Abstract
Vascular development is a dynamic process that relies on the coordinated expression of numerous genes, but the factors that regulate gene expression during blood vessel development are not well defined. ATP-dependent chromatin-remodeling complexes are gaining attention for their specific temporal and spatial effects on gene expression during vascular development. Genetic mutations in chromatin-remodeling complex subunits are revealing roles for the complexes in vascular signaling pathways at discrete developmental time points. Phenotypic analysis of these models at various stages of vascular development will continue to expand our understanding of how chromatin remodeling impacts new blood vessel growth. Such research could also provide novel therapeutic targets for the treatment of vascular pathologies.
Keywords: ATP-dependent chromatin-remodeling complex, Embryo, Angiogenesis, Blood vessel, SWI/SNF, NuRD
Introduction
Developmental programs require exquisite temporal and spatial regulation of gene expression. In the developing vascular system, for example, a carefully orchestrated series of signal transduction events leads to the formation of a primary vascular plexus, which further differentiates into an arborized vascular network comprised of vessels with different sizes and functions. Mutations of single genes within these signaling cascades can result in dysmorphic blood vessels and embryonic lethality [1]. What regulates and coordinates expression of the many genes required for building a functional vascular system? Several transcription factors have been identified that influence expression of genes essential for blood vessel development [2]. In addition, epigenetic regulators are gaining recognition for the important roles they play in modulating gene expression during vascular development [3–6].
Epigenetic regulators affect developmental gene expression by influencing the ability of general transcriptional machinery, transcription factors, and coregulatory proteins to access genomic DNA. Almost 2 m of genomic DNA are tightly packaged into a three-dimensional chromatin structure in order to fit within the confines of each eukaryotic cell nucleus. Nucleosomes, which are the basic subunits of chromatin, consist of 147 base pairs of DNA wrapped around a scaffold of histone proteins. As nucleosomes are further compacted into a final chromatin structure, DNA becomes inaccessible for critical cellular processes like transcription. Epigenetic regulators such as histone-modifying enzymes and ATP-dependent chromatin-remodeling complexes impact DNA accessibility by altering local chromatin structure. Histone-modifying enzymes generate or remove chemical moieties from histone tails, thereby changing the biochemical properties of chromatin and affecting its condensation. ATP-dependent chromatin-remodeling complexes transiently disrupt DNA-histone contacts and slide nucleosomes closer together or further apart. Both types of epigenetic regulators influence the binding of additional regulatory proteins to DNA or histones within promoter or enhancer regions and thereby impact transcription. This review will focus on the role of ATP-dependent chromatin-remodeling complexes in coordinating temporally and spatially specific transcriptional events and will examine their influence on embryonic vascular development.
ATP-dependent chromatin-remodeling complexes
ATP-dependent chromatin-remodeling complexes are comprised of varying numbers of proteins, but each complex contains a catalytic ATPase that drives displacement or compaction of nucleosomes [7]. These catalytic ATPases are related to the yeast Swi2/Snf2 ATPase, which is essential for transcription of genes involved in mating type switching and nutrient metabolism [8–10]. Through ATP hydrolysis, they generate energy required for altering chromatin accessibility by breaking histone-DNA contacts, sliding nucleosomes along DNA, and removing or exchanging nucleosomes. The biological mechanisms underlying these processes are described in several other reviews [7, 11–14].
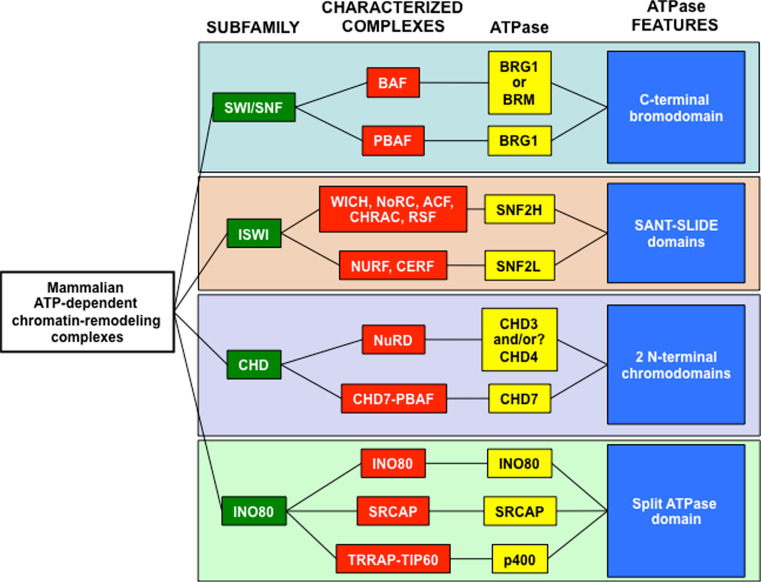
The human genome contains almost 30 Swi2/Snf2-like ATPases, which all share an evolutionarily conserved DEAD/H (Asp-Glu-Ala-Asp/His) box domain [7, 15]. While chromatin-remodeling activity has not been assigned to all of these ATPases, several of them have been described in protein complexes with chromatin-remodeling capacity. These chromatin-remodeling complexes are subclassified into four groups based on the presence of additional functional domains within the ATPases: mating type switching/sucrose nonfermenting (SWI/SNF), imitation switch (ISWI), chromodomain helicase DNA-binding (CHD), and inositol autotroph 80 (INO80) (Fig. 1).
Fig. 1.
