Abstract
Coordinated gene expression changes across the CNS are required to produce the mammalian maternal phenotype. Lateral septum (LS) is a brain region critically involved with aspects of maternal care, and we recently examined gene expression of whole septum (LS and medial septum) in selectively bred maternal mice. Here, we expand on the prior study by 1) conducting microarray analysis solely on LS in virgin and postpartum mice, 2) using outbred mice, and 3) evaluating the role of sensory input on gene expression changes. Large scale changes in genes related to neuronal signaling were identified, including four GABAA receptor subunits. Subunits α4 and δ were downregulated in maternal LS, likely reflecting a reduction in the extrasynaptic, neurosteroid-sensitive α4/δ containing receptor subtype. Conversely, subunits ε and θ were increased in maternal LS. Fifteen K+ channel related genes showed altered expression, as did dopamine receptors Drd1a and Drd2 (both downregulated), hypocretin receptor 1 (Hcrtr1), kappa opioid receptor 1 (Oprk1), and transient receptor potential channel 4 (Trpc4). Expression of a large number of genes linked to developmental processes or cell differentiation were also altered in postpartum LS, including chemokine (C-X-C) motif ligand 12 (Cxcl12), fatty acid binding protein 7 (Fabp7), plasma membrane proteolipid (Pllp), and suppressor of cytokine signaling 2 (Socs2). Additional genes that are linked to anxiety, such as glutathione reductase (Gsr), exhibited altered expression. Pathway analysis also identified changes in genes related to cyclic nucleotide metabolism, chromatin structure, and the Ras gene family. The sensory presence of pups was found to contribute to the altered expression of a subset of genes across all categories. This study suggests that both large changes in neuronal signaling and the possible terminal differentiation of neuronal and/or glial cells play important roles in producing the maternal state.
Introduction
The establishment of the maternal phenotype requires a coordinated suite of changes in numerous biological pathways, from endocrine signaling and metabolic activity to nervous system properties and adaptive behaviors [1]–[3]. Maternal behavior in many mammals is critical for the survival of offspring. In mice, this includes behaviors such as nest building, nursing, and protection of offspring [4]. The generation of effective maternal behavior also involves modulation of pathways related to bond formation and sociability, as the mother-infant relationship is the primary social bond in all mammalian species [5]. Additional emotional pathways altered in the postpartum state include fear, stress, and anxiety. The transition from a virgin to lactating state provides a unique and powerful opportunity to examine the fundamental neurophysiology of a range of emotional traits because the observed changes are naturally occurring.
Lateral septum (LS) is a brain region that is centrally featured in a network of structures known to influence social and parental behavior and emotional states [6], [7]. It has connections to the medial preoptic area, hypothalamus, amygdala, ventral tegmental area, periaqueductal gray, and receives input from medial prefrontal cortex [7]–[9]. The goal of this study was to identify gene expression changes occurring naturally in the LS of lactating outbred mice that may be important markers of the maternal phenotype. LS has been linked to certain aspects of maternal care, including offspring protection. Pharmacological manipulations of GABAA receptors in LS alter offspring protection [10] and it has recently been demonstrated that the production of GABA is increased in the LS of postpartum mice [11]. The heteropentameric, ionotropic GABAA receptor is assembled from a pool of 16 known subunits, resulting in a diversity of receptor subtypes with unique properties, pharmacological profiles, and distributions throughout the brain. This diversity provides a high degree of flexibility in signal transduction and allosteric modulation [12]–[14], but the dynamic regulation of GABAA receptor subunits in LS of maternal mice has yet to be studied. This study therefore has a particular focus on investigating expression changes in GABAA receptors themselves as a possible mechanism of modulating GABA signaling in the maternal LS.
We recently performed a gene expression study in the whole septum of maternal mice selectively bred for high offspring protection [15]. The present study used a similar microarray approach and quantitative real-time PCR to expand on that line of work by 1) utilizing a more specific dissection exclusively of LS, 2) using outbred mice to yield more natural and broadly applicable results, and 3) evaluating the effects of sensory input from interaction with pups on gene expression. While one component of the maternal phenotype is established initially by the actions of hormones during pregnancy, sensory inputs from both changes within the mother and from interactions with offspring additionally shape the maternal brain and parental behavior. This study evaluated the degree to which expression changes in the maternal LS require the continued presence of pups.
The transition from a virgin to lactating state involves many genes across numerous biological pathways. To address this, we utilized system level approaches to analyze microarray data and detect enrichment in sets of genes with related functions. Using these methods, we were able to identify functional themes in the expression profile of the maternal LS.
Results
Using Probe Logarithmic Intensity Error (PLIER) analysis, we identified 1,001 targets with significant expression differences (FDR-adjusted p<0.25) between the LS (Fig. 1) of lactating maternal and virgin mice. Of these, 809 corresponded to well-annotated genes. Table 1 presents the 100 most significant genes from this comparison, categorized by basic functional data from the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/) and GeneCards compendium (http://www.genecards.org). This table serves as a representative sample to highlight the microarray results, but many genes of biological interest and statistical significance cannot be included in such a short list. The full list of all 35,557 targets, their relative expression, and significance is presented in Table S1. Genes of interest reported in the microarray results include Gabra1 (FDR-adjusted p = 0.32, fold change 1.06), Gabra4 (FDR-adjusted p = 0.21, fold change 0.88), Gabrd (FDR-adjusted p = 0.11, fold change 0.75), Gabre (FDR-adjusted p = 0.12, fold change 1.38), Gabrq (FDR-adjusted p = 0.42, fold change 1.26), Cxcl12 (FDR-adjusted p = 0.25, fold change 1.14), Fabp7 (FDR-adjusted p = 0.20, fold change = 0.71), Gsr (FDR-adjusted p = 0.37, fold change 0.92), Pllp (FDR-adjusted p = 0.25, fold change 0.89), and Socs2 (FDR-adjusted p = 0.30, fold change 1.27).
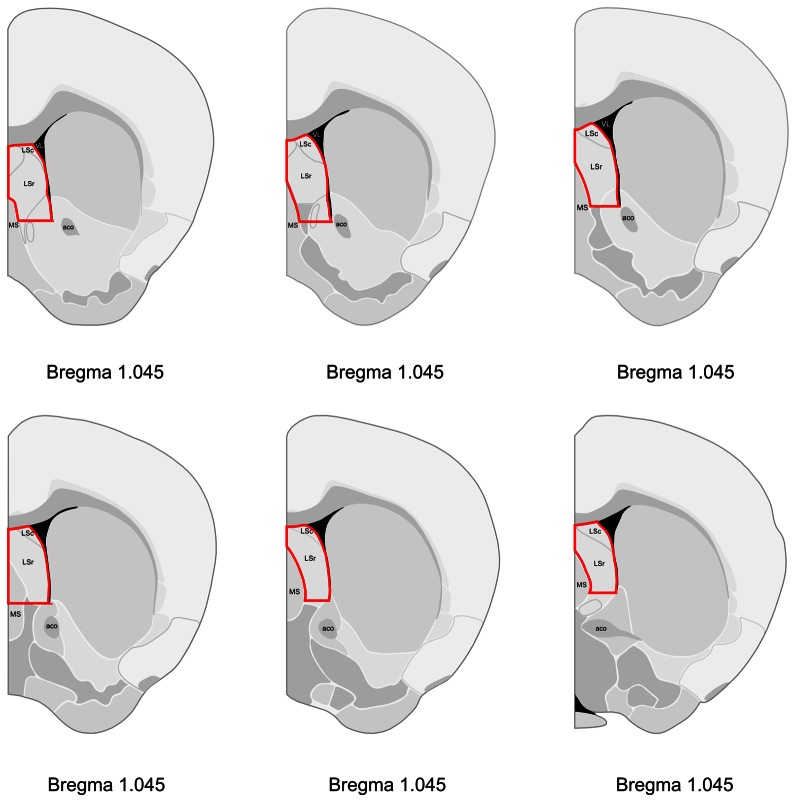
Figure 1. Schematic representation of the brain regions dissected for microarray and qPCR experiments.
Dissections are outlined in red. Distance from Bregma in the rostrocaudal plane is indicated. Reprinted and modified from The Allen Mouse Brain Atlas (reference atlas version 1, 2008). Abbreviations: LSc, caudal part of the lateral septal nucleus; LSr, rostral part of the lateral septal nucleus; MS; medial septal nucleus; VL, lateral ventricle; aco, anterior commissure, olfactory limb.
