Abstract
Perfringolysin O (PFO), a bacterial cholesterol-dependent cytolysin, binds to a mammalian cell membrane, oligomerizes into a circular prepore complex (PPC), and forms a 250-Å transmembrane β-barrel pore in the cell membrane. Each PFO monomer has two sets of 3 short α-helices that unfold and ultimately refold into two transmembrane β-hairpin (TMH) components of the membrane-embedded β-barrel. Inter-strand disulfide bond scanning revealed that β-strands in a fully assembled PFOβ-barrel were strictly aligned and tilted at 20 ° to the membrane perpendicular. In contrast, in a low temperature-trapped PPC intermediate, the TMHs were unfolded and had sufficient freedom of motion to interact transiently with each other; yet the TMHs were not aligned or stably hydrogen-bonded. The PFO PPC-to-pore transition therefore converts TMHs in a dynamic folding intermediate far above the membrane into transmembrane β-hairpins that are hydrogen bonded to those of adjacent subunits in the bilayer-embedded β-barrel.
Cholesterol-dependent cytolysins (CDCs) are a large family of secreted bacterial pore-forming toxins that specifically bind to cholesterol-containing mammalian membranes1. While the pore size (250–300 Å) and number of monomers per pore (35–50) can vary somewhat for different CDCs, the mechanism of pore formation involves membrane binding of the monomer and their oligomerization into circular prepore complexes (PPCs), followed by significant secondary and tertiary structural changes as the PPC becomes a membrane-spanning β-barrel pore2.
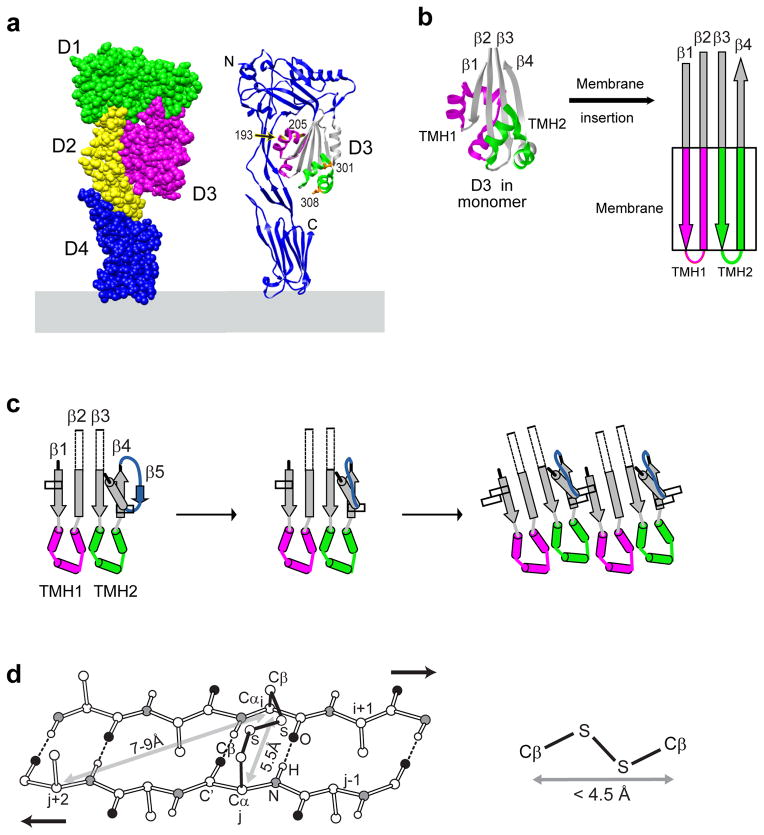
The mechanism of CDC pore formation has been studied most extensively using the Clostridium perfringens CDC, perfringolysin O (PFO). PFO is an elongated four-domain protein3 that binds to the membrane at the tip4 of domain 4 (D4) and projects approximately perpendicularly from the surface5,6 (Fig. 1a). During the PPC to pore transition, two α-helical bundles in D3 of each PFO monomer are converted to two transmembrane β-hairpins (TMHs) that extend from the core β-sheet in D3 and contribute to the formation of theβ-barrel pore7,8 (Fig. 1a,b). In addition, D1 and D3 move closer to the membrane surface5,6,9, with D3 moving more than 60 Å to reach the membrane surface and insert its TMHs5.
Figure 1. PFO structure and structural alterations.
(a) Individual domains of the elongated PFO monomer bound to a membrane surface (pale blue) are depicted in different colors (left), while the ribbon representation (right) shows the locations of TMH1 (magenta) and TMH2 (green) when folded into α-helices within domain 3 in monomeric PFO. The core β-sheet (gray) and four individual residues are also shown. The image was generated using Chimera (http://www.cgl.ucsf.edu/chimera). (b) The transition from helical TMHs in the PFO monomer to β-hairpins in the β-barrel pore is depicted. Four antiparallel β-strands of the D3 core that ultimately extend into the membrane (box) are identified as β1-β4 as shown. (c) When monomeric PFO binds to a membrane, β5 first rotates away from β4, and this then allowsβ4 to form hydrogen bonds with β1 from another PFO. β4 and β1 alignment relative to each other involves the π-stacking of an aromatic residue found in each strand (open rectangles). (d) The arrangement of atoms in anti-parallel β-strands is fixed by inter-strand hydrogen bonding. A Cα atom on one side of the β-sheet is separated by 5.5 Å from the juxtaposed Cα to which it is hydrogen bonded and by 7–9 Å from the next-nearest Cα atom on the adjacent β-strand. Since the two carbon atoms in a disulfide bond are separated by less than 4.5 Å, a disulfide bond can only be formed between the Cβ atoms directly opposite each other in adjacent anti- parallel β-strands.
No crystals of CDC pore complexes have yet been reported, but crystallography of more than 15 bacterial outer membrane proteins showed that the tilt of β-strands relative to the pore axis was 37° or more in each case10. Yet cryoelectron microscope images of a pore complex formed by pneumolysin, a CDC homologue of PFO, indicate that the TMHs in a CDC β-barrel pore are oriented perpendicular to the plane of the membrane and parallel to the pore axis9 (tilt = 0°). Molecular modeling of the TMH orientation in giant β-barrels is compatible with an alignment perpendicular to the membrane, but the modeling data are best fit with a tilted alignment in which the shear or stagger number (S) equals one-half of the total number of βstrands (n) in the β-barrel11 (S = n/2). However, no examples of the predicted S = n/2 β-barrel have been documented experimentally.
