Abstract
Translational control is a vital aspect of gene expression. Message specific translational repressors have been known of decades. Recent evidence, however, suggest that a general machinery exists that dampens the translational capacity of the majority of mRNAs. This activity has been best ascribed to a conserved family of RNA helicases called the DHH1 / RCKp54 family. The function of these helicases is to promote translational silencing. By transitioning mRNA into quiescence, DHH1 / RCKp54 helicases promote either mRNA destruction or storage. In this review we describe the known roles of these helicases and propose a mechanistic model to explain their mode of action.
Translational repression is a critical node of gene expression
The control of mRNA translation is vital to maintain cellular homeostasis. Just as the process of transcription is reiterative, so is translation. Each RNA message can be translated hundreds of times, thereby encoding hundreds of polypeptide. Therefore, the effects of nuclear changes in expression would not be felt without additional control of cytoplasmic transcripts, which must stop translating in a regulated fashion if protein expression is to be downregulated, for example. It therefore makes sense that regulatory mechanisms exist to enforce translational regulation, ensuring the mRNA doesn’t translate ad infinitium. While down-regulating mRNA translation is often coupled to robust degradation of the transcript, it is also possible to store messages as stable species so that they can be reutilized. Arresting translation and maintaining transcripts in a stable, inert form provides the cell with a complex poised to respond quickly to biological cues. For example, translationally silenced mRNAs are found at neuronal synapses(1), where rapid expression is critical for cellular function. Additionally, translational silencing mediated by changes in polyadenylation has been shown to be important in gene expression control during oocyte maturation and embryo development(2). For most mRNA transcripts, however, translational silencing feeds the message into decay(3). The regulated destruction of the mRNA is the ultimate mechanism to stop a transcript’s translation. Despite the importance of the processes, the interplay between mRNA translational control and mRNA decay is not well understood.
In this review we discuss the events that occur at the interface of translational control pathways and mRNA decay, and the importance thereof. Specifically we focus on the DEAD box helicase protein DHH1/RCKp54 which appears to transition mRNA from a state that is favorable for translation to a state of quiescence that can lead to either storage of an mRNA or entry into degradation. We hypothesize that this family of helicases is part of a basal machinery that is exploited for both general and mRNA specific translational control.
DEAD-box helicases and translation
The DEAD-box family of proteins is a large group of proteins generally involved in RNP remodeling(4). They are classified as members of helicase superfamily 2 (SF2)(5), along with the related DEAH and similar helicases (collectively referred to as the DExD/H helicase family). They are defined by a set of 9 conserved sequence motifs that have been identified by homology to the founding member of the family, eIF4A – an RNA helicase involved in translation initiation. These motifs permit DEAD-box proteins to bind and hydrolyze ATP as well as interact with RNA. Structural data, including the crystal structure of eIF4A alone(6) and D. melanogaster Vasa with RNA and ATP(7), have supported the involvement of the identified critical residues. While DEAD box proteins are classified as helicases, this is somewhat of a misnomer as no members of the family have been found to carry out processive unwinding ascribed to classical helicases. Instead, the known members of the family function in a variety of roles related to RNA metabolism, with functions ranging from local disassociation of short duplexes to RNA binding(8). eIF4A, for example, was found to change the sensitivity of RNA to nuclease, presumably by rearranging its structure(9). In addition, eIF4III-A appears to clamp tightly to RNA in response to ATP binding and hydrolysis(8). Members of this family have been found to be involved at every step in RNA processing, particularly ribosome biogenesis, splicing, and translation.
The DHH1/RCKp54 family of helicases
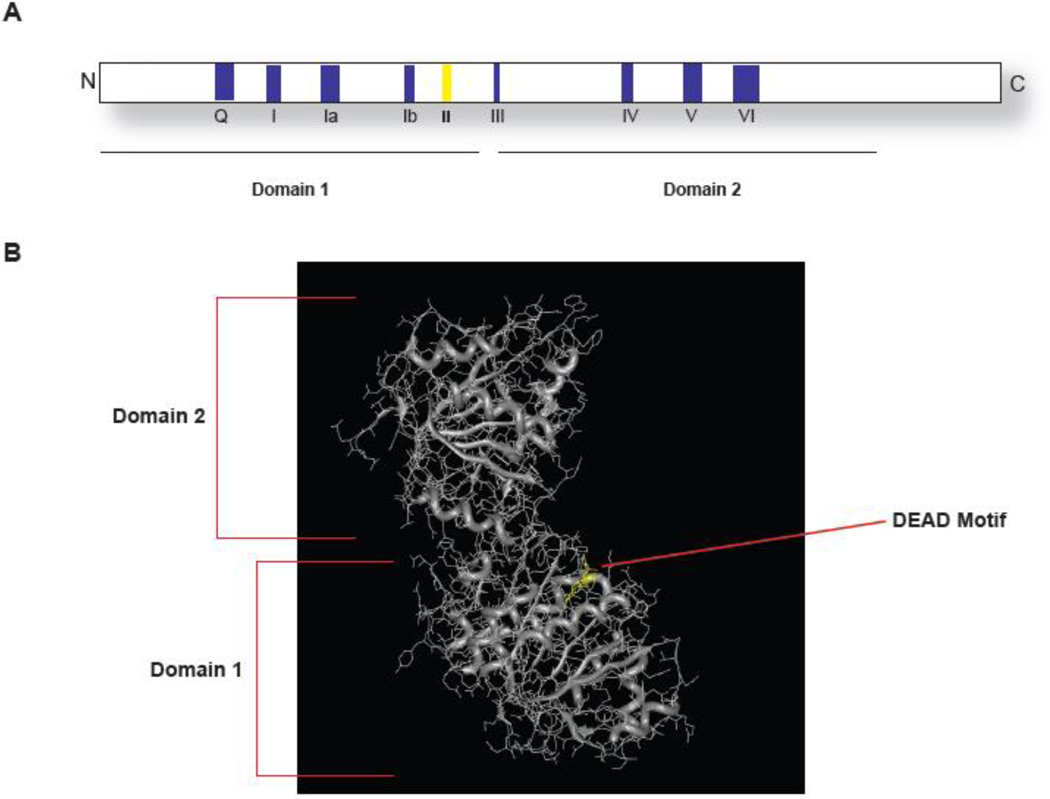
DHH1/RCKp54 proteins are stereotypic members of the DEAD-box helicase family, with 9 conserved motifs and characterized ATP binding and hydrolysis activities (Figure 1). This family of helicases was first identified in Drosophila as Me31b (maternally expressed from chromosomal locus 31b): a gene important for embryonic development(10). Since then, homologs have been identified in most eukaryotes and its functions have been described to be at the interface between mRNA translation and decay. Specifically, DHH1/RCKp54 helicases have been implicated in promoting maternal mRNA silencing in oocytes, as well as developmental regulation in the embryo, controlling cell-cycle checkpoints, maintaining neuronal synaptic plasticity, promoting stress responses, allowing viral replication, and facilitation of mRNA decapping and decay(11). In this review, we hypothesize that these diverse functions are a result of its core conserved function in promoting translational repression.