Subfamilies of mammalian SWI2/SWF2-like chromatin-remodeling complexes. The four subfamilies of mammalian SWI2/SNF2-like chromatin-remodeling complexes (green) contain well-characterized complexes (red). Each complex contains at lease one ATPase protein (yellow) that is critical for its function. The subfamilies are defined by distinct protein domains contained within the ATPases (blue), some of which mediate interactions with other epigenetic factors. SWI/SWF ATPases posses a bromodomain that binds acetylated histones [84]. ISWI ATPases have a SANT–SLIDE domain that binds histone tails [85]. CHD ATPases contain tandem amino-terminal chromodomains that interact with DNA, RNA, and methylated histone tails [86–88], and INO80 ATPases have a highly conserved ATPase domain that is split by a spacer region [89]
In addition to their catalytic ATPases, mammalian chromatin-remodeling complexes contain varying numbers of non-catalytic proteins. Some complexes contain invariant core proteins that enhance the nucleosome-remodeling activity of the ATPases [16]. However, other non-catalytic proteins are encoded by multiple genes and are mutually exclusive within a given complex [17, 18]. Based on the potential combinations of these alternative proteins, it is estimated that hundreds of mammalian SWI/SNF, CHD, ISWI, and INO80 complexes exist [18].
ATP-dependent chromatin-remodeling complexes play critical roles throughout mammalian development [19–23]. Their influence on transcriptional regulation impacts cellular differentiation in a variety of tissues from the single-cell gamete stage through postnatal developmental processes. Many of these influences have been elucidated through genetic studies on chromatin-remodeling complex proteins in vertebrate organisms. Global knockouts of the ATPases and non-catalytic subunits frequently have early and lethal consequences for developing mouse embryos [24–40]. Conditional knockouts, however, yield tissue-specific phenotypes that shed light on the mechanisms by which chromatin-remodeling complexes regulate distinct developmental processes [41–60]. In the following section, we will highlight the phenotypes that have emerged from the deletion of chromatin-remodeling complex components in developing embryonic vasculature.
Chromatin-remodeling complexes and vascular development
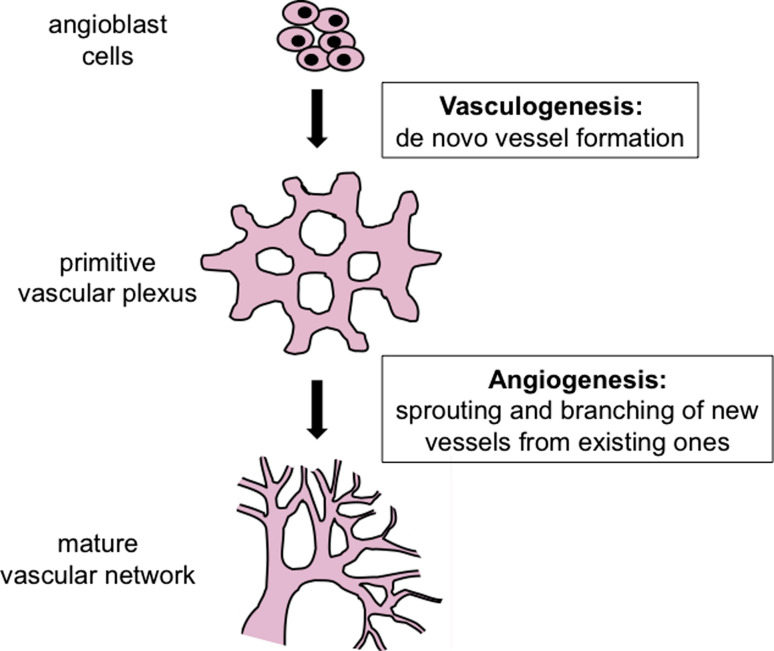
The circulatory system is the first functional organ system to develop in vertebrate embryos. It is required for embryonic survival because blood vessels carry oxygen and nutrients necessary for sustaining rapidly developing tissues. Vascular development is a highly dynamic process that can be subdivided into two stages: vasculogenesis and angiogenesis (Fig. 2). During vasculogenesis, blood vessels arise from progenitor cells that coalesce into a primitive vascular plexus. Angiogenesis refers to the growth of new vessels from pre-existing ones and provides a mechanism by which a primitive vascular plexus expands and differentiates into a mature vessel system.
Fig. 2.
Model of blood vessel development. Blood vessel formation initiates when precursor angioblast cells coalesce to form a primitive vascular plexus through the process of vasculogenesis. A mature vascular network forms through the process if angiogenesis which involves sprouting and branching of pre-existing vessels to form new ones. Both vasculogenesis and angiogenesis are influenced by known genes and signaling pathways [1], and epigenetic mechanisms for regulating those genes are currently those genes are currently being investigated
Both vasculogenesis and angiogenesis are orchestrated by regulatory factors that coordinate cell signaling events and gene expression changes. Genetic studies in mice and zebrafish have confirmed the importance of multiple growth factors (i.e., VEGF, FGF, TGFβ, and PDGFβ) and their receptors in vascular development [1]. Although these and many other genes and signaling pathways are recognized for their roles in developing vessels, little is known about what governs their temporal and spatial expression patterns. Mounting evidence indicates that epigenetic regulators—including chromatin-remodeling complexes—influence the transcriptional regulation of vascular development. For this review, we have catalogued the embryonic blood vessel phenotypes resulting from mutation of chromatin-remodeling complex components (Table 1). We do not summarize the growing number of developmental cardiac phenotypes associated with chromatin-remodeling complex mutants but instead refer the reader to recent reviews [3, 4, 6].
Table 1.