PFO pore formation occurs spontaneously on membranes containing sufficient cholesterol12–16. Cholesterol binding17 initiates an obligatory and tightly coupled sequence of conformational changes in PFO13,18–20. The timing and progression of these changes are regulated by structural elements such as the short β-strand (β5) that forms part of the core β-sheet in D3 (Fig. 1c). In the soluble monomer, the hydrogen bonding of β5 to β4 blocks oligomerization by preventing high affinity association with another PFO21. However, D4 binding to the membrane initiates conformational changes in monomer structure2 that cause β5 to rotate away from the core β-sheet in D321,22 and exposeβ4 for hydrogen bonding with the always-exposed core β1 strand of another membrane-bound PFO (Fig. 1c).
Oligomerization then proceeds after the core β4 and β1 strands of adjacent monomers align with their hydrogen-bonding partners. Proper alignment is ensured by the formation of an intermolecular π-stacking interaction. As the coreβ4 and β1 strands of adjacent monomers scan different alignments in search of the correct interstrand hydrogen bonding, the stacking of the aromatic side chains of single aromatic residues in β4 and β121 (indicated by open rectangles in Fig. 1c) dictate a particular set of core β4-β1 hydrogen bonding partners. However, TMH structure is unknown in the PPC oligomer.
Here we have used disulfide scanning to examine the hydrogen bonding and alignment of TMHs in both the fully assembled pore β-barrel and the PPC. These studies revealed that adjacent membrane-spanning regions of β4 and β1 in the pore β-barrel were locked into a specific alignment that introduced a 20° tilt in theβ-strands of the β-barrel. In sharp contrast, in the PPC, the α-helical bundles in D3 were partially unfolded and moving dynamically so that pairing of β4 to β1 was variable, transient, and restricted to the residues in the central regions of the TMHs. These studies therefore revealed the nature and extent of TMH interactions between PFO monomers in both the PPC and the pore β-barrel.
RESULTS
Detecting β-strand alignment
A PFO pore β-barrel is formed by ~140 anti-parallel β–strands6 that position amino acids and their side chains as depicted in Figure 1d. In theβ-barrel, the alignment of adjacentβ–strands relative to each other is fixed: a residue in one strand is always juxtaposed and hydrogen-bonded to the same residue in an adjacent strand. The most direct approach for determining the alignment of two β-strands relative to each other is to chemically crosslink a residue in one chain to its nearest neighbor in the opposite strand. Such crosslinks must be formed via the amino acid side chains to avoid disrupting the β-sheet. The hydrogen-bonded Cα atoms of opposing residues in adjacent β-strands are separated by 5.5 Å, while Cα atoms in opposite strands are separated by 7–9 Å when offset by two residues (the side chain from a one-residue offset is directed to the other side of the plane formed by the β–sheet and hence is unavailable; Fig. 1d). A Cys residue in a β-barrel can form a disulfide bond with a second Cys only if the two Cys residues are directly juxtaposed in adjacent β-strands because the Cβ-Cβ separation in Cys-S-S-Cys is less than 4.5 Å23 (Fig. 1d). Thus, if Cβ atoms on adjacent strands are crosslinked by a disulfide bond, then the crosslinked residues must be positioned directly across from each other in the β-sheet.
Mono-cysteine PFO derivatives were constructed from rPFO, a Cys-free version of wild-type PFO7, by substituting a Cys for one of the seven residues in either the TMH1β1 or the TMH2β4 strand whose side-chains faced the aqueous pore in the β-barrel. These mutants, with Cys locations spanning the bilayer (β1: K189C, Q191C, S193C, A195C, N197C, N199C, and K201C; β4: N300C, D302C, K304C, S306C, Q308C, K310C, and I312C)7,8, are active in pore formation: β1 mutants are 88–104% as active as wild-type in hemolytic assays with red blood cells7, and even after Cys modification with a fluorescent dye,β4 mutants were more than 70% as active as wild-type, except for 302 which was 47–60% as active8. Eachβ4 mutant was separately mixed with eachβ1 mutant in the presence of cholesterol-containing liposomes; the pairs of substituted Cys residues that formed a disulfide bond would therefore unambiguously identify the intermolecular alignment ofβ1 and β4 in adjacent monomers within the oligomeric PFO β-barrel.
Detection of covalent rPFO dimers
PFO binds to membranes with sufficient cholesterol12–16 and assembles into an oligomeric PPC at either 4°C or 37°C24,25. Pore formation proceeds readily at 37°C, but pore formation at 2–4°C (detected by monitoring either fluorescent-labeled TMH insertion into the nonpolar membrane interior or by the release of liposome-encapsulated small fluorophores) is insignificant after 30 min24,26. Thus, PFO samples are trapped in the PPC intermediate state at 4°C because the rate of β-barrel pore formation is extremely slow at 4°C.
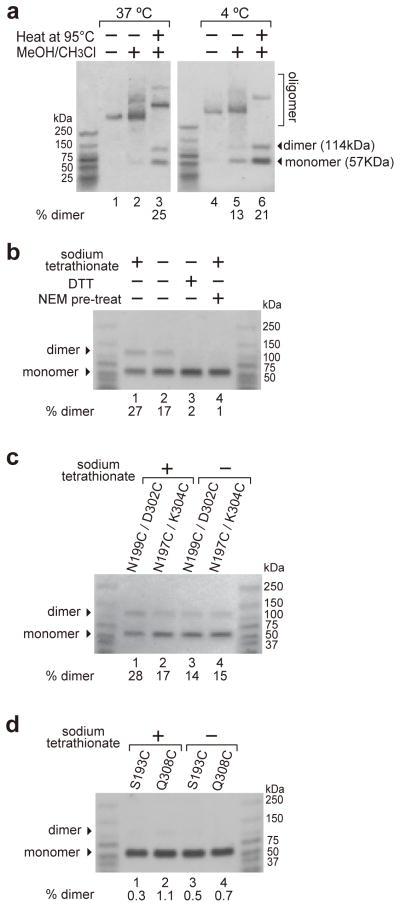
When equimolar rPFOS193C and rPFOQ308C, with Cys residues near the middle of β1 andβ4, respectively, were incubated with cholesterol-rich liposomes at 37°C or 4°C, the resulting oligomers were intact after agarose gel electrophoresis in SDS (SDS-AGE; Fig. 2a). Oligomer dissociation was improved slightly if methanol/chloroform was added27 to remove lipids and concentrate proteins by precipitation prior to SDS-AGE. But when methanol/chloroform- treated samples were dried, resuspended in sample buffer, and heated at 95°C, oligomer dissociation increased dramatically. This approach was then used throughout to maximize the dissociation of PPCs and pore complexes.
Figure 2. Detection of disulfide-bonded PFO dimers in PPC and pore complexes.
