Abstract
Chaperone-mediated autophagy (CMA) contributes to cellular quality control and the cellular response to stress through the selective degradation of cytosolic proteins in lysosomes. Decrease in CMA activity occurs in aging and in age-related disorders (for example, neurodegenerative diseases and diabetes). Although prevention of this age-dependent decline through genetic manipulation in mouse has proven beneficial, chemical modulation of CMA is not currently possible, due in part to the lack of information on the signaling mechanisms that modulate this pathway. In this work, we report that signaling through the retinoic acid receptor alpha (RARα) inhibits CMA and apply structure-based chemical design to develop synthetic derivatives of all-trans-retinoic acid (ATRA) to specifically neutralize this inhibitory effect. We demonstrate that chemical enhancement of CMA protects cells from oxidative stress and from proteotoxicity, supporting a potential therapeutic opportunity when reduced CMA contributes to cellular dysfunction and disease.
Keywords: all-trans-retinoic acid, lysosomes, oxidative stress, proteotoxicity, retinoic acid receptor
Introduction
Autophagy is the process by which intracellular components undergo degradation in lysosomes1,2, contributing to the maintenance of cellular homeostasis and to cellular quality control. Autophagy is upregulated as a cellular defense against aggressors or to allow cellular adaptation to changing environmental conditions3. Alterations of the autophagic process underlie the pathogenesis of severe human diseases1,4.
The best characterized autophagic pathways are macroautophagy, microautophagy and CMA1. The characteristic of CMA that distinguishes it from other forms of autophagy is the degradation of a specific subset of cytosolic proteins upon direct translocation across the lysosomal membrane into the lysosomal lumen5. Substrates for this pathway all have a targeting motif in their amino acid sequence6 that once recognized by the cytosolic chaperone hsc70, mediates substrate delivery to the surface of lysosomes7. Once there, substrates bind to the lysosome-associated membrane protein type 2A (LAMP-2A) and promote its multimerization into a high molecular weight complex that is required for substrate translocation8. A variant of hsc70, resident in the lysosomal lumen, assists substrates to complete translocation into lysosomes.
CMA is maximally activated in response to stressors such as prolonged nutritional deprivation, oxidative stress or exposure to toxic compounds5. Malfunction of CMA has been described in neurodegenerative conditions such as familial forms of Parkinson’s disease (PD)9,10 and tauopathies11, in metabolic disorders such as diabetes12 and in lysosomal storage disorders13. Furthermore, a gradual decline in CMA activity with age has been proposed to aggravate multiple age-related disorders14. In fact, if the reduction in CMA activity with age is prevented through genetic manipulation in mice, cellular homeostasis and organ function can be preserved until late in life15. These findings, along with the growing number of connections between CMA and human diseases, justify the growing interest in developing efficient chemical modulators of this autophagic pathway.
Chemical compounds that impact CMA have lacked selectivity for this pathway16. For example, inhibition of protein synthesis or of lysosomal proteases results in reduced CMA degradation, but it also affects many other intracellular processes16. Inhibition of glucose-6-phosphate dehydrogenase or of the cytosolic chaperone hsp90 lead to higher CMA activity in some cell types but not in others16; in fact, later studies demonstrated that the effect was not direct but a consequence of compensatory upregulation of other CMA components8. One of the limitations for the development of CMA modulators is a lack of information on the cellular signaling mechanisms that activate this pathway.
In this work, we have investigated the effect of RAR signaling on CMA activity. The three types of RARs identified in mammals, RARα, RARβ and RARγ, are encoded by three different genes17. In contrast to the complex tissue-dependent expression of RARβ and RARγ, RARα is ubiquitously expressed. RARs act as transcriptional activators and repressors of a broad subset of genes, modulating cellular processes in which CMA has also been implicated, such as differentiation, proliferation and control of cellular homeostasis17. Furthermore, RAR loss or aberrant function has been described in many oncogenic processes, where CMA upregulation is a common feature required to sustain cancer cell growth18.
RAR are attractive druggable targets because their natural substrates ATRA and similar retinoids have been well characterized19. They efficiently traffic across lipid bilayers because of their small size and hydrophobic character20. This feature along with a growing understanding of the chemical modifications that retinoid derivatives undergo in cells21,22 makes ATRA byproducts and derivatives good candidates for therapeutic applications. We have designed and synthesized novel retinoid derivatives with distinct biological activities23,24 to generate RARα modulators that impact CMA activity. We have used structure-based chemical design to incorporate the critical regions of ATRA but have also introduced modifications that render these retinoids selective for particular RARα downstream pathways. We demonstrate that these compounds lead to the selective activation of CMA in the absence of detectable changes in other autophagic pathways. Furthermore, we show that chemical enhancement of CMA with these compounds renders cells more resistant to oxidative stress and proteotoxicity. These findings highlight the potential therapeutic applicability of these or related compounds in the treatment of chronic diseases that associate with loss of CMA activity.
Results
Opposing effects of RAR signaling in autophagic pathways
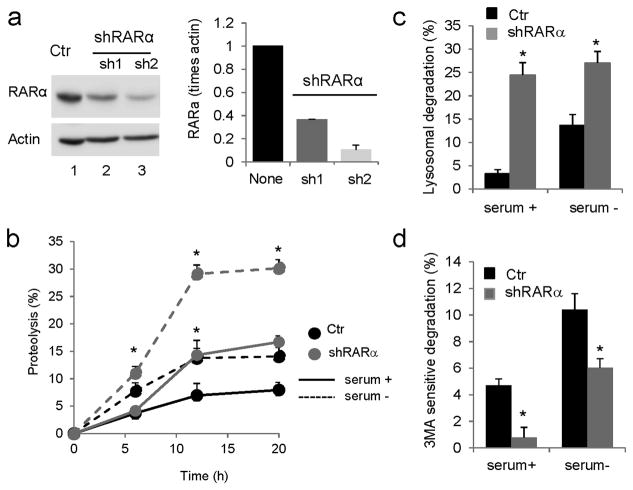
Retinoic acid (RA) exerts a cell-type dependent stimulatory effect on macroautophagy via Beclin-1 upregulation and inhibition of the mTOR pathway25–27 or by enhancing autophagosome maturation28, but the mechanistic relationship between RA and CMA has not been explored. To directly analyze the effect of RAR signaling on autophagic pathways, we knocked down RARα in mouse fibroblasts, the most abundant RAR in these cells. Lentiviral transduction with two different shRNA against RARα resulted in 75–90% stable knockdown of this receptor (Fig. 1a).
Figure 1. Effect of knockdown of RARα on intracellular turnover of long-lived proteins.
(a) Knockdown of RARα in NIH3T3 mouse fibroblasts was conducted using two different shRNAs, sh1 and sh2. Ctr: control. Left: Representative immunoblot. Actin is shown as loading control and full-length blots are shown in Supplementary Figure 21. Right: Levels of RARα in control and knockdown cells determined by densitometric quantification of immunoblots represented by the one shown on the left. Values are normalized for actin and expressed as times control (none) values. (n=3) (b) Rates of degradation of long-lived proteins in control and RARα-knockdown cells maintained in the presence or absence of serum for 12 h. Values are expressed as percentage of proteolysis. (n=3) (c, d) Percentage of lysosomal (c) and macroautophagy (d) degradation in cells assayed as in b, but treated with inhibitors of lysosomal proteolysis (c) or with 3-methyladenine to block macroautophagy (d). Values are expressed as percentage of total protein degradation sensitive to the lysosomal inhibitors (n=3). All values are mean±S.E. and differences with control are significant for *p<0.05.
Analysis of the rates of degradation of long-lived proteins, typical autophagy substrates, in control and knockdown cells revealed an increase in protein degradation in RARα(−) cells, when compared to control mouse fibroblasts evident both under basal conditions and when serum was removed from the culture media to upregulate autophagy. (Fig. 1b). Addition of lysosomal inhibitors demonstrated that most of the increase in basal and inducible protein degradation in RARα(−) cells originated in the lysosome (Fig. 1c). However, the amount of protein degradation sensitive to 3-methyladenine, a well-characterized inhibitor of macroautophagy, was significantly reduced in RARα(−) cells (Fig. 1d and Supplementary Results, Supplementary Fig. 1a, b), suggesting that the increase in lysosomal degradation was not attributable to macroautophagy.