Subunits of chromatin-remodeling complexes that influence embryonic blood vessel development
| Subfamily | Subunit | Mutation | Vascular phenotype | References |
|---|---|---|---|---|
| SWI/SNF | BRG1 | Endothelial cell knockout (Tie2-Cre +) | Yolk sac angiogenesis defects due to downregulated vascular Wnt signaling; hypotrabeculation of the heart due to overexpression of Adamts1; lethality between E10.5-11 | [44, 53, 64] |
| BAF180 | Global knockout | Defective coronary vascular development due to failed EMT and migration of epicardial cells | [66] | |
| BAF155 | Global knockout with partial transgenic rescue | Yolk sac angiogenesis defects | [65] | |
| CHD | CHD4 | Endothelial cell knockout (Tie2-Cre +) | Rescues Brg1 fl/fl :Tie2-Cre + yolk sac angiogenesis defects | [74] |
| ISWI | No blood vessel phenotypes currently described | |||
| INO80 | No blood vessel phenotypes currently described | |||
SWI/SNF
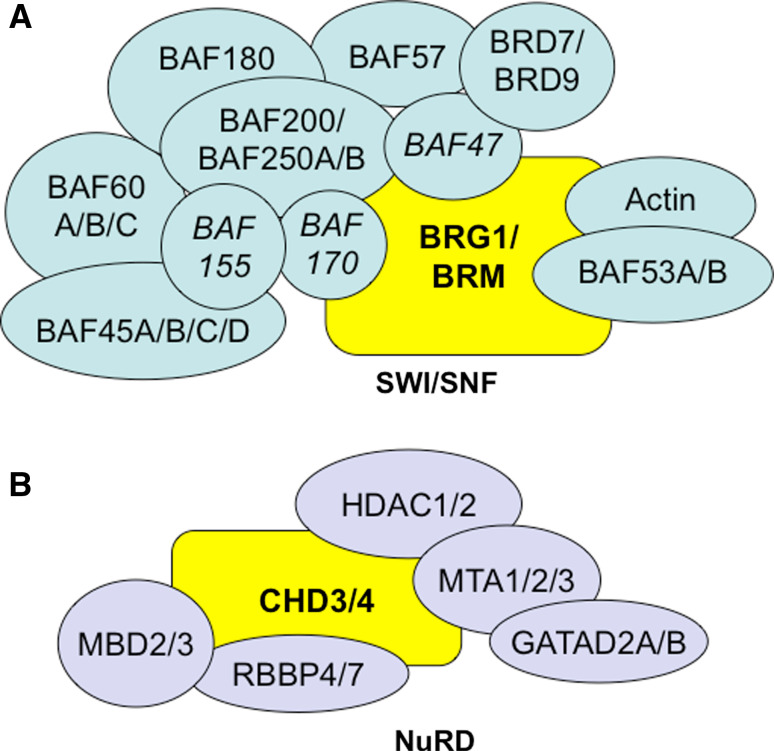
Mammalian SWI/SNF complexes contain one of two mutually exclusive ATPases: BRM and BRG1 (Fig. 3). When targeted for global deletion in mice, the functional distinctions between the ATPases become apparent. Brm −/− mice survive development and are overtly normal except for a slight increase in weight when compared to their control littermates [35]. Brg1 −/− embryos die around implantation, several days before blood vessel development begins [25]. Because BRG1 is upregulated in Brm −/− mice, it is hypothesized that BRG1 can functionally substitute for BRM in SWI/SNF complexes from Brm −/− mice [35]. However, Brg1 −/− lethality demonstrates that BRM cannot functionally substitute for BRG1 at the implantation stage of embryonic development.
Fig. 3.
Mammalian SWI/SNF and NuRD chromatin-remodeling complexes. a SWI/SNF complexes contain one of two core ATPase catalytic subunits, BRG1 or BRM (yelllow). Additional catalytic core subunits include BAF155, BAF170, and BAF47 (italics), which are sufficient for remodeling nucleosomes in vitro [16]. Other non-catalytic subunits, some of which have multiple isoforms, complex with the core subunits in differing cell types to provide additional regulatory functions. b NuRD complexes contain at least one ATPase catalytic subunit, CHD3 or CHD4 (yellow). Additional NuRD subunits include the histone deacetylases 1 and 2 (HDAC1/2) and the methyl-CpG-binding domain proteins 2 and 3 (MBD2/3), which interact with covalent epigenetic marks on histone tails and DNA, respectively
Evidence of a possible role for SWI/SNF complexes in vascular development first arose from expression data reported for their ATPases in developing embryos [61]. By immunofluorescence, BRM is expressed in the developing allantois, the inner layer of the yolk sac and amnion, the developing heart, and the vitelline artery and umbilical veins of the early midgestation embryo. BRM expression overlaps with that of PECAM-1, an endothelial cell marker, specifically in the yolk sac and vitelline artery but not in the dorsal aorta or heart. These data indicate that BRM expression predominates in extraembryonic rather than embryonic vasculature at midgestation (embryonic day 8.5 = E8.5). In the same study, BRG1 is ubiquitously expressed at midgestation. Since vascular development is essential for embryonic survival, and Brm −/− mice survive development, the expression data reported by Dauvillier and colleagues suggest that BRM plays an important role in extraembryonic vascular development but that BRG1 can functionally compensate for BRM in Brm −/− embryos.
To address this hypothesis, a comparison was made between embryos missing Brg1 in vascular endothelial cells and embryos missing both Brg1 and Brm in the vasculature [44]. Floxed Brg1 alleles were deleted from endothelial cells using a Tie2-Cre transgenic line, which is expressed in developing endothelial and hematopoietic cells [62]. Surprisingly, Brg1 fl/fl :Tie2-Cre + embryos display abnormal yolk sac angiogenesis, but concomitant deletion of Brm does not exacerbate the vascular phenotype in Brm −/− ;Brg1 fl/fl :Tie2-Cre + embryos [44]. These data indicate that BRG1 plays its own role in extraembryonic vascular development that is independent of BRM. Indeed, these genetic data do not indicate that BRM plays any recognizable role in midgestational vascular development, so the significance of its restricted expression pattern in developing vascular tissues is unclear.
Brg1 fl/fl :Tie2-Cre + embryos undergo normal yolk sac vasculogenesis; they develop a vascular plexus that is comparable to that seen in control embryos at embryonic day 8.5 (E8.5) [44]. However maturation of the vascular plexus is abnormal in Brg1 fl/fl :Tie2-Cre + yolk sacs. Mutant vessels are thin, disconnected, and display failed sprouting or regression. Notably, vascular patterning and maturation are grossly normal in Brg1 fl/fl :Tie2-Cre + embryos [44, 53], indicating that BRG1 is more important for angiogenesis in extraembryonic tissues than in the embryo proper.