(a) An equimolar mixture of S193C and Q308C PFO derivatives in pore complexes (incubated 40 min, 37°C) or PPCs (2 h, 4°C) were prepared in the presence of 100 μM Na-tetrathionate, extracted in some cases with methanol/chloroform, and analyzed by SDS-AGE with or without boiling prior to electrophoresis. The yield of disulfide-linked dimers is shown below the lane. (b) Dimer (193–308) formation in pore complexes was stimulated by 100 μM Na-tetrathionate or blocked by either 5 mM DTT or pre-treatment with 20 mM NEM. (c) Dimers were detected in pore complexes with other pairs of mono-cysteine PFO derivatives with or without tetrathionate. (d) No homodimers were observed in pore complexes formed with only a single mono-cysteine derivative.
However, when the 37°C sample of rPFOS193C and rPFOQ308C was reduced with DTT, the dimer band disappeared (Fig. 2b). This result suggested that a disulfide bond covalently linked the monomers. Consistent with this interpretation, addition of the oxidizing agent tetrathionate increased the number of dimers. Furthermore, no dimers formed, even in the presence of tetrathionate, when rPFO thiols were reacted with N-ethylmaleimide (NEM) before mixing. Thus, our analysis conditions separated disulfide-linked dimers from monomers. The amount of residual oligomer in individual samples varied, but there was no evidence that this variability correlated with the extent of dimer formation. We therefore concluded that the percentage of rPFOs in covalent dimers in a sample was given by [dimer/(dimer + monomer)] × 100.
Covalent dimers were also detected in equimolar mixtures of liposome-bound rPFON199C/rPFOD302C and rPFON197C/rPFOK304C at 37°C, and tetrathionate again stimulated disulfide bond formation, though to different extents (Fig. 2c). Since oxidative conditions increased covalent dimer production to the theoretical maximum, 25%, for some pairs of rPFO derivatives (e.g., Fig. 2b,c), tetrathionate was routinely added to samples to maximize the detection of covalent dimers. In contrast, no homodimers were detected when samples contained only a single rPFO derivative, whether or not tetrathionate was present (Fig. 2d). Thus, dimers were not formed by random collisions between thiol-containing rPFOs on the membrane surface, even in the presence of tetrathionate. Instead, covalent dimer formation in β-barrel pores required the close juxtaposition of a Cys in β1 and a Cys on β4 of adjacent monomers in a fully assembled rPFO pore, and three such pairs of Cys residues are identified in Fig. 2.
Disulfide bond formation in the β-barrel pore
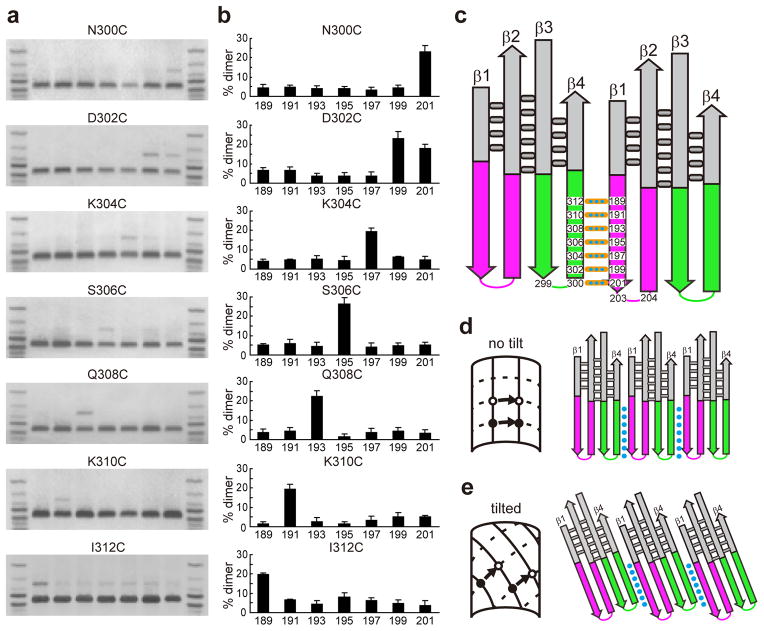
Each rPFO with a Cys in β4 was incubated with an equimolar amount of each rPFO derivative with a Cys in β1 in the presence of tetrathionate, and covalent dimer formation was measured in each sample. If β4 and β1 from adjacent monomers are locked into a specific alignment, then each residue inβ4 should form a disulfide with only its nearest neighbor residue in β4. For example, rPFON300C formed dimers only with rPFOK201C (Fig. 3a). The adjacent panel in Figure 3b quantifies the percentage of dimers formed with each pair of rPFO derivatives in three or more independent experiments. Residue 300 in β4 is therefore positioned directly across from, and is hydrogen bonded to, residue 201 in β1 of the adjacent monomer in the poreβ-barrel. In addition, residue pairs 300-201, 304-197, 306-195, 308-193, 310-191, and 312-189 are hydrogen bonded to each other.
Figure 3.
β4-β1 cross-linking and β-strand alignment in pore complex.
(a) Eachβ1 mutant was mixed with an equimolar amount of each β4 mutant in the presence of tetrathionate, and then incubated (40 min, 37°C) with cholesterol-rich liposomes to form pore complexes. Dimer formation was detected by SDS-AGE. (b) The % yield of disulfide-linked dimers was calculated from the intensities of the bands shown in (a): 100 × dimer/(dimer + monomer). The average yields (± S.D.) are shown for three or more independent experiments with each pair of β1 and β4 mutants. (c) Seven specific β1-β4 pairs formed a disulfide bond in the β-barrel pores with high efficiency. Aligning β1 and β4 of adjacent monomers based on these crosslinks reveals a two residue-offset in the tips of the two β-strands. (d) An untilted (α = 0°) parallel alignment of strands in a β-barrel is created when adjacent β-strands and TMHs are not offset. (e) When TMHs are offset by 2 residues as in panel c, the closed ends of the TMHs must tilt to form a circular β-barrel in the plane of the membrane.
Given this invariant pattern of crosslinking, one would have expected rPFOD302C to form disulfide-linked dimers only with rPFON199C. However, we observed that rPFOD302C also formed dimers with rPFOK201C nearly as efficiently as with rPFON199C (Fig. 3a,b). Similarly, in the complementary experiment, we found that rPFOK201C formed dimers with both rPFOD302C and rPFON300C (Supplementary Results, Supplementary Figure 1). However, in the absence of tetrathionate, no 302-201 dimers formed. The tetrathionate dependence of dimer formation indicates that 201 and 302 are not stably paired, but are transiently close enough to each other for tetrathionate to catalyze disulfide bond formation. This conformational ambiguity presumably occurs because the reversal of the β1 backbone orientation at the TMH tip positions the sulfhydryls of Cys residues at 201 and 302 in proximity, at least dynamically.
β-strand tilt in the β-barrel pore
The residues at the tips of the TMHs were identified as 203 and 204 for TMH1 and 299 and 300 for TMH27,8. When the residues in β4 were aligned with the hydrogen-bonding partners in β1 of the adjacent monomer identified above, the β1 residues were located two residues further from the TMH tip than were the β4 residues. This difference is easily visualized by aligning β4 and β1 of adjacent monomers vertically and opposite their hydrogen bonding partners (Fig. 3c). The resulting two-residue stagger of β-strands dictates how the TMHs are oriented when the β-barrel pore is embedded in the membrane.