Table 1. List of the 100 most significant gene expression differences between LS of lactating maternal mice and age-matched virgins.
| Accession No. | Gene Symbol | Gene Title | Fold Change | |
| Apoptosis, anti-apoptosis | ||||
| NM_001079883 | Bcl11b | B-cell leukemia/lymphoma 11B | 0.79 | |
| NM_007537 | Bcl2l2 | BCL2-like 2 | 1.10 | |
| NM_009668 | Bin1 | bridging integrator 1 | 1.11 | |
| NM_011636 | Plscr1 | phospholipid scramblase 1 | 1.13 | |
| Cell cycle, adhesion, division, death, differentiation and proliferation | ||||
| NM_183187 | Fam107a | family with sequence similarity 107, member A | 1.17 | |
| NM_183183 | Gprin3 | GPRIN family member 3 | 0.79 | |
| NM_008258 | Hn1 | hematological and neurological expressed sequence 1 | 0.90 | |
| NM_001161535 | Islr2 | immunoglobulin superfamily containing leucine-rich repeat 2 | 0.85 | |
| NM_010660 | Krt10 | keratin 10 | 0.91 | |
| NM_178714 | Lrfn5 | leucine rich repeat and fibronectin type III domain containing 5 | 1.10 | |
| NM_177725 | Lrrc8a | leucine rich repeat containing 8A | 1.10 | |
| NM_028190 | Luc7l | Luc7 homolog (S. cerevisiae)-like | 1.08 | |
| NM_146061 | Prr5 | proline rich 5 (renal) | 1.13 | |
| NM_011386 | Skil | SKI-like | 0.83 | |
| NM_011418 | Smarcb1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | 0.92 | |
| NM_133786 | Smc4 | structural maintenance of chromosomes 4 | 0.84 | |
| NM_001033272 | Spata13 | spermatogenesis associated 13 | 0.79 | |
| Metabolic | ||||
| NM_139306 | Acer2 | alkaline ceramidase 2 | 1.32 | |
| NM_177872 | Adamts3 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3 | 0.74 | |
| NM_009625 | Adcyap1 | adenylate cyclase activating polypeptide 1 | 1.13 | |
| NM_018737 | Ctps2 | cytidine 5′-triphosphate synthase 2 | 1.10 | |
| NM_016699 | Exosc10 | exosome component 10 | 0.96 | |
| NM_021896 | Gucy1a3 | guanylate cyclase 1, soluble, alpha 3 | 0.81 | |
| NM_008509 | Lpl | lipoprotein lipase | 0.65 | |
| NM_172948 | Mgat5b | mannoside acetylglucosaminyltransferase 5, isoenzyme B | 0.91 | |
| NM_019840 | Pde4b | phosphodiesterase 4B, cAMP specific | 0.91 | |
| NM_172267 | Phyhd1 | phytanoyl-CoA dioxygenase domain containing 1 | 1.20 | |
| NM_029614 | Prss23 | protease, serine, 23 | 1.17 | |
| NM_027997 | Serpina9 | serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 | 0.52 | |
| Phosphorylation, dephosphorylation | ||||
| NM_177407 | Camk2a | calcium/calmodulin-dependent protein kinase II alpha | 0.95 | |
| NM_146125 | Itpka | inositol 1,4,5-trisphosphate 3-kinase A | 0.86 | |
| NM_175171 | Mast4 | microtubule associated serine/threonine kinase family member 4 | 1.13 | |
| NM_008587 | Mertk | c-mer proto-oncogene tyrosine kinase | 1.24 | |
| NM_016891 | Ppp2r1a | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | 0.95 | |
| Protein ubiquitination, deubiquitination | ||||
| NM_172835 | Peli3 | pellino 3 | 1.09 | |
| NM_144873 | Uhrf2 | ubiquitin-like, containing PHD and RING finger domains 2 | 1.14 | |
| NM_001033173 | Usp31 | ubiquitin specific peptidase 31 | 1.14 | |
| NM_025830 | Wwp2 | WW domain containing E3 ubiquitin protein ligase 2 | 0.94 | |
| Regulation of transcription | ||||
| NM_010155 | Erf | Ets2 repressor factor | 0.88 | |
| NM_175660 | Hist1h2ab | histone cluster 1, H2ab | 0.68 | |
| NM_013550 | Hist1h3a | histone cluster 1, H3a | 0.78 | |
| NM_178203 | Hist1h3b | histone cluster 1, H3b | 0.77 | |
| NM_178204 | Hist1h3d | histone cluster 1, H3d | 0.78 | |
| NM_178204 | Hist1h3e | histone cluster 1, H3d | 0.78 | |
| NM_145073 | Hist1h3g | histone cluster 1, H3g | 0.77 | |
| NM_178206 | Hist1h3h | histone cluster 1, H3h | 0.77 | |
| NM_178207 | Hist1h3i | histone cluster 1, H3i | 0.78 | |
| NM_175662 | Hist2h2ac | histone cluster 2, H2ac | 0.88 | |
| NM_001013813 | Maml2 | mastermind like 2 (Drosophila) | 0.87 | |
| NM_001033713 | Mef2a | myocyte enhancer factor 2A | 0.91 | |
| NM_001136072 | Meis2 | Meis homeobox 2 | 0.88 | |
| NM_172788 | Sh3rf3 | SH3 domain containing ring finger 3 | 1.11 | |
| NM_172913 | Tox3 | TOX high mobility group box family member 3 | 0.86 | |
| Signal transduction | ||||
| NM_133237 | Apcdd1 | adenomatosis polyposis coli down-regulated 1 | 0.88 | |
| NM_207655 | Egfr | epidermal growth factor receptor | 0.85 | |
| NM_008072 | Gabrd | gamma-aminobutyric acid (GABA) A receptor, subunit delta | 0.75 | |
| NM_175668 | Gpr4 | G protein-coupled receptor 4 | 1.19 | |
| NM_146072 | Grik1 | glutamate receptor, ionotropic, kainate 1 | 1.21 | |
| NM_181850 | Grm3 | glutamate receptor, metabotropic 3 | 0.87 | |
| NM_174998 | Hpcal4 | hippocalcin-like 4 | 0.89 | |
| NM_013568 | Kcna6 | potassium voltage-gated channel, shaker-related, subfamily, member 6 | 1.14 | |
| NM_010597 | Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | 0.86 | |
| NM_008427 | Kcnj4 | potassium inwardly-rectifying channel, subfamily J, member 4 | 0.72 | |
| NM_001081027 | Kcnt2 | potassium channel, subfamily T, member 2 | 1.15 | |
| NM_001081298 | Lphn2 | latrophilin 2 | 0.86 | |
| NM_024200 | Mfn1 | mitofusin 1 | 1.15 | |
| NM_152229 | Nr2e1 | nuclear receptor subfamily 2, group E, member 1 | 0.85 | |
| NM_178751 | Orai2 | ORAI calcium release-activated calcium modulator 2 | 0.86 | |
| NM_177411 | Rab5b | RAB5B, member RAS oncogene family | 0.91 | |
| NM_011243 | Rarb | retinoic acid receptor, beta | 0.66 | |
| NM_009107 | Rxrg | retinoid X receptor gamma | 0.71 | |
| NM_011313 | S100a6 | S100 calcium binding protein A6 (calcyclin) | 0.81 | |
| NM_030889 | Sorcs2 | sortilin-related VPS10 domain containing receptor 2 | 0.91 | |
| NM_016908 | Syt5 | synaptotagmin V | 0.94 | |
| NM_021344 | Tesc | tescalcin | 0.82 | |
| NM_011648 | Tshr | thyroid stimulating hormone receptor | 0.82 | |
| NM_031877 | Wasf1 | WASP family 1 | 0.91 | |
| Translation | ||||
| NM_178627 | Poldip3 | polymerase (DNA-directed), delta interacting protein 3 | 0.94 | |
| NM_024212 | Rpl4 | ribosomal protein L4 | 0.92 | |
| NM_007475 | Rplp0 | ribosomal protein, large, P0 | 0.90 | |
| NM_177214 | Snrnp200 | small nuclear ribonucleoprotein 200 (U5) | 0.92 | |
| Transport | ||||
| NM_017391 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | 1.16 | |
| NM_194355 | Spire1 | spire homolog 1 (Drosophila) | 1.07 | |
| Other | ||||
| NM_133729 | 2610018G03Rik | RIKEN cDNA 2610018G03 gene | 0.80 | |
| ENSMUST00000055842 | Armcx4 | armadillo repeat containing, X-linked 4 | 1.15 | |
| NM_027560 | Arrdc2 | arrestin domain containing 2 | 1.13 | |
| NM_028386 | Asphd2 | aspartate beta-hydroxylase domain containing 2 | 0.87 | |
| BC096543 | B930095G15Rik | RIKEN cDNA B930095G15 gene | 1.14 | |
| NM_027909 | C2cd2l | C2 calcium-dependent domain containing 2-like | 0.90 | |
| NM_030179 | Clip4 | CAP-GLY domain containing linker protein family, member 4 | 0.88 | |
| NM_146067 | Cpped1 | calcineurin-like phosphoesterase domain containing 1 | 0.88 | |
| NM_198866 | Dbpht2 | DNA binding protein with his-thr domain | 0.85 | |
| NM_001134457 | Fam55c | family with sequence similarity 55, member C | 1.10 | |
| NM_178673 | Fstl5 | follistatin-like 5 | 1.16 | |
| NM_207222 | Lmo3 | LIM domain only 3 | 0.81 | |
| NM_001164805 | Thsd7a | thrombospondin, type I, domain containing 7A | 0.81 | |
| NM_198627 | Vstm2l | V-set and transmembrane domain containing 2-like | 0.90 | |
| NM_001081382 | Zfp777 | zinc finger protein 777 | 0.92 | |
| NM_013859 | Znhit2-ps | zinc finger, HIT domain containing 2, pseudogene | 0.90 | |
| NM_145456 | Zswim6 | zinc finger, SWIM domain containing 6 | 0.84 | |
All expression changes in Table 1 have FDR-adjusted p-values less than 0.11. Expression is given as fold change in lactating maternal LS relative to virgin; numbers greater than 1.0 indicate increases in maternal mice.
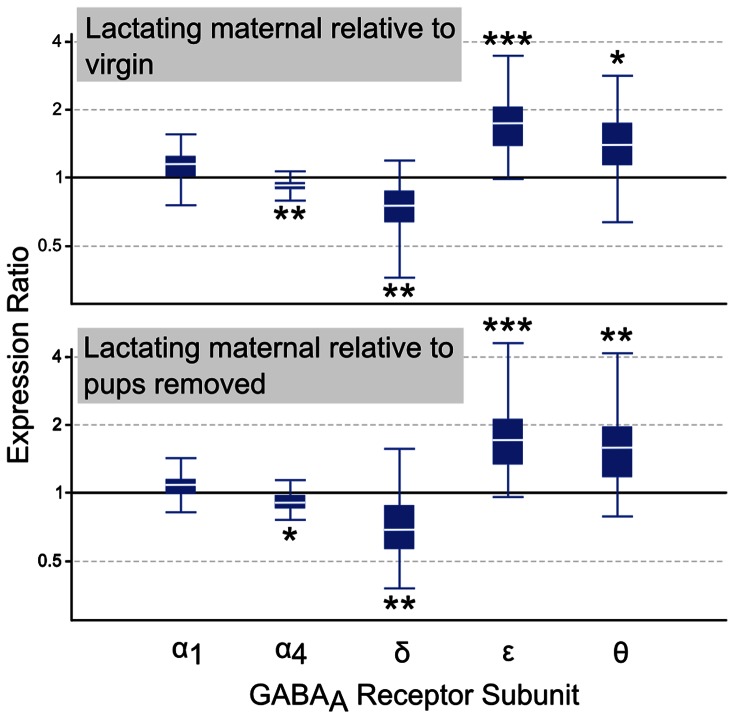
Numerous expression changes in GABAA receptor subunits were confirmed by quantitative real-time PCR analysis (Fig. 2). For each subunit that exhibited significant expression changes, differences were found to be significant in lactating maternal LS relative to both virgin and pups removed groups. The α4 subunit was downregulated (p = 0.004 and p = 0.046), as was the δ subunit (p = 0.003 and p = 0.01). The ε subunit expression was elevated (p<0.001 and p = 0.001), along with the θ subunit (p = 0.023 and p = 0.001). The observed increase in α1 subunit expression did not reach significance (p = 0.077 and p = 0.092).