Brg1 fl/fl :Tie2-Cre + embryos eventually die from failed primitive erythropoiesis by E11.5 [44, 53]. Tie2-Cre is expressed in hematopoietic cells as well as endothelial cells, and deletion of Brg1 in developing erythroblasts leads to downregulated embryonic globin expression and subsequent apoptosis [44]. Since yolk sac angiogenesis is partially subject to regulation through hemodynamic forces [63], it is possible that Brg1 fl/fl :Tie2-Cre + yolk sac vascular abnormalities are secondary to failed primitive hematopoiesis. However, more recent work demonstrates that misregulated vascular Wnt signaling plays a key role in the Brg1 mutant yolk sac vascular phenotype and is independent of the hematopoietic phenotype [64]. BRG1 modulates Wnt signaling in yolk sac vasculature at two levels: it promotes transcription of multiple frizzled (Fzd) receptors, and it directly coactivates transcription of certain Wnt target genes. When Wnt signaling is pharmacologically stimulated in vivo, vascular patterning is significantly rescued in Brg1 fl/fl :Tie2-Cre + yolk sacs, although primitive hematopoiesis is still compromised. These data indicate that Brg1 fl/fl :Tie2-Cre + yolk sac vascular abnormalities are predominantly driven by downregulated Wnt signaling rather than reduced hemodynamic forces.
Further support for the role of mammalian SWI/SNF complexes in extraembryonic vascular development comes from deletion of the core non-catalytic subunit Srg3 (Baf155). Global deletion of Srg3 results in embryonic lethality around the implantation stage [30], which is the same stage at which Brg1 −/− embryos die. However, partial rescue of Srg3 −/− embryos with an exogenous, global Srg3 transgene allows the embryos to survive until E11.5 [65]. Srg3 −/− :Tg + embryos are developmentally delayed beginning at E8.5 and harbor numerous morphological defects. Among these, the Srg3 −/− :Tg + yolk sacs have defective vascular development. While Srg3 −/− :Tg + yolk sac vessels undergo normal vasculogenesis to form a primitive plexus, the plexus fails to develop into a mature vascular network. Several genes involved in angiogenesis are downregulated in Srg3 −/− :Tg + yolk sacs by RT-PCR, including members of the Angiopoietin/Tie2, Hedgehog, Eph/ephrin, and Notch signaling pathways. However, genes analyzed within the VEGF pathway are expressed at normal levels, which is consistent with the normal vasculogenesis observed in the mutant yolk sacs. Because the Srg3 −/− :Tg + mutations are global and the mutant embryos display numerous developmental defects, including aberrant yolk sac visceral endoderm morphology, it is difficult to know whether the yolk sac vascular defects are primary or secondary in nature. A conditional deletion of Srg3 in embryonic vasculature would be important for determining whether SRG3 plays a direct role in yolk sac vascular development. Nevertheless, the similar defects in yolk sac (but not embryonic vasculature) in both Srg3 −/− :Tg + and Brg1 fl/fl :Tie2-Cre + mutants suggest that SWI/SNF complexes predominantly impact extraembryonic vascular development at midgestation.
Although this review does not summarize the many intriguing cardiac phenotypes that have been documented from mutations in SWI/SNF complex members (see [3, 4, 6] for reviews), it is worth mentioning a specific cardiac defect that is seen in Brg1 fl/fl :Tie2-Cre + mutants because it definitively stems from loss of BRG1 in endocardial cells. Brg1 fl/fl :Tie2-Cre + embryos undergo hypotrabeculation of the heart ventricles beginning at E9.5 due to a deficit of extracellular matrix material (cardiac jelly) that separates the ventricular endocardium from the myocardium [53]. This phenotype arises when Brg1 is deleted from endothelial/endocardial cells and not from myocardial cells. The hypotrabeculation results from upregulation of the extracellular protease ADAMTS1 in Brg1 fl/fl :Tie2-Cre + endocardial cells. BRG1 binds the Adamts1 promoter in endocardial cells and represses its transcription at midgestation; loss of Brg1 results in overproduction of ADAMTS1 and aberrant degradation of the cardiac jelly that supports trabeculation. This work supports a role for BRG1 in embryonic (cardiac) vasculature and demonstrates how chromatin remodeling in the vasculature can affect morphogenesis of adjacent tissues.
Another example of a role for SWI/SNF in embryonic vessels comes from mutation of the non-catalytic subunit Baf180. Global deletion of Baf180 results in placental defects, hypoplastic ventricle development in the heart, and embryonic lethality by E15.5 [40]. Furthermore, Baf180 −/− embryos have defective coronary vessel development around the ventricle and within the myocardium of mutant hearts [66]. Since coronary vessels derive from epicardial cells surrounding the developing heart [67], the coronary abnormalities in Baf180 −/− embryos are believed to stem from defective epithelial-to-mesenchymal-transition and migration of epicardial cells. These data reveal an additional role for SWI/SNF complexes in specific sites of embryonic vascular development and demonstrate how chromatin remodeling in a progenitor tissue can impact the downstream development of blood vessels.
Finally, since SWI/SNF complexes modify expression of smooth muscle-specific genes in vitro [68, 69], it is important to address the impact of these complexes on vascular smooth muscle development in the embryo. Brg1 was conditionally deleted using a transgenic line in which Cre recombinase is expressed under the control of the transgelin (SM22α) promoter [45, 53]. This SM22α-Cre transgene is expressed in adult arterial smooth muscle cells and in E10.5 myocardium, aortic vascular smooth muscle cells, and aortic endothelial cells [53, 70, 71]. The extent of its expression in embryonic vascular smooth muscle cells apart from those associated with the developing aorta is unclear. Brg1 fl/fl :SM22α-Cre + embryos die by E11.5 with thin myocardium and septation defects, presumably due to excision of Brg1 in cardiomyocytes [45]. Since no hemorrhage or vascular dilation were reported for these mutants, BRG1 is either unnecessary for smooth muscle cell function in developing blood vessels or this SM22α-Cre line fails to excise Brg1 in vascular smooth muscle cells outside of the heart at midgestation. Brg1 was also excised later in development using a transgenic line in which Cre recombinase is driven by the smooth muscle myosin heavy-chain promoter (smMHC) [56]. This smMHC-Cre line is first expressed around the aorta and major airway at E12.5 and becomes more fully penetrant in differentiated smooth muscle cells by E16.5 [72]. However, no vascular abnormalities were noted in E17.5 Brg1 fl/fl :smMHC-Cre + embryos or in double-deficient Brg1 fl/fl :smMHC-Cre + :Brm −/− embryos [56]. Therefore, SWI/SNF does not appear to play a critical role in maintaining differentiated vascular smooth muscle cells, but its role in proliferating smooth muscle cells during embryonic blood vessel development requires further elucidation.
