Abstract
Deposition of amyloid-β (Aβ) in the cerebral cortex is thought to be a pivotal event in Alzheimer’s disease (AD) pathogenesis with a significant genetic contribution. Molecular imaging can provide an early noninvasive phenotype but small samples have prohibited genome-wide association studies (GWAS) of cortical Aβ load until now. We employed florbetapir (18F) positron emission tomography (PET) imaging to assess brain Aβ levels in vivo for 555 participants from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). More than six million common genetic variants were tested for association to quantitative global cortical Aβ load controlling for age, gender, and diagnosis. Independent genome-wide significant associations were identified on chromosome 19 within APOE (rs429358, p = 5.5 × 10−14) and on chromosome 3 upstream of BCHE (rs509208, p = 2.7 × 10−8) in a region previously associated with serum butyrylcholinesterase activity. Together, these loci explained 15% of the variance in cortical Aβ levels in this sample (APOE 10.7%, BCHE 4.3%). Suggestive associations were identified within ITGA6, near EFNA5, EDIL3, ITGA1, PIK3R1, NFIB, and ARID1B, and between NUAK1 and C12orf75. These results confirm the association of APOE with Aβ deposition and represent the largest known effect of BCHE on an AD-related phenotype. Butyrylcholinesterase has been found in senile plaques and this new association of genetic variation at the BCHE locus with Aβ burden in humans may have implications for potential disease-modifying effects of butyrylcholinesterase-modulating agents in the AD spectrum.
Keywords: Alzheimer’s disease (AD), amyloid, apolipoprotein E (APOE), butyrylcholinesterase (BCHE), florbetapir (AV-45), genome-wide association study (GWAS)
INTRODUCTION
Cortical deposition of amyloid-β (Aβ) peptide is thought to be a crucial early step in the cascade of events leading to Alzheimer’s disease (AD).1 The presence of cortical neuritic plaques, consisting of Aβ fibrils surrounded by degenerating neuronal processes, is a hallmark feature for pathologic diagnosis of AD.2 Aβ plaques have also been identified in individuals meeting clinical criteria for mild cognitive impairment (MCI)3 and have exhibited subtle relationships with cognition among older individuals without dementia or MCI symptoms.4 In addition, pathogenic mutations in genes related to Aβ processing, including the amyloid precursor protein (APP) and presenilin-1 and -2 (PSEN1, PSEN2) genes, have been discovered in patients with the rare, autosomal dominant form of AD.5 As a result, Aβ accumulation is increasingly proposed as a major antecedent ultimately leading to incident AD.6
The fundamental biological influences on brain Aβ levels are not yet fully understood. The strongest known genetic risk factor for AD is presence of the apolipoprotein E (APOE) ε4 allele,7 and in vitro and murine studies have proposed plausible links between the encoded ApoE E4 isoform and aberrant Aβ mechanisms.8 However, APOE ε4 is neither necessary nor sufficient for development of AD pathology, suggesting that the biology underlying Aβ accumulation involves contributions from other genes and pathways, as well as the environment.
The ongoing search for genetic modulators of brain Aβ deposition in humans has been bolstered by advances in imaging methods for noninvasive detection of fibrillar Aβ in vivo. While existing genetic studies of brain Aβ have focused on candidate genes due to moderate sample sizes, the enhanced stability of recently-developed 18F-labeled positron emission tomography (PET) imaging Aβ tracers such as florbetapir (also known as AV-45 or Amyvid) allows for more widespread evaluation to facilitate the acquisition of larger cohorts for analysis.9 Importantly, florbetapir PET data has demonstrated strong relationships with pathologically-verified assessments of fibrillar Aβ burden,10, 11 and thus represents a novel and robust quantitative phenotype that can be assessed in samples with heightened power for discovery of genes influencing Aβ neuropathology.
We used quantitative florbetapir PET data from 555 participants enrolled in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) to perform the first genome-wide association study (GWAS) of cortical Aβ burden in humans. We hypothesized that GWAS would confirm the association of APOE as well as identify other genetic modulators of brain Aβ deposition.
METHODS
Study participants
This report utilized data from ADNI12 (http://adni.loni.ucla.edu/), a multi-site longitudinal study that was launched in 2004 as a public-private partnership. The initial phase (ADNI-1) enrolled individuals aged 55–90 years who were recruited from over 50 sites across the United States and Canada and followed at 6-to 12-month intervals for 2–3 years. These individuals included approximately 200 healthy controls (HC), 400 patients with late MCI (LMCI), and 200 patients clinically diagnosed with probable AD. Subsequent phases (ADNI-GO and ADNI-2) have extended follow-up for existing participants and have enrolled additional individuals, including those meeting criteria for early MCI (EMCI). All participants provided written informed consent and study protocols were approved by each site’s institutional review board. Further information about ADNI, including full study protocols, complete inclusion and exclusion criteria, and data collection and availability can be found at http://www.adni-info.org/.