Figure 2. Quantitative real-time PCR analysis of GABAA receptor subunit expression in lateral septum.
Relative expression distribution (Y-axis) represented as a ratio of lactating maternal versus virgin (top panel, n = 8/group) and lactating maternal versus pups removed (bottom panel, n = 8/group), was normalized against two reference genes, Ppia and Ywhaz, and shown by box-and-whisker plots as medians (white lines), interquartile ranges (boxes), and ranges (whiskers). Ratios over one indicate genes that are more highly expressed in lactating maternal LS than in virgin or pups removed LS. *p<0.05; **p<0.01; ***p<0.001.
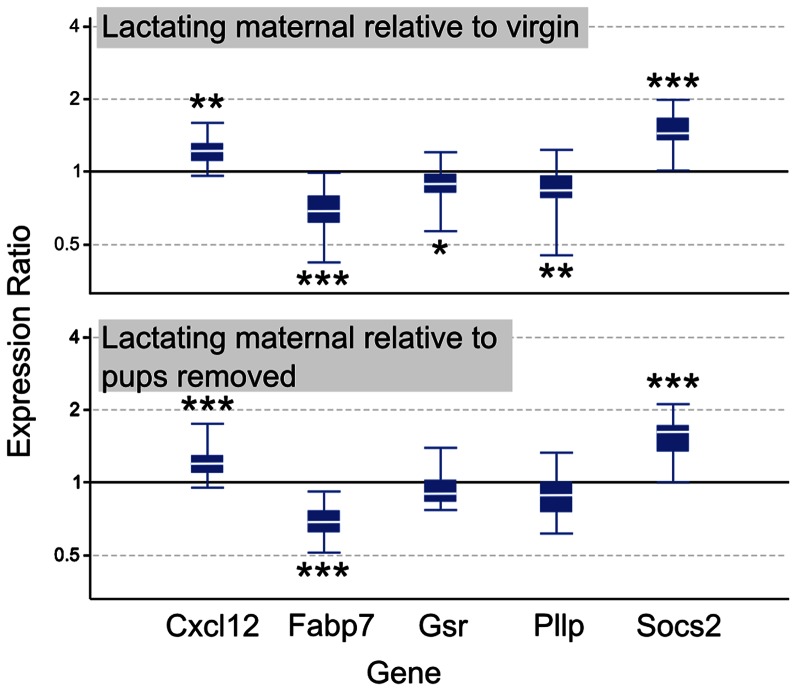
Quantitative real-time PCR analysis was used to confirm expression changes observed in microarray analysis for several genes of interest (Fig. 3). These genes were selected not only for their statistical significance in the current microarray experiment, but because they have appeared with regularity in numerous sets of significant expression results conducted in our laboratory across various regions of the maternal mouse brain ([15], unpublished observations). In lactating maternal LS relative to virgin, Cxcl12 and Socs2 were upregulated (p = 0.002 and p<0.001), while Fabp7, Gsr, and Pllp were downregulated (p<0.001, p = 0.04, and p = 0.018). In lactating maternal LS relative to pups removed, expression of Cxcl12 and Socs2 were again significantly elevated (p = 0.001 and p<0.001). Fabp7 was downregulated (p<0.001), but decreases in Gsr and Pllp did not reach significance (p = 0.125 and p = 0.074).
Figure 3. Quantitative real-time PCR analysis of gene expression in lateral septum.
Relative expression distribution (Y-axis) represented as a ratio of lactating maternal versus virgin (top panel, n = 8/group) and lactating maternal versus pups removed (bottom panel, n = 8/group), was normalized against two reference genes, Ppia and Ywhaz, and shown by box-and-whisker plots as medians (white lines), interquartile ranges (boxes), and ranges (whiskers). Ratios over one indicate genes that are more highly expressed in lactating maternal LS than in virgin or pups removed LS. *p<0.05; **p<0.01; ***p<0.001.
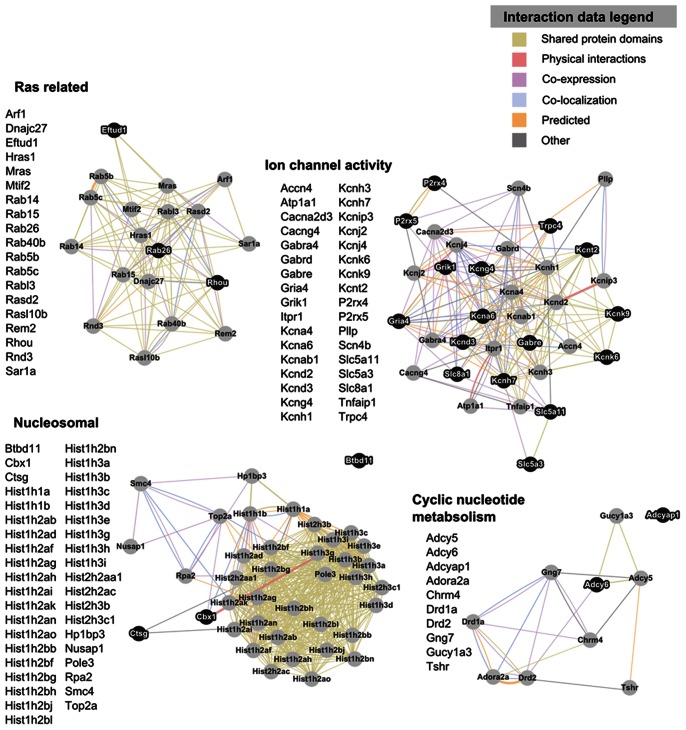
NIH DAVID’s functional annotation clustering tool identified enriched biological pathways among the 809 significant expression changes in lactating maternal LS relative to virgin (765 IDs recognized by DAVID). Figure 4 visualizes four gene clusters of interest with particularly high enrichment scores as networks of nodes connected by a variety of annotated interaction data. The ion channel activity network consists of 34 genes, of which 15 were upregulated and 19 were downregulated. The 19-gene network related to the Ras superfamily of small GTPases was primarily downregulated. The cyclic nucleotide metabolism network is comprised of 10 genes that influence the activity of adenylyl cyclase and the biogenesis of cyclic nucleotides. The nucleosome network is a large group of histone and histone-related proteins. It exhibits downregulation of the core histones H2a, H2b, H3, as well as two linker H1 histones. No H4 histones displayed significant expression changes. A more detailed expression data summary for enriched clusters is provided in Table S2.
Figure 4. Gene clusters found to be enriched in the LS of lactating maternal mice relative to virgin mice as interaction networks.
Gene lists for each cluster are presented to the left of their respective network visualization. Gene symbols in bold text are upregulated in lactating maternal LS relative to virgin, and are represented in the network images as dark nodes with white text. Non-bold gene symbols and grey nodes with dark text correspond to genes that are downregulated. The nature of the interaction data linking any two nodes is encoded by color. Distance between nodes is proportional to the strength of evidence for their interactions.
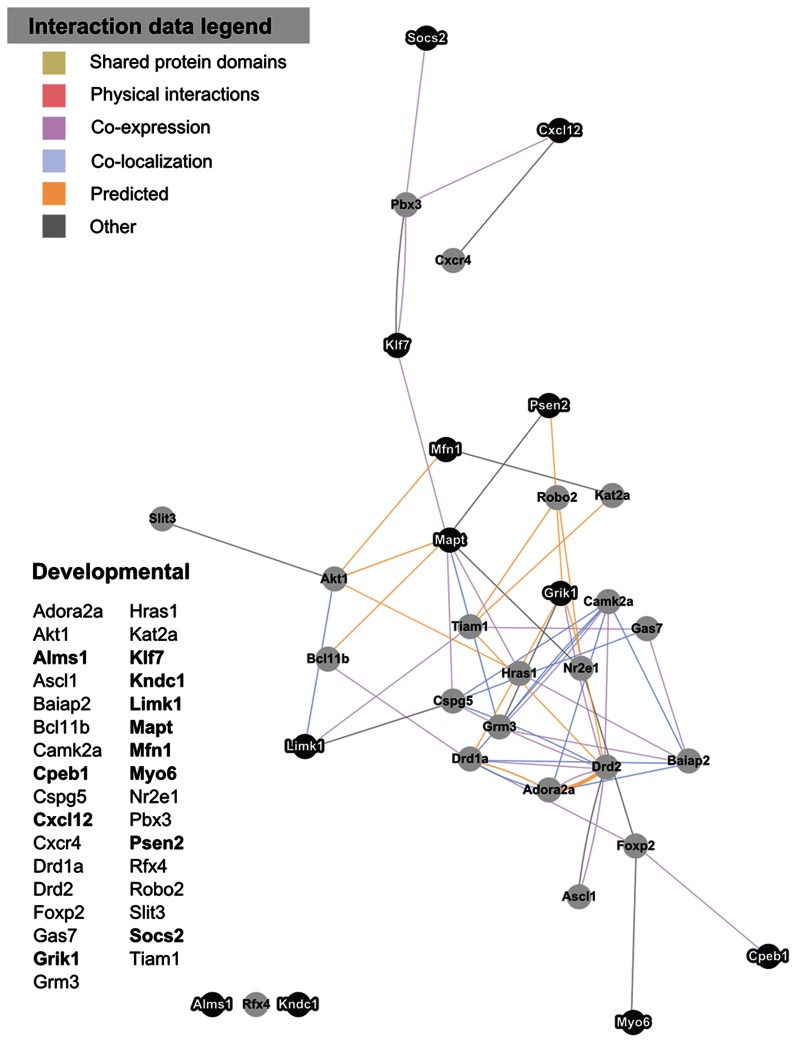
In the 809 significant expression changes in maternal LS relative to virgin, NIH DAVID’s functional annotation clustering tool detected enrichment in numerous, small gene sets that each included genes linked to developmental processes and neuron/glial differentiation. We consolidated these developmentally related genes into a single, larger cluster that is visualized in Figure 5. Expression data for this cluster can be viewed in Table S2.
