematics, metabolism, molecular biology, and cell biology (49). The emerging field of free radical and redox biology should be at the foundation of this approach. Presently, no single mathematical approach is able to provide an integrative cellular model (50). Within the multi-algorithm framework a key task will be to address the dynamic redoxome. The elements of the redoxome do not function in isolation rather each is a part of an interacting network. Systems biology can help us integrate the redoxome into all the domains of molecular cell biology.
6. Illuminating Drug Discovery and Understanding of Toxins
Systems biology holds the promise to significantly impact the process of drug discovery (51). A systems approach is needed to identify pathways and pose interventions for disease. It could also alert to secondary effects of drugs that can be harmful. For example, the state of the redoxome can also be influenced by a wide range of toxins, especially those with redox cycling quinone/semiquinone/hydroquinone triads as a component of their toxicity (52). These triads can redox cycle producing superoxide and hydrogen peroxide and as a consequence consume GSH (46). Understanding and including the redoxome in systems biology will lead to new interventions to address disease and enhance health.
These concepts on the nature and the role of the redoxome in the fundamental biology of the cell are informing our approach to treat cancer with pharmacological doses of ascorbate (53, 54). The oxidation of ascorbate produces hydrogen peroxide; this flux of hydrogen peroxide appears to affect a subset of cancer cells more than normal cells, providing a potential treatment advantage.
The redoxome is being recognized as a cornerstone for biological processes. The integration of the young, rapidly advancing field of redox biology in conjunction with the new mathematical biology into the traditional fields of genetics and protein biochemistry will allow researchers to meet the demands to improve human health in the 21st century.
Acknowledgments
Supported by Grants R01GM073929 from the NIGMS and P42ES013661 from the NIEHS. The content is solely the responsibility of the authors and does not represent views of the NIGMS, NIEHS, or the NIH. The University of Iowa ESR Facility provided invaluable support.
References
- 1.Russell R, Superti-Furga G. Systems biology: understanding the biological mosaic. FEBS Lett. 2005;579:1771. 2005. [Google Scholar]
- 2.Forman HJ, Torres M, Fukuto J. Signal transduction by reactive oxygen and nitrogen species: pathways and chemical principles. Kluwer Academic Publishers; Dordrecht, Netherlands: 2003. [Google Scholar]
- 3.Nathan C. Specificity of a third kind: reactive oxygen and nitrogen intermediates in cell signaling. Journal of Clinical Investigation. 2003;111:769–778. doi: 10.1172/JCI18174. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper CE, Patel RP, Brookes PS, Darley-Usmar VM. Nanotransducers in cellular redox signaling: modification of thiols by reactive oxygen and nitrogen species. Trends in Biochemical Sciences. 2002;27:489–492. doi: 10.1016/s0968-0004(02)02191-6. 2002. [DOI] [PubMed] [Google Scholar]
- 5.Buettner GR. Superoxide dismutase in redox biology: The roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 2011;11:341–346. doi: 10.2174/187152011795677544. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schafer FQ, Buettner GR. Redox state of the cell as viewed though the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. 2001. [DOI] [PubMed] [Google Scholar]
- 7.Wagner BA, Venkataraman S, Buettner GR. The rate of oxygen utilization by cells. Free Radic Biol Med. 2011;51:700–712. doi: 10.1016/j.freeradbiomed.2011.05.024. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maulik N, Das DK. Redox signaling in vascular angiogenesis. Free Radic Biol Med. 2002;33:1047–60. doi: 10.1016/s0891-5849(02)01005-5. 2002. [DOI] [PubMed] [Google Scholar]