CHD
The best-characterized complexes within the CHD subfamily of ATP-dependent chromatin-remodeling complexes are the nucleosome remodeling and deacetylase (NuRD) complexes (Fig. 3). NuRD complexes contain the ATPases CHD3 (Mi-2α) or CHD4 (Mi-2β), and it is not yet clear whether these proteins are mutually exclusive within a single complex. NuRD complexes also contain histone deacetylases (HDAC1 or HDAC2) and methyl-CpG-binding domain proteins (MBD2 or MBD3) in addition to other non-catalytic proteins. Because histone deacetylation and DNA methylation are typically associated with transcriptional repression, NuRD has historically been considered a repressive complex. However, mounting evidence supports a role for NuRD in mediating transcriptional activation in certain contexts, so NuRD—like SWI/SNF—can promote or repress transcription at specific target genes [73].
No global or conditional mutations of Chd3 have yet been described in the mouse. However, a floxed allele of Chd4 has been generated and used for deleting the gene in thymocytes, bone marrow, and epidermis [48, 54, 55]. Very little information is available about the expression patterns or function of CHD4 in early embryonic development; however, evidence for its participation in extraembryonic vascular development comes from its deletion with the Tie2-Cre transgene [74]. Chd4 fl/fl :Tie2-Cre + yolk sacs have normal vascular development and patterning through E10.5, as assessed by whole mount immunostaining for the endothelial cell marker PECAM-1. However, deletion of both Chd4 and the SWI/SNF ATPase Brg1 in endothelial cells rescues the vascular abnormalities seen in Brg1 fl/fl :Tie2-Cre + yolk sacs. These genetic data indicate that CHD4 does play a role in yolk sac vascular patterning, although this role is not revealed by mutation of the ATPase alone.
Importantly, the defective primitive erythropoiesis associated with Brg1 fl/fl :Tie2-Cre + mutants is not rescued in Brg1 fl/fl ;Chd4 fl/fl :Tie2-Cre + mutants, providing further evidence that the vascular phenotypes in Brg1 fl/fl :Tie2-Cre + yolk sacs are not secondary to anemia. Instead, since Brg1 fl/fl :Tie2-Cre + yolk sac vascular abnormalities are largely attributed to downregulated Wnt signaling [64], it was hypothesized that deletion of Chd4 upregulates Wnt signaling to rescue vascular patterning and morphology in Brg1 fl/fl ;Chd4 fl/fl :Tie2-Cre + yolk sacs. Indeed, deletion of Chd4 results in upregulation of several Wnt target genes in yolk sac vascular endothelium [74]. This phenotype is explained by the finding that CHD4 directly inhibits expression of the Wnt-responsive transcription factor Tcf7 as well as a subset of Wnt target genes. Although BRG1 and CHD4 have been shown to act in opposition on specific target genes in non-vascular cells [75, 76], these data provide the first evidence that the enzymes can antagonistically regulate the same signaling pathway via different target genes. Further work will be required to determine if BRG1 and CHD4 maintain their antagonistic relationship with the Wnt signaling pathway in other vascular beds and at later developmental time points.
Chromatin-remodeling complexes and target gene specificity
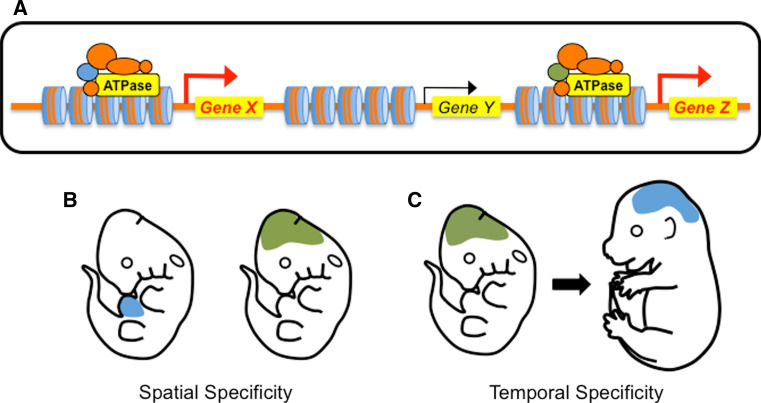
Chromatin-remodeling complexes are not indiscriminate mediators of transcription but rather exercise precise temporal and spatial control over gene expression. Given that the core subunits of the complexes—including the ATPases—are often ubiquitously expressed, how do the complexes impart such specific regulation over their targets? Typically, the alternative non-catalytic subunits have been attributed with this role [18] (Fig. 4). These proteins are often mutually exclusive and many contain protein domains that allow them to differentially bind DNA, proteins, or posttranslational histone/DNA modifications, which help direct the complexes to distinct target genes [77]. In addition, the non-catalytic subunits can impart temporal and spatial specificity on the function of a chromatin-remodeling complex if their expression patterns are restricted. For example, the mammalian SWI/SNF subunits BAF45a and BAF53a are expressed in neural stem and progenitor cells at various sites and times during development but are replaced by the subunits BAF45b, BAF45c, and BAF53b in postmitotic neurons [50]. This switch in the expression and utilization of SWI/SNF non-catalytic subunits is required for neuronal differentiation and presumably for targeting distinct genes in progenitor and postmitotic neurons.