Figure 5. Cluster of genes enriched in maternal LS related to developmental processes and cell differentiation.
Gene lists for the cluster is presented to the left of the network visualization. Gene symbols in bold text are upregulated in lactating maternal LS relative to virgin, and are represented in the network images as dark nodes with white text. Non-bold gene symbols and grey nodes with dark text correspond to genes that are downregulated. The nature of the interaction data linking any two nodes is encoded by color. Distance between nodes is proportional to the strength of evidence for their interactions.
To assess which gene changes may be most strongly associated with continued sensory input from pups, we identified genes that exhibited significance in lactating maternal LS when compared to both virgin and pups removed groups. Table 2 presents the 69 genes for which expression changes relative to maternal were significant (FDR-adjusted p-values for both comparisons <0.25) and were in the same direction. This list therefore represents genes for which expression changes detected between lactating maternal and virgin LS were most closely mirrored in lactating maternal relative to pups removed. A full listing of all gene comparisons of maternal versus the pups removed group is also provided in Table S1.
Table 2. List of 68 genes that displayed significant expression changes in the LS of lactating maternal mice compared to both virgin and pups removed groups.
| Accession No. | Gene Symbol | Gene Title | Fold Change (vs.pups removed) | Fold Change(vs. virgin) | |
| Cell cycle, adhesion, division, death, differentiation and proliferation | |||||
| NM_170597 | Creg2 | cellular repressor of E1A-stimulated genes 2 | 0.90 | 0.92 | |
| NM_001166273 | Cspg5 | chondroitin sulfate proteoglycan 5 | 0.90 | 0.92 | |
| NM_183187 | Fam107a | family with sequence similarity 107, member A | 1.26 | 1.17 | |
| NM_011812 | Fbln5 | fibulin 5 | 1.24 | 1.19 | |
| NM_010218 | Fjx1 | four jointed box 1 (Drosophila) | 0.84 | 0.87 | |
| NM_008538 | Marcks | myristoylated alanine rich protein kinase C substrate | 0.90 | 0.88 | |
| NR_033728 | Nit1 | nitrilase 1 | 1.06 | 1.08 | |
| NM_011386 | Skil | SKI-like | 0.87 | 0.83 | |
| Metabolic | |||||
| NM_001081204 | B3galtl | beta 1,3-galactosyltransferase-like | 0.91 | 0.92 | |
| NM_028979 | Cyp2j9 | cytochrome P450, family 2, subfamily j, polypeptide 9 | 1.18 | 1.14 | |
| NM_172948 | Mgat5b | mannoside acetylglucosaminyltransferase 5, isoenzyme B | 0.88 | 0.91 | |
| NM_172267 | Phyhd1 | phytanoyl-CoA dioxygenase domain containing 1 | 1.27 | 1.20 | |
| NM_029614 | Prss23 | protease, serine, 23 | 1.17 | 1.17 | |
| NM_001101430 | Psmg4 | proteasome (prosome, macropain) assembly chaperone 4 | 0.90 | 0.91 | |
| NM_133670 | Sult1a1 | sulfotransferase family 1A, phenol-preferring, member 1 | 1.34 | 1.30 | |
| Phosphorylation, dephosphorylation | |||||
| NM_009652 | Akt1 | thymoma viral proto-oncogene 1 | 0.93 | 0.92 | |
| NM_008587 | Mertk | c-mer proto-oncogene tyrosine kinase | 1.26 | 1.24 | |
| NM_016891 | Ppp2r1a | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | 0.95 | 0.95 | |
| Protein ubiquitination, deubiquitination | |||||
| NM_173784 | Ubtd2 | ubiquitin domain containing 2 | 0.87 | 0.90 | |
| NM_144873 | Uhrf2 | ubiquitin-like, containing PHD and RING finger domains 2 | 1.13 | 1.14 | |
| Regulation of transcription | |||||
| BC024814 | BC024814 | cDNA sequence BC024814 | 0.87 | 0.90 | |
| NM_023876 | Elp4 | elongation protein 4 homolog (S. cerevisiae) | 0.91 | 0.93 | |
| NM_133658 | Ercc3 | excision repair cross-complementing rodent repair deficiency, complementation group 3 | 0.93 | 0.93 | |
| NM_178213 | Hist2h2ab | histone cluster 2, H2ab | 0.89 | 0.87 | |
| NM_175662 | Hist2h2ac | histone cluster 2, H2ac | 0.90 | 0.88 | |
| NM_001033713 | Mef2a | myocyte enhancer factor 2A | 0.93 | 0.91 | |
| NR_030470 | Mir669a-2 | microRNA 669a-2 | 1.24 | 1.20 | |
| NM_181650 | Prdm4 | PR domain containing 4 | 0.93 | 0.95 | |
| NM_001166410 | Rbm3 | RNA binding motif protein 3 | 0.70 | 0.76 | |
| NM_001024918 | Rfx4 | regulatory factor X, 4 (influences HLA class II expression) | 0.86 | 0.89 | |
| NM_029949 | Snapc3 | small nuclear RNA activating complex, polypeptide 3 | 1.06 | 1.07 | |
| NM_001168578 | Tceal8 | transcription elongation factor A (SII)-like 8 | 0.88 | 0.86 | |
| NM_172913 | Tox3 | TOX high mobility group box family member 3 | 0.87 | 0.86 | |
| NM_001164578 | Tsr2 | TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) | 0.94 | 0.91 | |
| NM_144546 | Zfp119a | zinc finger protein 119a | 0.87 | 0.89 | |
| Signal transduction | |||||
| NM_133237 | Apcdd1 | Adenomatosis polyposis coli down-regulated 1 | 0.83 | 0.88 | |
| NM_008533 | Cd180 | CD180 antigen | 0.86 | 0.84 | |
| NM_009895 | Cish | cytokine inducible SH2-containing protein | 1.20 | 1.20 | |
| NM_010108 | Efna3 | ephrin A3 | 0.91 | 0.89 | |
| NM_207655 | Egfr | epidermal growth factor receptor | 0.89 | 0.85 | |
| NM_053072 | Fgd6 | FYVE, RhoGEF and PH domain containing 6 | 1.09 | 1.08 | |
| NM_175668 | Gpr4 | G protein-coupled receptor 4 | 1.18 | 1.19 | |
| NM_001081298 | Lphn2 | latrophilin 2 | 0.89 | 0.86 | |
| NM_001083897 | Mpzl1 | myelin protein zero-like 1 | 0.91 | 0.91 | |
| NM_019515 | Nmu | neuromedin U | 1.35 | 1.42 | |
| NM_152229 | Nr2e1 | nuclear receptor subfamily 2, group E, member 1 | 0.87 | 0.85 | |
| NM_178751 | Orai2 | ORAI calcium release-activated calcium modulator 2 | 0.86 | 0.86 | |
| NM_027571 | P2ry12 | purinergic receptor P2Y, G-protein coupled 12 | 0.88 | 0.87 | |
| NM_001042499 | Rabl3 | RAB, member of RAS oncogene family-like 3 | 0.92 | 0.91 | |
| NM_007706 | Socs2 | suppressor of cytokine signaling 2 | 1.32 | 1.30 | |
| NM_133789 | Strn4 | striatin, calmodulin binding protein 4 | 0.93 | 0.92 | |
| Transport | |||||
| NM_027560 | Arrdc2 | arrestin domain containing 2 | 1.11 | 1.13 | |
| NM_010634 | Fabp5 | fatty acid binding protein 5, epidermal | 0.79 | 0.81 | |
| NM_021272 | Fabp7 | fatty acid binding protein 7, brain | 0.69 | 0.71 | |
| NM_175112 | Rae1 | RAE1 RNA export 1 homolog (S. pombe) | 0.94 | 0.92 | |
| NM_009155 | Sepp1 | selenoprotein P, plasma, 1 | 1.17 | 1.08 | |
| NM_178639 | Sfxn5 | sideroflexin 5 | 0.93 | 0.93 | |
| NM_146198 | Slc5a11 | solute carrier family 5 (sodium/glucose cotransporter), member 11 | 1.14 | 1.21 | |
| NM_138599 | Tomm70a | translocase of outer mitochondrial membrane 70 homolog A (yeast) | 0.90 | 0.88 | |
| Other | |||||
| NM_025675 | Atpbd4 | ATP binding domain 4 | 0.86 | 0.91 | |
| BC090976 | BC022687 | cDNA sequence BC022687 | 0.89 | 0.90 | |
| NM_001166164 | Ccdc74a | coiled-coil domain containing 74A | 0.89 | 0.89 | |
| NM_030179 | Clip4 | CAP-GLY domain containing linker protein family, member 4 | 0.91 | 0.88 | |
| NM_153507 | Cpne2 | copine II | 0.91 | 0.92 | |
| NM_146067 | Cpped1 | calcineurin-like phosphoesterase domain containing 1 | 0.86 | 0.88 | |
| NM_026062 | Fam69a | family with sequence similarity 69, member A | 0.87 | 0.87 | |
| NM_001033550 | Lrrc8b | leucine rich repeat containing 8 family, member B | 0.92 | 0.92 | |
| NM_026845 | Ppil1 | peptidylprolyl isomerase (cyclophilin)-like 1 | 0.87 | 0.83 | |
| NM_024196 | Tbc1d20 | TBC1 domain family, member 20 | 0.90 | 0.89 | |
The genes in Table 2 have FDR-adjusted p-values for each individual comparison under 0.25. Expression is given as fold change in lactating maternal LS relative to pups removed or virgin groups.