- 9.Forman HJ, Torres M, Fukuto J. Redox signaling. Mol Cell Biochem. 2002;234-235:49–62. 2002. [PubMed] [Google Scholar]
- 10.Oh JI, Kaplan S. Redox signaling: globalization of gene expression. EMBO Journal. 2000;19:4237–4247. doi: 10.1093/emboj/19.16.4237. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sen CK. Cellular thiols and redox-regulated signal transduction. Current Topics in Cellular Regulation. 2000;36:1–30. doi: 10.1016/s0070-2137(01)80001-7. 2000. [DOI] [PubMed] [Google Scholar]
- 12.Rhee SG. Redox signaling: hydrogen peroxide as intracellular messenger. Experimental & Molecular Medicine. 1999;31:53–59. doi: 10.1038/emm.1999.9. 1999. [DOI] [PubMed] [Google Scholar]
- 13.Cai J, Jones DP. Mitochondrial redox signaling during apoptosis. Journal of Bioenergetics & Biomembranes. 1999;31:327–334. doi: 10.1023/a:1005423818280. 1999. [DOI] [PubMed] [Google Scholar]
- 14.Powis G, Gasdaska JR, Baker A. Redox signaling and the control of cell growth and death. Advances in Pharmacology. 1997;38:329–359. doi: 10.1016/s1054-3589(08)60990-4. 1997. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Oberley LW. Redox regulation of transcriptional activators. Free Radic Biol Med. 1996;21:335–348. doi: 10.1016/0891-5849(96)00109-8. 1996. [DOI] [PubMed] [Google Scholar]
- 16.Menon SG, Sarsour EH, Spitz DR, Higashikubo R, Sturm M, Zhang H, Goswami PC. Redox regulation of the G1 to S transition in the mouse embryo fibroblast cell cycle. Cancer Research. 2003;63:2109–2117. 2003. [PubMed] [Google Scholar]
- 17.Menon SG, Sarsour EH, Kalen AL, Venkataraman S, Oberley LW, Goswami PC. Superoxide signaling mediates N-acetyl-L-cysteine induced G1 arrest: Regulatory role of manganese superoxide dismutase and cyclin D1. Cancer Research. 2007;67:6392–6399. doi: 10.1158/0008-5472.CAN-07-0225. 2007. [DOI] [PubMed] [Google Scholar]
- 18.McCord JM, Fridovich I. The reduction of cytochrome c by milk xanthine oxidase. J Biol Chem. 1968;243:5753–5760. 1968. [PubMed] [Google Scholar]
- 19.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. 1969. [PubMed] [Google Scholar]
- 20.Oberley LW, Oberley TD, Buettner GR. Cell division in normal and transformed cells: The possible role of superoxide dismutase and hydrogen peroxide. Med Hypotheses. 1981;7:21–42. doi: 10.1016/0306-9877(81)90018-9. 1981. [DOI] [PubMed] [Google Scholar]
- 21.Gechev TS, Hille J. Hydrogen peroxide as a signal controlling plant programmed cell death. Journal of Cell Biology. 2005;168:17–20. doi: 10.1083/jcb.200409170. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel RP, Moellering D, Murphy-Ullrich J, Jo H, Beckman JS, Darley-Usmar VM. Cell signaling by reactive nitrogen and oxygen species in atherosclerosis. Free Radic Biol Med. 2000;28:1780–1794. doi: 10.1016/s0891-5849(00)00235-5. 2000. [DOI] [PubMed] [Google Scholar]
- 23.Moriarty-Craige SE, Jones DP. Extracellular thiols and thiol/disulfide redox in metabolism. Annu Rev Nutr. 2004;24:481–509. doi: 10.1146/annurev.nutr.24.012003.132208. 2004. [DOI] [PubMed] [Google Scholar]
- 24.Menon SG, Goswami PC. A redox cycle within the cell cycle: ring in the old with the new. Oncogene. 2007;26:1101–1109. doi: 10.1038/sj.onc.1209895. 2007. [DOI] [PubMed] [Google Scholar]
- 25.Sarsour EH, Kumar MG, Chaudhuri L, Kalen AL, Goswami PC. Redox control of the cell cycle in health and disease. Antioxid Redox Signal. 2009;11:2985–3011. doi: 10.1089/ars.2009.2513. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarsour EH, Venkataraman S, Kalen AL, Oberley LW, Goswami PC. Manganese superoxide dismutase activity regulates transitions between quiescent and proliferative growth. Aging Cell. 2008;7:405–417. doi: 10.1111/j.1474-9726.2008.00384.x. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burhans WC, Heintz NH. The cell cycle is a redox cycle: linking phase-specific targets to cell fate. Free Radic Biol Med. 2009;47:1282–1293. doi: 10.1016/j.freeradbiomed.2009.05.026. 2009. [DOI] [PubMed] [Google Scholar]