If PFO TMHs are inserted perpendicularly into the plane of the membrane, a closed circle (β-barrel) can only form in the bilayer if the adjacent β4 and β1 strands are exactly in register and not offset (Fig. 3d); a 2-residue offset would prevent the formation of a closed βbarrel becauseβ1 and β4 would no longer be aligned horizontally. But since the TMHs in adjacent PFO monomers are offset, a closed circular β-barrel with each TMH tip at the membrane surface can only be formed if the TMHs are tilted (Fig. 3e). Specifically, the 2-residue offset revealed by intermolecular disulfide crosslinking experiments demonstrates that the anti-parallelβ-strands are tilted relative to the pore axis with the right-handed twist favored by the conformational energetics of β-sheets28. Furthermore, the 2-residue offset showed experimentally and conclusively via covalent bonds that S = n/2 for PFO (Supplementary Figure 2).
The angle of the strands relative to the barrel axis, α, was calculated using α = tan−1 (aS/bn), where a = the Cα-Cα distance along the strands and b = the inter-strand distance29. Published a and b values for anti-parallel β-strands have ranged from 3.3 Å and 4.4 Å29, respectively, to 3.48 Å and 4.83 Å11 to 3.53 Å and 4.87 Å30, but all three pairs yield α values of 20° since we have shown that S = n/2 for PFO. The individual β-strands in the PFO β-barrel are therefore tilted at an angle of 20° to the pore axis.
PPC TMHs are unfolded, flexible, and moving
To create samples of PPCs that can be examined, one needs to block the prepore to pore transition of PFO, but retain the ability to form a pore in the absence of reducing agents. Since those requirements can only be met by trapping PFO in PPCs at low temperature24,26, we prepared PPCs at 4°C to monitor the TMHs of functional toxins at an intermediate stage of pore formation.
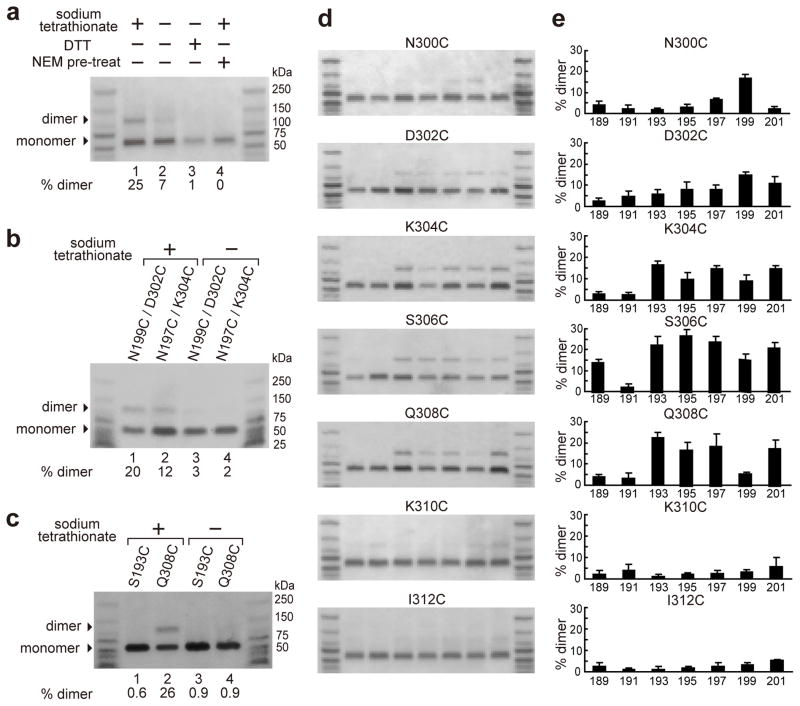
When PPCs were formed at 4°C on liposomes with equimolar mixtures of rPFOS193C and rPFOQ308C in the absence of tetrathionate, only a small amount of disulfide-linked dimer was observed (Fig. 4a). As was seen with the pore complexes (Fig. 2b), adding tetrathionate to the sample maximized dimer formation (Fig. 4a). However, adding DTT to the sample or preincubating the proteins with NEM eliminated dimer formation. Thus, disulfide crosslinks were formed between Cys residues at 193 and 308 in adjacent PPC subunits. In addition, other PPCβ4-β1 crosslinks were detected, but only when tetrathionate was present (Fig. 4b). Since TMH1 and TMH2 are folded and located on opposite sides of the core β-sheet in PFO monomers (Fig. 1b), the two TMHs must be unfolded in PPCs to get β4-β1 crosslinks. Yet the low yield of dimer formation in the absence of tetrathionate (Fig. 4a,b) suggests that β4 and β1 are not stably hydrogen bonded in PPCs.
Figure 4. Dimer formation in PPCs.
(a) An equimolar mixture of S193C and Q308C rPFO mutants were incubated for 2 h at 4°C to form PPCs, and the effects of 100 μM sodium tetrathionate, 5 mM DTT, and pre-treatment with 20 mM NEM on disulfide crosslinking of PFO monomers are shown. (b) The tetrathionate dependence of disulfide crosslinking is shown for two other pairs of β4 and β1 mutants in PPCs. (c) In PPCs, the Q308C rPFO mutant forms homodimers in the presence of tetrathionate, while the S193C mutant does not. (d) Samples were prepared as in Figure 3a in the presence of tetrathionate, and then incubated (2 h, 4°C) to form PPCs. Dimer formation was detected by SDS-AGE. (e) The % yield of disulfide-linked dimers for each pair of β1 and β4 mutants was determined as in Figure 3b. Mean values (± S.D.) are shown for three or more independent experiments with each pair of β1 and β4 mutants.
This conclusion was confirmed when a sample containing a single derivative, rPFOQ308C, was found to form dimers in the presence, but not in the absence, of tetrathionate (Fig. 4c). In a β-barrel, Cys residues located at the same β4 site in adjacent subunits are separated by more than 20 Å and cannot form a disulfide bond. The existence of homodimers therefore reveals that the β4 strands in adjacent PPC subunits are free to move and sufficiently flexible to bring two β4 Cys residues together. The transient nature of this β4-β4 proximity is indicated by the absolute dependence of disulfide bond formation on tetrathionate catalysis. Moreover, the high homodimer content in such samples attests to the high frequency of β4-β4 encounters in PPCs. Interestingly, the absence of rPFOS193C homodimers, even in the presence of tetrathionate, suggests that TMH1 is less unfolded and/or more restricted in its movement than TMH2 (Fig. 4c), presumably because TMH1 is initially buried at the D2–D3 interface in the monomer (Fig. 1a).