Discussion
This study used Affymetrix microarray and quantitative real-time PCR to identify gene expression changes occurring naturally in LS of mice in association with the transition from a virgin to postpartum state at multiple levels of analysis, from single genes to enriched networks and biological pathways. Additionally, we evaluated the degree to which expression changes are dependent on the continued sensory input from pups. The results reveal numerous changes in genes of interest that may be of particular importance as markers of the maternal phenotype, including differential regulation of GABAA receptor subtypes.
Dynamic Regulation of GABAA Receptor Subunits in Maternal LS
The GABAA receptor was selected as a protein of interest based on previous studies suggesting the importance of GABA signaling in the LS for maternal behavior. Site-directed application of GABAA receptor antagonist was shown to inhibit offspring protection in lactating maternal mice [10], and it has recently been demonstrated that glutamic acid decarboxylase (GAD) 65 and 67, the rate-limiting enzymes in the production of GABA, are upregulated in rostral LS of postpartum maternal mice [11]. The present study explores whether changes in GABAA receptor expression might be a mechanism of regulating GABA signaling in LS.
In the microarray analysis, three GABAA receptor subunits showed significant expression differences in lactating maternal LS relative to virgin, including δ, ε, and α4 (FDR-adjusted p<0.25). Several additional subunits, such as α1, ρ2, γ1, α5, θ, and β2, exhibited possible significance (PLIER p<0.05). The most ubiquitous and abundant GABAA receptor subtype in the brain is composed of two α1, two β2, and one γ2 subunit [12], [14]. α1 and β2 displayed relatively small fold changes (1.06 and 1.05), while much more robust alterations were detected for δ (0.75), ε (1.39), α4 (0.88), and θ (1.26 (Table S1)). qPCR confirmation of the α1 result showed an increase that did not reach significance (p = 0.077). This suggests that the commonly found α1β2γ2 subtype is not dynamically regulated (at least at high levels) in the LS during the transition from a virgin to lactating maternal state. Conversely, qPCR confirmed significant expression changes for δ, ε, α4, and θ subunits (Fig. 2).
δ assembles with α4 endogenously to form a benzodiazepine insensitive receptor that mediates a tonic, rather than phasic, form of inhibitory current. α4/δ containing receptors have a high affinity for GABA, resist desensitization, and exhibit some degree of agonist-independent activity. They are located extrasynaptically and are positively modulated by the binding of neurosteroids [16]–[18]. Expression of the δ subunit has been shown to be dynamically downregulated in several brain regions in pregnant mice and is proposed to be a mechanism of maintaining a steady level of inhibition in the face of increasing progesterone associated with the prepartum period [19]. Our results provide evidence that a reduced sensitivity to neurosteroid influence on GABA signaling in the LS may also be important for maintaining the maternal phenotype after parturition. Neurosteroids can be synthesized in peripheral locations before crossing the blood brain barrier and are also produced locally in some regions of the brain by certain neurons and glia that exert autocrine and paracrine-like effects [20]. It is also possible that a reduction in tonic GABA inhibition could allow for the emergence of more precise rapid, synaptic transmission. One study has shown that transgenic mice expressing more extrasynaptic GABAA receptors than their wild type counterparts exhibited greater tonic currents and smaller GABA-mediated mIPSCs [21]. If an inverse relationship between these two modes of signaling exists, then regulation of the relative amounts of synaptic versus extrasynaptic GABAA receptors could have significant effects on the nature of net neuronal activity in the maternal LS.
Additional Changes in Genes Related to Neuronal Signaling in Maternal LS
A large and diverse group of genes related to neuronal signaling was enriched in the maternal LS relative to virgin, including a large number of potassium channel related genes (Fig. 4). These channels are involved with many physiological functions, including regulation of neurotransmitter release and neuronal excitability. They also play a crucial role in shaping the action potential [22]–[24]. Regulation of potassium channel subunits appears to be complex; approximately half of potassium channel genes in this enriched cluster were upregulated in maternal LS compared to virgin, while the other half were downregulated. The ion channel cluster also includes genes involved in purinergic signaling, calcium channels, and glutamate signaling. The transient receptor potential cation channel, subfamily C, member 4 (Trpc4) was detected by microarray to be elevated by 13% in maternal LS compared to both virgin and pups removed groups (Fig. 4; Table S1). Trpc4 is interesting because it has a very restricted distribution in the brain and is most highly expressed in LS. Trpc4 and Trpc1 are the predominant Trpcs in LS, and may form heteromeric channels responsible for maintaining plateau potentials that can cause epileptic burst firing and even cell death in LS neurons [25]. Trpc4 knockout rats exhibit a phenotype of reduced social exploration and heightened social anxiety [26]. It has additionally been shown that Trpc4 is expressed in tyrosine hydroxylase positive dopamine neurons in the ventral tegmental area [27], demonstrating a possible link to reward circuitry. It is therefore possible that the dynamic regulation of Trpc4 expression in the postpartum LS could be a central mechanism of altering sociability in maternal mammals.
The modulatory neurotransmitter dopamine contributes to learning motivation and reward associated behaviors [28]. Dopamine receptors 1a (Drd1a) and 2 (Drd2) were downregulated in maternal LS relative to virgin (by 21% and 17%, respectively), but the functional significance of the changes is still be to be evaluated. There is growing evidence that Drd2 interacts with the adenosine A2a receptor (Adora2a) [29], which is also downregulated in the maternal LS. Hypocretin (orexin) receptor 1 (Hcrtr1) was 24% higher in maternal LS relative to virgin in the microarray results. Hypocretin is produced in lateral hypothalamic neurons that project to numerous brain regions, including the septum, and these neurons show altered activity in the postpartum state [30]. Hypocretin acting on Hcrtr1 influences arousal, vigilance, and feeding behavior [31]–[33] as well as maternal behaviors [34] .
The kappa opioid receptor (Oprk1) displayed a 17% decrease in maternal LS relative to virgin. Some studies report that site specific manipulations of Oprk1 can mediate anxiety-like behavior, the effects of social defeat stress, and the extent to which social play is rewarding [35]–[37]. Plasmolipin (Pllp, also known as plasma membrane proteolipid) is a proteolipid expressed in oligodendrocytes [38] and is a component of myelin, representing up to nearly 5% of the membrane [39]. Changes in Pllp expression, including the 17% reduction in maternal LS compared to virgin as detected by our qPCR analysis (Fig. 3) could influence myelination and action potential conductance. Pllp also makes up 1–2% of clathrin-coated vesicles which appear to target specifically to the synaptic plasma membrane [40]. Additionally, Pllp has been observed to increase dramatically in primary cultures during differentiation for embryonic rat neurons and neonatal rat glia [41]. Therefore, it appears to have roles in development, myelination, and synaptic function in the LS. Collectively, the striking enrichment in neuronal signaling and ion channel activity represents a significant alteration in basic signaling properties and neuronal excitability in the maternal LS.
Large Expression Changes of Genes Related to Development and Cell Differentiation in Maternal LS
A number of studies have indicated plasticity of the maternal brain, and it is possible that the maternal state represents an endpoint in neuronal or glial differentiation. It could be that some cells do not fully differentiate until the postpartum state, and this contributes to the emergence of the maternal phenotype. We identified changes in a number of genes with strong ties to developmental processes and neuronal/glial differentiation in the maternal LS (Fig. 5). A subset of these genes were of interest (and confirmed by qPCR) because they had also been highlighted as showing altered expression in prior and ongoing maternal brain studies ([15], unpublished observations). Thus, these genes could provide key support for long lasting developmental changes in the maternal LS. A 21% increase in Chemokine (C-X-C motif) ligand 12 (Cxcl12, also known as SDF1-1) was detected by qPCR in the LS of lactating maternal mice relative to virgin (Fig. 3). Cxcl12 has been shown to be essential for the guidance of neuronal and glial stem cells in embryonic development [42], and has also been linked to angiogenesis in both embryo and tumor formation [43], [44]. Cxcl12 is unique among chemokines, which are commonly promiscuous in their receptor binding, in that it is highly specific to its receptor, Cxcr4. Interestingly, our microarray analysis reports that Cxcr4 is significantly downregulated in lactating maternal LS compared to virgin (Fold change 0.82, FDR-adjusted p = 0.13 (Fig. 5; Table S1)), opposite to the observed upregulation of the Cxcl12 ligand. While Cxcl12 and Cxcr4 are known to be stably expressed and function in mature cells, their apparent dynamic regulation in postpartum LS suggests that developmental processes with which they are involved could be of importance in shaping the maternal phenotype.
A highly significant 32% decrease in fatty acid binding protein 7 (Fabp7) was observed by qPCR in maternal LS relative to virgin (Fig. 3). Fabp7 is a brain-specific member of a family of long chain polyunsaturated fatty acid binding proteins. Polyunsaturated fatty acids, such as arachidonic acid and docosahexaenoic acid are important structural components of the developing brain [45]. Fabp7 is responsible for transporting these hydrophobic molecules through cytoplasmic environments to their ultimate membranous destination [46]. In addition to its role in development, it has been shown that a null mutation in Fabp7 results in a rodent phenotype characterized by elevated anxiety and fear memory [47]. The robust Fabp7 decrease in lactating maternal LS relative to virgin detected in the present study is in agreement with a previous experiment carried out with lactating maternal whole septum of a mouse strain previously selected for high offspring protection [15] and in hypothalamus of outbred mice [48]. This confirmation is notable because it demonstrates that the observed phenomenon is not likely strain specific and may occur in multiple brain regions. These results suggest that Fabp7 may play a role in developmental processes involved with plasticity in the maternal LS and actively mediate emotional changes associated with the postpartum period.