Fig. 4.
The protein composition of chromatin-remodeling complexes influences temporal and spatial expression of target genes. The functional diversity of mammalian chromatin-remodeling complexes is often attributed to their non-catalytic proteins. a In this schematic a non-catalytic protein (blue) is critical for expression of Gene X while an alternative protein (green) is required for expression of Gene Z. These proteins may impart target gene specificity by recognizing different DNA sequences, epigenetic tags, or chromatin-binding proteins at the two loci. b If the alternative non-catalytic proteins are expressed in different tissues at a particular developmental time point, they can influence the spatial specificity of target gene expression. c If the alternative non-catalytic proteins are expressed at different developmental stages, they can impact the temporal specificity of target gene expression
However, evidence from vascular mutations of chromatin-remodeling complex ATPases suggest that these broadly expressed proteins also play instructive roles in dictating the complexes’ specificity for target genes. Conditional deletions of SWI/SNF and NuRD ATPases in the mouse result in distinct phenotypes that are not only temporally and spatially restricted but are also attributed to a surprisingly small group of target genes. For example, BRG1 and CHD4 appear to regulate vascular Wnt signaling in the yolk sac but not the embryo proper at midgestation, since Brg1 and Chd4 mutant embryos do not have vascular phenotypes resembling those seen in embryos with a deficit or overabundance of Wnt signaling [78, 79]. Although the enzymes are broadly expressed, their specificity for Wnt signaling targets appears to be spatially restricted at midgestation. This specificity could still be due to spatial restriction of non-catalytic subunits in the complexes. For example, a blood-vessel specific SWI/SNF component could be restricted in expression to the yolk sac at midgestation where it mediates interactions between BRG1 and Wnt pathway target genes. A precedent for this scenario lies in the developing heart where the SWI/SNF component BAF60c is specifically expressed and mediates interactions between BRG1 and cardiac transcription factors [80]. Alternatively, tissue-specific transcription factors may play a critical role in recruiting chromatin-remodeling complexes to their target genes in the vasculature. Precise combinations of transcription factor binding sites are found in the promoters of many vascular-specific genes [81], so combinatorial transcription factors could influence the temporal and spatial specificity of the chromatin-remodeling complexes with which they interact. Finally, functional redundancy with other chromatin-remodeling complexes could factor into the spatially restricted phenotypes seen in Brg1 and Chd4 vascular mutants. It is possible that BRG1-containing and CHD4-containing complexes target Wnt pathway genes in the embryo as well as the yolk sac but that other complexes can functionally compensate for their loss in the embryo. This idea was genetically addressed by simultaneous deletion of both SWI/SNF ATPases Brg1 and Brm in developing vessels [44]. The double-mutants did not display additive or exacerbated phenotypes over those seen in Brg1 mutants alone, indicating that BRM does not functionally compensate for BRG1 in embryonic vascular Wnt signaling. However, other known or novel complexes may regulate Wnt signaling in the embryo proper at midgestation, with or without a coordinated effort from BRG1- or CHD4-containing complexes.
Another indication of chromatin-remodeling complex specificity for vascular target genes comes from the surprising lack of early vascular phenotypes in Brg1 fl/fl :Tie2-Cre + or Chd4 fl/fl :Tie2-Cre + mutants [44, 53, 74]. Both embryonic and yolk sac vascular plexus formation appears normal in these mutants, indicating that neither of these ATPases mediates transcription of genes—such as those in the vascular endothelial growth factor (VEGF) signaling pathway—required for vasculogenesis. Unless VEGF pathway genes are immune to transcriptional regulation by chromatin-remodeling complexes, this begs the question of why vasculogenesis is unaffected in Brg1 and Chd4 mutants. One possibility is that Brg1 and Chd4 are not excised early or efficiently enough by the Tie2-Cre transgene to impact transcription of VEGF pathway genes. Alternatively, it is possible that BRG1 and CHD4 do not belong to complexes that mediate VEGF pathway gene transcription. In this case, mutational analysis of more chromatin-remodeling complex ATPases will be required to identify complexes involved in early vascular development. Likewise, genetic analyses using temporally inducible Cre recombinases will help elucidate the role of chromatin-remodeling enzymes at later stages of vascular development.
Implications for chromatin-remodeling complexes and pathological vascular development
Angiogenesis has been scrutinized as a therapeutic target for combating cancer ever since it was first proposed to play a critical role in tumor growth [82]. In addition to cancer, other diseases associated with misregulated vascular growth include diabetic retinopathy, rheumatoid arthritis, and obesity. To date, multiple anti-angiogenic therapies—primarily targeted against growth factors and their receptors—have been introduced into clinical practice, although the efficacy of these therapies has been limited [83]. We propose that epigenetic machinery—such as chromatin-remodeling complexes—could serve as novel targets for anti-angiogenesis therapy if the complexes are utilized during pathological angiogenesis in the adult as they are during physiological angiogenesis in the embryo. One example of this type of reutilization of chromatin-remodeling complexes under embryonic and postnatal pathological conditions occurs in developing myocardium and stressed adult hearts [45]. The SWI/SNF ATPase BRG1 mediates expression of β-myosin heavy chain (β-MHC) and promotes proliferation of embryonic cardiomyocytes. BRG1 is typically turned off in adult cardiomyocytes, and α-MHC replaces β-MHC expression. However, postnatal cardiac stresses cause BRG1 to reactivate, initiate a pathological shift from α-MHC to β-MHC expression, and drive hypertrophic proliferation of cardiomyocytes. We hypothesize that BRG1 and other chromatin-remodeling enzymes may play analogous roles in promoting angiogenesis in embryonic vessels and postnatal pathological vessels. Future work will require genetic analysis of temporally specific vascular mutants for chromatin-remodeling complex subunits to determine if vascular phenotypes are discernable under postnatal pathological angiogenic conditions.