TMHs are not aligned in PPCs
D3 in PPCs is sufficiently high above the membrane in PPCs to allow the unfolding and alignment of TMHs into a hydrogen-bonded β-barrel above the membrane5. If a β-barrel were to form in PPCs, then the PPCβ4-β1 disulfide bond crosslinking pattern would be identical to that of the poreβ-barrel. However, the disulfide crosslinking patterns are very different for prepore (Fig. 4d,e) and pore (Fig. 3a,b) complexes. Whereas only a single crosslinking partner was identified for each β4 residue in the β-barrel pore, disulfide bond formation in PPCs was residue dependent: Cys at 310 and 312 formed few disulfide bonds with β1 Cys, while Cys residues at 304, 306, and 308 crosslinked to 4 or more different β1 Cys residues. Thus, β4 and β1 of adjacent monomers were not stably aligned relative to each other in the PPC, and a pre-formed β-barrel was not poised to puncture the bilayer in the PFO PPC.
Instead, β1 and β4 residues in the middle regions of the hairpins were transiently proximal to multiple residues of the complementary strand, as evidence by the PPC crosslinking yields (Fig. 4e; Supplementary Figure 3a). This region of the TMH is therefore in flux, dynamically sampling accessible space with a high collisional frequency between β4 andβ1. The extent of TMH unfolding was substantial, as evidenced by the large separation between residues in the monomer that are disulfide-linked in the PPC (Supplementary Figure 3b). But the absence of anyβ4-β1 crosslinks to residues near the core β-sheet (189, 191, 310, 312) shows that these segments of β4 and β1 do not have access to any β1 and β4 residues, respectively, in PPCs. This restriction suggests that TMHs in PPCs adopt a partially folded intermediate state in PPCs, with sufficient TMH flexibility and freedom of motion to dynamically sample potential hydrogen-bonding partners in the PPC folding intermediate.
Fluorescence-detected environment of TMH residues in PPCs
The state of the TMHs during the monomer to PPC to pore complex transitions was also examined after covalently attaching a small and uncharged fluorescent dye (7-nitrobenz-2-oxa-1,3-diazole = NBD) to a Cys that was substituted for a residue in rPFO. NBD fluorescence differs dramatically in aqueous and nonpolar environments: NBD emission intensity is 5-to-10-fold higher in nonpolar than aqueous milieus, and its wavelength of maximum emission decreases by 15–20 nm (a blue shift) upon moving from an aqueous to a nonpolar environment7,8,31,32. Thus, NBD’s spectral properties reveal directly the nature of its microenvironment.
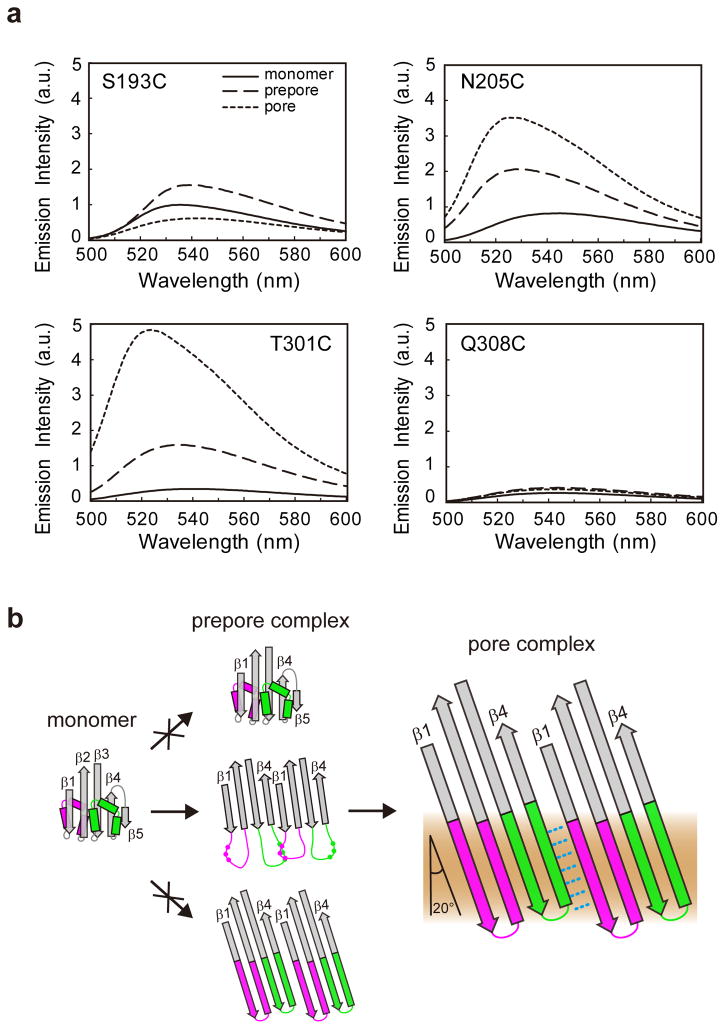
NBD was positioned at each of four different locations: the side chains of residues 193 and 308 in the middle of β1 and β4, respectively, face the aqueous pore in the β-barrel, while the side chains of residues 205 and 301 near the tips of β1 and β4 face the lipids in the membrane-inserted barrel. In monomeric PFO, the low emission intensities and the red-shifted maximum wavelengths of NBDs at 301 and 308 reveal that these probes are in an aqueous environment (Fig. 5a). The slightly higher monomeric NBD emission intensities at 193 and 205 indicate that these dyes are in a less polar milieu, consistent with the locations of these residues in the crystal structure of the PFO monomer3 where TMH1 is folded inside the core β-sheet of D3, and TMH2 is exposed on the D3 surface (Fig. 1a). Pore formation had little effect on the emission of NBDs at 308 (Fig. 5a), while 193 NBD emission intensity decreased and red-shifted as the dyes moved into the aqueous pore. In contrast, the large increases in the emission intensities of the NBDs at 205 and 301 and the large blue shifts of their emission maxima showed that they moved into the nonpolar core of the lipid bilayer during pore formation (Fig. 5a). These conclusions agree with our earlier results7,8, but the focus here is on how these TMH environments compare to those in the PPC.
Figure 5. Fluorescence-detected changes in TMH environment reveal stages in TMH unfolding and alignment during pore formation.
(a) Fluorescence emission spectra are shown for NBDs positioned at each of four different sites in D3 (see Fig. 1a). Each sample was sequentially transitioned through three states: soluble monomer; membrane-bound PPC; and membrane-embedded pore complex. The spectra are normalized to accurately reflect the relative emission intensities, with one arbitrary unit (a.u.) defined as the peak intensity of the S193C monomer. (b) TMH1 (magenta) and TMH2 (green) helices folded around the core β-sheet (gray) in PFO monomers are converted to membrane-inserted hairpins in the pore complex β-barrel through any of several possible PPC structures. Since PPC TMHs are partially unfolded and not aligned, intermediate states with the TMHs folded into helices (top) or aligned in a β-sheet (bottom) do not occur. A few of the β4-β1 disulfide crosslinking sites observed in the presence of sodium tetrathionate are indicated by the dots in the TMHs shown in the center panel.