Cytokine signaling in the CNS influences how stem cells respond to hormones and plays an important role in the differentiation of neural progenitor cells into either glia or neurons [49]. A family of genes called “suppressors of cytokine signaling” (Socs) is known to be a negative regulator of such pathways [50]. Socs2 is the most abundant of these proteins in the CNS [51], and it is thought to mediate a negative feedback loop on messaging pathways downstream of growth hormone binding in the central nervous system. Stem cells cultured from Socs2 knockout mice produced 50% fewer neurons when induced to differentiate, and generated more astrocytic glial cells [52]. Conversely, Socs2 overexpressing stem cells yielded a higher than normal neuron to astrocyte ratio after differentiation. Socs2 was dramatically upregulated in maternal LS compared to both virgin and pups removed (46% and 53%; Fig. 3). The strength of developmental relevance in these microarray data provides some support for the idea that maternity can be viewed as another stage in the mammalian life cycle characterized by terminal differentiation of the CNS. If indeed these gene changes reflect developmental activity in the maternal LS, further anatomical and histological studies may be able to provide more direct evidence by visualizing structural changes involved in shaping the maternal brain.
Additional Enrichment Findings in Maternal LS
Functional annotation clustering revealed a small cluster of genes influencing the synthesis of cyclic nucleotides (Fig. 4), with most members exhibiting downregulation in the maternal LS compared to virgin. Cyclic nucleotides play a central role in a variety of intracellular signaling pathways as second messengers [53], [54] and can modulate neuronal excitability via the binding of cyclic nucleotide gated (CNG) channels. CNG channels are well known for their role in sensory transduction in retinal and olfactory cells, but are also expressed widely in the mammalian CNS and are likely involved with synaptic plasticity and development [55], [56]. The enrichment of this gene cluster indicates that, in addition to altering ion channel activity directly through expression of the channels themselves, there may be an additional level of regulation facilitated by fluctuating levels of cyclic nucleotides available to cells in the maternal LS.
There was a large degree of enrichment in a nucleosomal gene cluster primarily composed of histone genes. These genes were almost exclusively downregulated, but it is not clear what implications this has on the function of the maternal LS. It is possible that a downregulation of histone mRNA may reflect changes in post-transcriptional processes that influence the stability of histone mRNA transcripts [57], [58]. The visualized cluster in Figure 4 shows a strong interaction between chromobox homolog 1 (Cbx1, also known as heterochromatin protein 1 β) and the H3 histone. Cbx1 plays a major role in regulating higher order chromatin structure and gene transcription [59]. Additionally, Cbx1−/− knockout mice exhibit a lethal phenotype characterized by aberrant neocortical development with reduced proliferation of neuronal precursors, demonstrating developmental relevance [60]. While the significance of the nucleosomal gene cluster is not clear, the robustness and consistency of its enrichment is profound, and may influence chromatin remodeling in LS in the establishment of the maternal brain.
The Ras related gene cluster includes members of the Ras family, which is involved with many different cellular processes. It has been widely linked to tumor formation and has been shown to contribute in conjunction with thymoma viral proto-oncogene (Akt) signaling to glioblastoma formation in the brain [61]. All three members of the Akt family exhibited indications of altered expression in maternal LS compared to virgin in our microarray results (Table S1). In addition to cancer, certain Ras members of this enriched gene cluster also influence exocytosis and vesicle trafficking [62], [63].
Expression Changes of Anxiety Related Genes in Maternal LS
A number of genes, such as the GABAA receptors and other neuronal signaling genes have been linked to anxiety. Glutathione reductase (Gsr) is also linked to anxiety, and qPCR confirmed a 12% decrease in Gsr in the maternal LS compared to virgin (Fig. 3). Lentiviral in vivo overexpression of Gsr in the cingulate cortex of C57BL/6J mice results in significant increases in anxiety-related behavior [64]. The same study also showed that, across several strains of mice, Gsr activity was highest in the most anxious strains and lowest in the least anxious strains, suggesting relevance of Gsr activity to normal variation in anxiety. The transition from virgin to lactating maternal states involves a natural change in anxiety, in which postpartum mice respond less to general stressors [65]. The Gsr mRNA reduction we measured in maternal mice is in agreement with these observations, but whether these changes in LS are causally linked to altered anxiety is not known.
Sensory Input Contributions to Gene Expression in Maternal LS
The present study revealed that a subset of genes was strongly influenced by the presence of pups. The maternal state can involve both short term and long term changes. Studies indicate that maternal characteristics are more strongly and stably expressed with increasing numbers of pregnancies and that this occurs with long lasting gene expression changes [66]–[68]. Among the significant 809 gene expression changes between maternal and virgin LS, 69 were found to also be significantly different (FDR-adjusted p<0.25) between maternal and pups removed LS (Table 2). For these genes, the removal of pups most fully restored virgin-like levels of mRNA even after the experience of mating, pregnancy, and parturition. We interpret these genes as being more dependent on the continued presence of pups, in contrast to genes that were differentially expressed between maternal and virgin LS but not between maternal and pups removed LS. This list represents an interesting subset of genes that are more environmentally malleable to the social aspects of maternity. While formal pathway analysis cannot be reliably conducted on such a small set of genes, it can be seen from the basic categorization in Table 2 that they span numerous functional groupings. Fabp7 and Socs2 are notable members of this subset, which suggests that the developmental processes with which they are involved may be ongoing and driven in the postpartum period by social interaction.
Materials and Methods
Animals
Outbred hsd:ICR female mice (Mus domesticus) (Harlan, Madison WI) were used for all experiments. Nulliparous animals were split into three age-matched groups (∼70 days of age at time of dissection), designated as lactating maternal, pups removed, and virgin. For mating, females in the lactating maternal and pups removed groups were housed in polypropylene cages with a breeder male for 2 weeks. Virgin females were concurrently co-housed with one another to provide similar levels of exposure to social stimuli. After the separation of breeder males, all females (pregnant and virgin) were housed individually and provided precut nesting material until dissections. Under this schedule, all females experienced similar levels of co-housing and single housing to minimize potential effects of isolation-induced stress. Cages were changed once per week until pups were born (postpartum Day 0), after which cages were not changed again for all animals until dissection. On Day 0, pups were culled, if necessary, to standardize litter size to eleven. For females in the pups removed group, pups were removed from the cage on postpartum Day 2. The pups removed group was included in the experimental design to provide insight as to whether or not continued sensory input from pups is required, in addition to parturition, to generate expression changes characteristic of the maternal phenotype. All animals were housed in the same room with cages of each experimental group positioned in an alternating fashion on the same shelves. A 12∶12 light/dark cycle with lights on at 06∶00 h CST was used. Female mice were provided with ad lib access to breeder chow (Harlan) and tap water. Procedures were performed in strict accordance with the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and all studies were approved by the University of Wisconsin Animal Care and Use Committee.
Tissue Collection and RNA Extraction
On postpartum Day 7, brains were removed from females in the lactating maternal and pups removed groups between 10∶00 and 12∶00 h. Brains from age-matched virgin females were collected on the same day, during the same time period. Dissections were alternated among groups so that an equal number of dissections from each group were performed. Animals were lightly anesthetized with isoflurane and decapitated. After decapitation, vaginal lavage was performed on virgin and pups removed females to determine their estrous state. All females in the pups removed group were diestrous, while virgin females exhibited variance in estrous states. To control for effects of estrous cycling on gene expression, only diestrous virgins were used for analysis [69], [70]. Tissue collection was performed as previously described [15]. Briefly; the whole brain was removed, snap frozen in isopentane on dry ice, and stored at −80°C until sliced. Brain sections were sliced in a cryostat (Leica, CM1850, Bannockburn, IL, USA) at 200 micron thickness and mounted on glass slides. Target tissue was extracted by micropunch technique [71]. Microdissection of frozen brain sections was performed with the Brain Punch Set (Stoelting, Wood Dale, IL, USA) under a dissecting microscope. LS, including caudal and rostral portions, was collected bilaterally from Bregma 1.10 to 0.14 mm (Fig. 1) and consolidated such that each animal yielded one sample of LS. Microdissections from eight animals in each group were flash frozen on dry ice and stored at −80°C until processing for gene array analysis or quantitative real-time PCR. Total RNA was extracted and prepared in matched triplets, one sample from each experimental group, using the Aurum Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad, Hercules, CA, USA) in accordance with manufacturer’s instructions. After isolation, RNA integrity was assessed using Agilent RNA 6000 Nano Chips and the Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). The purity and yield of RNA samples were determined with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Purified total RNA was stored at −80°C until processing.
High-Density Oligonucleotide Array Hybridization
Six out of eight samples collected for each group (n = 6 per group) were randomly selected for use in the microarray experiment. Microarray analysis was performed with the GeneChip Mouse Gene 1.0 ST array (Affymetrix, Santa Clara, CA, USA) using targets derived from total RNA isolated from LS as described above. cDNA for array hybridization was reverse transcribed from 200 ng of total RNA using an Ambion GeneChip WT Expression Kit (Ambion, Austin, TX, USA) in accordance with the manufacturer’s specifications. In short, total RNA was used to synthesize double-stranded cDNA, which was then used as a template to synthesize single-stranded cRNA. This cRNA was subsequently used as a template for one round of single-stranded cDNA synthesis, and the resulting DNA-RNA hybrids were then degraded with RNase H. Amplified single-stranded cDNA was fragmented and biotinylated with an Affymetrix WT Terminal Labeling Kit (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s instructions. Fragmented, labeled cDNA samples were hybridized with the arrays for 16 hours at 45°C. Hybridized arrays were washed, stained, and subsequently scanned at 570 nm on an Affymetrix GC3000 G7 Scanner. The Affymetrix Command Console v. 3.1.1.1229 was used to extract and process data from scans. cDNA synthesis, fragmentation, labeling, array hybridization, and scanning were performed by the Gene Expression Center at the University of Wisconsin-Madison.