Figure 1. Structure of DHH1/RCKp54 helicases.
DHH1/RCKp54 helicases are traditional members of the SF2 family of helicases. They all have the nine characteristic helicase domains and are similar to SF2 helicases in structure (B).
Structural and biochemical characterization of DHH1/RCKp54 helicases
The biochemical characterization of proteins in this family is relatively lacking, with the best characterized members being Xp54 and RCKp54. Xp54 was shown to have unwinding activity towards a 46-bp duplex with 5’ overhangs. The protein used was purified from cell lysates using salt elution from an oligo(dT)-cellulose column and appeared pure by coomassie staining. Additionally, the unwinding activity was blocked specifically by the addition of α-Xp54 antibodies(12). These results strongly suggest that Xp54 is able to unwind RNA, though the requirement for a cofactor cannot be ruled out. The activity of RCK/p54 was assayed via electron microscopy. Recombinant RCKp54 was mixed with in vitro transcribed RNA and observed for 30 minutes. The protein was observed to coat and straighten the RNA, which tended to have a structured appearance in the absence of the protein. The RNA appeared to demonstrate thinner single-stranded regions in these assays, though these are hard to interpret due to the relatively low resolution of the microscopy(13). This experimentation clearly shows that RCKp54 is able to bind mRNA without modification or cofactors; however, attempts to elicit unwinding activities from other members of the family (e.g. DHH1(14)) have been unsuccessful. Importantly, however, work from the Reese lab has shown that DHH1 has weak RNA-dependent ATPase activity and that this activity is restricted by inter domain interactions. This suggests that co-factors might exist within the cell to help modulate DHH1/RCKp54 enzymatic activity(14).
While it remains unclear whether all members of the DHH1 subfamily have bona fide unwinding activity, or whether some members carry out other activities, genetic studies have shown that mutants that are predicted to affect ATP binding or helicase activity can nullify DHH1 function or even create a dominant negative effect(14, 15). These results argue that whatever catalytic activity DHH1 may have is important to its function. Other evidence of activity includes the fact that DHH1 preferentially cross-links to ATP and exhibits tight binding to RNA in vitro(14). The activity could be something that is distinct from traditional helicase activity, such as RNA structural conversion, RNA clamping, or RNP displacement. It has been shown that DexH/D proteins can possess activities of this sort alongside or instead of traditional unwinding(8).
Most of the details of the molecular function of this helicase subfamily have been elucidated in the yeast system. The DHH1 protein is a 506 aa polypeptide with 52% identity and 63% similarity to the human homolog. These figures improve to 69% and 83% respectively if only the core 400aa domain is considered. It possesses the traditional DEAD RNA helicase motifs in the core conserved region and doesn’t have any other recognizable sequence elements.
The structures of several subfamily members have been solved, including DHH1(15) and RCKp54(16–18). These have shown that like previously solved DEAD-box proteins (eIF4A(19) and mjDEAD(20)), the DHH1 core contains two RecA-like domains containing previously described motifs. Each individual domain overlaid with its counterparts from previous structures gives a maximum RMSD value of 1.3 Ǻ. Importantly, the orientation of the domains is different, with the two forming extensive interactions with each other, with many of the conserved motifs clustered near the interface. Evidence for the interaction of the domains was found in the observation that addition of ATP and RNA to DHH1 greatly increased its resistance to trypsin, suggesting that the protein adopted a more closed conformation. A mutant predicted to sterically hinder the domain packing largely negated this effect, as well as creating a dominant negative effect in vivo(15). These results indicate that DHH1 has a typical twin Rec-A domain structure, but adopts a unique closed conformation that is important to the function of the protein.
Evidence for function in translational repression across many organisms
DHH1/RCKp54 helicases have been ascribed many different functions, at different developmental stages, in many different cell types, and in many different organisms; however, we hypothesize that one unifying theme unites these functions. In this review, we propose that each function of the protein hinges on a more general role of this helicase family in translational repression. As a general decay factor in yeast, it represses translation and allows decay to overtake the message. As a component of oocyte granules, it serves to keep RNAs silenced during oocyte development and maturation. It serves similar roles in neuronal granules, stress granules, as well as other systems and functions. In the following sections, we will present details about the specific roles of several key members of this family and then propose a unifying model.