Florbetapir PET scans
PET imaging using the 18F-labeled Aβ tracer florbetapir was performed for participants enrolled in ADNI-GO or ADNI-2. Participants were administered a bolus injection of approximately 370 MBq florbetapir intravenously. Fifty minutes later, a 20-minute continuous cranial PET scan was initiated. Images were reconstructed immediately following the scan using iterative algorithms, and repeat scans were acquired if motion artifact was detected. Preprocessing of the scans was performed as previously described.13 Briefly, image frames were averaged, aligned to a standard space (AC-PC), resampled to a standard image and voxel size, smoothed to a uniform resolution, and normalized to an atlas-based bilateral and symmetric cerebellar reference region. This cerebellar reference region consisted largely but not exclusively of grey matter and was expected to exhibit nonspecific binding, ultimately resulting in standardized uptake value ratio (SUVR) images. These preprocessed scans were downloaded from the ADNI database (http://adni.loni.ucla.edu/) for 621 participants. For each scan, mean regional SUVR values were extracted for the frontal, parietal, temporal, limbic, and occipital lobes using the MarsBaR toolbox implemented in the Statistical Parametric Mapping 8 (SPM8) software (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/). The average SUVR for these 5 regions was then calculated to represent a global cortical measure of Aβ deposition to be used as a quantitative phenotype for GWAS. Overall, 19 participants (11 HC, 6 MCI, 2 AD) were excluded due to missing scan data or failed processing, leaving data for 602 individuals available for further analysis.
Genotyping and imputation
A blood draw for genomic DNA extraction was obtained at the screening or baseline visit for all study participants.14 Genotyping on these samples was performed according to manufacturer’s protocol (Illumina, Inc., San Diego, CA) using the Human610-Quad BeadChip (for subjects initially enrolled during ADNI-1) or the Human OmniExpress BeadChip (for subjects initially enrolled in ADNI-GO or ADNI-2). In addition, the two SNPs characterizing APOE ε2/ε3/ε4 status (rs429358 and rs7412) were genotyped separately as previously described14 and merged with the array data sets. All genotype data underwent stringent quality control procedures14 (sample exclusion for call rate < 95% or failed identity or gender check, and SNP exclusion for call rate < 95%, Hardy-Weinberg equilibrium test p < 1 × 10−6, or minor allele frequency (MAF) < 1%) using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/), version 1.07.15 In addition, to limit possible effects of population stratification, multidimensional clustering analysis was used to select only participants with non-Hispanic Caucasian (CEU or TSI) ancestry based on HapMap3 reference populations. Overall, one individual was excluded due to a failed gender check and 42 individuals were excluded based on ancestry.
Next, haplotype patterns from the 1000 Genomes Project reference panel were used to impute genotypes for markers not directly assayed. Prior to imputation, the orientation of all genotyped markers in relation to the plus strand alignment of the reference panel genome (NCBI build 37 coordinates) was verified and monomorphic variants from the reference panel were excluded. Minimac16 was used to impute samples within groups based on the genotyping platform employed (Illumina 610-Quad or OmniExpress). Following imputation, SNPs with r2 < 0.5 between imputed and assayed genotypes were removed.17 The remaining array SNPs demonstrated > 99.9% concordance between imputed and assayed genotypes. The independently-imputed data sets were then merged to generate a common set of more than 10 million SNPs for the full ADNI sample. Following quality control (SNP call rate < 95%, Hardy-Weinberg p < 1 × 10−6) and frequency filtering (MAF < 5%), 6,108,668 SNPs were included in the GWAS. Of the 602 participants with Aβ PET data, 559 individuals were included in the resulting genetic data set. Among these individuals, four pairs exhibited significant relatedness (PLINK identity by descent PI_HAT > 0.5) and therefore one individual from each pair was randomly selected for exclusion (2 HC, 1 EMCI, 1 LMCI), leaving 555 participants for the final GWAS sample.
Statistical analysis
For the GWAS, linear regression was performed using PLINK to determine the association of each SNP to global cortical Aβ levels. An additive genetic model was specified and age, gender, and diagnosis (through a set of binary dummy variables indicating HC, EMCI, LMCI, or AD) were applied as covariates. To account for multiple comparisons, we employed a conservative threshold for genome-wide significant association (p < 5 × 10−8) based on a Bonferroni correction of one million independent tests.18 Haploview19 (http://www.broad.mit.edu/mpg/haploview/) was used to generate Manhattan and Q-Q plots and SNAP20 (http://www.broadinstitute.org/mpg/snap/) was used to obtain regional association plots for selected loci. Post-hoc analyses, including hierarchical linear regression, effect size calculations, and exploratory correlation and interaction studies using Bonferroni corrections for multiple comparisons, were performed using IBM SPSS Statistics for Windows, Version 20.0 (Armonk, NY).
RESULTS
This study analyzed data from 555 ADNI participants with non-Hispanic Caucasian ancestry. Participants were diagnosed at the time of the PET scan as HC, EMCI, LMCI, or AD. EMCI subjects met clinical criteria for amnestic MCI21 but exhibited milder (between 1.0 and 1.5 standard deviations below age-associated norms) memory impairment (Table 1). In this sample, the EMCI group was younger than the other groups (p < 0.001). All groups displayed comparable levels of education but the LMCI group included fewer female participants compared to the HC group (Chi-square p = 0.029).
Table 1.