The similarities in the monomer, pore, and PPC emission scans of the NBDs at 308 reveal that this residue was in an aqueous environment throughout the transitions from monomer to PPC to pore (Fig. 5a). Intriguingly, the slightly nonpolar environment of NBD at poisition 193 in monomers became even more nonpolar in PPCs, but upon pore formation, the emission intensity of the 193 NBD decreased and was red shifted as the NBDs moved into the aqueous pore during the PPC to pore transition. Residue 193 therefore occupied a more nonpolar milieu within the PPC than it did in either the monomer or the pore.
The emission spectra of PPC NBDs at 205 and 301 lie between the aqueous (monomer) and nonpolar (pore) extremes (Fig. 5a), thereby indicating that the NBD environments at 205 and 301 had an intermediate nonpolarity in PPCs. The substantial differences in the spectral properties of the 205 and 301 NBDs in monomers, PPCs, and pores shows that these residues move through different locales as pore formation proceeds.
Thus, the fluorescence data reveal that individual TMH residues have distinctly different average conformations in monomers, PPCs, and pores. The progression of PFO from monomer to PPC to pore therefore involves the controlled transition of TMH conformation from α-helices to a specific partially folded intermediate state to a β-barrel.
Discussion
Several important structural features of the PFO TMH structures in the prepore and pore complexes were revealed by systematically examining the extent of inter-β-strand disulfide bond crosslinking. This approach identified, chemically and unambiguously, the pairs of TMH residues that were nearest neighbors and hydrogen bonded to each other in the β-barrel, and thereby showed that the hydrogen-bonded β1 and β4 strands of adjacent monomers were offset by two residues in fully assembled β-barrel pores. When this amount of stagger is applied to the 140 anti-parallel β-strands in the giant PFO β-barrel, the individual strands orient in a right-handed twist with a 20° tilt relative to the pore axis.
This new chemical approach provides detailed structural information about the oligomeric CDCβ-barrels with diameters of 250–300 Å1 that is not available using other methods. No crystals of CDC pore complexes have been reported, perhaps because of heterogeneity in the number of monomers per oligomer. CryoEM images suggest that the tilt of the pneumolysin β-barrel strands is 0°9 instead of the 20° observed here for the PFO β-barrel, but it remains to be seen to what extent the β-strand tilts in CDC pore complexes vary. In any case, PFO β-barrels clearly differ from the many β-barrels with smaller pores that have tilt angles ≥ 37° and S values between n and 2n10,11,33.
The variable yield and identity of disulfide crosslinks in PPCs revealed that the TMHs are unfolded, extended, and in the initial stages of β-barrel formation. Specifically, the multiplicity of crosslinking partners for someβ1 and β4 residues showed that the β4 and β1 strands of adjacent PPC subunits were moving and transiently sampling potential hydrogen-bonding alignments and partners without forming any stable β1-β4 associations between adjacent PPC subunits. Yet the TMHs did not have complete freedom of motion because other TMH residues did not form any β1-β4 disulfide crosslinks in PPCs and hence were not accessible for reaction even in the presence of tetrathionate.
The fluorescence data also showed that the dynamic movement of unfolded TMHs in the PPC is not random. Two residues destined to face the membrane interior in the fully assembled pore were segregated into partially nonpolar milieus in the PPC, as was one of the aqueous-facing residues examined. These environments were distinct from the environments of those residues in both pore complexes and monomers, thereby revealing that individual TMH residues were reproducibly directed to specific locales within the PPC intermediate.
Since TMHs in thermally trapped PFO PPCs are partially unfolded and not aligned, the extreme possibilities for PPC intermediate states, with the TMHs completely folded or completely aligned (Fig. 5b), do not occur. Instead, PPC TMHs are partially unfolded, extended, and transiently proximal to nearby TMHs that are also in flux and dynamically sampling the available space. Although the TMHs in PPCs (Fig. 5b, center) are not as compact or tightly folded as in the native monomer and β-barrel structures, TMH movement is somewhat restricted within the oligomeric PPC by residual folding and/or spatial constraints. TMH freedom of motion within the PPC is therefore controlled, presumably to ensure that the major changes in TMH alignment, location, and environment during the temperature- dependent conversion from PPC to membrane-embedded β-barrel occur in the proper sequence and/or to ensure that the unfolded TMHs retain on-pathway folding conformations, consistent with our observation that a thermally trapped PPC readily forms a pore upon warming.
The disulfide scanning approach we have introduced here is based on the recognition that inter-β-strand disulfide bond formation can only occur between two Cys residues that are directly juxtaposed in adjacent β-strands. This structural feature allows us to quantify important aspects of β-barrel structure, as well as to directly monitor β-barrel assembly. Using this approach, we have shown experimentally that PFO forms an unprecedented β-barrel structure with S = n/2, a folding paradigm that is likely to describe the giantβ-barrels of other CDCs. Similarly, this approach will prove invaluable for identifying the details of anyβ-barrel structure and/or the folding and assembly of proteins that form aβ-barrel, no matter what their size. In particular, this approach will prove useful in identifying the mechanisms by which various protein complexes promote the folding of β-barrel proteins as they are inserted into the outer membranes of mitochondria by the SAM (sorting and assembly machinery)/TOB (topogenesis of β-barrel proteins) complex34–36 or into bacterial outer membranes by the Bam (β-barrel assembly machine) complex37.
Online Methods
PFO
Single-site mutations were introduced into a pRSETB plasmid (Invitrogen) coding for rPFO, a PFO derivative in which wild-type Cys459 was replaced by Ala7, using the QuikChange procedure (Agilent Technologies) and confirmed by DNA sequencing. PFO species were expressed with an amino-terminal hexahistidine tag in E. coli BL21(DE3)pLysS (Invitrogen) and purified as follows7,38. The IPTG-induced cells were harvested, suspended in Buffer C [10 mM MES (pH 6.5), 150 mM NaCl], and then lysed by passage through a French press (Thermo Scientific) twice. Cell debris was removed by centrifugation (30,000g, 15 min, 4°C) and filtering through a 0.45 μm syringe filter (Millipore). The cell lysate was loaded onto Chelating Sepharose FF (GE Healthcare) pre-treated with Co2+ equilibrated with Buffer C. After washing the column with 100 mL of Buffer C, PFO was eluted with a linear gradient from 0 to 1 M imidazole in Buffer C at 4°C. Fractions containing PFO were pooled and dialyzed overnight at 4°C against 4 L of Buffer B [10 mM MES (pH 7.5), 100 mM NaCl, 0.5 mM DTT]. After centrifugation as above, the supernatant was loaded onto SP Sephariose HP equilibrated in Buffer B and then eluted with a linear gradient from 0 to 1 M NaCl in Buffer B. Fractions containing PFO were pooled and dialyzed overnight at 4°C against 4 L of Buffer A [50 mM HEPES (pH 7.5), 100 mM NaCl, 0.5 mM DTT]. After another 2-hr dialysis against Buffer A in the morning, PFO was mixed and stored with 10% (v/v) glycerol and 5 mM DTT at −80°C.