DHH1: a regulator of decapping functions by altering mRNA translation
In yeast, the investigation of DHH1 has been tied to its effects on mRNA decay(21, 22). It was originally shown to function as a general decay factor, interacting with the multiple proteins involved in the process, including deadenylase components POP2, NOT1, and CCR4(23, 24), the decapping cofactors LSM1 and PAT1, and even the decapping holoenzyme through DCP1(21). Furthermore, it was shown that its deletion leads to general stabilization of mRNA, inability to repress translation, and poor formation of P-bodies (processing bodies potentially involved in storage of mRNA or decay factors)(25). Additionally, there are reports that DHH1 can function in regulatory roles, changing expression of specific factors in several cellular processes, specifically in cell cycle regulation(26–28), mating(29), iron metabolism(30), and mitochondrial function(31). In many of these cases, it was found to function with other proteins acting as adapters, recruiting DHH1 to specific messages, where it could carry out its function. Unsurprisingly, these studies have shown that protein output is controlled by DHH1, as are mRNA levels. However, there has been little evidence until recently that would allow us to distinguish whether the level of translation was being reduced by removal of mRNA or whether the level of mRNA was being reduced by the inhibition of translation. Recently, however, two studies have shown that DHH1 represses translation in the absence of an active decapping pathway, indicating that the protein is able to impact translation without reducing mRNA levels(32, 33). This confirms that DHH1 can function to reduce the level of translation directly, likely leading to an increase in the decay rates in its target genes. Taken together, these experiments identify DHH1 as a versatile translation repressor, able to function in general decay as well as targeted functions with specific adaptor proteins.
In early studies, DHH1 was suspected to have a role in steps of mRNA processing relating to deadenylation, as it had been identified in complexes found to be related to that process(24). Subsequently, DHH1 was identified as a factor important in decapping, as mutant cells showed accumulation of capped transcripts, similar to mutants in the decapping complex(21, 22). However, it was ruled out that DHH1 could simply be a decapping cofactor, as nonsense-mediated decay (another process that depends on decapping) appears unaffected(21). Furthermore, DHH1 was found to associate with both the deadenylase complex and the decapping complex in an RNA independent manner. Bringing the ideas together, the growth phenotype of a DHH1 deletion was found to be exacerbated by overexpressing Caf20p, a known antagonist of cap-dependent translation. As these effects were unique, and did not replicate with mutants in the deadenylase or decapping enzymes, it was posited to directly tie DHH1 function to translational regulation(21).
Further studies showed that DHH1, along with another factor PAT1, were capable of repressing translation(25). The overexpression of these proteins leads to a loss of polysomes and accumulation of these proteins in cytoplasmic foci called P-bodies as well as loss of reporter mRNAs from heavy polysomes. In contrast, overexpression of other decapping components like DCP1 does not duplicate this effect. Complementary experiments in vitro showed that recombinant DHH1 could inhibit translation in a manner dependent on its ATP hydrolysis activity. Finally, it was shown that deletion of DHH1 (which stabilizes normal messages) failed to stabilize translation-inhibited reporters, unlike general decapping factors. This again directly linked DHH1 with translational repression rather than direct decapping control(25).
It has been shown in more recent experimentation that mRNA decays in the context of translation, with ribosomes still associated with the message at the time of decay(34). In this state, where the message is accessible to both the translational machinery and the decay proteins, translational control is critical to directing individual mRNAs into the decay pathway rather than further rounds of translation. This is illustrated by the conclusion that mutation of translation initiation factors, which inhibits translation, increases the rate of decay(35). DHH1 appears to function at this junction between the two processes.
From a molecular standpoint, it has been shown that DHH1 inhibits the formation of the 48S pre-initiation complex in vitro, implying that DHH1 could have a similar function in vivo, which would be consistent with the observed function in translational repression(25). More recently, however, it was shown that DHH1 functions even in cells where translation initiation is limited via initiation mutants. While this is not conclusive evidence, the fact that limiting initiation does not prevent DHH1 function implies that it may function at another step. Additionally, it was observed that artificially tethering DHH1 to a reporter resulted in the saturation of message with slowly moving ribosomes(33). Consistent with these observations, endogenous DHH1 was found to preferentially associate with slow-moving ribosomes in a manner dependent on its ATPase activity(33). Furthermore, destabilization of messages impeded for translation elongation was entirely DHH1 dependent. Taken together, these pieces of evidence indicate that DHH1 functions at steps later than initiation, associating with translating ribosomes and responding to changes in elongation rate. Together, the in vitro and in vivo data suggest DHH1 impacts some aspect of ribosome function. We hypothesize that the context of DHH1’s interaction with ribosomes dramatically impacts the effect on translation observed (see below).
Xp54, cgh-1, Me31b: Activities in reproductive and developmental pathways
In Xenopus, Xp54 was first identified as a component of maternal storage particles. It was shown to have bona fide unwinding activity, the first of the DHH1/RCKp54 family of helicases to do so(12). In Xenopus oocyte systems, tethered Xp54 was shown to repress translation in oocyte systems lacking decay(36), supporting similar evidence in yeast indicating a role primarily in translational control. Interestingly, similarly tethered inactive versions of Xp54 show an upregulation of protein output, suggesting that the mutant proteins function in translation somehow, but fail to fulfill their normal role(37).
Further studies found that during active transcription in oocyte development, Xp54 appears to shuttle between the nucleus and the cytoplasm, associating with newly synthesized RNA in the nuclei and participating in mRNP storage body formation in the cytoplasm. Upon arrest of transcription during maturation, Xp54 becomes restricted to the cytoplasm and remains there until transcription is reactivated(12). Further studies have shown that Xp54 forms higher order structure in an RNA dependent manner, which may contribute to the formation of these storage granules(37). These findings suggest a role for Xp54 in promoting and maintaining the translational silencing of stored mRNAs in oocytes, which is vital to embryonic development.
In drosophila, Me31B has been shown to have several functions in widely varying cell types. It functions in silencing of oocyte-specific genes in transport to the oocyte(38). Interestingly, Me31B has been shown to be present in synaptic foci in neurons which lack degradation factors. Knockdown of Me31B in this context leads to elevation in levels of postsynaptic proteins known to be translationally regulated(1). Similarly to other systems, it has been shown associate with general decay factors, including Pacman and DCP1(39).