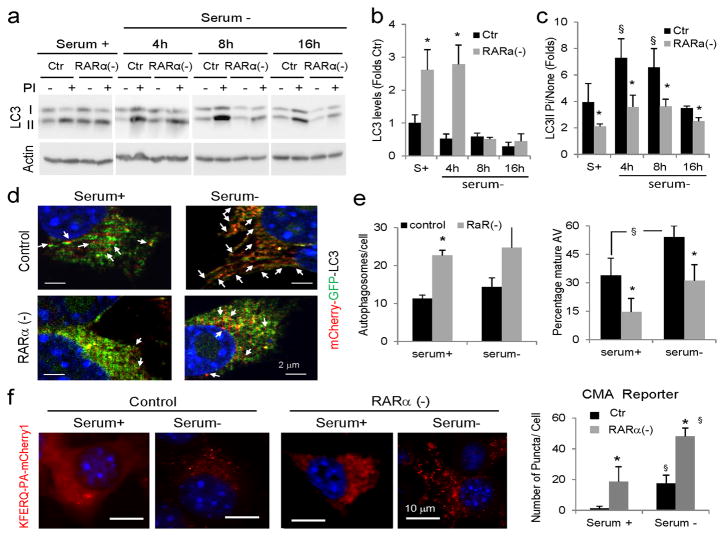
We applied a widely accepted assay based on the analysis of the intracellular levels and degradation of the lipid-conjugated form of the light chain protein type 3 (LC3-II), a constitutive component of autophagosomes29, to directly measure macroautophagy. Steady-state levels of LC3-II provide information on the amount of autophagosomes present in a cell at a given time, whereas the amount of LC3-II that accumulates upon blockage of lysosomal proteolysis provides a measure of the efficiency of fusion and degradation of autophagic vacuoles by lysosomes (autophagic flux). RARα(−) cells displayed significantly higher levels of LC3-II both under basal and inducible conditions when compared with control cells (Fig. 2a–c). Immunofluorescence for LC3 also revealed higher content of LC3-positive vesicles in the cells defective for RARα (Supplementary Fig. 1c). We confirmed that these vesicles were autophagosomes using a double-tagged form of LC3 (mCherry-GPF-LC3) that due to quenching of GFP fluorescence at low pH highlights autophagosomes in yellow and lysosomes in red. As shown in Figure 2d and Supplementary Figure 2a, the content of double-labeled vesicles was significantly higher in RARα(−) cells when compared with control cells. An increase in autophagosome content can result from increased formation or reduced clearance by lysosomes. To differentiate between these possibilities, we compared the autophagic flux in control and RARα(−) cells. Immunoblot (Fig. 2a, c) and immunofluorescence for endogenous LC3 (Supplementary Fig. 1c) revealed significantly reduced increase in autophagosome-associated form of this protein upon blockage of lysosomal degradation, and we also observed a reduction in the number of single-labeled vesicles (lysosomes) with the double-tagged form of LC3 (Fig. 2d, e and Supplementary Fig. 2a). These results suggest that elimination of signaling through the RARα reduces macroautophagy, in agreement with the previously described stimulatory effect of RA on this pathway25,27,28.
Figure 2. Effect of knockdown of RARα on autophagic pathways.
(a) Immunoblot for LC3-II of mouse fibroblasts control (ctr) or knocked-down for RARα (RARα(−)) maintained in the presence or absence of serum for the indicated times. Where indicated, protease inhibitors (PI) against lysosomal proteolysis were added. Actin is shown as loading control. (b) Levels of LC3-II determined by densitometric quantification of immunoblots. Values are expressed as folds values in serum supplemented control cells (n=4). (c) Ratio of levels of LC3-II in cells treated with PI compared to untreated cells. Values are expressed as fold untreated (n=4). (d,e) Autophagic flux in the same cells expressing mCherry-GFP-LC3 and maintained in the presence or absence of serum: (d) representative merged channels images. Arrows: autolysosomes (red); (e) Quantification of number of autophagosomes (mCherry and GFP positive) per cell (left) and percentage of autolysosomes (mCherry positive and GFP negative; right) in > 50 cells in at least 4 different fields. (f) Control and RARα(−) cells were transfected with the KFERQ-mcherry1 photoactivable reporter and after photoactivation were maintained in media with or without serum. Left: representative images. Inset show higher magnification images. Right: Quantification of the number of puncta per cell in > 50 cells in at least 4 different fields. Nuclei are labeled with DAPI. All values are mean±S.E. Differences with control (*) or with serum supplemented cells (§) are significant for p<0.05. Full-field fluorescence images and full-length blots are shown in Supplementary Figures 2 and 21, respectively.
Because the downregulation of macroautophagy upon RARα knockdown cannot explain the increase in lysosomal degradation observed in these cells (Fig. 1c), we next measured the effect of this intervention on CMA using a photoactivable (PA) reporter fused to the CMA-targeting motif (KFERQ-PA-mcherry1)30. Activation of CMA mobilizes this artificial CMA substrate from the cytosol to lysosomes, which can be tracked as a change in the reporter fluorescence from a diffuse to a punctate pattern30. Upon transfection with the CMA reporter, RARα(−) cells had a significantly higher number of fluorescent puncta per cell both in the presence or absence of serum when compared to control cells (Fig. 2f and Supplementary Fig. 2b). These results suggest that the increase in lysosomal degradation observed upon RARα blockage was, for the most part, a consequence of CMA upregulation, and support an inhibitory effect of RARα signaling on basal and inducible CMA.
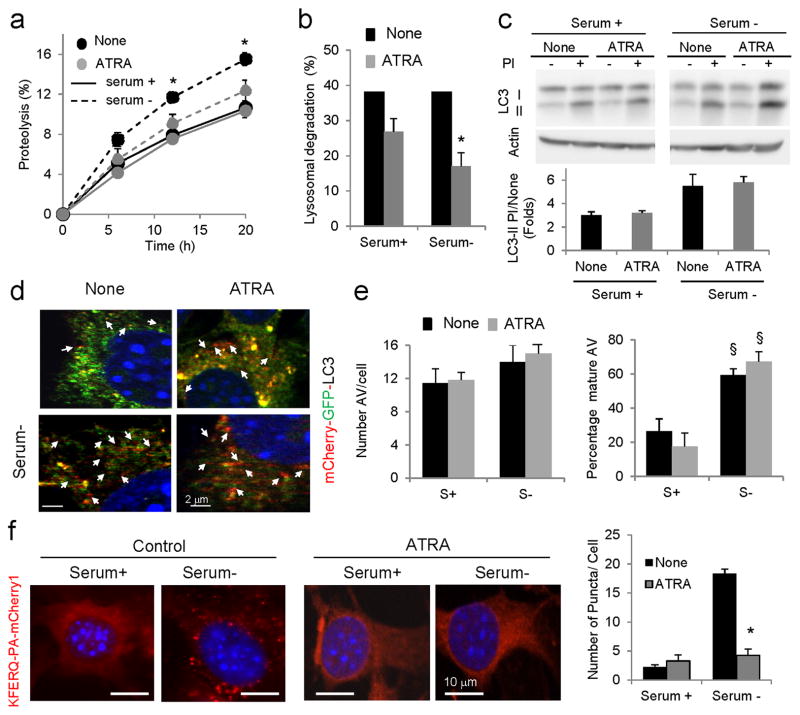
To further confirm the opposing effects of RARα signaling on autophagic pathways and the inhibitory effect on CMA, we performed similar experiments in cells treated or not with ATRA, a potent activator of RARα signaling. ATRA supplementation of mouse fibroblasts did not affect total rates of protein degradation under basal conditions, but significantly reduced the increase in protein degradation normally observed in response to prolonged serum removal (Fig. 3a). Addition of inhibitors of lysosomal proteolysis confirmed that both basal and inducible lysosomal degradation were significantly compromised after ATRA supplementation (Fig. 3b). In contrast to reports in other cell types25,27,28, we did not find significant changes in steady-state levels or lysosomal flux of LC3 (Fig. 3c–e and Supplementary Fig. 3a), supporting the previously proposed cell-type dependent stimulatory effect of retinoids on macroautophagy. Treatment with ATRA did not have a noticeable effect on basal CMA but significantly reduced the activation of this pathway in response to serum removal (Fig. 3f and Supplementary Fig. 3b). Overall our findings suggest that ATRA and RARα signaling exert an inhibitory effect on CMA activity.
Figure 3. Effect of ATRA on autophagy.
(a) Rates of degradation of long-lived proteins in mouse fibroblasts untreated (None) or treated with (40μM) ATRA and maintained in the presence or absence of serum. Values are expressed as a percentage of proteolysis. (n=3) (b) Percentage of lysosomal degradation calculated after treatment with inhibitors of lysosomal proteolysis for 12 h (n=3). (c) Immunoblot for LC3-II of the same cells maintained in the presence or absence of serum and protease inhibitors (PI). Left: representative immunoblot. Actin is shown as loading control. Bottom: Ratio of levels of LC3-II in cells treated with PI compared to untreated cells. Values are expressed as fold untreated (n=4). (d, e) Autophagic flux in untreated and ATRA-treated cells expressing mCherry-GFP-LC3 and maintained in the presence or absence of serum: (d) representative merged channels images. Arrows: autolysosomes (red); (e) Number of autophagosomes (left) and percentage of autolysosomes (right) after quantification of > 50 cells. (f) Mouse fibroblasts expressing the KFERQ-mcherry1 photoactivable reporter with or without ATRA and after photoactivation maintained in the presence or absence of serum. Left: Representative images. Nuclei are labeled with DAPI. Insets: high magnification images. Right: Quantification of the number of puncta per cell in > 50 cells. All values are mean±S.E. and differences with untreated (*) or with serum-supplemented cells (§) are significant for p<0.01. Full-field fluorescence images and full-length blots are shown in Supplementary Figures 3 and 21, respectively.