Selected demographic and clinical characteristics of ADNI participants at the time of PET scan
| HC (n=179) | EMCI (n=190) | LMCI (n=115) | AD (n=71) | |
|---|---|---|---|---|
| Age (years) | 76.68 (6.25) | 71.04 (7.41) | 75.61 (8.14) | 75.87 (8.15) |
| Women | 87 (49%) | 83 (44%) | 41 (36%) | 27 (38%) |
| Education (years) | 16.27 (2.72) | 15.89 (2.65) | 16.11 (2.90) | 16.04 (2.87) |
| APOE ε4 allele present | 41 (23%) | 77 (41%) | 49 (43%) | 45 (64%) |
| CDR-SOB | 0.07 (0.29) | 1.22 (0.73) | 1.73 (1.18) | 5.63 (2.70) |
| Mini Mental Status Examination | 29.07 (1.25) | 28.39 (1.52) | 27.74 (1.84) | 21.68 (4.24) |
| Logical memory immediate recall (WMS-R) | 14.94 (3.36) | 10.93 (2.81) | 8.74 (4.35) | 4.20 (3.10) |
| Logical memory delayed recall (WMS-R) | 14.08 (3.64) | 8.87 (1.73) | 6.13 (4.38) | 1.67 (2.50) |
Data are number (%) or mean (SD).
CDR-SOB = Clinical Dementia Rating-Sum of Boxes; WMS-R = Wechsler Memory Scale-Revised.
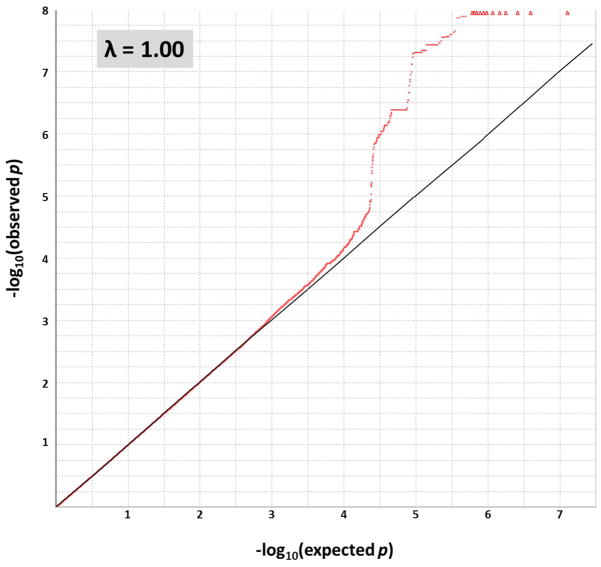
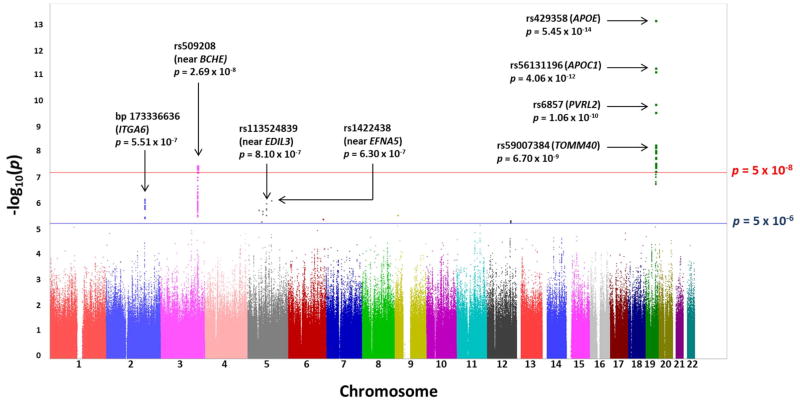
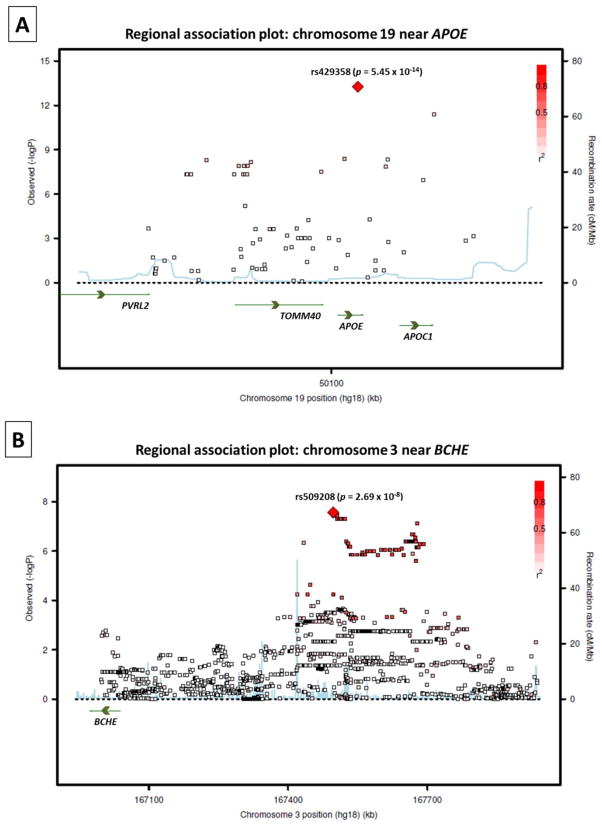
The GWAS results did not indicate evidence of spurious inflation of association test statistics (λ = 1.00) due to population stratification or other confounding factors (Figure 1). Loci on two chromosomes exhibited genome-wide significant association (p < 5 × 10−8) to cortical Aβ levels (Figure 2). As expected, the peak association originated on chromosome 19 from rs429358 (p = 5.45 × 10−14), which codes for the APOE ε4 allele.7 While other SNPs within APOE and in the region of its adjacent genes APOC1, TOMM40, and PVRL2 also displayed significant association to cortical Aβ levels in the primary GWAS model (Figure 2 and Figure 3A), their association signals disappeared (p > 0.05) when APOE ε4 status (absence = 0, presence = 1) was included as a covariate.