In C. elegans, cgh-1 has been suggested to have distinct functions in somatic cells and germ cells. In somatic cells, it appears to form cytoplasmic granules consistent with processing bodies (P-bodies) containing other decay factors(40, 41). In germ cells, however it is essential for male gametogenesis, as well as oocyte development, triggering an apoptosis pathway that destroys developing oocytes upon knockdown(40). In these cells, the protein is found in distinct cytoplasmic granules devoid of decay factors. Strikingly, RIP-chip analysis revealed that in this situation, as many as 85% of mRNAs bound by cgh-1 is identified as maternal mRNAs present in embryos before zygotic transcription. Knockdown of cgh-1 led to specific mislocalization and destabilization of these mRNAs(41).
RCKp54: Translational silencing in multiple scenarios
In humans, the study of RCKp54 (also called DDX6) has largely focused on the involvement of the gene in disease processes. It was first described in 1992 as a target of a particular t(11;14) translocation in the RC-K8 lymphoma cell line from which it took its name(42). It has been found to be upregulated in cancer cell lines(43) and implicated in regulation of oncogenes such as c-myc(13). These roles were confirmed by knockdown studies that showed RCKp54 was important to the proliferation of certain cancer lines(44). Studies of hepatocellular carcinoma led to a connection with the viral field by showing that RCKp54 may be contributing to the cancer process in part by facilitating the progression of the Hepatitis C virus(45) in that system. Since then, it has also been shown to be involved in the replication and maintenance of HCV(46, 47) and replication and encapsidation of other viruses in humans(48, 49), including HIV(50), as well as other systems(51). These studies theorize that the same activity that functions to modulate translation and decay in other systems may function to remodel the viral mRNP so that it is properly recognized and packaged. Another explanation would be that viral replication and packaging requires the viral RNA to be clear of ribosomes, so the translational repression function of the protein may function in this process as well. It is also possible that viruses take advantage of the ability of RCKp54 to repress expression of host proteins to facilitate these functions.
Specific functions in humans have also been described outside of viral systems. The protein is required during erythroid development as it is required to store silenced hr15-LOX mRNA from the point of enucleation until the latest stages, at which point the product of that message destroys the mitochondria of the mature erythrocyte(52). This functions similarly to the translational repression seen in oocyte systems, where mRNA is stored for future use under circumstances that require it. Furthermore, there is evidence that RCKp54 carries out general translational repression functions in this system also, including repression mediated by miRNA mechanisms(53). In this case, the RCKp54 protein was found to physically interact with the miRISC machinery, specifically Ago2. Depletion of the protein led to significant changes in localization of Ago2 and a significant reduction in its ability to repress protein production from a miRNA-specific reporter, but not a siRNA-specific reporter. Importantly, knockdown of another protein known to function in mRNA decay (Lsm1) did not recapitulate the effects. These findings show that RCKp54 is important to carrying out the translational repression effects seen in miRNA silencing. The protein is not required by the siRNA pathway as that pathway involves cleavage of the mRNA and is not dependent on translational repression in this case. It should be noted that other proteins in this family have since been shown to have functions in miRNA function, such as Me31b(1).
Granularization: A putative mechanism for translational control?
In many systems, the function of DHH1 and related proteins is connected with aggregation of protein and RNA into large cytoplasmic complexes termed granules. These are generally large enough to be viewed microscopically and vary in composition according to the organism and the situation that leads to their formation. These can be broadly divided into four classes: germ granules, which are complexes formed during oogenesis which carry maternal mRNA into the embryo; P-bodies, which may serve as processing or storage areas for silenced RNAs in somatic cell; stress granules, which serve to sequester mRNAs during severe changes in transcription in response to stress; and specialized granules, such as neuronal granules, which exist in specific cell types and carry out functions unique to those cells.
DEAD-box proteins have long been known to participate in granule formation. The family member vasa was identified as a key component of germ granules in drosophila and was found to be highly similar to eIF4A very early in the study of granule formation(54). Similarly, DHH1 family members are integral components of germ granules, functioning to silence maternal mRNAs until necessary(12, 38, 40). In neuronal granules, DHH1/RCKp54 also functions to maintain silencing(1). P-bodies and stress granules are two sets of closely related granules containing similar sets of translation and decay regulatory factors. They vary largely by the fact that PBs are present in growing cells whereas SGs are only present under stress and the fact that PBs are devoid of ribosomal components and translation factors, whereas SGs contain small ribosomal subunits as well as eIF3/4 initiation factors and poly(A) binding protein(55). DHH1 has been shown to be a found in both types of bodies, though it is much more heavily represented in P-bodies(56).
One problem that confounds the interpretation of DHH1 function in the context of P-bodies and stress granules is the fact the function of these bodies has not been clearly elucidated. It is clear that P-bodies form in cells under normal conditions as well as stress and that stress granules form under many different kinds of cellular stresses. Some functions are assumed through the observations that stress granules form at the same time as overall translational downregulation at the onset of stress. These assumptions are strengthened by the findings that proteins like DHH1 are found to associate with miRNA machinery and mediate the function of this machinery(53). Furthermore, Ago2 was found to be directly localized to P-bodies, further adding to the idea that these were central sites of mRNA regulation and decay(57). However, it has been shown that disruption of P-bodies through the deletion of EDC3(58) has no effect on either decay or translation in yeast. Additional evidence has demonstrated that translational repression can be induced without formation of stress granules(59) and miRNA-mediated repression can be carried out without P-bodies(60) in other organisms. Thus it remains to be established if P-bodies and stress granules are storage bodies for repressed mRNAs or if they have other functions within the cell.
Summary and model
Many different functions for the DHH1/RCKp54 subfamily of proteins have been discussed. They function in organisms ranging from yeast to mammals, during development and adulthood, in both somatic cells and germ lines. They carry out functions from general regulation of decay to cell-specific functions such as maternal mRNA maintenance, neurotransmitter regulation, and miRNA silencing. The adaptability and flexibility of these proteins showcases the versatility of this protein family. However, one thread unites all of the functions of these proteins. Whatever the context may be, they seem to function in repressing translation of messages, whether the outcome of that repression is rapid decay in the yeast system or maintenance and protection in oocytes. In many of these systems, the mechanism that these proteins use to carry out their functions has not been assayed directly, but all of the effects discussed above are consistent with the idea that these proteins carry out a translational repression function in each case.