Design and synthesis of RARα antagonist
Previous reports have revealed that ATRA does not affect macroautophagy via RAR signaling, as changes in macroautophagy occurred in the presence or absence of these receptors28. We confirmed that signaling through RARα did not lead to the inhibition of autophagic degradation of cytosolic proteins, because it was still detectable when RARα( −) cells were supplemented with ATRA (Supplementary Fig. 4a). In contrast, the inhibitory effect of ATRA on CMA was dependent on the RARα, as ATRA treatment did not inhibit CMA activity in RARα( −) cells (Supplementary Fig. 4b). The marked upregulation of CMA when RARα was eliminated (Fig. 2f), the opposite effects of this intervention on macroautophagy activity (Fig. 2d, e), and the fact that part of the effect of ATRA on macroautophagy was not mediated through RARα signaling (Supplementary Fig. 4a) led us to propose that it may be possible to design RARα– targeted compounds capable of upregulating CMA without affecting other autophagic pathways.
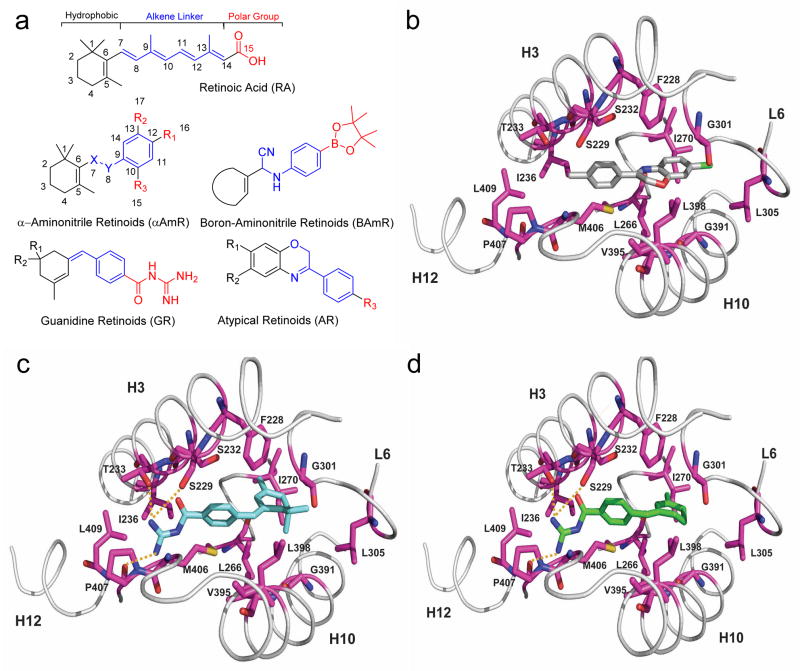
To this effect, we used structure-based chemical-design strategies and novel chemistry to generate a small library of RA derivatives. We introduced chemical changes to protect the regions of ATRA most prone to intracellular modifications and to enhance ATRA reactive properties with RARα. Figure 4 depicts the three basic domains common to all retinoid molecules: a hydrophobic component, an all-trans-configured alkene linker and a polar group (the carboxylic acid moiety)22. The alkene linker region is sensitive to photochemical changes21 and to oxidation at the allylic C4 position in the trimethylcyclohexyl ring by enzymes such as cellular isomerases and cytochrome P45022.
Figure 4. Design, synthesis and molecular docking of RARα-targeting compounds.
(a) Molecular structure of ATRA (top) highlighting three different regions: the hydrophobic ring (black), the polyene linker “connector” (blue), and the carboxylic acid moiety (red). The basic structure of the four families of compounds generated through modifications of ATRA using structure-based chemical design strategies are shown. Numbering is shown in the retinoic acid and the α-aminonitrile retinoid backbone to indicate how these positions have been conserved in the new molecules. (b–d) Molecular docking of AR7 (b), GR1 (c) and GR2 (d) in the RARα-binding pocket. A close view of the RARα-binding pocket in ribbon (gray) and interacting residues in stick (magenta) for each compound docked in the lowest-energy conformation for docking pose I is shown. Compounds are docked to a hydrophobic region of the RARα-binding pocket formed by h3, h10 and h12. Hydrogen bonds are formed from the guanidinium group of GR1 and GR2 to side-chain hydroxyls of Ser229 and Thr233, and backbone carbonyl oxygen of Pro407 (yellow dotted lines).
Through chemical modifications using structure-based chemical-design approach, we generated a library of 29 RA-related compounds grouped in four families: α-aminonitrile retinoids (αAmR)31, boron-aminonitrile retinoids (BAmR) (Supplementary Note I), guanidine retinoids (GR)32 and atypical retinoids (AR)33 protected in particular positions (Fig. 4a). These families contain modifications at the C4 position of the hydrophobic ring, to protect it from possible oxidation to 4-oxo, and in general shortened alkene linkers, which are susceptible to photoisomerization. The newly designed alkene linker conserves its aromaticity in the ring-constrained form. In addition, by incorporating groups such as –CN, -NH or -B, we aimed to enhance ATRA-reactive properties (for example, sp2 or sp3 hybridization of B can facilitate formation of hydrogen or covalent bonds). The structures of all the compounds generated for the library and their synthetic schemes are shown in Supplementary Table 1.
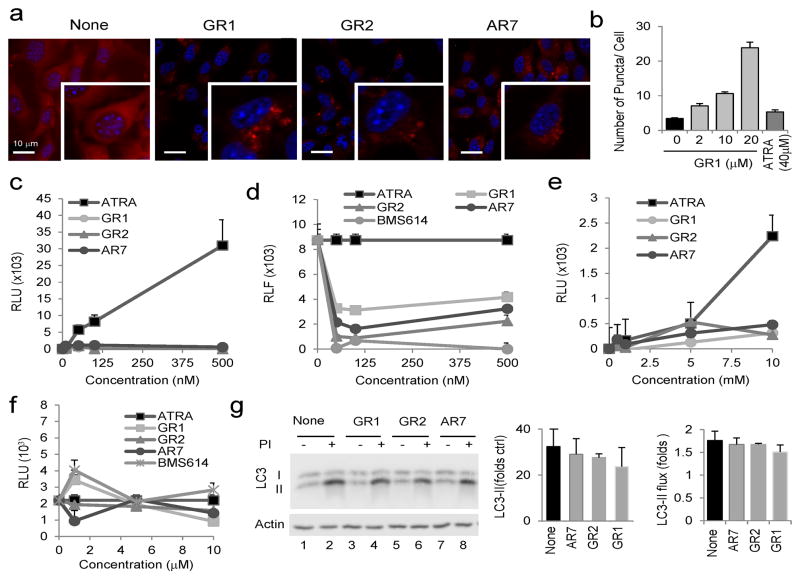
We first assessed dose-dependent effects of the compounds on cellular viability (Supplementary Fig. 5a) and found that for most compounds, toxicity was not clearly manifested until concentrations ≥50μM. Consequently, for all subsequent testing, compounds were used at 20μM, a concentration where we observed less than 20% decrease in cellular viability and did not detect apoptosis (annexin V labeling; Supplementary Fig. 5b). We then screened the library for an effect on CMA (Supplementary Table 2), using mouse fibroblasts expressing the CMA reporter (Fig. 5a and Supplementary Fig.6). For those compounds showing a positive effect (≥ 2.5-fold increase in the number of fluorescent puncta in serum supplemented cells), we validated changes in total protein degradation using metabolic labeling. These combined analyses revealed marked dose-dependent activation of CMA activity in cells treated with compounds AR7 (1), GR1 (2) and GR2 (3) (Fig. 5b and Supplementary Fig. 7). NMR data showed that GR1 was a mixture of isomers with E- and Z-stereoselectivity in 2:1 ratio, whereas in GR2 the major isomer was E and the minor Z in a 1:0.2 ratio (Supplementary Fig. 8).
Figure 5. Effect of the chemical activators of CMA on RARα activity.