Conclusions
A dynamic developmental process such as angiogenesis that requires rapid and coordinated gene expression changes is well suited to regulation by ATP-dependent chromatin-remodeling complexes. However, we have only just begun to comprehend the impact of these complexes on vascular development. An important goal for this field is to generate more vascular mutations in chromatin-remodeling enzymes and non-catalytic subunits. Such mutations will reveal vascular phenotypes in developing embryos, which can be exploited for identifying misregulated target genes of chromatin-remodeling complexes (Fig. 5). This approach serves two important purposes: it distinguishes genes that are epigenetically modulated by chromatin-remodeling complexes and it defines the function of these genes in developing blood vessels. Importantly, this unbiased approach can greatly expand our understanding of vascular development because it provides an opportunity for identifying genes with previously unrecognized roles in vascular development.
Fig. 5.
Strategy for identifying chromatin-remodeling complex target genes and their function in developing blood vessels. Left Panel In wild-type vascular cells, chromatin-remodeling complexes modulate expression of selective target genes and thereby regulate the function of those genes during vascular development. Right Panel When chromatin-remodeling complex components are genetically mutated in vascular cells, misregulated target genes can produce abnormal vascular phenotypes. Once vascular phenotypes are identified in mutant embryos (1), comparison of transcripts from mutant and wild-type embryos can reveal genes whose misregulation mediates the phenotypes (2). To verify candidate target genes of chromatin-remodeling complexes, vascular cells isolated from wild-type embryos at the developmental stage corresponding to the onset of the mutant phenotypes can be subjected to chromatin immunoprecipitation (ChIP) assays to reveal whether the chromatin-remodeling complex of interest binds regulatory regions for the candidate target gene
So far, the available vascular mutants for chromatin-remodeling complex components have yielded spatially specific phenotypes, with extraembryonic blood vessels affected more visibly than embryonic vessels. Future work will likely reveal which chromatin-remodeling complexes mediate vascular development in the embryo proper and in postnatal models of physiological and pathological vascular development. A combination of genetic mutations and biochemical assays can provide powerful tools for assessing the temporal and spatial influence of chromatin-remodeling complexes on vascular development. New insight may require simultaneous mutations of subunits from two or more complexes to reveal functional interactions between different complexes on specific genes or signaling pathways in blood vessels. Importantly, isolation of vascular cells from genetic mutants will allow for functional analysis of vascular gene expression, nucleosome density, covalent chromatin modifications, and recruitment of core transcriptional machinery at specific developmental time points. Such analysis on physiologically relevant cells rather than a static cell line will elucidate how chromatin-remodeling complexes impact their target genes during blood vessel development in vivo.
Acknowledgments
This work was supported by grants from the National Institutes of Health: the National Center for Research Resources and the National Institute of General Medical Sciences through grant number P20GM103441 and the National Heart, Lung and Blood Institute through grant number R00HL087621.
Abbreviations
- SWI/SNF
Mating type switching/sucrose nonfermenting
- NuRD
Nucleosome remodeling and deacetylase
- ISWI
Imitation switch
- CHD
Chromodomain helicase DNA-binding
- INO80
Inositol autotroph 80
- BRM
Brahma
- BRG1
Brahma-related gene 1
- PECAM-1
Platelet endothelial cell adhesion molecule-1
- HDAC
Histone deacetylase
- MBD
Methyl-CpG-binding domain
- BAF
BRG/BRM-associated factor
- VEGF
Vascular endothelial growth factor
References
- 1.Patel-Hett S, D’Amore PA. Signal transduction in vasculogenesis and developmental angiogenesis. Int J Dev Biol. 2011;55:353–363. doi: 10.1387/ijdb.103213sp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang CP, Bruneau B. Epigenetics and cardiovascular development. Annu Rev Physiol. 2011;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 4.Vallaster M, Vallaster CD, Wu SM. Epigenetic mechanisms in cardiac development and disease. Acta Biochim Biophys Sin (Shanghai) 2012;44:92–102. doi: 10.1093/abbs/gmr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan MS, Matouk CC, Marsden PA. Epigenetics of the vascular endothelium. J Appl Physiol. 2010;109:916–926. doi: 10.1152/japplphysiol.00131.2010. [DOI] [PubMed] [Google Scholar]
- 6.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circ Res. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 9.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae . Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-F. [DOI] [PubMed] [Google Scholar]
- 11.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 12.Johnson CN, Adkins NL, Georgel P. Chromatin-remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol. 2005;83:405–417. doi: 10.1139/o05-115. [DOI] [PubMed] [Google Scholar]
- 13.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 14.Clapier CR, Cairns BR. The biology of chromatin-remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 15.Laurent BC, Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- 16.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin-remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/S1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 18.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown E, Malakar S, Krebs JE. How many remodelers does it take to make a brain? Diverse and cooperative roles of ATP-dependent chromatin-remodeling complexes in development. Biochem Cell Biol. 2007;85:444–462. doi: 10.1139/O07-059. [DOI] [PubMed] [Google Scholar]
- 20.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 21.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Wu JI. Diverse functions of ATP-dependent chromatin-remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 2012;44:54–69. doi: 10.1093/abbs/gmr099. [DOI] [PubMed] [Google Scholar]