The differences in all of these situations are the proteins that bind and recruit DHH1/RCKp54. The system appears to be very flexible and modular. In the case of general regulation of translation and decay DHH1 functions in concert with other translational repressors and decay activators, such as CCR4/NOT. These proteins complement DHH1 function and may serve to recruit it or make certain messages better substrates. In the example of CCR4/NOT, the deadenylase function of the complex serves to repress translation and create conditions amenable to decay. Even in this simple system, the PUF regulatory proteins are known to interact with members of the CCR4/NOT complex and may promote recruitment of DHH1 to the targeted mRNA(61). In similar fashion, factors involved in miRNA regulation such as Ago1 and Ago2 interact with RCKp54 in active miRISC complexes. These serve to target and deliver RCKp54 to its target mRNAs, where it can carry out its function and create the silencing effect observed with miRNA regulation.
Its function in storage bodies like the maternal and neuronal bodies is also defined by its interactions with its binding partners. It has a complex role in Xenopus oocytes, with a nuclear component, where Xp54 binds newly synthesized RNAs in the nucleus a cytoplasmic function where Xp54 maintains those maternal mRNAs in a silenced state. Though its binding partners in the nucleus remain unknown, it interacts with CPEB and FRGY2 in the cytoplasm, both factors implicated in maintenance and regulation of the maternal granules(62). In the neuronal system, Me31b has been found to associate with FMRP in neuronal granules. A deletion of Me31b in that system has been shown to lead to loss of FMRP-associated phenotypes, implying that Me31b is involved in FMRP-mediated repression(1).
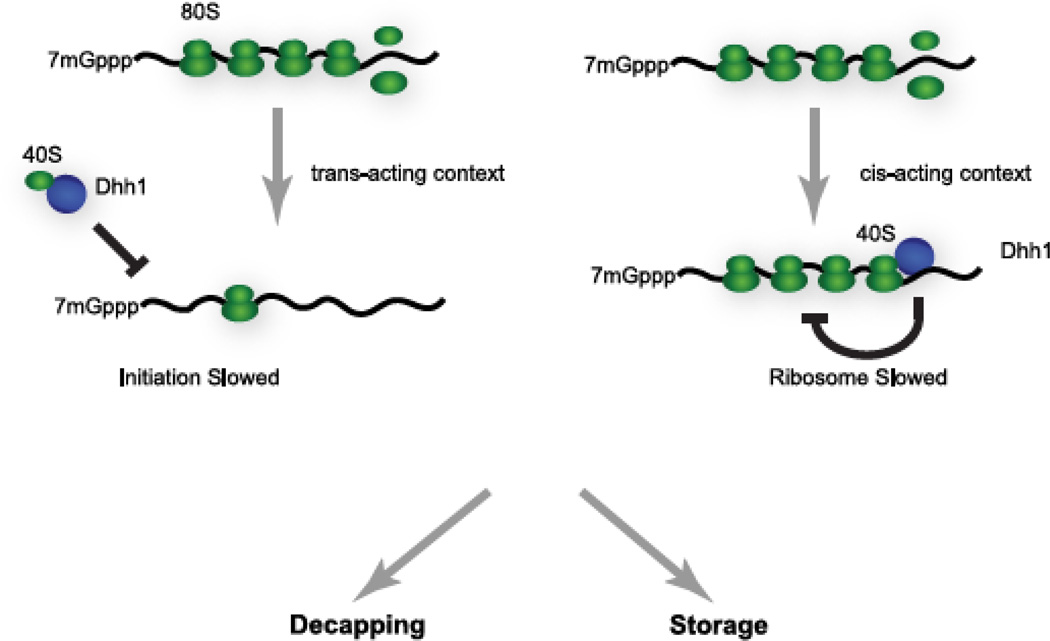
Mechanistically, it has been proposed that DHH1/RCKp54 helicases repress translation by acting on translational initiation(25). More recently, we have observed that DHH1 in yeast can slow translational elongation/termination when tethered(33). One potentially unifying theory of the data is that DHH1 directly affects the function of the 40S ribosomal subunit itself and the context of this interaction dictates the mRNA’s fate. Indeed, it has been observed that Dhh1 binds ribosomes(33, 63). Moreover, Dhh1 represses translation in vitro of an mRNA harboring the Cricket Paralysis Virus IRES, which requires only the 40S ribosomes to initiate translation(25). The context upon which Dhh1 binds to the 40S ribosomal subunit might affect which step in translation that appears to be inhibited (Figure 2). Interaction between Dhh1 and free 40S subunits could influence translation at early steps and manifest as an initiation block. Such might be occurring both in vitro and during DHH1 over-expression in cells(25). In the context of an actively translating mRNA, however, DHH1 interaction with 40S subunits might impede ribosome movement on mRNA as we have observed and implies a role for DHH1 in inhibiting translation either during elongation, termination, or ribosome recycling. Additional experiments will be needed to define precisely how DHH1 functions mechanistically.
Figure 2. A novel function of Dhh1 is to repress a late step in translation.
We hypothesize that DHH1/RCKp54 helicases may function directly on 40S ribosomal subunits. If Dhh1 were to function on free 40S subunits, the consequence would be repression of translation at initiation (depicted in the left side of the figure). If DHH1/RCKp54 helicases act on assembled polyribosomes in vivo this would lead to repression of translation at a late, post-initiation step (depicted in the right side of the figure). Repression of ribosome movement could either be direct repression of ribosomes or possibly further consolidation of already slowed ribosomes. Repressed polyribosomal mRNA can then either be decapped or stored depending on the biological conditions.