Figure 1. Quantile-quantile (Q-Q) plot of observed −log10 p-values from the GWAS of cortical Aβ load versus those expected under the null hypothesis.
The Q-Q plot exhibits no evidence of genomic inflation (PLINK-calculated λ = 1.00) or population stratification in the GWAS. An additive genetic model was used and age, gender, and diagnosis were applied as covariates. Analyses were restricted to subjects with non-Hispanic Caucasian (CEU or TSI) ancestry as determined by genetic clustering. Observed −log10 p-values > 8 are represented along the top of the plot as red triangles, while all other values are represented as red dots.
Figure 2. Manhattan plot of observed −log10 p-values from the GWAS of cortical Aβ load.
More than six million SNPs were tested for association to global cortical Aβ burden under an additive genetic model and applying age, gender, and diagnosis as covariates. Genome-wide significant associations (exceeding the threshold represented by the red line) were identified on chromosome 19 within APOE and its neighboring genes and on chromosome 3 at the BCHE locus. Suggestive associations (exceeding the threshold represented by the blue line) were identified on five additional chromosomes. Annotations are provided for genome-wide significant associations and for the top three suggestive associations.
Figure 3. Regional association plots for the loci exhibiting genome-wide significant association to cortical Aβ burden.
Magnified association plots are displayed for the regions around A) rs429358 within APOE and B) rs509208 at the BCHE locus. SNPs are plotted based on their GWAS −log10 p-values (left vertical axis) and their genomic position (NCBI build 36). Genes in these regions are labeled with arrows denoting their 5′-to-3′ orientation, and the red color scale of r2 values is used to label SNPs based on their degree of linkage disequilibrium with the annotated peak SNP. Recombination rates calculated from 1000 Genomes Project reference data are also displayed in a blue line corresponding to the right vertical axis.
Multiple SNPs on chromosome 3 near BCHE (butyrylcholinesterase) also displayed genome-wide significant association to cortical Aβ load (Figure 2). The peak association signal at this locus originated from rs509208 (p = 2.69 × 10−8), which is approximately 450 kb upstream (5′) of BCHE (Figure 3B). This association remained strong with the inclusion of APOE ε4 status as a covariate (p = 1.94 × 10−7).
Several additional loci exhibited suggestive association (p < 5 × 10−6) to cortical Aβ levels (Figure 2 and Supplementary Table 1). These loci included SNPs within or near the cell adhesion genes of ITGA6 (integrin, alpha 6; chromosome 2) and ITGA1 (integrin, alpha 1; chromosome 5) as well as SNPs near the insulin signaling pathway gene PIK3R1 (phosphoinositide-3-kinase, regulatory subunit 1; chromosome 5), among others. The association signals at these loci were not reduced after the inclusion of APOE ε4 allele status as a covariate (data not shown).
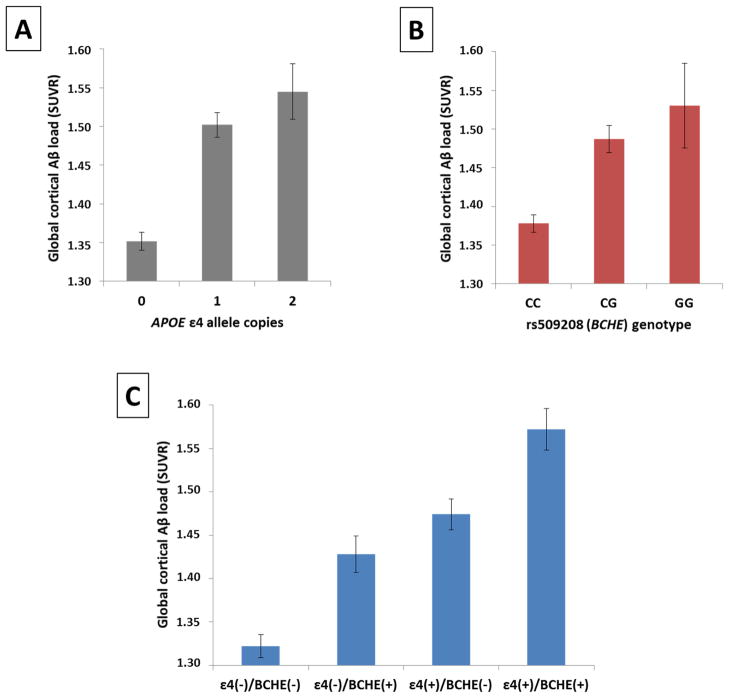
Following the GWAS, we performed post-hoc analyses to further assess the impact of the APOE and BCHE loci on Aβ burden. While both the APOE ε4 allele and the minor allele (G) of rs509208 conferred increases in cortical Aβ levels (Figure 4A and 4B), there was no evidence of epistasis modeled as an interaction between these factors (p = 0.871). Instead, these factors appeared to exert independent and additive effects on Aβ burden (Figure 4C), with comparable effect size associated with presence of at least one copy of the minor allele at rs509208 whether APOE ε4 allele status was included as a covariate (Cohen’s d = 0.50) or not (Cohen’s d = 0.52). Exploratory analyses did not reveal significant interactions (p > 0.05) of the APOE or BCHE risk loci with age, diagnosis, education, or gender.
Figure 4. APOE ε4 and rs509208 (BCHE) appear to exhibit independent, additive effects on cortical Aβ levels.