(a) Mouse fibroblasts expressing the KFERQ-mcherry1 photoactivable reporter with or without the indicated compounds (20μM) imaged 16 h after photoactivation. Insets: higher magnification images. Nuclei are labeled with DAPI. (b) Quantification of the effect of increasing concentrations of GR1 on the same cells. Untreated cells and cells treated with 40μM ATRA are also shown. Representative images are shown in Supplementary Figure 7. Graph shows the average number of fluorescent puncta per cell, quantified in > 50 cells. All values are mean±S.E. (c–f) Mouse fibroblasts were co-transfected with the hRARα receptor (c, d) or the hRXR receptor (e,f), a relevant reporter luciferase plasmid and the non-retinoid regulated renilla reporter to control for transfection. Values show luciferase units detected in cells subjected to: (c, e) the indicated concentrations of ATRA and the three retinoid derivatives for 12 h. (d, f) 100 nM (d) or 10 μM (f) ATRA alone (ATRA) or in the presence of the indicated concentrations of the three retinoid derivatives or the antagonist BMS614. Values show luciferase intensity expressed as percentage of that in cells treated only with ATRA and Kis are shown on the right (n=4–6). (g) Immunoblot for LC3 of cells treated with 20μM of the retinoid derivatives and protease inhibitors (PI), as labeled. Actin is shown as loading control and full-length blots are shown in Supplementary Figure 21. Levels of LC3-II in untreated cells (left) and increase after PI treatment (LC3-II flux) (right) were calculated from the densitometric quantification of immunoblots. Values are mean±S.E. (n=3).
To determine the effect of these molecules on RARα signaling, we co-transfected cells with a plasmid coding for RARα fused to the Gal4 DNA-binding domain and a Gal4-dependent luciferase reporter. In contrast to the dose-dependent activation observed in cells treated with ATRA, similar doses of the three compounds that activate CMA did not have any effect on luciferase activity (Fig. 5c), whereas some compounds in the other families in our library (αAmR and BAmR; 4–9) displayed discrete activity (Supplementary Fig. 9a, b). When administered in combination with ATRA, these compounds had a marked inhibitory effect on the ATRA-dependent activation of luciferase (Fig. 5d). In fact, GR2 and AR7 were among the most potent antagonist compounds in the library (Supplementary Fig. 9c). Using a similar luciferase-based reporter for RXR we found that although some compounds in the library (αAmR family) exhibited activity through this receptor (Supplementary Fig.10), none of the three leading compounds had significant agonist or antagonist activity on this receptor (Fig. 5e, f).
The RAR antagonistic potency of the three leading molecules was close to that of the bona fide antagonist BMS614 (Fig. 5d and Supplementary Fig. 11a). In fact, BMS614 also enhanced CMA activity (Supplementary Fig. 11b), but whereas this antagonist inhibited macroautophagy (Supplementary Fig. 11c), none of the new compounds significantly affected autophagosome content or their clearance by lysosomes (Fig. 5g). These results confirm that the novel retinoid derivatives act as RARα antagonists and are capable of upregulating CMA without affecting macroautophagy.
Retinoid derivatives stimulate CMA
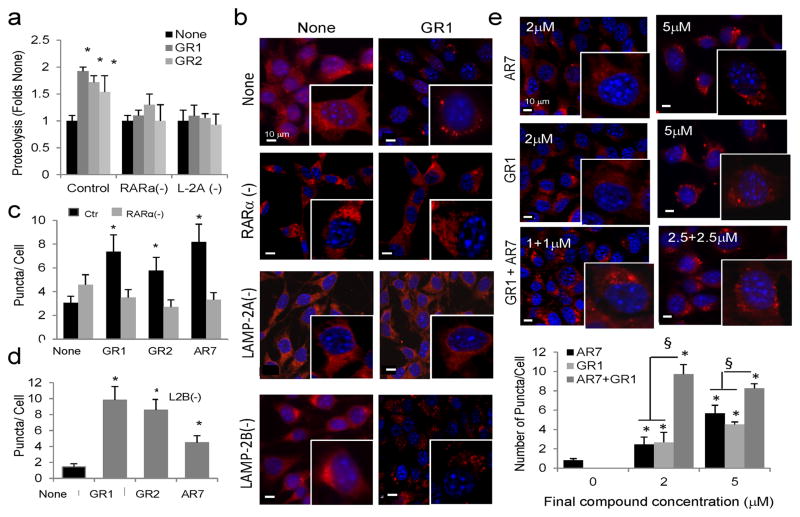
We further characterized the effect on autophagy of the three RARα antagonist compounds generated in this study. Using RARα knockdown cells, we confirmed that the stimulatory effect of the new derivatives on total protein degradation (Fig. 6a and Supplementary Fig. 12) and on CMA (Fig. 6b, c and Supplementary Fig. 13a) was dependent on the presence of RARα. Thus, although both activities were higher in the RARα( −) cells, addition of the retinoid derivatives no longer had a stimulatory effect. We also confirmed that the increase in protein degradation (Fig. 6a) and in the amount of CMA-positive puncta per cell (Fig. 6b and Supplementary Fig. 13b) induced by the retinoid derivates was abolished in cells knocked down for LAMP-2A and consequently incapable of carrying on CMA34. However, the compound-induced increase in fluorescent puncta was still present in cells knocked down for LAMP-2B, a protein with 85% homology to LAMP-2A but that does not participate in CMA (Fig. 6d and Supplementary Fig. 13c).
Figure 6. Characterization of the effect of the retinoid derivatives on CMA.
(a) Rates of degradation of long-lived proteins in mouse fibroblasts control or knocked-down (−) for RARα or for LAMP-2A and left untreated (None) or treated with (20μM) the indicated compounds. Values are expressed as fold-change in the proteolytic rate compared to untreated cells for each group. (n=3) (b–d) Mouse fibroblasts control (Ctr) knocked-down (−) for RARα, LAMP-2A or LAMP-2B were transfected with the KFERQ-mcherry1 photoactivable reporter with or without the indicated compounds (20μM). (b) Representative fields and high magnification insets for GR1. Nuclei are labeled with DAPI. Representative fields for GR2 and AR7 are shown in Supplementary Figure 13. (c, d) Average number of fluorescent puncta per cell quantified in > 50 cells in at least 4 different fields. No puncta were detected in LAMP-2A(−) cells. (e) Mouse fibroblasts transfected with the KFERQ-mcherry1 photoactivable reporter with or without the indicated concentrations of AR7, GR1 or with both compounds to reach the same final concentration, as indicated. Top: Representative fields and high magnification insets. Nuclei are labeled with DAPI. Bottom: Quantification of the number of fluorescent puncta per field in each condition. Values are mean±s.em. (n > 50 cells). All values are mean±S.E. Differences with untreated samples (*) or between single and combined treatments (§) are significant for p<0.01.
To elucidate the basis for the differences in the antagonistic effect on RARα activity of our new retinoid derivatives when compared with other well-characterized antagonist molecules, we performed molecular docking and molecular dynamics simulations of the three leading compounds using the RARα X-ray crystal structure in the inactive conformation31. Docking studies suggest that our three leading compounds may bind with two different orientations (pose I and pose II) in the ligand-binding site of RARα (Supplementary Fig. 14). In both docking orientations, none of the three compounds interact or are close to the catalytic Arg272, a feature common to ATRA and other known agonists and antagonists. The binding site in docking pose I is formed by residues of helices 3 (h3), h10 and h12 and in pose II by residues of h3, h10 and h5 (Supplementary Fig. 14). As a result, the compounds adopt opposite orientations in poses I and II and the interacting residues overlap. Docking also revealed that both E- and Z-isomers of GR1 and GR2 can bind in poses I and II and that isomerization has a minimal impact on the docked structure of these compounds, suggesting that both isomers might be active RARα antagonists (Supplementary Fig. 15). However, to determine if they are both active and have comparable potency, future studies with purified isomers are needed.
Analysis of docking and molecular dynamics simulations (Supplementary Fig. 14) suggests that the compounds adopt the binding mode shown in Figure 4b–d. The compounds are at the junction of h12, h3 and h10 in orientations that mimic a portion of BMS614 when it is bound to RARα13. The compounds form extensive hydrophobic interactions with the site, stabilizing the open conformation of h12 that regulates recruitment of RARα coregulators. As shown in Supplementary Figure 16, compounds would have steric clashes with h12 in the closed conformation when RARα is active. Additionally, docking suggests that the guanidinium group of compounds GR1 and GR2 can form hydrogen bonds with the carbonyl backbone of Pro407 in h12 and the hydroxyl group of Thr233 in h3. Thus, docking positions the compounds in a critical region of the RARα-binding site where they likely stabilize its inactive conformation. This binding region is distinct from the ATRA-binding site, which may account for the selectivity of the compounds for RARα.
The small size of our lead molecules compared to the RARα-binding site and the complementarity of hydrophobic interactions between the compounds and the RARα residues in the two docking orientations led us to hypothesize that AR7 and the GR compounds could bind simultaneously to the RARα-binding site. We found a marked increase in CMA-activating potency when AR7 and GR1 were combined, supporting their cooperative effect (Fig. 6e).