NBD labeling
Stock solutions containing 2 mg of cysteine-substituted PFO derivatives were passed through a PD-10 desalting columen (GE Healthcare) equilibrated in Buffer A to remove excess DTT. After concentration to ~1 mg PFO/mL using Amicon Ultra (Millipore), guanidinium hydrochloride was added to 3 M. A ten-fold molar excess of N,N′-dimethyl-N- (iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine was then added to the PFO. After 2 hr at room temperature, the reaction was quenched by adding DTT to 5 mM. NBD-labeled PFO was separated from free dye by gel filtration through Sephadex G-50 (GE Healthcare)(1.5 cm i.d. × 25 cm) equilibrated in Buffer A before storage in 10% (v/v) glycerol at −80°C. Labeling efficiency was determined by using the molar absorptivity coefficients of 74,260 M−1 at 280 nm and 25,000 M−1 at 478 nm for PFO and NBD, respectively7,38
Liposomes
A lipid mixture composed of 45 mol% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; Avanti Polar Lipids) and 55 mol% cholesterol (Steraloids) was dried under a stream of nitrogen, and then further dried under vacuum for 3 hr. After the lipid film was resuspended by vortexing in Buffer A, the lipid was bath sonicated for 5 min. The resulting suspension was passed 21 times through 100-nm and 200-nm pore polycarbonate membranes using a Liposofast extruder (Avestin)26.
Disulfide crosslinking
In most experiments, 60 pmoles of a β1 mutant were mixed with 60 pmoles of a β4 mutant on ice before liposomes were added to 1.2 mM in a final volume of 100μl of 50 mM Hepes (pH 7.5), 100 mM NaCl (Buffer A) plus 5 mM DTT. Samples were incubated for 40 min at 37°C or 2 h at 4°C to form pore or prepore complexes, respectively. PPC and pore complexes were then purified by flotation in a sucrose density step gradient to remove PFO monomers, aggregates, and DTT by adding 250 μl of a 70% (w/v) sucrose in buffer A solution to the PFO. Following transfer of the resulting PFO in 50% (w/v) sucrose solution to a SW55Ti centrifuge tube (Beckman), the PFO was overlaid with 2 ml of 40% (w/v) and then 1.5 ml of 5% (w/v) sucrose in buffer A. After sedimentation at 87,000g for 1 h at 4°C, liposomes with PPCs or pore complexes were collected from the interface between 40% and 5% layers. In many cases, the purified membrane-bound PPCs or pore complexes were then incubated with 100 μM sodium tetrathionate for 10 min on ice to stimulate disulfide bond formation. The unreacted thiol groups in every sample were blocked by incubation (0°C, 5 min) with 20 mM NEM prior to protein precipitation and lipid extraction in methanol/chloroform27. After dried samples were resuspended in SDS sample buffer and boiled, PFO oligomer content was analyzed by SDS-AGE using a 1.8% (w/v) gel (100 V, 40 min) as before24. Gels were dried, stained with Coomassie Blue, re-dried, and scanned with a flatbed scanner (Epson) before the data were quantified with ImageJ (NIH). The ratio of dimer to (monomer + dimer) PFO revealed which pairs of β1 and β4 cysteine residues were adjacent.
Spectral measurements
Steady-state fluorescence emission spectra (λex = 470 nm, 4 nm bandpass; λem = 500–600 nm, 4 nm bandpass) were obtained as before7 with continuous stirring using 1 cm × 1 cm quartz cuvettes coated with POPC vesicles to minimize protein absorption39. Each sample of 50 nM NBD-labeled PFO in buffer A was examined: (i) by itself at 4°C; (ii) after addition of 50 mM cholesterol-containing liposomes and incubation at 4°C for 2 hr to allow PPC formation; and (iii) after incubation at 37°C for 30 min to allow pore formation, followed by 5 min on ice and 5 min of stirring and equilibration to 4°C in the instrument. An equivalent NBD-free sample prepared with unmodified PFO was examined in parallel and its signal was subtracted from the sample signal to obtain the net NBD emission by correcting for light scattering and background signals. Each depicted net emission scan is the average obtained from three separate samples.
Trypsin digestion
Samples containing prepore or pore complexes were isolated by sedimentation in a sucrose density gradient as described above. Parallel samples of PFO (~7 μg) in the monomer, PPC, or β-barrel pore complex state were treated with porcine trypsin (Sigma) in a final volume of 200 μL of Buffer A at 4°C for various incubation times and concentrations of trypsin (specified in Supplementary Figure 8). After digestions were halted by the addition of TLCK (Sigma) to 5 mM, PFO was precipitated by methanol/chloroform27 and analyzed by SDS-PAGE (12.5% gels, 20 mA, 250V, 40 min).
Supplementary Material
Acknowledgments
Support was provided by NIH grant AI-37657 (RKT) and Robert A. Welch Foundation Chair grant BE-0017 (AEJ).
Footnotes
Author contributions:
TKS did experiments and contributed to experimental design and writing text, AEJ and RWT contributed to experimental design and writing text.
Competing financial interests
The authors have no competing financial interests relevant to this work.