Table 1.
| Protein | System | Function | Ref |
|---|---|---|---|
| DHH1 | Yeast | Decay/decapping activation, p-body formation, translational repression, cell cycle regulation, mating | 21, 25–29, 33 |
| RCKP54 | Human | Oncogene, viral functions, erythroid maturation, repression by miRNA | 42–53 |
| Me31b | Drosophila | Oocyte transport silencing, repression by miRNA, neuronal granules, general decay | 1, 38, 39 |
| Cgh-1 | C. elegans | Somatic granules, gametogenesis, oocyte development | 40, 41 |
| Xp54 | Xenopus | Maternal granules, in vitro unwinding, tethered repression, granule formation | 12, 36, 37 |
Highlights.
-
-
DHH1 / RCKp54 helicases are a conserved family of translational regulators
-
-
DHH1 / RCKp54 helicases function in diverse biological contexts
-
-
Translational control links to mRNA decay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006 Dec 21;52(6):997–1009. doi: 10.1016/j.neuron.2006.10.028. PubMed PMID: WOS:000243115100008. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Moor CH, Richter JD. Translational control in vertebrate development. International review of cytology. 2001;203:567–608. doi: 10.1016/s0074-7696(01)03017-0. PubMed PMID: 11131527. [DOI] [PubMed] [Google Scholar]
- 3.Coller J, Parker R. Eukaryotic mRNA decapping. Annual review of biochemistry. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. PubMed PMID: 15189161. [DOI] [PubMed] [Google Scholar]
- 4.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006 Feb 15;367:17–37. doi: 10.1016/j.gene.2005.10.019. PubMed PMID: 16337753. [DOI] [PubMed] [Google Scholar]
- 5.Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic acids research. 2006;34(15):4168–4180. doi: 10.1093/nar/gkl468. PubMed PMID: 16936318. Pubmed Central PMCID: 1616962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proceedings of the National Academy of Sciences of the United States of America. 2000 Nov 21;97(24):13080–13085. doi: 10.1073/pnas.97.24.13080. PubMed PMID: 11087862. Pubmed Central PMCID: 27181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006 Apr 21;25(2):287–300. doi: 10.1016/j.cell.2006.01.054. PubMed PMID: 16630817. [DOI] [PubMed] [Google Scholar]
- 8.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nature reviews Molecular cell biology. 2011 Aug;12(8):505–516. doi: 10.1038/nrm3154. PubMed PMID: 21779027. [DOI] [PubMed] [Google Scholar]
- 9.Ray BK, Lawson TG, Kramer JC, Cladaras MH, Grifo JA, Abramson RD, et al. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. The Journal of biological chemistry. 1985 Jun 25;260(12):7651–7658. PubMed PMID: 3838990. [PubMed] [Google Scholar]
- 10.de Valoir T, Tucker MA, Belikoff EJ, Camp LA, Bolduc C, Beckingham K. A second maternally expressed Drosophila gene encodes a putative RNA helicase of the "DEAD box" family. Proceedings of the National Academy of Sciences of the United States of America. 1991 Mar 15;88(6):2113–2117. doi: 10.1073/pnas.88.6.2113. PubMed PMID: 1900936. Pubmed Central PMCID: 51179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic acids research. 2006;34(10):3082–3094. doi: 10.1093/nar/gkl409. PubMed PMID: 16769775. Pubmed Central PMCID: 1477856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladomery M, Wade E, Sommerville J. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic acids research. 1997 Mar 1;25(5):965–973. doi: 10.1093/nar/25.5.965. PubMed PMID: WOS:A1997WM30000006. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akao Y, Yoshida H, Matsumoto K, Matsui T, Hogetu K, Tanaka N, et al. A tumour-associated DEAD-box protein, rck/p54 exhibits RNA unwinding activity toward c-myc RNAs in vitro. Genes Cells. 2003 Aug;8(8):671–676. doi: 10.1046/j.1365-2443.2003.00665.x. PubMed PMID: WOS:000184333400002. English. [DOI] [PubMed] [Google Scholar]
- 14.Dutta A, Zheng ST, Jain D, Cameron CE, Reese JC. Intermolecular Interactions within the Abundant DEAD-box Protein Dhh1 Regulate Its Activity in Vivo. Journal of Biological Chemistry. 2011 Aug 5;286(31):27454–27470. doi: 10.1074/jbc.M111.220251. PubMed PMID: WOS:000293268700041. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng ZH, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. Rna. 2005 Aug;11(8):1258–1270. doi: 10.1261/rna.2920905. PubMed PMID: WOS:000230856100011. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsui T, Hogetsu K, Akao Y, Tanaka M, Sato T, Kumasaka T, et al. Crystallization and X-ray analysis of the N-terminal core domain of a tumour-associated human DEAD-box RNA helicase, rck/p54. Acta Crystallogr D. 2004 Jan;60:156–159. doi: 10.1107/s0907444903024223. PubMed PMID: WOS:000187399200031. English. [DOI] [PubMed] [Google Scholar]
- 17.Matsui T, Hogetsu K, Usukura J, Sato T, Kumasaka T, Akao Y, et al. Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes. Genes Cells. 2006 Apr;11(4):439–452. doi: 10.1111/j.1365-2443.2006.00951.x. PubMed PMID: WOS:000236244500010. English. [DOI] [PubMed] [Google Scholar]
- 18.Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural Basis for the Mutually Exclusive Anchoring of P Body Components EDC3 and Tral to the DEAD Box Protein DDX6/Me31B. Mol Cell. 2009 Mar 13;33(5):661–668. doi: 10.1016/j.molcel.2009.02.014. PubMed PMID: WOS:000264237800012. English. [DOI] [PubMed] [Google Scholar]
- 19.Benz J, Trachsel H, Baumann U. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae - the prototype of the DEAD box protein family. Struct Fold Des. 1999 Jun 15;7(6):671–679. doi: 10.1016/s0969-2126(99)80088-4. PubMed PMID: WOS:000080967100010. English. [DOI] [PubMed] [Google Scholar]