Mean cortical Aβ levels (adjusted for age, gender, and diagnosis) ± standard errors are displayed based on A) the number of APOE ε4 allele copies and B) rs509208 genotype. Presence of at least one copy of the ε4 allele was significantly associated with increased Aβ burden (Cohen’s d = 0.71), as was presence of at least one copy of the minor allele (G) of rs509208 (Cohen’s d = 0.52). These loci appeared to exert additive effects on Aβ levels (panel C): subjects having both risk factors exhibited significantly greater Aβ burden than subjects having either factor in isolation, and no significant epistasis modeled as an interaction was identified.
Finally, we performed hierarchical linear regression to assess the variance in Aβ levels uniquely explained by these genetic factors (ΔR2). Age, gender, and diagnosis were entered as the first block in the model, and collectively accounted for 2.7% of the variance in cortical Aβ levels in this sample. APOE ε4 allele status (+/−) and BCHE rs509208 allele status (+/−) were included in the second block for stepwise entry into the model. APOE ε4 was found to explain an additional 10.7% of the variance, while BCHE rs509208 accounted for 4.3% of variance over and above that explained by the previously entered variables.
Discussion
Using florbetapir PET and GWAS, this study confirmed the association of APOE and identified a novel and independent association of BCHE to cerebral Aβ deposition. Together, the risk variants at these loci explained 15% of the variation in cortical Aβ levels, a substantial effect for two genes in a study of complex disease of this size. Additional loci, including both new genes and others previously studied in relation to AD, also displayed suggestive association to Aβ burden and provide further targets for follow-up.
Florbetapir PET allows for noninvasive detection of brain Aβ plaques,10, 11 a hallmark pathologic feature of AD. It also serves as a quantitative endophenotype that can provide increased statistical power for discovery using GWAS compared to case-control designs.22 Although the heritability of Aβ deposition, a dynamic process captured by PET at one time point, is unknown and not a direct proxy for AD heritability, implicated markers using this approach may be more closely related to underlying processes impacting disease risk and progression.23
For late-onset AD, the largest known genetic risk factor is the APOE ε4 allele. Extensive prior studies have suggested a number of mechanistic roles for APOE ε4 on Aβ burden, including hindering clearance of soluble Aβ from the brain,24 favoring Aβ aggregation into fibrils,25 and promoting neurodegeneration by directing toxic Aβ oligomers to synapses.26 Among the genes neighboring APOE, we found no significant associations with Aβ burden after including APOE ε4 status as a covariate. While this suggests that these genes did not exhibit independent effects on Aβ deposition, the extensive linkage disequilibrium (LD) structure around APOE makes this difficult to definitively determine.
BCHE has previously been proposed as an AD risk gene.27 Butyrylcholinesterase is known to be enriched within Aβ plaques in post-mortem human AD brains,28 and its increased presence has been suggested as a critical factor in the formation of the neuritic plaques of dementia.29 The most commonly studied BCHE SNP is the K-variant (rs1803274), which is approximately 500 kb downstream of and not in LD with rs509208. The BCHE K-variant has demonstrated synergistic effects with APOE ε4 on incidence of pathologically-confirmed late-onset AD27 and on risk of progression from MCI to AD.30 Nevertheless, the present study is the first to implicate genetic variation at the BCHE locus in brain Aβ burden in humans and represents the largest reported effect for this gene on an AD-related phenotype.
There are several mechanisms which might explain the effect of BCHE on Aβ plaque burden. Genetic variation at BCHE has been associated with increased cortical butyrylcholinesterase activity in autopsy tissue from elderly individuals with dementia.31 Increased enzyme activity, leading to decreased acetylcholine levels, may disrupt synaptic functioning, glial cell activation, and myelin maintenance to favor Aβ plaque formation and neurodegeneration.32 Indeed, cholinesterase inhibitors are presently first-line symptomatic therapies for AD,33 and drugs such as rivastigmine which inhibit butyrylcholinesterase are suggested to have potential disease-modifying effects in certain individuals compared to exclusive acetylcholinesterase inhibitors.34 Alternatively, through non-enzymatic functions butyrylcholinesterase may promote Aβ fibrillogenesis from soluble precursors35 or may interact with Aβ and ApoE to alter the cerebrospinal fluid environment36, 37 and to render developing plaques more resistant to clearance.38 Interestingly, butyrylcholinesterase activity has been associated with insulin resistance,39 and rs509208 in particular has been associated in a separate GWAS with diabetes-related traits,40 suggesting a broader role of the BCHE locus in disorders characterized by amyloidogenic protein accumulation.
Although rs509208 is approximately 450 kb upstream (5′) of BCHE and not within conventional gene boundaries, the next closest genes (ZBBX, SERPINI2) are nearly 1 Mb in the opposite direction (Supplementary Figure 3). Of note, SNPs as far as 800 kb upstream of BCHE have previously demonstrated genome-wide significant association with serum butyrylcholinesterase activity in a population sample of nearly 9000 individuals from several Australian twin and family studies.41 These SNPs included a peak signal 250 kb from the gene (rs2034445) and other non-independent signals from SNPs in high linkage disequilibrium with rs509208 (e.g., rs6443374 and rs13314077), suggesting that variants upstream of the gene may exert regulatory effects on BCHE expression with consequences to the activity of its encoded enzyme. Converging evidence in genomics indicates that this kind of regulation is quite common and may involve mechanisms influencing chromatin structure, transcription factor binding, and splicing component recognition sequences, among others.42 Molecular characterization in brain tissue and cell cultures will be important to determine the complex functional architecture of the BCHE locus and the genes and other DNA elements surrounding it.