CMA activators protect against proteotoxicity
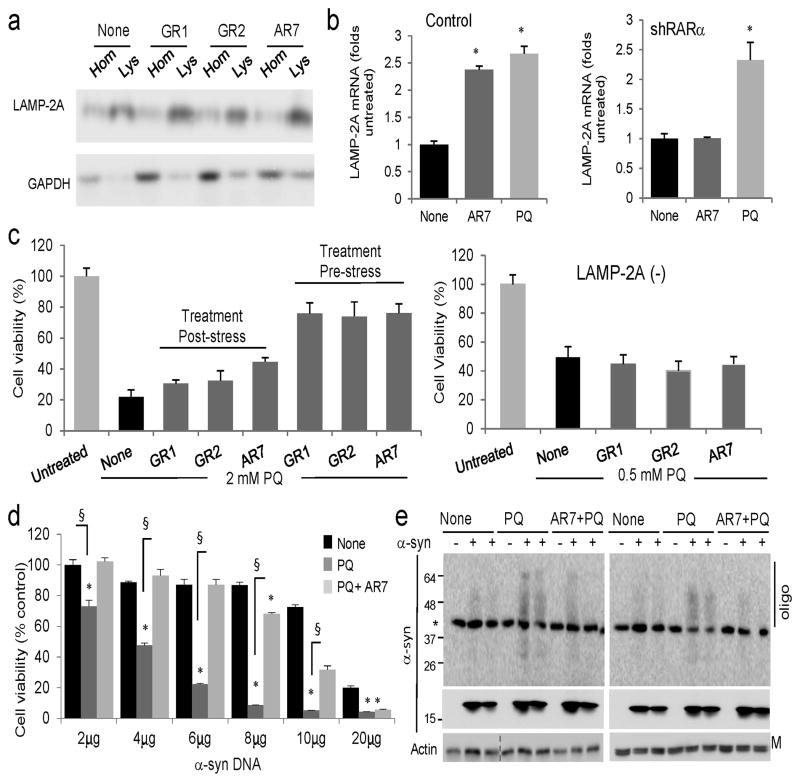
To gain insight into the mechanism by which the retinoid derivatives activate CMA, we analyzed their effect on the cellular oxidative status, because RA exerts antioxidant effects in a variety of cellular settings35. Inhibition of RARα signaling could enhance oxidative stress, a condition known to activate CMA 36. However, we did not find differences in the amount of oxidized proteins after treatment with the different retinoid derivatives (Supplementary Fig. 17a). We also ruled out a direct effect of the compounds on the lysosomal compartment, because pretreatment of intact lysosomes isolated from rat liver with the retinoid derivatives did not increase binding or uptake of the well-characterized CMA substrate glyceraldhehyde-3-phosphate dehydrogenase (GADPH) in these lysosomes in a standard in vitro assay for CMA (Supplementary Fig. 17b). In contrast, lysosomes isolated from cells treated with the retinoid derivatives had a higher content of endogenous GAPDH, suggesting that the compounds activate CMA through RARα signaling and not by direct interaction with CMA components in lysosomes (Fig. 7a).
Figure 7. Effect of the retinoid derivatives in the cellular response against different stressors.
(a) Immunoblot for the indicated proteins in homogenates (Hom) and lysosomes (Lys) isolated from cells untreated (none) or treated for 12 h with 20μM of the indicated compounds. (b) mRNA levels of LAMP-2A in mouse fibroblasts control (Ctr) or knocked down (−) for RARα, and treated with AR7 or paraquat (PQ) as in a (n=4–5). (c) Cellular viability of control (left) or LAMP-2A(−) (right) fibroblasts exposed to 2 mM or 0.5 mM PQ, respectively, and treated with the indicated compounds for 12 h before or after the PQ treatment. (n=3). (d) Viability of mouse fibroblasts transfected with the indicated concentrations of a plasmid encoding α-synuclein and left untreated (none) or treated with 1 mM PQ alone or in the presence of 20μM AR7. (n=3). (e) Immunoblot for α-synuclein in the same cells as d. Top: higher exposure blot to highlight oligomeric (Oligo) species. *nonspecific band. M: monomer; All values are mean±S.E. Differences with cells untreated (*) or treated only with PQ (§) were significant for p<0.001. Full-length blots are shown in Supplementary Figure 21.
We did not observe significant differences in RARα nuclear translocation in response to ATRA in the presence or absence of our retinoic derivatives (Supplementary Fig. 18), ruling out a possible inhibitory effect. Treatment with the transcriptional repressor Actinomycin D partially reduced the stimulatory effect of AR7 on CMA, consistent with transcriptional changes contributing to the upregulation of CMA (Supplementary Fig. 19a).
In our search for CMA targets modulated by the retinoid derivatives, we focused on the lysosomal receptor LAMP-2A, as it is limiting for CMA. We found higher levels of this receptor in the lysosomes from the treated cells (Fig. 7a) along with a discrete but significant transcriptional activation of LAMP-2A in these cells, comparable to that previously described in conditions of maximal activation of CMA such as paraquat (PQ)-induced oxidative stress (Fig. 7b). This transcriptional upregulation was not observed for other lysosomal membrane proteins (Supplementary Fig. 19b), was partially inhibited by treatment with Actinomycin D (Supplementary Fig. 19c) and was no longer observed in the absence of the RARα (Fig. 7b). We did not find changes in LAMP-2A mRNA levels upon stimulation of RARα activity with ATRA under basal conditions (Supplementary Fig. 19dd). However, LAMP-2A mRNA significantly decreased when ATRA was added to cells deprived of serum (Supplementary Fig. 19d), a condition in which ATRA treatment reduced CMA activity (Fig. 3f). We anticipate that the stimulatory effect of the retinoid derivatives on CMA results from the combination of multiple effects that can be elucidated in future investigations. However, it was reassuring to identify that an essential component of CMA, LAMP-2A is one of the possible targets.
Lastly, we determined the possible beneficial effect of chemically enhancing CMA with the retinoid derivatives in cellular homeostasis and resistance to stress. We compared sensitivity of cells to the pro-oxidant PQ when they were treated with the retinoid derivatives before or right after the oxidative insult. As shown in Figure 7c addition of retinoid derivatives to cells exposed to PQ for 4 hours had only a very discrete positive effect on cellular viability. In contrast, when the compounds were added before inducing oxidative stress, we observed a marked improvement in cellular viability (Fig. 7c). The enhanced resistance to the oxidative insult in retinoid-treated cells was completely abolished in cells unable to carry out CMA (knocked down for LAMP-2A; Fig. 7c: note that one-fourth of the concentration of PQ used in control cells was used in the CMA-incompetent cells because of their sensitivity to oxidative stress).
Sustained chronic oxidation is a common feature of aging and an aggravating factor in multiple degenerative disorders. To model the detrimental effect of oxidative stress on proteotoxicity and analyze the possible beneficial effect of enhancing CMA activity in these conditions, we used cells transfected with α-synuclein, a protein that accumulates as intracellular aggregates in both familial and idiopatic forms of PD and was previously identified as a bona fide CMA substrate9,10. Different intracellular factors and aggressors, including oxidative stress, have been proposed to contribute toα-synuclein deposition. Transfection of cultured cells with increasing concentrations of cDNA coding for α-synuclein did not result in toxicity until concentrations above 10μg (Fig. 7d), but the toxicity-induced by a fixed concentration of PQ clearly depended on the amount of α-synuclein in a cell. We adjusted the concentration of PQ so that it decreased viability in cells expressing low levels of α-synuclein no more than 20%. The same concentrations of PQ reduced cell viability up to 80% in cells expressing higher concentrations of α-synuclein (Fig. 7d). The toxic effect in our experimental paradigm resulted from the combination of the oxidative stress and the proteotoxicity associated withα-synuclein, rather than to DNA toxicity, because the effect of PQ remained constant in cells transfected with increasing concentrations of an empty plasmid (Supplementary Fig. 20). Activation of CMA, by pre-treatment of α-synuclein-transfected cells with the retinoid derivatives (Fig. 7d) before addition of PQ, increased cellular viability, even in cells expressing very high levels of α-synuclein. Pretreatment with the compounds also reduced the formation of oligomeric species of α-synuclein in the cells expressing α-synuclein and treated with PQ (Fig. 7e).
These results are consistent with the upregulation of CMA having a beneficial effect in the defense against cellular oxidative stress and proteotoxicity and confirm the ability of the retinoid derivatives to activate CMA even under pathological conditions.