- 20.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proceedings of the National Academy of Sciences of the United States of America. 2001 Feb 13;98(4):1465–70. doi: 10.1073/pnas.98.4.1465. PubMed PMID: WOS:000166949200031. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. Rna. 2001 Dec;7(12):1717–1727. doi: 10.1017/s135583820101994x. PubMed PMID: 11780629. Pubmed Central PMCID: 1370212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. Embo J. 2002 Jun 3;21(11):2788–2797. doi: 10.1093/emboj/21.11.2788. PubMed PMID: WOS:000175912500028. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, et al. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998 Feb;148(2):571–9. doi: 10.1093/genetics/148.2.571. PubMed PMID: 9504907. Pubmed Central PMCID: 1459828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maillet L, Collart MA. Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. The Journal of biological chemistry. 2002 Jan 25;277(4):2835–2842. doi: 10.1074/jbc.M107979200. PubMed PMID: 11696541. [DOI] [PubMed] [Google Scholar]
- 25.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005 Sep 23;122(6):875–886. doi: 10.1016/j.cell.2005.07.012. PubMed PMID: 16179257. Pubmed Central PMCID: 1853273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westmoreland TJ, Olson JA, Saito WY, Huper G, Marks JR, Bennett CB. Dhh1 regulates the G1/S-checkpoint following DNA damage or BRCA1 expression in yeast. The Journal of surgical research. 2003 Jul;113(1):62–73. doi: 10.1016/s0022-4804(03)00155-0. PubMed PMID: 12943812. [DOI] [PubMed] [Google Scholar]
- 27.Bergkessel M, Reese JC. An essential role for the Saccharomyces cerevisiae DEAD-box helicase DHH1 in G1/S DNA-damage checkpoint recovery. Genetics. 2004 May;167(1):21–33. doi: 10.1534/genetics.167.1.21. PubMed PMID: 15166134. Pubmed Central PMCID: 1470881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo XA, Talarek N, De Virgilio C. Initiation of the yeast G(0) program requires Igo1 and Igo2, which antagonize activation of decapping of specific nutrient-regulated mRNAs. Rna Biol. 2011 Jan-Feb;8(1):14–17. doi: 10.4161/rna.8.1.13483. PubMed PMID: WOS:000288492200004. English. [DOI] [PubMed] [Google Scholar]
- 29.Ka M, Park YU, Kim J. The DEAD-box RNA helicase, Dhh1, functions in mating by regulating Ste12 translation in Saccharomyces cerevisiae. Biochem Bioph Res Co. 2008 Mar 14;367(3):680–686. doi: 10.1016/j.bbrc.2007.12.169. PubMed PMID: WOS:000253013000027. English. [DOI] [PubMed] [Google Scholar]
- 30.Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, Parker R, Thiele DJ, Puig S. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. Journal of Biological Chemistry. 2008 Oct 17;283(42):28527–28535. doi: 10.1074/jbc.M804910200. PubMed PMID: WOS:000259969300056. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang LC, Lee FJS. The RNA helicase Dhh1p cooperates with Rbp1p to promote porin mRNA decay via its non-conserved C-terminal domain. Nucleic acids research. 2012 Feb;40(3):1331–1344. doi: 10.1093/nar/gkr803. PubMed PMID: WOS:000300422400040. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol. 2011 Aug 22;194(4):527–537. doi: 10.1083/jcb.201007151. PubMed PMID: WOS:000294158200004. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sweet T, Kovalak C, Coller J. The DEAD-Box Protein Dhh1 Promotes Decapping by Slowing Ribosome Movement. Plos Biol. 2012 Jun;10(6) doi: 10.1371/journal.pbio.1001342. PubMed PMID: WOS:000305945600004. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu WQ, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009 Sep 10;461(7261) doi: 10.1038/nature08265. 225-U96. PubMed PMID: WOS:000269654600036. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999 Aug;19(8):5247–5256. doi: 10.1128/mcb.19.8.5247. PubMed PMID: WOS:000081513800002. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minshall N, Thom G, Standart N. A conserved role of a DEAD box helicase in mRNA masking. Rna-a Publication of the Rna Society. 2001 Dec;7(12):1728–1742. doi: 10.1017/s135583820101158x. PubMed PMID: WOS:000172966300006. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic acids research. 2004 Feb;32(4):1325–1334. doi: 10.1093/nar/gkh303. PubMed PMID: WOS:000220179700017. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001 Sep;128(17):3233–3242. doi: 10.1242/dev.128.17.3233. PubMed PMID: WOS:000171478600002. English. [DOI] [PubMed] [Google Scholar]
- 39.Zabolotskaya MV, Grima DP, Lin MD, Chou TB, Newbury SF. The 5'-3' exoribonuclease Pacman is required for normal male fertility and is dynamically localized in cytoplasmic particles in Drosophila testis cells. Biochem J. 2008 Dec 15;416:327–335. doi: 10.1042/BJ20071720. PubMed PMID: WOS:000261781500002. English. [DOI] [PubMed] [Google Scholar]
- 40.Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C-elegans. Development. 2001 Sep;128(17):3221–3232. doi: 10.1242/dev.128.17.3221. PubMed PMID: WOS:000171478600001. English. [DOI] [PubMed] [Google Scholar]
- 41.Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol. 2008 Aug 11;182(3):543–557. doi: 10.1083/jcb.200801183. PubMed PMID: WOS:000258529100014. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akao Y, Seto M, Yamamoto K, Iida S, Nakazawa S, Inazawa J, et al. The Rck Gene Associated with T(11-14) Translocation Is Distinct from the Mll/All-1 Gene with T(4-11) and T(11-19) Translocations. Cancer Res. 1992 Nov 1;52(21):6083–6087. PubMed PMID: WOS:A1992JV49600034. English. [PubMed] [Google Scholar]