Probeset Level Summarization and Microarray Statistical Analysis
Probeset level summarization and data normalization were performed with the PLIER algorithm and GC bin background correction using Affymetrix Power Tools v. 1.12.0. The raw and summarized microarray data presented in this publication have been deposited in NCBI’s Gene Expression Omnibus [72], and are accessible through GEO Series accession number GSE43627. GEO reporting meets the requirements of the Minimum Information About a Microarray Experiment (MIAME). An array-specific, empirical Bayesian implementation of ANOVA was used in the BioConductor package limma v.3.6.9 [73] to perform inferential statistics on gene expression changes between groups (lactating maternal vs. virgin, and lactating maternal vs. pups removed). Nominal and false discovery rate (FDR) corrected p-values were calculated, and fold changes for each gene were calculated in Excel by dividing the limma-calculated average lactating maternal expression by the limma-calculated average expression of virgin or pups removed groups.
Quantitative Real-time PCR (qPCR)
To confirm expression changes detected by microarray analysis, qPCR was performed on genes of interest (n = 8 per group). Target genes included five GABAA receptor subunits; α1 (Gabra1), α4 (Gabra4), δ (Gabrd), ε (Gabre), and θ (Gabrq), as well as five genes currently being evaluated as potential markers of the maternal phenotype; chemokine (C-X-C) motif ligand 12 (Cxcl12), fatty acid binding protein 7 (Fabp7), glutathione reductase (Gsr), plasma membrane proteolipid (Pllp), and suppressor of cytokine signaling 2 (Socs2). Two stable reference genes were used to normalize relative expression results in genes of interest; Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (Ywhaz), and peptidylprolyl isomerase A (Ppia). Primer information can be viewed in Table S3. A SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA) was used to reverse transcribe 100 ng of RNA to cDNA in an Eppendorf MasterCycler Personal PCR machine (Eppendorf, Hamburg, Germany) with poly-T 20mer primers.
Functional Annotation Clustering and Interaction Mapping
DAVID’s functional annotation clustering is a tool which analyzes enrichment in a variety of precompiled gene sets assembled by shared ontology, protein domains, and biological pathways within a given list of input genes. DAVID then groups related gene sets into clusters. Enrichment scores are generated by detecting overrepresentation of a cluster within the input list compared to a reference background [74]. Functional annotation clustering with high classification stringency was performed on the 809 genes with FDR-adjusted p-values less than 0.25 between the lactating maternal and virgin groups, as reported by the probeset level summarization. Of these 809 genes, DAVID recognized 765 of them in its analysis. For several significant clusters, the non-redundant list of gene members comprising them were used as input in GeneMANIA to visualize each cluster as a network of nodes connected by lines representing known interactions and similarities.
Supporting Information
Full list of all microarray targets, their relative expression, and FDR-adjusted p-values.
(XLSX)
Expression data summary for enriched gene clusters found in NIH DAVID functional annotation clustering.
(XLSX)
Primers for genes of interest and reference genes used in real-time quantitative PCR experiments.
(DOCX)
Acknowledgments
The authors wish to thank Sharon Stevenson for managerial support, Wayne Davis and the University of Wisconsin-Madison Gene Expression Center for microarray technical assistance, and Kate Skogen and Jeff Alexander for animal care.
Funding Statement
This work was supported by United States National Institutes of Health Grant R01 MH 085642 to S.G. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Montagnese CM, Poulain DA, Vincent JD, Theodosis DT (1987) Structural plasticity in the rat supraoptic nucleus during gestation, post-partum lactation and suckling-induced pseudogestation and lactation. J Endocrinol 115: 97–105. [DOI] [PubMed] [Google Scholar]
- 2. Levy F, Gheusi G, Keller M (2011) Plasticity of the parental brain: a case for neurogenesis. J Neuroendocrinol 23: 984–993. [DOI] [PubMed] [Google Scholar]
- 3. Lisk RD (1971) Oestrogen and progesterone synergism and elicitation of maternal nest-building in the mouse (Mus musculus). Anim Behav 19: 606–610. [DOI] [PubMed] [Google Scholar]
- 4. Kuroda KO, Tachikawa K, Yoshida S, Tsuneoka Y, Numan M (2011) Neuromolecular basis of parental behavior in laboratory mice and rats: with special emphasis on technical issues of using mouse genetics. Prog Neuropsychopharmacol Biol Psychiatry 35: 1205–1231. [DOI] [PubMed] [Google Scholar]
- 5. Broad KD, Curley JP, Keverne EB (2006) Mother-infant bonding and the evolution of mammalian social relationships. Philos Trans R Soc Lond B Biol Sci 361: 2199–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slotnick BM, Nigrosh BJ (1975) Maternal behavior of mice with cingulate cortical, amygdala, or septal lesions. J Comp Physiol Psychol 88: 118–127. [DOI] [PubMed] [Google Scholar]
- 7. Sheehan TP, Chambers RA, Russell DS (2004) Regulation of affect by the lateral septum: implications for neuropsychiatry. Brain Res Brain Res Rev 46: 71–117. [DOI] [PubMed] [Google Scholar]
- 8. Risold PY, Swanson LW (1997) Chemoarchitecture of the rat lateral septal nucleus. Brain Res Brain Res Rev 24: 91–113. [DOI] [PubMed] [Google Scholar]
- 9. Risold PY, Swanson LW (1997) Connections of the rat lateral septal complex. Brain Res Brain Res Rev 24: 115–195. [DOI] [PubMed] [Google Scholar]
- 10. Lee G, Gammie SC (2009) GABA(A) receptor signaling in the lateral septum regulates maternal aggression in mice. Behav Neurosci 123: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao C, Driessen T, Gammie SC (2012) Glutamic acid decarboxylase 65 and 67 expression in the lateral septum is up-regulated in association with the postpartum period in mice. Brain Res 1470: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whiting PJ, Bonnert TP, McKernan RM, Farrar S, Le Bourdelles B, et al. (1999) Molecular and functional diversity of the expanding GABA-A receptor gene family. Ann N Y Acad Sci 868: 645–653. [DOI] [PubMed] [Google Scholar]
- 13. Levitan ES, Schofield PR, Burt DR, Rhee LM, Wisden W, et al. (1988) Structural and functional basis for GABAA receptor heterogeneity. Nature 335: 76–79. [DOI] [PubMed] [Google Scholar]
- 14. Olsen RW, Sieghart W (2009) GABA A receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology 56: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao C, Saul MC, Driessen T, Gammie SC (2012) Gene expression changes in the septum: possible implications for microRNAs in sculpting the maternal brain. PLoS One 7: e38602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farrant M, Nusser Z (2005) Variations on an inhibitory theme: phasic and tonic activation of GABA(A) receptors. Nat Rev Neurosci 6: 215–229. [DOI] [PubMed] [Google Scholar]
- 17. Sinkkonen ST, Vekovischeva OY, Moykkynen T, Ogris W, Sieghart W, et al. (2004) Behavioural correlates of an altered balance between synaptic and extrasynaptic GABAAergic inhibition in a mouse model. Eur J Neurosci 20: 2168–2178. [DOI] [PubMed] [Google Scholar]
- 18. Maguire JL, Stell BM, Rafizadeh M, Mody I (2005) Ovarian cycle-linked changes in GABA(A) receptors mediating tonic inhibition alter seizure susceptibility and anxiety. Nat Neurosci 8: 797–804. [DOI] [PubMed] [Google Scholar]
- 19. Maguire J, Ferando I, Simonsen C, Mody I (2009) Excitability changes related to GABAA receptor plasticity during pregnancy. J Neurosci 29: 9592–9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lambert JJ, Cooper MA, Simmons RD, Weir CJ, Belelli D (2009) Neurosteroids: endogenous allosteric modulators of GABA(A) receptors. Psychoneuroendocrinology 34 Suppl 1S48–58. [DOI] [PubMed] [Google Scholar]
- 21. Wisden W, Cope D, Klausberger T, Hauer B, Sinkkonen ST, et al. (2002) Ectopic expression of the GABA(A) receptor alpha6 subunit in hippocampal pyramidal neurons produces extrasynaptic receptors and an increased tonic inhibition. Neuropharmacology 43: 530–549. [DOI] [PubMed] [Google Scholar]
- 22. Pongs O (1992) Molecular biology of voltage-dependent potassium channels. Physiol Rev 72: S69–88. [DOI] [PubMed] [Google Scholar]
- 23. Armstrong CM, Hille B (1998) Voltage-gated ion channels and electrical excitability. Neuron 20: 371–380. [DOI] [PubMed] [Google Scholar]
- 24. Simmons ML, Chavkin C (1996) k-Opioid receptor activation of a dendrotoxin-sensitive potassium channel mediates presynaptic inhibition of mossy fiber neurotransmitter release. Mol Pharmacol 50: 80–85. [PubMed] [Google Scholar]
- 25. Phelan KD, Mock MM, Kretz O, Shwe UT, Kozhemyakin M, et al. (2012) Heteromeric canonical transient receptor potential 1 and 4 channels play a critical role in epileptiform burst firing and seizure-induced neurodegeneration. Mol Pharmacol 81: 384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmus K, Wang J-G, Varnell A, Ostertag E, Cooper D (2011) Sociability is decreased following deletion of the trpc4 gene. Nature Precedings.