The current study has several limitations. Although this represents the largest genetic study of Aβ PET, the sample size has limited power for a GWAS. With a larger sample, the suggestive loci we highlighted might have achieved genome-wide significance. Among these included ITGA6 which encodes a component of a receptor complex proposed to mediate the pro-inflammatory interaction of microglia with Aβ fibrils,43 PIK3R1 which may contribute to disruptions in insulin signaling in the AD brain,44 and other genes with potential relationships to AD pathogenesis. Similarly, given the novelty of the present Aβ PET GWAS dataset, a comparable replication sample is not yet available.
In addition, the ADNI cohort represents a sample typical of a clinical trial for MCI/AD and the analyses here were restricted to non-Hispanic Caucasians. The extent to which the present findings can be generalized to other populations and to the community setting of MCI/AD individuals remains to be determined. Future multi-center and international collaborations are expected to yield larger samples with greater power for analyses of potential interactions of genetic risk factors with each other and with clinical variables such as gender, ethnicity, family history, age of onset, and rate of disease progression, which could not be appropriately addressed with presently available data. Florbetapir also does not bind soluble forms of Aβ45 which may exhibit dynamic relationships with deposited, fibrillar Aβ to drive neurodegeneration in AD.1 Finally, while this study employed imputed (probabilistically-predicted) genotype data to provide deep coverage of the genome, higher-density genotyping arrays and sequencing will eventually provide direct assays for a similar number of variants.
Despite these limitations, the present findings point to several intriguing extensions for follow-up. First, GWAS of longitudinal change in florbetapir PET Aβ burden is likely to elucidate additional genes modulating the rate of progression of AD neuropathology. Complementary analytical strategies, including pathway- and epistasis-based approaches, may also reveal functional influences on Aβ deposition that are not easily observed through standard GWAS methodologies. In addition, whole genome sequencing will dramatically enhance the granularity of coverage for GWAS-implicated loci and could be particularly valuable for discovering additional novel loci, rare variants, copy number variants, and DNA regulatory elements. Finally, the approval for widespread use of both florbetapir PET imaging and butyrylcholinesterase inhibitors creates a unique opportunity to prospectively assess the effects of these drugs on Aβ deposition over time, particularly among individuals at early stages in the AD spectrum where clinical efficacy would likely be most valuable.
This study highlights the power of pairing targeted molecular imaging with genome-wide analytical strategies to elucidate mechanisms underlying AD pathophysiology. The association of the BCHE locus to Aβ deposition merits further investigation and may have significant implications for risk stratification, biomarker interpretation, and therapeutic development.
Supplementary Material
The initial sample and the number of participants remaining after key data processing steps are displayed (black boxes) along with exclusion criteria at these steps (red boxes).
Global cortical Aβ levels are displayed for all GWAS participants stratified by diagnosis (HC, EMCI, LMCI, AD) and by APOE ε4 allele status (top 4 panels) and BCHE locus rs509208 minor allele (G) status (bottom 4 panels). Group means are indicated by red lines.
Supplementary Figure 3: Regional association plot (wide view) around rs509208 at the BCHE locus
SNPs within BCHE and the region spanning approximately 1.5 Mb upstream of BCHE are plotted based on their GWAS −log10 p-values (left vertical axis), NCBI build 36 genomic position (horizontal axis), and recombination rates calculated from 1000 Genomes Project reference data (right vertical axis). Genes are labeled with arrows denoting their 5′-to-3′ orientation. The color scale of r2 values is used to label SNPs based on their degree of linkage disequilibrium with rs509208. As displayed, rs509208 is 450 kb upstream of BCHE (blue arrows) and is approximately 1 Mb away from the next closest genes (orange arrows). The association plot was generated using the Locus Zoom software (http://csg.sph.umich.edu/locuszoom).
Supplementary Table 1: Peak association signals (p < 5 × 10−6) from unique genes or intergenic loci in the GWAS of global cortical Aβ load
Acknowledgments
Data collection and sharing for this project was supported by the ADNI National Institutes of Health (NIH) grant U01 AG024904 (PI: Michael W. Weiner, M.D., VA Medical Center and University of California-San Francisco). Funding sources for ADNI include the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering, the U.S. Food and Drug Administration, the nonprofit partners of the Alzheimer’s Association, the Alzheimer’s Drug Discovery Foundation, and the Dana Foundation, and the following private sector contributors: Abbott, AstraZeneca AB, Amorfix, Bayer Schering Pharma AG, Bioclinica Inc., Biogen Idec, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, Innogenetics, IXICO, Janssen Alzheimer Immunotherapy, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Meso Scale Diagnostic & LLC, Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Servier, Synarc, Inc., and Takeda Pharmaceuticals. Private sector contributions to ADNI are facilitated by the Foundation for the NIH (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education. The study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego, and ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. Additional ADNI support comes from the NIH grants P30 AG010129, K01 AG030514, and U24 AG21886. Data management and the specific analyses reported here were supported by NSF IIS-1117335 and NIH R01 AG19771, P30 AG10133, R01 LM011360, K24 AG027841, K99 LM011384.