- 24.Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang ZQ. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet. 2001;29:206–211. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- 25.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/S1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 26.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, Confalonieri S, Cesaroni M, Marchesi F, Gasco M, Scanziani E, Capra M, Mai S, Nuciforo P, Crook T, Lough J, Amati B. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 29.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, Park SD, Seong RH. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS, Kuehn MR, Yamaguchi TP, Wu C. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marino S, Nusse R. Mutants in the mouse NuRD/Mi2 component P66alpha are embryonic lethal. PLoS One. 2007;2:e519. doi: 10.1371/journal.pone.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohan KN, Ding F, Chaillet JR. Distinct roles of DMAP1 in mouse development. Mol Cell Biol. 2011;31:1861–1869. doi: 10.1128/MCB.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci USA. 2003;100:14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tominaga K, Kirtane B, Jackson JG, Ikeno Y, Ikeda T, Hawks C, Smith JR, Matzuk MM, Pereira-Smith OM. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda T, Watanabe-Fukunaga R, Ogawa H, Fukuyama H, Higashi Y, Nagata S, Fukunaga R. Critical role of the p400/mDomino chromatin-remodeling ATPase in embryonic hematopoiesis. Genes Cells. 2007;12:581–592. doi: 10.1111/j.1365-2443.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin-remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/S1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 43.Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008;68:3689–3696. doi: 10.1158/0008-5472.CAN-07-6652. [DOI] [PubMed] [Google Scholar]
- 44.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He S, Pirity MK, Wang WL, Wolf L, Chauhan BK, Cveklova K, Tamm ER, Ashery-Padan R, Metzger D, Nakai A, Chambon P, Zavadil J, Cvekl A. Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics Chromatin. 2010;3:21. doi: 10.1186/1756-8935-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Indra AK, Dupe V, Bornert JM, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005;132:4533–4544. doi: 10.1242/dev.02019. [DOI] [PubMed] [Google Scholar]
- 48.Kashiwagi M, Morgan BA, Georgopoulos K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development. 2007;134:1571–1582. doi: 10.1242/dev.001750. [DOI] [PubMed] [Google Scholar]
- 49.Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin-remodeling complex NURF regulates thymocyte maturation. Genes Dev. 2011;25:275–286. doi: 10.1101/gad.2007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin-remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 52.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/S1535-6108(02)00185-X. [DOI] [PubMed] [Google Scholar]
- 53.Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, Georgopoulos K. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang M, Chen M, Kim JR, Zhou J, Jones RE, Tune JD, Kassab GS, Metzger D, Ahlfeld S, Conway SJ, Herring BP. SWI/SNF complexes containing Brahma or Brahma related gene 1 play distinct roles in smooth muscle development. Mol Cell Biol. 2011;31:2618–2631. doi: 10.1128/MCB.01338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aguilera C, Nakagawa K, Sancho R, Chakraborty A, Hendrich B, Behrens A. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 2011;469:231–235. doi: 10.1038/nature09607. [DOI] [PubMed] [Google Scholar]
- 58.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dauvillier S, Ott MO, Renard JP, Legouy E. BRM (SNF2alpha) expression is concomitant to the onset of vasculogenesis in early mouse postimplantation development. Mech Dev. 2001;101:221–225. doi: 10.1016/S0925-4773(00)00560-8. [DOI] [PubMed] [Google Scholar]
- 62.Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA. Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. J Exp Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lucitti JL, Jones EA, Huang C, Chen J, Fraser SE, Dickinson ME. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development. 2007;134:3317–3326. doi: 10.1242/dev.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc Natl Acad Sci USA. 2011;108:2282–2287. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han D, Jeon S, Sohn DH, Lee C, Ahn S, Kim WK, Chung H, Seong RH. SRG3, a core component of mouse SWI/SNF complex, is essential for extra-embryonic vascular development. Dev Biol. 2008;315:136–146. doi: 10.1016/j.ydbio.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 66.Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P, Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol. 2008;319:258–266. doi: 10.1016/j.ydbio.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 67.Perez-Pomares JM, Pompa JL. Signaling during epicardium and coronary vessel development. Circ Res. 2011;109:1429–1442. doi: 10.1161/CIRCRESAHA.111.245589. [DOI] [PubMed] [Google Scholar]
- 68.Zhang M, Fang H, Zhou J, Herring BP. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282:25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 69.Zhou J, Zhang M, Fang H, El-Mounayri O, Rodenberg JM, Imbalzano AN, Herring BP. The SWI/SNF chromatin-remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol. 2009;29:921–928. doi: 10.1161/ATVBAHA.109.187229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holtwick R, Gotthardt M, Skryabin B, Steinmetz M, Potthast R, Zetsche B, Hammer RE, Herz J, Kuhn M. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proc Natl Acad Sci USA. 2002;99:7142–7147. doi: 10.1073/pnas.102650499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, Tallquist MD, Iruela-Arispe ML. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xin HB, Deng KY, Rishniw M, Ji G, Kotlikoff MI. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol Genomics. 2002;10:211–215. doi: 10.1152/physiolgenomics.00054.2002. [DOI] [PubMed] [Google Scholar]
- 73.Ramirez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- 74.Curtis CD, Griffin CT. The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Mol Cell Biol. 2012;32:1312–1320. doi: 10.1128/MCB.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao H, Lukin K, Ramirez J, Fields S, Lopez D, Hagman J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin-remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci USA. 2009;106:11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erdel F, Krug J, Langst G, Rippe K. Targeting chromatin remodelers: signals and search mechanisms. Biochim Biophys Acta. 2011;1809:497–508. doi: 10.1016/j.bbagrm.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 78.Cattelino A, Liebner S, Gallini R, Zanetti A, Balconi G, Corsi A, Bianco P, Wolburg H, Moore R, Oreda B, Kemler R, Dejana E. The conditional inactivation of the beta-catenin gene in endothelial cells causes a defective vascular pattern and increased vascular fragility. J Cell Biol. 2003;162:1111–1122. doi: 10.1083/jcb.200212157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corada M, Nyqvist D, Orsenigo F, Caprini A, Giampietro C, Taketo MM, Iruela-Arispe ML, Adams RH, Dejana E. The Wnt/beta-catenin pathway modulates vascular remodeling and specification by upregulating Dll4/Notch signaling. Dev Cell. 2010;18:938–949. doi: 10.1016/j.devcel.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 81.De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197108122850711. [DOI] [PubMed] [Google Scholar]
- 83.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marmorstein R, Berger SL. Structure and function of bromodomains in chromatin-regulating complexes. Gene. 2001;272:1–9. doi: 10.1016/S0378-1119(01)00519-4. [DOI] [PubMed] [Google Scholar]
- 85.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 86.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. BioEssays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 87.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 88.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]