Discussion
Despite recent findings showing compromise of CMA in neurodegenerative diseases, diabetes and lysosomal storage disorders9,11,12, and the pronounced beneficial effect observed when the age-dependent decline of this pathway is prevented15, chemical modulators of CMA have for the most part been lacking. One of the main limitations has been the absence of chemical targets for this pathway since most of the key components in this pathway are multifunctional proteins that participate in many cellular processes, which makes targeting them for specific modulation of CMA challenging. LAMP-2A, the most unique component for CMA, is a difficult target due to its high homology (almost 85% identity) with the other spliced variants of the lamp2 gene, which are known to participate in other cellular functions such as macroautophagy, lysosomal biogenesis and cholesterol trafficking37.
We have identified the RARα receptor as a novel regulator of CMA activity amenable for chemical targeting. Disruption of signaling through this receptor has a stimulatory effect on CMA but it leads to inhibition of macroautophagy. Using structure-based chemical design, we have been able to dissociate the opposing effects of RARα on macroautophagy and CMA and to generate compounds capable of antagonizing only the CMA inhibitory effect, without affecting macroautophagy.
The antagonistic effect of the retinoid derivatives likely results from a combination of tight binding to RARα (favored by the multiplicity of contact sites within the binding pocket) and high stability (obtained by protecting the sites in retinoids usually amenable to intracellular modification). For example, in GR1 and GR2, the polyene linker and the hydrophobic ring are predominantly surrounded by hydrophobic side chains that protect this region, and the interaction with RARα residues Thr233, Ser229 and Pro407 may contribute to the resonance stabilization of the guanidine type polar moiety (Fig. 4d, e). Similarly, in the case of AR7, the bulky aromatic rings offer excellent interaction contacts with the surrounding hydrophobic RARα residues such as Met406, Leu266, Leu398 and Ile270 (Fig. 4g). Future structural and mutagenesis studies on RARα will help to identify the precise binding orientation of each compound, the bound RARα conformation and the contribution of each RARα residue to the antagonistic effect of the retinoid derivatives generated in this study.
Both gene activation and gene repression have been described to occur through the RAR family, and examples of redundant and type-specific functions for each of the members of this family have been reported17. This functional diversity suggests that the interaction of RAR molecules with their targets is probably modulated by multiple factors, including unique characteristics of the ligands to this receptor. We have exploited this last property and introduced chemical modifications in the ligands to favor the effect of RARα on a particular subset of targets that in turn leads to the selective activation of CMA.
Reciprocal crosstalk between macroautophagy and CMA has been previously reported, and this relationship is responsible for the compensatory activation of one of these pathways when the other malfunctions5. However, activation of CMA by the retinoid derivatives generated in this study is not reactive to blockage of macroautophagy or to a highly oxidative intracellular environment. Instead, upregulation of CMA occurs selectively, and in contrast to the receptor-independent effects described for ATRA on macroautophagy, CMA activation depends on a functional RARα. The identification of the specific mechanism behind CMA activation is complex and goes beyond the scope of the current study. However, we present evidence that LAMP-2A is one of the downstream targets of this pathway. The facts that the LAMP-2A gene does not contain a recognizable RA-response element and that our retinoid derivatives suppressed rather than activated a reporter with that sequence suggest that transcriptional activation of LAMP-2A is under negative control by RARα. It is noteworthy that although most lysosomal proteins are under a common transcriptional program controlled by TFEB38, LAMP-2A is one of the few exceptions. RARα signaling is thus the first signaling mechanism shown to regulate this lysosomal receptor. Interestingly, ChIP-Seq analysis have revealed that one of the two largest functional classes of RAR target genes was related to proteolysis39. Our study further reinforces the importance of RAR signaling in protein degradation.
The protective effect against oxidation and proteotoxicity observed in response to the retinoid derivatives supports a possible therapeutic potential of these or related compounds in chronic age-related diseases. Maintenance of protein homeostasis is achieved through a tightly coordinated balance between chaperones and proteolytic systems. It is thus essential to develop interventions that can separately affect one of these processes without compromising the functionality of the other. The efficient upregulation of CMA observed with the retinoid derivatives and their lack of noticeable effects on macroautophagy makes them suitable for the selective modulation of CMA in contexts where this pathway is primarily compromised, such as neurodegeneration and aging.
Online Methods
Animals, cells and reagents
Adult male Wistar rats (Charles River Laboratories) fasted for 48h before sacrifice were used for isolation of lysosomes from liver. All animal work was approved and performed according to the guidelines set by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee. Mouse fibroblasts (NIH3T3) from the American Type Culture Collection were cultured in Dulbecco’s Modified Eagle’s Medium (Sigma) in the presence of 10% newborn calf serum. Serum removal was performed by thoroughly washing the cells with Hanks’ Balanced Salt Solution (Invitrogen) and placing them in serum-free complete medium. Where indicated, cells were treated with the macroautophagy inhibitor 3-methyladenine (Sigma) at a final concentration of 10mM or with 20mM NH4Cl and 100μM leupeptin (Fisher BioReagents) to inhibit lysosomal proteolysis. Where indicated, paraquat was added directly to the culture media to induce oxidative stress. Stable knock-down of LAMP-2A, LAMP-2B or RARα was obtained using vector-mediated stable RNA interference (RNAi) directed specifically against the LAMP-2A or LAMP-2B exon as described previously34 or against the two following regions of RARα: GAAAGTCTACGTCCGGAAA and GCAGCAGTTCCGAAGAGAT. The plasmid encoding for mCherry-GFP-LC3 was from Addgene and for α-synuclein was a generous gift from Dr. Esther Wong (Nanyang Technological University, Singapore). Sources of chemicals and antibodies were as described previously8,9,30,36. ATRA was purchased from Sigma, BMS614 and AM580 were from Tocris Bioscience and the antibody against RARα was from Cell Signaling.
Autophagic measurements
1) Intracellular protein degradation was measured by metabolic labeling and pulse chase experiments40. Briefly, confluent cells labelled with [3H]leucine (2μCi/ml) (NEN-PerkinElmer Life Sciences) for 48h were extensively washed and maintained in medium with an excess of unlabeled leucine. Aliquots of the medium taken at different times were precipitated in trichloroacetic acid and proteolysis measured as the amount of acid-precipitable radioactivity transformed in acid-soluble radioactivity at each time. The amount of lysosomal proteolysis was calculated by treating parallel wells with 20mM NH4Cl and 100μM leupeptin during the chase period. 2) Autophagic flux was measured as changes in levels of LC3-II upon inhibition of lysosomal proteolysis29 and as the ratio mcherry-positive puncta to double labelled (mCherry-GFP) puncta in cells transfected with the mCherry-GFP-LC3 reporter41. 3) CMA activity was determined using the photoactivable KFERQ-PA-mCherry1 reporter that allows visualization of CMA activation as an increase in the number of fluorescent puncta per cell 30. Briefly, cells transduced with a lentivirus carrying the KFERQ-bearing constructs were photoactivated by exposure to a 3.5mA (current constant) light emitting diode (LED: Norlux, 405nm) for 10min and at the desired times fixed in 3% formaldehyde and images were captured with an Axiovert 200 fluorescence microscope (Zeiss) with apotome and equipped with a 63x 1.4 NA oil objective lens and red (ex. 570/30 nm, em. 615/30 nm), cyan (ex. 365/50 nm and em. 530/45 nm) and green (ex. 475/40 nm and em. 535/45 nm) filter sets (Chroma). All images were acquired with a high-resolution CCD camera after optical sectioning through the apotome. Images were prepared using Adobe Photoshop 6.0 software (Adobe Systems). Quantification was performed in individual frames after deconvolution and thresholding using ImageJ software (NIH) in a minimum of 50 cells. 4) Analysis of CMA in isolated lysosomes was performed using a previously developed in vitro assay to measure the ability of intact lysosomes to take up and degrade well-characterized CMA substrate proteins40. Briefly, intact lysosomes treated or not with a pool of protease inhibitors for 10 min on ice, were incubated with glyceraldheyde-3-phosphate dehydrogenase (GAPDH) for 20 min at 37oC. At the end of the incubation, lysosomes were recovered by centrifugation, washed, and subjected to SDS-PAGE and immunoblot for GAPDH. Binding was calculated after densitometric analysis as the amount of GAPDH recovered in the lysosomes not treated with protease inhibitors, and uptake as the difference in the amount of GAPDH recovered in treated minus untreated lysosomes. Lysosomes active for CMA were isolated from rat liver using differential centrifugation and floatation in discontinuous density metrizamide gradients following a previously optimized procedure42. Lysosomes from cultured cells were isolated by similar procedures but using instead discontinuous metrizamide/percoll gradients after rupture of the plasma membrane through nitrogen cavitation34.