- 28.Watson WH, Cai J, Jones DP. Diet and apoptosis. Annu Rev Nutr. 2000;20:485–505. doi: 10.1146/annurev.nutr.20.1.485. 2000. [DOI] [PubMed] [Google Scholar]
- 29.Watson WH, Chen Y, Jones DP. Redox state of glutathione and thioredoxin in differentiation and apoptosis. Biofactors. 2003;17:307–314. doi: 10.1002/biof.5520170130. 2003. [DOI] [PubMed] [Google Scholar]
- 30.Jiang S, Moriarty-Craige SE, Orr M, Cai J, Sternberg P, Jr, Jones DP. Oxidant-induced apoptosis in human retinal pigment epithelial cells: dependence on extracellular redox state. Invest Ophthalmol Vis Sci. 2005;46:1054–1061. doi: 10.1167/iovs.04-0949. 2005. [DOI] [PubMed] [Google Scholar]
- 31.Allen RG, Newton RK, Sohal RS, Shipley GL, Nations C. Alterations in superoxide dismutase, glutathione, and peroxides in the plasmodial slime mold physarum polycephalum during differentiation. J. Cell Physiol. 1985;125:413–419. doi: 10.1002/jcp.1041250308. 1985. [DOI] [PubMed] [Google Scholar]
- 32.Schafer FQ, Buettner GR. Redox state and redox environment in biology. In: Forman HJ, Torres M, Fukuto J, editors. Signal Transduction by Reactive Oxygen and Nitrogen Species: Pathways and Chemical Principles. Kluwer Academic Publishers; Dordrecht, Netherlands: 2003. pp. 1–14. [Google Scholar]
- 33.Nkabyo YS, Ziegler TR, Gu LH, Watson WH, Jones DP. Glutathione and thioredoxin redox during differentiation in human colon epithelial (Caco-2) cells. American Journal of Physiology - Gastrointestinal & Liver Physiology. 2002;283:G1352–G1359. doi: 10.1152/ajpgi.00183.2002. 2002. [DOI] [PubMed] [Google Scholar]
- 34.Mannery YO, Ziegler TR, Park Y, Jones DP. Oxidation of plasma cysteine/cystine and GSH/GSSG redox potentials by acetaminophen and sulfur amino acid insufficiency in humans. J Pharmacol Exp Ther. 2010;333:939–947. doi: 10.1124/jpet.110.166421. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyer SS, Accardi CJ, Ziegler TR, Blanco RA, Ritzenthaler JD, Rojas M, Roman J, Jones DP. Cysteine redox potential determines pro-inflammatory IL-1beta levels. PLoS One. 2009;4(3):e5017. doi: 10.1371/journal.pone.0005017. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DP, Liang Y. Measuring the poise of thiol/disulfide couples in vivo. Free Radic Biol Med. 2009;47(10):1329–38. doi: 10.1016/j.freeradbiomed.2009.08.021. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shyntum Y, Iyer SS, Tian J, Hao L, Mannery YO, Jones DP, Ziegler TR. Dietary sulfur amino acid supplementation reduces small bowel thiol/disulfide redox state and stimulates ileal mucosal growth after massive small bowel resection in rats. J Nutr. 2009;139(12):2272–2278. doi: 10.3945/jn.109.105130. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antunes F, Salvador A, Marinho HS, Alves R, Pinto RE. Lipid peroxidation in mitochondrial inner membranes. I. An integrative kinetic model. Free Radic Biol Med. 1996;21(7):917–943. doi: 10.1016/s0891-5849(96)00185-2. 1996. [DOI] [PubMed] [Google Scholar]
- 39.Marinho HS, Antunes F, Pinto RE. Role of glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase in the reduction of lysophospholipid hydroperoxides. Free Radic Biol Med. 1997;22(5):871–883. doi: 10.1016/s0891-5849(96)00468-6. 1997. [DOI] [PubMed] [Google Scholar]
- 40.Johnson RM, Ho YS, Yu DY, Kuypers FA, Ravindranath Y, Goyette GW. The effects of disruption of genes for peroxiredoxin-2, glutathione peroxidase-1, and catalase on erythrocyte oxidative metabolism. Free Radic Biol Med. 2010;48(4):519–525. doi: 10.1016/j.freeradbiomed.2009.11.021. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adimora NJ, Jones DP, Kemp ML. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxidants & Redox Signaling. 2010:731–743. doi: 10.1089/ars.2009.2968. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DP. Disruption of mitochondrial redox circuitry in oxidative stress. Chem Biol Interact. 2006;163(1-2):38–53. doi: 10.1016/j.cbi.2006.07.008. 2006. [DOI] [PubMed] [Google Scholar]