Footnotes
CONFLICT OF INTEREST
Dr. Jagust has served as a consultant to TauRx Therapeutics LTD, GE Healthcare, Siemens, Synarc, and Janssen Alzheimer Immunotherapy. Dr. Weiner has served on scientific advisory boards for Eli Lilly, Araclon, Institut Catala de Neurociencies Aplicades, the Gulf War Veterans Illnesses Advisory Committee, Biogen Idec, and Pfizer; has served as a consultant to Astra Zeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics LDT, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, Servier, Synarc, Janssen, Harvard University, and KLJ Associates; has received funding for travel from NeuroVigil, Inc., CHRU Hôpital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Eli Lilly, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Ipsen, Clinical Trials on Alzheimer’s Disease, the AD/PD Meeting, Paul Sabatier University, Novartis, Tohoku University, Fundacio ACE, and Travel eDreams, Inc.; has received honoraria from NeuroVigil, Inc., Institut Catala de Neurociencies Aplicades, PMDA/Japanese Ministry of Health, Labour, and Welfare, Tohoku University, and the Alzheimer’s Drug Discovery Foundation; has received research support from Merck and Avid; and has stock options for Synarc and Elan. Dr. Saykin has received investigator-initiated research funding from Welch Allyn and Siemens Healthcare and has served as a consultant or advisory board member for Siemens Healthcare and Eli Lilly. The other authors declare no conflict of interest.
References
- 1.Karran E, Mercken M, De Strooper B. The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nat Rev Drug Discov. 2011;10(9):698–712. doi: 10.1038/nrd3505. [DOI] [PubMed] [Google Scholar]
- 2.Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ, et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. Journal of neuropathology and experimental neurology. 2012;71(5):362–381. doi: 10.1097/NEN.0b013e31825018f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen RC, Parisi JE, Dickson DW, Johnson KA, Knopman DS, Boeve BF, et al. Neuropathologic features of amnestic mild cognitive impairment. Arch Neurol. 2006;63(5):665–672. doi: 10.1001/archneur.63.5.665. [DOI] [PubMed] [Google Scholar]
- 4.Bennett DA, Schneider JA, Arvanitakis Z, Kelly JF, Aggarwal NT, Shah RC, et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66(12):1837–1844. doi: 10.1212/01.wnl.0000219668.47116.e6. [DOI] [PubMed] [Google Scholar]
- 5.Sleegers K, Lambert JC, Bertram L, Cruts M, Amouyel P, Van Broeckhoven C. The pursuit of susceptibility genes for Alzheimer’s disease: progress and prospects. Trends in genetics: TIG. 2010;26(2):84–93. doi: 10.1016/j.tig.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63(3):287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furst AJ, Kerchner GA. From Alois to Amyvid: Seeing Alzheimer disease. Neurology. 2012 doi: 10.1212/WNL.0b013e3182662084. [DOI] [PubMed] [Google Scholar]
- 10.Clark CM, Schneider JA, Bedell BJ, Beach TG, Bilker WB, Mintun MA, et al. Use of florbetapir-PET for imaging beta-amyloid pathology. JAMA: the journal of the American Medical Association. 2011;305(3):275–283. doi: 10.1001/jama.2010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM, et al. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-β plaques: a prospective cohort study. The Lancet Neurology. 2012;11(8):669–678. doi: 10.1016/S1474-4422(12)70142-4. [DOI] [PubMed] [Google Scholar]
- 12.Weiner MW, Aisen PS, Jack CR, Jr, Jagust WJ, Trojanowski JQ, Shaw L, et al. The Alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimers Dement. 2010;6(3):202–211. e207. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement. 2010;6(3):221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44(5):552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen RC. Clinical practice. Mild cognitive impairment. The New England journal of medicine. 2011;364(23):2227–2234. doi: 10.1056/NEJMcp0910237. [DOI] [PubMed] [Google Scholar]
- 22.Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DB, et al. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cognitive neuropsychiatry. 2009;14(4–5):391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kendler KS, Neale MC. Endophenotype: a conceptual analysis. Mol Psychiatry. 2010;15(8):789–797. doi: 10.1038/mp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, et al. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med. 2011;3(89):89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3(77):77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koffie RM, Hashimoto T, Tai HC, Kay KR, Serrano-Pozo A, Joyner D, et al. Apolipoprotein E4 effects in Alzheimer’s disease are mediated by synaptotoxic oligomeric amyloid-beta. Brain. 2012;135(Pt 7):2155–2168. doi: 10.1093/brain/aws127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehmann DJ, Johnston C, Smith AD. Synergy Between the Genes for Butyrylcholinesterase K Variant and Apolipoprotein E4 in Late-Onset Confirmed Alzheimer’s Disease. Human Molecular Genetics. 1997;6(11):1933–1936. doi: 10.1093/hmg/6.11.1933. [DOI] [PubMed] [Google Scholar]
- 28.Darvesh S, Hopkins DA, Geula C. Neurobiology of butyrylcholinesterase. Nat Rev Neurosci. 2003;4(2):131–138. doi: 10.1038/nrn1035. [DOI] [PubMed] [Google Scholar]
- 29.Guillozet AL, Smiley JF, Mash DC, Mesulam MM. Butyrylcholinesterase in the life cycle of amyloid plaques. Ann Neurol. 1997;42(6):909–918. doi: 10.1002/ana.410420613. [DOI] [PubMed] [Google Scholar]