Design and synthesis of novel-retinoid derivatives
Preparation of atypical retinoids (Supplementary Fig. 8 and Table 1)33: The 2H-benzo[b][1,4] oxazines were synthesized by modifying the existing methods43. 2-bromo-4-chloroacetophenone (0.01mol) in dichloromethane was added drop-wise to a solution of 2-aminophenol in dichloromethane, aqueous potassium carbonate (20% w/v) and tetrabutylammonium hydrogen sulphate (0.0005 mol). The resulting mixture was refluxed till completion for 4–6 h and the organic layer was extracted with dichloromethane and dried over sodium sulphate evaporated in vacuum to give a crude solid product. The solid was then recrystallized with hot ethanol to obtain pure yield 87–95%. Compound AR7 (1): 1H NMR (300 MHz, CDCl3): δ = 2.44 (s, 3H), 5.09 (s, 2H), 6.91–6.98 (m, 2H), 7.12 (d, J = 12Hz, 2H), 7.33–7.38 (d, J = 8 Hz, 1H), 7.82–7.84 (d, J = 12 Hz, 2H); 13C NMR (75 MHz, CDCl3): δ = 21.56, 62.82, 76.58, 115.94, 122.46, 126.41, 128.34, 132.15, 132.44, 133.02, 141.97, 146.82, 158.94; HR FT-ICR MS: calcd for C15H12ClNO ([M+H]+) 258.0641 Found: 258.0702.
Preparation of guanidine retinoids (Supplementary Fig. 8 and Table 1)32: Solid sodium chloride obtained from reacting guanidine hydrochloride (2mmol) in DMF:dioxane (1:1) and sodium tert-butoxide (2mmol) under nitrogen at 50–55 °C for 30 min was filtered and the filtrate was added to a solution of retinoid and CDI in DMF stirred at room temperature for 1h. The progress of the reaction was monitored by TLC, the solid product was collected by filtration and washed with cold water to remove excess guanidine. Compound GR1 (2): N-[4-(3,5,5-trimethyl-cyclohex-2-enylidenemethyl)-benzoyl]-guanidine. 1H NMR (300 MHz, CDCl3): δ = 0.88 (s, 6H), 1.78 (s, 3H), 1.91 (s, 2H), 2.33 (s, 2H), 2.48–2.53 (m, 4H), 2.74 (s, 1H), 2.90 (s, 1H), 3.52 (s, 1H), 6.01 (s, 1H), 6.32 (s, 1H), 7.24–7.28 (d, J = 12 Hz, 2H), 8.01–8.05 (d, J = 12 Hz, 2H); 13C NMR (75 MHz, CDCl3): δ = 25.0, 29.1, 31.2, 45.1, 126.0, 127.3, 128.8, 129.5, 137.6, 137.9, 138.0, 140.6, 163.2, 176.5; HR-MS: (C18H24N3O) calcd ([M+H]+) 298.1921; found 298.1928. Compound GR2 (3): N-[4-(3-methyl-cyclohex-2-enylidenemethyl)-benzoyl]-guanidine. 1H NMR (300 MHz, CDCl3): δ = 1.57–1.1.70 (m, 2H), 1.80 (s, 3H), 2.03–2.13 (m, 2H), 2.48–2.60 (m, 6H), 6.01 (s, 1H), 6.20 (s, 1H), 7.23–7.27 (d, J = 12 Hz, 2H), 7.98–7.02 (d, J = 12 Hz, 2H); 13C NMR (75 MHz, CDCl3): δ = 23.2, 24.7, 26.8, 30.7, 124.5, 127.9, 128.8, 129.3, 137.4, 139.3, 140.2, 140.7, 142.7, 163.8, 176.4; HR-MS: (C17H20N3O) calcd ([M+H]+) 270.1606; found 270.1614. In the NOE analysis, E/Z isomers of GR1 were assigned based on a pair of weak vinyl peaks at 6.4ppm and 6.12ppm (Z-isomer) and another pair of strong vinyl peaks at 6.27ppm and 5.98ppm (E-isomer). For GR2, E/Z isomers were assigned based on a pair of weak vinyl peaks at 6.43ppm and 6.12ppm (Z-isomer) and another pair of strong vinyl peaks at 6.19ppm and 6.00ppm (E-isomer).
Preparation ofα-aminonitrile functionalized constrained retinoids (Supplementary Table 1)31: β-cyclocitral (1.0 mmol), amine (1.0 mmol), TMSCN (1.2 mmol), H2O (2 mL), and InCl3 (0.1 mmol) were added sequentially into a 10 ml round-bottomed flask. The reaction mixture was stirred vigorously for 4–6 h at room temperature and the progress of the reaction was monitored by TLC. The resulting solid was filtered and washed with water and hexane to yield the desired product retinoids (αAmR1 -αAmR11).
Preparation of boron-α-aminonitrile functionalized constrained retinoids (Supplementary Table 1 and note I): aldehyde (1.0 mmol), amine (1.0 mmol), TMSCN (1.2 mmol), H2O (2 mL), and InCl3 (0.1 mmol) were added sequentially into a 10 ml round-bottomed flask. The reaction mixture was stirred vigorously for 4–6 h at room temperature and the progress of the reaction was monitored by TLC. The resulting solid was filtered and washed with water and hexane to yield the desired product retinoids (BAmR1–BamR6; 4–9).
The characterization of the 29 compounds synthesized in this study matches the standards previously published 31–33 or is shown for the first time in Supplementary note I.
In silico docking
AR7, GR1 and GR2 structures were drawn in ChemDraw Ultra 12.0 and converted to three-dimensional all-atom structures from sdf format using LigPrep (Version 2.5, Schrödinger, LLC, New York, NY, 2011). For each ligand a maximum of 4 stereoisomers were generated, ionization states and tautomers were generated for pH 7 and pH 2 and geometries optimized and energy minimized before for docking. The structure of the RARα-RXR hetero-dimer in complex with the small molecule antagonist BM614 (PDB ID: 1DKF), was used for docking and molecular dynamics. The RARα-RXR structure was prepared using MAESTRO protein preparations module (Version 9.2, Schrödinger, LLC, New York, NY, 2011). The structure of the antagonist was removed from the RARα site, water molecules at a distance of more than 5 Å from heteroatoms were removed, all missing protons were generated, hydrogens were optimized for best hydrogen bonding network bonds and formal charges were assigned and structure was gently minimized by restrained energy minimization. The ligand-binding pocket was defined within 5 Å of the BMS614 pose and receptor grid size and center was generated based on the position and the size of the BMS614. To account for receptor flexibility in docking, scaling of van der Waals’ radii of non polar atoms with the absolute value of the partial atomic charge less than or equal to 0.25 for protein atoms was set to 1 and for ligand non polar atoms atoms with partial atomic charges less than or equal to 0.15 was set to 0.8. Docking was performed in ligand flexible mode using Glide 44–46 (Version 5.8, Schrödinger) using the extra precision (XP) mode. All three molecules were docked into the BMS614 binding site with and without rotatable binding site hydroxyl-groups. Structures were analyzed using MAESTRO and PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.)
Molecular Dynamics Simulations
The lowest-energy docked structures for each ligand were minimized using the Desmond molecular dynamics system 47–49 (Version 3.0, D. E. Shaw Research, New York, NY, 2011. Maestro-Desmond Interoperability Tools, version 3.1, Schrödinger, New York, NY, 2012). All minimization and simulation systems were set up using MAESTRO tools. The minimization protocol was performed using an OPLS-2005 force field with a cubic-TIP3P water-box, 150 mM NaCl and water-box boundaries set to a minimum of 10 Å from the proteins surface in all directions. Minimization was performed with a maximum of 2000 iterations with a convergence threshold of 1 kcal/mol/Å, a steepest descent method was used initially until a gradient threshold of 25 kcal/mol/Å is reached. Simulated annealing and molecular dynamics in explicit water were performed on the refined lowest energy docked structures, using the Desmond molecular dynamics system and OPLS-2005 force field. A five stage simulated annealing protocol was performed with 40 ps intervals where the temperature was linearly interpolated between each time point with temperature steps of 10, 100, 300, 400, and 300 K. An NVT ensemble class was employed with a Berendsen thermostat with a relaxation time of 1 ps. A 6 ns molecular dynamics simulation was performed with and without simulated annealing for each of the minimized lowest-energy docking possess. Molecular dynamics simulations were performed using an NPT ensemble class at 300 K and 1.01325 bar. A Nose-Hoover chain thermostat method with a 1 ps relaxation time and a Martyna-Tobias-Klein Barostat Method with a 2ps relaxation time were used. All docking and molecular dynamic simulations were analyzed using tools within MAESTRO and figures were prepared using PyMOL.
Measurement of RARa and RXR activity
A RAR-responsive luciferase construct was utilized to monitor the activity of RARα in cultured cells. Cells were co-transfected with pCMX-Gal-L-hRARα and a construct encoding the firefly luciferase reporter gene under the control of a minimal promoter and tandem repeats of the retinoic-acid response element (tk-px3-luc). Co-transfection with a renilla luciferase reporter plasmid was performed to control for efficiency of transfection. RXR activity was measured by similar procedures using co-transfection with pCMX-mRXR and tk-apoA1-luc. Compounds were added 24h after transfection and 48h later cells were lysed and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Progmega). Luciferase values were normalized to the renilla luciferase reporter.