- 43.Akao Y, Marukawa O, Morikawa H, Nakao K, Kamei M, Hachiya T, et al. The Rck/P54 Candidate Protooncogene Product Is a 54-Kilodalton D-E-a-D Box Protein Differentially Expressed in Human and Mouse-Tissues. Cancer Res. 1995 Aug 1;55(15):3444–3449. PubMed PMID: WOS:A1995RL49300039. English. [PubMed] [Google Scholar]
- 44.Lin F, Wang R, Shen JJ, Wang X, Gao P, Dong K, et al. Knockdown of RCK/p54 expression by RNAi inhibits proliferation of human colorectal cancer cells in vitro and in vivo. Cancer Biol Ther. 2008 Oct;7(10):1669–1676. doi: 10.4161/cbt.7.10.6660. PubMed PMID: WOS:000260263600026. English. [DOI] [PubMed] [Google Scholar]
- 45.Miyaji K, Nakagawa Y, Matsumoto K, Yoshida H, Morikawa H, Hongou Y, et al. Overexpression of a DEAD box/RNA helicase protein, rck/p54, in human hepatocytes from patients with hepatitis C virus-related chronic hepatitis and its implication in hepatocellular carcinogenesis. J Viral Hepatitis. 2003 Jul;10(4):241–248. doi: 10.1046/j.1365-2893.2003.00447.x. PubMed PMID: WOS:000183685400001. English. [DOI] [PubMed] [Google Scholar]
- 46.Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, et al. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proceedings of the National Academy of Sciences of the United States of America. 2009 Aug 11;106(32):13517–13522. doi: 10.1073/pnas.0906413106. PubMed PMID: WOS:000268877300067. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jangra RK, Yi M, Lemon SM. DDX6 (Rck/p54) Is Required for Efficient Hepatitis C Virus Replication but Not for Internal Ribosome Entry Site-Directed Translation. J Virol. 2010 Jul;84(13):6810–6824. doi: 10.1128/JVI.00397-10. PubMed PMID: WOS:000278551900051. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu SF, Lujan P, Jackson DL, Emerman M, Linial ML. The DEAD-box RNA Helicase DDX6 is Required for Efficient Encapsidation of a Retroviral Genome. Plos Pathog. 2011 Oct;7(10) doi: 10.1371/journal.ppat.1002303. PubMed PMID: WOS:000296734300033. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward AM, Bidet K, Ang YL, Ler SG, Hogue K, Blackstock W, et al. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3' UTR structures. Rna Biol. 2011 Nov-Dec;8(6):1173–1186. doi: 10.4161/rna.8.6.17836. PubMed PMID: WOS:000298630600024. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, et al. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J Cell Biol. 2012 Aug 6;198(3):439–456. doi: 10.1083/jcb.201111012. PubMed PMID: WOS:000307412200018. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alves-Rodrigues I, Mas A, Diez J. Xenopus Xp54 and human RCK/p54 helicases functionally replace yeast Dhh1p in brome mosaic virus RNA replication. J Virol. 2007 Apr;81(8):4378–4380. doi: 10.1128/JVI.02246-06. PubMed PMID: WOS:000245692900073. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naarmann IS, Harnisch C, Muller-Newen G, Urlaub H, Ostareck-Lederer A, Ostareck DH. DDX6 recruits translational silenced human reticulocyte 15-lipoxygenase mRNA to RNP granules. Rna-a Publication of the Rna Society. 2010 Nov;16(11):2189–2204. doi: 10.1261/rna.2211110. PubMed PMID: WOS:000283047900015. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. Plos Biol. 2006 Jul;4(7):1122–1136. doi: 10.1371/journal.pbio.0040210. PubMed PMID: WOS:000238974300006. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hay B, Jan LY, Jan YN. A Protein-Component of Drosophila Polar Granules Is Encoded by Vasa and Has Extensive Sequence Similarity to Atp-Dependent Helicases. Cell. 1988 Nov 18;55(4):577–587. doi: 10.1016/0092-8674(88)90216-4. PubMed PMID: WOS:A1988R075200007. English. [DOI] [PubMed] [Google Scholar]
- 55.Anderson P, Kedersha N. Stress granules: The Tao of RNA triage. Trends Biochem Sci. 2008 Mar;33(3):141–150. doi: 10.1016/j.tibs.2007.12.003. PubMed PMID: WOS:000254451400006. English. [DOI] [PubMed] [Google Scholar]
- 56.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008 Nov 3;183(3):441–455. doi: 10.1083/jcb.200807043. PubMed PMID: WOS:000261060200012. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu JD, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005 Jul;7(7) doi: 10.1038/ncb1274. 719-U118. PubMed PMID: WOS:000230190800017. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007 Nov 5;179(3):437–449. doi: 10.1083/jcb.200704147. PubMed PMID: WOS:000250705200011. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mokas S, Mills JR, Garreau C, Fournier MJ, Robert F, Arya P, et al. Uncoupling Stress Granule Assembly and Translation Initiation Inhibition. Mol Biol Cell. 2009 Jun 1;20(11):2673–2683. doi: 10.1091/mbc.E08-10-1061. PubMed PMID: WOS:000266950900004. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007 Jun;27(11):3970–3981. doi: 10.1128/MCB.00128-07. PubMed PMID: WOS:000246698300008. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Bio. 2008 Apr;9(4):337–344. doi: 10.1038/nrm2370. PubMed PMID: WOS:000254274300019. English. [DOI] [PubMed] [Google Scholar]
- 62.Standart N, Minshall N. Translational control in early development: CPEB, P-bodies and germinal granules. Biochemical Society transactions. 2008 Aug;36(Pt 4):671–676. doi: 10.1042/BST0360671. PubMed PMID:18631138. [DOI] [PubMed] [Google Scholar]
- 63.Drummond SP, Hildyard J, Firczuk H, Reamtong O, Li N, Kannambath S, et al. Diauxic shift-dependent relocalization of decapping activators Dhh1 and Pat1 to polysomal complexes. Nucleic acids research. 2011 Sep 1;39(17):7764–7774. doi: 10.1093/nar/gkr474. PubMed PMID: 21712243. Pubmed Central PMCID: 3177209. [DOI] [PMC free article] [PubMed] [Google Scholar]