- 27. Sergeeva OA, Korotkova TM, Scherer A, Brown RE, Haas HL (2003) Co-expression of non-selective cation channels of the transient receptor potential canonical family in central aminergic neurones. J Neurochem 85: 1547–1552. [DOI] [PubMed] [Google Scholar]
- 28. Schultz W (2010) Dopamine signals for reward value and risk: basic and recent data. Behav Brain Funct 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Childs E, Hohoff C, Deckert J, Xu K, Badner J, et al. (2008) Association between ADORA2A and DRD2 polymorphisms and caffeine-induced anxiety. Neuropsychopharmacology 33: 2791–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Espana RA, Berridge CW, Gammie SC (2004) Diurnal levels of Fos immunoreactivity are elevated within hypocretin neurons in lactating mice. Peptides 25: 1927–1934. [DOI] [PubMed] [Google Scholar]
- 31. Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, et al. (1999) Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98: 437–451. [DOI] [PubMed] [Google Scholar]
- 32. Hagan JJ, Leslie RA, Patel S, Evans ML, Wattam TA, et al. (1999) Orexin A activates locus coeruleus cell firing and increases arousal in the rat. Proc Natl Acad Sci U S A 96: 10911–10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, et al.. (1998) Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92: 1 page following 696. [DOI] [PubMed]
- 34. D’Anna KL, Gammie SC (2006) Hypocretin-1 dose-dependently modulates maternal behaviour in mice. J Neuroendocrinol 18: 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bruchas MR, Land BB, Lemos JC, Chavkin C (2009) CRF1-R activation of the dynorphin/kappa opioid system in the mouse basolateral amygdala mediates anxiety-like behavior. PLoS One 4: e8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McLaughlin JP, Li S, Valdez J, Chavkin TA, Chavkin C (2006) Social defeat stress-induced behavioral responses are mediated by the endogenous kappa opioid system. Neuropsychopharmacology 31: 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vanderschuren LJ, Niesink RJ, Spruijt BM, Van Ree JM (1995) Mu- and kappa-opioid receptor-mediated opioid effects on social play in juvenile rats. Eur J Pharmacol 276: 257–266. [DOI] [PubMed] [Google Scholar]
- 38. Fischer I, Cochary EF, Konola JT, Romano-Clark G (1991) Expression of plasmolipin in oligodendrocytes. J Neurosci Res 28: 81–89. [DOI] [PubMed] [Google Scholar]
- 39. Cochary EF, Bizzozero OA, Sapirstein VS, Nolan CE, Fischer I (1990) Presence of the plasma membrane proteolipid (plasmolipin) in myelin. J Neurochem 55: 602–610. [DOI] [PubMed] [Google Scholar]
- 40. Sapirstein VS, Nolan C, Stern R, Ciocci M, Masur SK (1988) Identification of the plasma membrane proteolipid protein as a constituent of brain coated vesicles and synaptic plasma membrane. J Neurochem 51: 925–933. [DOI] [PubMed] [Google Scholar]
- 41. Shea TB, Fischer I, Sapirstein V (1986) Expression of a plasma membrane proteolipid during differentiation of neuronal and glial cells in primary culture. J Neurochem 47: 697–706. [DOI] [PubMed] [Google Scholar]
- 42. Tran PB, Miller RJ (2003) Chemokine receptors: signposts to brain development and disease. Nat Rev Neurosci 4: 444–455. [DOI] [PubMed] [Google Scholar]
- 43. Li M, Ransohoff RM (2009) The roles of chemokine CXCL12 in embryonic and brain tumor angiogenesis. Semin Cancer Biol 19: 111–115. [DOI] [PubMed] [Google Scholar]
- 44. Banisadr G, Skrzydelski D, Kitabgi P, Rostene W, Parsadaniantz SM (2003) Highly regionalized distribution of stromal cell-derived factor-1/CXCL12 in adult rat brain: constitutive expression in cholinergic, dopaminergic and vasopressinergic neurons. Eur J Neurosci 18: 1593–1606. [DOI] [PubMed] [Google Scholar]
- 45. Gordon N (1997) Nutrition and cognitive function. Brain Dev 19: 165–170. [DOI] [PubMed] [Google Scholar]
- 46. Gerstner JR, Bremer QZ, Vander Heyden WM, Lavaute TM, Yin JC, et al. (2008) Brain fatty acid binding protein (Fabp7) is diurnally regulated in astrocytes and hippocampal granule cell precursors in adult rodent brain. PLoS One 3: e1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Owada Y, Abdelwahab SA, Kitanaka N, Sakagami H, Takano H, et al. (2006) Altered emotional behavioral responses in mice lacking brain-type fatty acid-binding protein gene. Eur J Neurosci 24: 175–187. [DOI] [PubMed] [Google Scholar]
- 48. Gammie SC, Hasen NS, Awad TA, Auger AP, Jessen HM, et al. (2005) Gene array profiling of large hypothalamic CNS regions in lactating and randomly cycling virgin mice. Brain Res Mol Brain Res 139: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Turnley AM (2005) Role of SOCS2 in growth hormone actions. Trends Endocrinol Metab 16: 53–58. [DOI] [PubMed] [Google Scholar]
- 50. Wang J, Campbell IL (2002) Cytokine signaling in the brain: putting a SOCS in it? J Neurosci Res 67: 423–427. [DOI] [PubMed] [Google Scholar]
- 51. Ransome MI, Goldshmit Y, Bartlett PF, Waters MJ, Turnley AM (2004) Comparative analysis of CNS populations in knockout mice with altered growth hormone responsiveness. Eur J Neurosci 19: 2069–2079. [DOI] [PubMed] [Google Scholar]
- 52. Turnley AM, Faux CH, Rietze RL, Coonan JR, Bartlett PF (2002) Suppressor of cytokine signaling 2 regulates neuronal differentiation by inhibiting growth hormone signaling. Nat Neurosci 5: 1155–1162. [DOI] [PubMed] [Google Scholar]
- 53. Vandamme J, Castermans D, Thevelein JM (2012) Molecular mechanisms of feedback inhibition of protein kinase A on intracellular cAMP accumulation. Cell Signal 24: 1610–1618. [DOI] [PubMed] [Google Scholar]
- 54. Jordan JD, Landau EM, Iyengar R (2000) Signaling networks: the origins of cellular multitasking. Cell 103: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bradley J, Zhang Y, Bakin R, Lester HA, Ronnett GV, et al. (1997) Functional expression of the heteromeric “olfactory” cyclic nucleotide-gated channel in the hippocampus: a potential effector of synaptic plasticity in brain neurons. J Neurosci 17: 1993–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zufall F, Shepherd GM, Barnstable CJ (1997) Cyclic nucleotide gated channels as regulators of CNS development and plasticity. Curr Opin Neurobiol 7: 404–412. [DOI] [PubMed] [Google Scholar]
- 57. Sittman DB, Graves RA, Marzluff WF (1983) Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A 80: 1849–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Graves RA, Pandey NB, Chodchoy N, Marzluff WF (1987) Translation is required for regulation of histone mRNA degradation. Cell 48: 615–626. [DOI] [PubMed] [Google Scholar]
- 59. Lomberk G, Wallrath L, Urrutia R (2006) The Heterochromatin Protein 1 family. Genome Biol 7: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aucott R, Bullwinkel J, Yu Y, Shi W, Billur M, et al. (2008) HP1-beta is required for development of the cerebral neocortex and neuromuscular junctions. J Cell Biol 183: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, et al. (2000) Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 25: 55–57. [DOI] [PubMed] [Google Scholar]
- 62. Fischer von Mollard G, Stahl B, Li C, Sudhof TC, Jahn R (1994) Rab proteins in regulated exocytosis. Trends Biochem Sci 19: 164–168. [DOI] [PubMed] [Google Scholar]
- 63. Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525. [DOI] [PubMed] [Google Scholar]
- 64. Hovatta I, Tennant RS, Helton R, Marr RA, Singer O, et al. (2005) Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature 438: 662–666. [DOI] [PubMed] [Google Scholar]
- 65. Slattery DA, Neumann ID (2008) No stress please! Mechanisms of stress hyporesponsiveness of the maternal brain. J Physiol 586: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mann PE, Bridges RS (1992) Neural and endocrine sensitivities to opioids decline as a function of multiparity in the rat. Brain Res 580: 241–248. [DOI] [PubMed] [Google Scholar]
- 67. Mann PE, Kinsley CH, Ronsheim PM, Bridges RS (1989) Long-term effects of parity on opioid and nonopioid behavioral and endocrine responses. Pharmacol Biochem Behav 34: 83–88. [DOI] [PubMed] [Google Scholar]
- 68. Nephew BC, Bridges RS, Lovelock DF, Byrnes EM (2009) Enhanced maternal aggression and associated changes in neuropeptide gene expression in multiparous rats. Behav Neurosci 123: 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Romano GJ, Harlan RE, Shivers BD, Howells RD, Pfaff DW (1988) Estrogen increases proenkephalin messenger ribonucleic acid levels in the ventromedial hypothalamus of the rat. Mol Endocrinol 2: 1320–1328. [DOI] [PubMed] [Google Scholar]
- 70. Arosh JA, Parent J, Chapdelaine P, Sirois J, Fortier MA (2002) Expression of cyclooxygenases 1 and 2 and prostaglandin E synthase in bovine endometrial tissue during the estrous cycle. Biol Reprod 67: 161–169. [DOI] [PubMed] [Google Scholar]
- 71. Makino S, Gold PW, Schulkin J (1994) Effects of corticosterone on CRH mRNA and content in the bed nucleus of the stria terminalis; comparison with the effects in the central nucleus of the amygdala and the paraventricular nucleus of the hypothalamus. Brain Res 657: 141–149. [DOI] [PubMed] [Google Scholar]
- 72. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- 74. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full list of all microarray targets, their relative expression, and FDR-adjusted p-values.
(XLSX)
Expression data summary for enriched gene clusters found in NIH DAVID functional annotation clustering.
(XLSX)
Primers for genes of interest and reference genes used in real-time quantitative PCR experiments.
(DOCX)