General methods
Protein concentration was measured by the Lowry method using bovine serum albumin as a standard. Cell viability was determined using the Cell Titer Blue Kit (Promega). Carbonyl groups were detected with the OxyBlot Oxidized Protein Detection Kit (Chemicon International). Apoptosis was determined with Annexin V-PE apoptosis detection kit (BD Pharmingen). Lentiviral particles were generated by co-transfection with the lentiviral transfer vector carrying the hairpin sequence against the desired mRNA and the third-generation packaging constructs pMDLg/pRRE and pRSV-REV, and as envelope the G glycoprotein of the VSV (pMD2.G) into HEK293T cells as described before34. Cultured cells were transduced by addition of packed virus at a titter of 2.63×106 units/ml. After SDS-PAGE and immunoblotting, the proteins recognized by the specific antibodies were visualized by chemiluminescence methods (Western Lightning; PerkinElmer) using peroxidase-conjugated secondary antibodies. Densitometric quantification of the immunoblotted membranes was performed using Image J (NIH). Quantitative real time PCR was used to determine changes in mRNA levels using the TaqMan One-Step RT-PCR Master Mix reagent (Applied Biosystem). Fluorescence was performed using conventional procedures and all images were captured with an Axiovert 200 fluorescence microscope (Zeiss) with apotome.
Statistical analysis
All numerical results are reported as mean + S.E., and represent data from a minimum of three independent experiments unless otherwise stated. Statistical significance of difference between groups was determined in instances of single comparisons by the two-tailed unpaired Student’s t-test of the means. In instances of multiple means comparisons, we used one-way analysis of variance (ANOVA) followed by the Bonferroni post-hoc test to determine statistical significance. Statistic analysis was performed in all the assays and significant differences are noted in the graphical representations.
Supplementary Material
Acknowledgments
We thank Dr. Rut Valdor for technical assistance with the luciferase assay, Dr. Roberta Kiffin for assistance with the quantitative RT-PCR, Dr. Fernando Macian for help with FACs procedures, Dr. Todd Evans and Ms. Ingrid Torregroza for advice with the RARα luciferase assay, Dr Clare-Louise Towse for advice on the simulated annealing and molecular dynamics simulations, and Dr. Susmita Kaushik for critically reviewing this manuscript. This work was supported by grants from the NIH/NIA AG021904 and AG031782 (to A.M.C.), Albert Einstein College of Medicine start-up funds and NIH/NHLBI HL095929 (to E.G.), NIH/NIAAA AA020630 (to B.D), and by the Rainwaters Foundation, the Beatrice and Roy Backus Foundation and a Robert and Renee Belfer gift (to A.M.C.).
Footnotes
Author Contributions
J.A. performed the experiments, analyzed the data and contributed to writing the paper; T.P.G. performed the in silico docking and molecular dynamic simulations; M.M. contributed to the synthesis of chemical compounds; B.D. designed the chemical compounds, analyzed the chemical data and revised the manuscript; E.G. designed and directed the in silico docking and molecular dynamic simulations, contributed the interpretation of the chemical data and to the writing and revising of the manuscript; A.M.C. designed the biological experiments, directed the study and wrote the manuscript.
Competing financial interests
The authors declare that they have no competing interests.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N. Autophagy in Protein and Organelle Turnover. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2011.76.011023. [DOI] [PubMed] [Google Scholar]
- 4.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nature Neuroscience. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2010;23:184–9. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 7.Chiang H, Terlecky S, Plant C, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 8.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–63. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–5. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 10.Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, Di Monte DA. Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem. 2010;285:13621–9. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–70. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–44. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 13.Venugopal B, et al. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–353. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 14.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–65. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn P, Mesires N, Vine M, Dice JF. Effects of small molecules on chaperone- mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 17.Duong V, Rochette-Egly C. The molecular physiology of nuclear retinoic acid receptors. From health to disease. Biochim Biophys Acta. 2011;1812:1023–31. doi: 10.1016/j.bbadis.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Kon M, et al. Chaperone-mediated autophagy is required for turmor growth. Sci Trans Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frolik CA, Roller PP, Roberts AB, Sporn MB. In vitro and in vivo metabolism of all-trans- and 13-cis-retinoic acid in hamsters. Identification of 13-cis-4-oxoretinoic acid. J Biol Chem. 1980;255:8057–62. [PubMed] [Google Scholar]
- 20.Rochette-Egly C, Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs) Nucl Recept Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Lera AR, Bourguet W, Altucci L, Gronemeyer H. Design of selective nuclear receptor modulators: RAR and RXR as a case study. Nat Rev Drug Discov. 2007;6:811–20. doi: 10.1038/nrd2398. [DOI] [PubMed] [Google Scholar]
- 22.Njar VC, et al. Retinoic acid metabolism blocking agents (RAMBAs) for treatment of cancer and dermatological diseases. Bioorg Med Chem. 2006;14:4323–40. doi: 10.1016/j.bmc.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 23.Das BC, et al. Design and Synthesis of 3,5-Disubstituted 1,2,4-Oxadiazole Containing Retinoids from a Retinoic Acid Receptor Agonist. Tetrahedron Lett. 2011;52:2433–2435. doi: 10.1016/j.tetlet.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das BC, McCartin K, Liu TC, Peterson RT, Evans T. A forward chemical screen in zebrafish identifies a retinoic acid derivative with receptor specificity. PLoS One. 2010;5:e10004. doi: 10.1371/journal.pone.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isakson P, Bjoras M, Boe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010;116:2324–31. doi: 10.1182/blood-2010-01-261040. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARalpha oncoprotein. Autophagy. 2011;7:401–11. doi: 10.4161/auto.7.4.14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trocoli A, et al. ATRA-induced upregulation of Beclin 1 prolongs the life span of differentiated acute promyelocytic leukemia cells. Autophagy. 2011;7:1108–14. doi: 10.4161/auto.7.10.16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajawat Y, Hilioti Z, Bossis I. Retinoic acid induces autophagosome maturation through redistribution of the cation-independent mannose-6-phosphate receptor. Antioxid Redox Signal. 2011;14:2165–77. doi: 10.1089/ars.2010.3491. [DOI] [PubMed] [Google Scholar]
- 29.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 is a Marker for Autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 30.Koga H, Martinez-Vicente M, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Comm. 2011;2:386. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das BC, Anguiano J, Mahalingam SM. Design and Synthesis of α-Aminonitrile Functionalized Novel Retinoids. Tetrahedron Lett. 2009;50:5670–5672. [Google Scholar]
- 32.Das BC, et al. Design and Synthesis of Potential New Apoptosis Agents: Hybrid Compounds Containing Perillyl Alcohol and New Constrained Retinoids. Tetrahedron Lett. 2010;51:1462–1466. doi: 10.1016/j.tetlet.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das BC, Madhukumar AV, Anguiano J, Mani S. Design, synthesis and biological evaluation of 2H-benzo[b][1,4] oxazine derivatives as hypoxia targeted compounds for cancer therapeutics. Bioorg Med Chem Lett. 2009;19:4204–6. doi: 10.1016/j.bmcl.2009.05.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahlemeyer B, et al. Retinoic acid reduces apoptosis and oxidative stress by preservation of SOD protein level. Free Radic Biol Med. 2001;30:1067–77. doi: 10.1016/s0891-5849(01)00495-6. [DOI] [PubMed] [Google Scholar]
- 36.Kiffin R, Christian C, Knecht E, Cuervo A. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eskelinen E, et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–68. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 39.Delacroix L, et al. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231–44. doi: 10.1128/MCB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaushik S, Cuervo AM. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 2009;452:297–324. doi: 10.1016/S0076-6879(08)03619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–15. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 43.Shridhar DR, Reddy CV, Sastry OP, Bansal OP, Rao PP. A convenient one-step synthesis of 3-Aryl-2H-1,4-benzoxazines. Synthesis. 1981;1981:912–913. [Google Scholar]
- 44.Friesner RA, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–96. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 45.Halgren TA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–9. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 46.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 47.Shivakumar D, et al. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J Chem Theory Comput. 2010;6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 48.Guo Z, et al. Probing the alpha-helical structural stability of stapled p53 peptides: molecular dynamics simulations and analysis. Chem Biol Drug Des. 2010;75:348–59. doi: 10.1111/j.1747-0285.2010.00951.x. [DOI] [PubMed] [Google Scholar]
- 49.Bowers KJ, Dror RO, Shaw DE. The midpoint method for parallelization of particle simulations. J Chem Phys. 2006;124:184109. doi: 10.1063/1.2191489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.