- 43.Wang M, Kirk JS, Venkataraman S, Domann FE, Zhang HJ, Schafer FQ, Flanagan SW, Weydert CJ, Spitz DR, Buettner GR, Oberley LW. Manganese superoxide dismutase suppresses hypoxic induction of hypoxia inducible factor-1α and vascular endothelial growth factor. Oncogene. 2005;24:8154–8166. doi: 10.1038/sj.onc.1208986. 2005. [DOI] [PubMed] [Google Scholar]
- 44.Kaewpila S, Venkataraman S, Buettner GR, Oberley LW. Manganese superoxide dismutase modulates Hypoxia Inducible Factor-1α induction via superoxide. Cancer Res. 2008;68(8):2781–2788. doi: 10.1158/0008-5472.CAN-07-2635. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buettner GR, Ng CF, Wang W, Rodgers VGJ, Schafer FQ. A new paradigm: manganese superoxide dismutase influences the production of H2O2 in cells and thereby their biological state. Free Radic Biol Med. 2006;41:1338–1350. doi: 10.1016/j.freeradbiomed.2006.07.015. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y, Buettner GR. Thermodynamic and kinetic considerations for the reaction of semiquinone radicals to form superoxide and hydrogen peroxide. Free Radic Biol Med. 2010;49:919–962. doi: 10.1016/j.freeradbiomed.2010.05.009. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funahashi A, Tanimura N, Morohashi M, Kitano H. CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. BIOSILICO. 2003;1:159–162. 2003. [Google Scholar]
- 48.Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H. CellDesigner 3.5: A Versatile Modeling Tool for Biochemical Networks. Proceedings of the IEEE. 2008;96(8):1254–1265. 2008. doi 10.1109/JPROC.2008.925458. [Google Scholar]
- 49.Aloy P, Russel EB. Structure-based systems biology: a zoom lens for the cell. FEBS Lett. 2005;579:1854–1858. doi: 10.1016/j.febslet.2005.02.014. 2005. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi K, Vel Arjunan SN, Tomita M. Space in systems biology signaling pathways - towards intracellular molecular crowding in silico. FEBS Lett. 2005;579:1783–1788. doi: 10.1016/j.febslet.2005.01.072. 2005. [DOI] [PubMed] [Google Scholar]
- 51.Apic G, Ignjatovic T, Boyer S, Russel RB. Illumiating drug discovery with biological pathways. FEBS Lett. 2005;579:1872–1877. doi: 10.1016/j.febslet.2005.02.023. 2005. [DOI] [PubMed] [Google Scholar]
- 52.O'Brien PJ. Molecular mechanisms of quinone cytotoxicity. Chem-Biol Interact. 1991;80:1–41. doi: 10.1016/0009-2797(91)90029-7. 1991. [DOI] [PubMed] [Google Scholar]
- 53.Chen Q, Espey MG, Sun AY, Lee JH, Krishna MC, Shacter E, Choyke PL, Pooput C, Kirk KL, Buettner GR, Levin M. Ascorbic acid in pharmacologic concentrations: a prodrug for selective delivery of ascorbate radical and hydrogen peroxide to extracellular fluid in vivo. Proc Natl Acad Sci USA. 2007;104:8749–8754. doi: 10.1073/pnas.0702854104. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du J, Martin SM, Levine M, Wagner BA, Buettner GR, Wang SH, Taghiyev AF, Du C, Knudson CM, Cullen JJ. Mechanisms of ascorbate-induced cytotoxicity in pancreatic cancer. Clin Cancer Res. 2010;16(2):509–520. doi: 10.1158/1078-0432.CCR-09-1713. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brigelius-Flohe R. Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. 1999. [DOI] [PubMed] [Google Scholar]
- 56.Watson WH, Yang X, Choi YE, Jones DP, Kehrer JP. Thioredoxin and its role in toxicology. Toxicological Sciences. 2004;78:3–14. doi: 10.1093/toxsci/kfh050. 2004. [DOI] [PubMed] [Google Scholar]
- 57.Nordberg J, Arner ES. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biol Med. 2001;31:1287–1312. doi: 10.1016/s0891-5849(01)00724-9. 2001. [DOI] [PubMed] [Google Scholar]
- 58.Powis G, Montfort WR. Properties and biological activities of thioredoxins. Annual Review of Pharmacology & Toxicology. 2001;41:261–295. doi: 10.1146/annurev.pharmtox.41.1.261. 2001. [DOI] [PubMed] [Google Scholar]
- 59.Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res. 2007;48(10):2306–23018. doi: 10.1194/jlr.M700299-JLR200. 2007. [DOI] [PubMed] [Google Scholar]