References
- 1.Tweten RK. Cholesterol-Dependent Cytolysins, a Family of Versatile Pore-Forming Toxins. Infect Immun. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotze EM, Tweten RK. Membrane Assembly of the Cholesterol-Dependent Cytolysin Pore Complex. Biochim Biophys Acta. 2012;1818:1028–1038. doi: 10.1016/j.bbamem.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossjohn J, Feil SC, Mckinstry WJ, Tweten RK, Parker MW. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 4.Ramachandran R, Heuck AP, Tweten RK, Johnson AE. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nature Struct Biol. 2002;9:823–827. doi: 10.1038/nsb855. [DOI] [PubMed] [Google Scholar]
- 5.Ramachandran R, Tweten RK, Johnson AE. The Domains of a Cholesterol-Dependent Cytolysin Undergo a Major FRET-Detected Rearrangement during Pore Formation. Proc Natl Acad Sci, USA. 2005;102:7139–7144. doi: 10.1073/pnas.0500556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. Vertical collapse of a cytolysin prepore moves its transmembrane β-hairpins to the membrane. EMBO J. 2004;23:3206–3215. doi: 10.1038/sj.emboj.7600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepard LA, et al. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an α-helical to β-sheet transition identified by fluorescence spectroscopy. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- 8.Shatursky O, et al. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell. 1999;99:293–299. doi: 10.1016/s0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- 9.Tilley SJ, Orlova EV, Gibert RJC, Andrew PW, Saibil HR. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 10.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 11.Reboul CF, Mahmood K, Whisstock JC, Dunstone MA. Predicting giant transmembrane β-barrel architecture. Bioinformatics. 2012;28:1299–1302. doi: 10.1093/bioinformatics/bts152. [DOI] [PubMed] [Google Scholar]
- 12.Ohno-Iwashita Y, Iwamoto M, Ando S, Iwashita S. Effect of lipidic factors on membrane cholesterol topology - mode of binding of θ-toxin to cholesterol in liposomes. Biochim Biophys Acta. 1992;1109:81–90. doi: 10.1016/0005-2736(92)90190-w. [DOI] [PubMed] [Google Scholar]
- 13.Heuck AP, Hotze EM, Tweten RK, Johnson AE. Mechanism of membrane insertion of a multimeric β-barrel protein: Perfringolysin O creates a pore using ordered and coupled conformational changes. Mol Cell. 2000;6:1233–1242. doi: 10.1016/s1097-2765(00)00119-2. [DOI] [PubMed] [Google Scholar]
- 14.Nelson L, Johnson AE, London E. How the interaction of Perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: insights into the origin of perfringolysin O-lipid raft interaction. J Biol Chem. 2008;283:4632–4642. doi: 10.1074/jbc.M709483200. [DOI] [PubMed] [Google Scholar]
- 15.Flanagan JJ, Tweten RK, Johnson AE, Heuck AP. Cholesterol Exposure at the Membrane Surface Is Necessary and Sufficient to Trigger Perfringolysin O Binding. Biochemistry. 2009;48:3977–3987. doi: 10.1021/bi9002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson LD, Chiantia S, London E. Perfringolysin O Association with Ordered Lipid Domains: Implications for Transmembrane Protein Raft Affinity. Biophys J. 2010;99:3255–3263. doi: 10.1016/j.bpj.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrand AJ, LaChapelle S, Hotze EM, Johnson AE, Tweten RK. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc Natl Acad Sci USA. 2010;107:4341–4346. doi: 10.1073/pnas.0911581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soltani CE, Hotze EM, Johnson AE, Tweten RK. Specific protein-membrane contacts are required for prepore and pore assembly by a cholesterol-dependent cytolysin. J Biol Chem. 2007;282:15709–15216. doi: 10.1074/jbc.M701173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soltani CE, Hotze EM, Johnson AE, Tweten RK. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc Natl Acad Sci, USA. 2007;104:20226–20231. doi: 10.1073/pnas.0708104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dowd KJ, Tweten RK. The Cholesterol-Dependent Cytolysin Signature Motif: A Critical Element in the Allosteric Pathway that Couples Membrane Binding to Pore Assembly. PLoS Pathogens. 2012;8:e1002787. doi: 10.1371/journal.ppat.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran R, Tweten RK, Johnson AE. Membrane-Dependent Conformational Changes Initiate Cholesterol-Dependent Cytolysin Oligomerization and Intersubunit β-Strand Alignment. Nature Struct Mol Biol. 2004;11:697–705. doi: 10.1038/nsmb793. [DOI] [PubMed] [Google Scholar]
- 22.Hotze EM, et al. Monomer-monomer interactions propagate structural transitions necessary for pore formation by the cholesterol-dependent cytolysins. J Biol Chem. 2012;287:24534–24543. doi: 10.1074/jbc.M112.380139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison PM, Sternberg MJ. Analysis and classification of disulphide connectivity in proteins. The entropic effect of cross-linkage. J Mol Biol. 1994;244:448–463. doi: 10.1006/jmbi.1994.1742. [DOI] [PubMed] [Google Scholar]
- 24.Shepard LA, Shatursky O, Johnson AE, Tweten RK. The mechanism of pore assembly for a cholesterol-dependent cytolysin: formation of a large prepore complex precedes the insertion of the transmembrane β-hairpins. Biochemistry. 2000;39:10284–10293. doi: 10.1021/bi000436r. [DOI] [PubMed] [Google Scholar]
- 25.Hotze EM, et al. Arresting pore formation of a cholesterol-dependent cytolysin by disulfide trapping synchronizes the insertion of the transmembrane β-sheet from a prepore intermediate. J Biol Chem. 2001;276:8261–8268. doi: 10.1074/jbc.M009865200. [DOI] [PubMed] [Google Scholar]
- 26.Heuck AP, Tweten RK, Johnson AE. Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J Biol Chem. 2003;278:31218–31225. doi: 10.1074/jbc.M303151200. [DOI] [PubMed] [Google Scholar]
- 27.Wessel D, Flügge UL. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 28.Chou KC, et al. Energetics of the structure and chain tilting of antiparallel β-barrels in proteins. Proteins: Structure, Function, and Genetics. 1990;8:14–22. doi: 10.1002/prot.340080105. [DOI] [PubMed] [Google Scholar]
- 29.Sansom MS, Kerr ID. Transbilayer pores formed by β-barrels: molecular modeling of pore structures and properties. Biophys J. 1995;69:1334–1343. doi: 10.1016/S0006-3495(95)80000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou KC, Scheraga HA. Origin of the right-handed twist of β-sheets of poly(LVal) chains. Proc Natl Acad Sci USA. 1982;79:7047–7051. doi: 10.1073/pnas.79.22.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- 32.Johnson AE. Fluorescence Approaches for Determining Protein Conformations, Interactions, and Mechanisms at Membranes. Traffic. 2005;6:1078–1092. doi: 10.1111/j.1600-0854.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 33.Song L, et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 34.Neupert W, Herrmann JM. Translocation of Proteins into Mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 35.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing Mitochondrial Proteins: Machineries and Mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endo T, Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta. 2010;1803:706–14. doi: 10.1016/j.bbamcr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Hagan CL, Silhavy TJ, Kahne D. β-Barrel Membrane Protein Assembly by the Bam Complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 38.Heuck AP, Savva CG, Holzenburg A, Johnson AE. Conformational changes that effect oligomerization and initiate pore formation are triggered throughout perfringolysin O upon binding to cholesterol. J Biol Chem. 2007;282:22629–22637. doi: 10.1074/jbc.M703207200. [DOI] [PubMed] [Google Scholar]
- 39.Ye J, Esmon NL, Esmon CT, Johnson AE. The active site of thrombin is altered upon binding to thrombomodulin: Two distinct structural changes are detected by fluorescence, but only one correlates with protein C activation. J Biol Chem. 1991;266:23016–23021. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.