- 30.Lane R, Feldman HH, Meyer J, He Y, Ferris SH, Nordberg A, et al. Synergistic effect of apolipoprotein E epsilon4 and butyrylcholinesterase K-variant on progression from mild cognitive impairment to Alzheimer’s disease. Pharmacogenetics and genomics. 2008;18(4):289–298. doi: 10.1097/FPC.0b013e3282f63f29. [DOI] [PubMed] [Google Scholar]
- 31.Tasker A, Ballard CG, Joachim C, Warden DR, Okello EJ, Perry RH, et al. Butyrylcholinesterase K variant associated with higher enzyme activity in the temporal cortex of elderly patients. Neurosci Lett. 2008;442(3):297–299. doi: 10.1016/j.neulet.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 32.Lane RM, He Y. Butyrylcholinesterase genotype and gender influence Alzheimer’s disease phenotype. Alzheimers Dement. 2012 doi: 10.1016/j.jalz.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Farlow MR, Miller ML, Pejovic V. Treatment options in Alzheimer’s disease: maximizing benefit, managing expectations. Dement Geriatr Cogn Disord. 2008;25(5):408–422. doi: 10.1159/000122962. [DOI] [PubMed] [Google Scholar]
- 34.Sabbagh MN, Farlow MR, Relkin N, Beach TG. Do cholinergic therapies have disease-modifying effects in Alzheimer’s disease? Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2006;2(2):118–125. doi: 10.1016/j.jalz.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Darvesh S, Cash MK, Reid GA, Martin E, Mitnitski A, Geula C. Butyrylcholinesterase is associated with beta-amyloid plaques in the transgenic APPSWE/PSEN1dE9 mouse model of Alzheimer disease. Journal of neuropathology and experimental neurology. 2012;71(1):2–14. doi: 10.1097/NEN.0b013e31823cc7a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darreh-Shori T, Forsberg A, Modiri N, Andreasen N, Blennow K, Kamil C, et al. Differential levels of apolipoprotein E and butyrylcholinesterase show strong association with pathological signs of Alzheimer’s disease in the brain in vivo. Neurobiology of Aging. 2011;32(12):2320.e2315–2320.e2332. doi: 10.1016/j.neurobiolaging.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 37.Darreh-Shori T, Modiri N, Blennow K, Baza S, Kamil C, Ahmed H, et al. The apolipoprotein E ε4 allele plays pathological roles in AD through high protein expression and interaction with butyrylcholinesterase. Neurobiology of Aging. 2011;32(7):1236–1248. doi: 10.1016/j.neurobiolaging.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Lane RM, He Y. Emerging hypotheses regarding the influences of butyrylcholinesterase-K variant, APOE ε4, and hyperhomocysteinemia in neurodegenerative dementias. Medical hypotheses. 2009;73(2):230–250. doi: 10.1016/j.mehy.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 39.Iwasaki T, Yoneda M, Nakajima A, Terauchi Y. Serum Butyrylcholinesterase is Strongly Associated with Adiposity, the Serum Lipid Profile and Insulin Resistance. Internal Medicine. 2007;46(19):1633–1639. doi: 10.2169/internalmedicine.46.0049. [DOI] [PubMed] [Google Scholar]
- 40.Meigs JB, Manning AK, Fox CS, Florez JC, Liu C, Cupples LA, et al. Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC medical genetics. 2007;8 (Suppl 1):S16. doi: 10.1186/1471-2350-8-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benyamin B, Middelberg RP, Lind PA, Valle AM, Gordon S, Nyholt DR, et al. GWAS of butyrylcholinesterase activity identifies four novel loci, independent effects within BCHE and secondary associations with metabolic risk factors. Hum Mol Genet. 2011;20(22):4504–4514. doi: 10.1093/hmg/ddr375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 43.Bamberger ME, Harris ME, McDonald DR, Husemann J, Landreth GE. A cell surface receptor complex for fibrillar beta-amyloid mediates microglial activation. J Neurosci. 2003;23(7):2665–2674. doi: 10.1523/JNEUROSCI.23-07-02665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bomfim TR, Forny-Germano L, Sathler LB, Brito-Moreira J, Houzel JC, Decker H, et al. An anti-diabetes agent protects the mouse brain from defective insulin signaling caused by Alzheimer’s disease- associated Abeta oligomers. The Journal of clinical investigation. 2012;122(4):1339–1353. doi: 10.1172/JCI57256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikonomovic MD, Klunk WE, Abrahamson EE, Mathis CA, Price JC, Tsopelas ND, et al. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer’s disease. Brain. 2008;131(Pt 6):1630–1645. doi: 10.1093/brain/awn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The initial sample and the number of participants remaining after key data processing steps are displayed (black boxes) along with exclusion criteria at these steps (red boxes).
Global cortical Aβ levels are displayed for all GWAS participants stratified by diagnosis (HC, EMCI, LMCI, AD) and by APOE ε4 allele status (top 4 panels) and BCHE locus rs509208 minor allele (G) status (bottom 4 panels). Group means are indicated by red lines.
Supplementary Figure 3: Regional association plot (wide view) around rs509208 at the BCHE locus
SNPs within BCHE and the region spanning approximately 1.5 Mb upstream of BCHE are plotted based on their GWAS −log10 p-values (left vertical axis), NCBI build 36 genomic position (horizontal axis), and recombination rates calculated from 1000 Genomes Project reference data (right vertical axis). Genes are labeled with arrows denoting their 5′-to-3′ orientation. The color scale of r2 values is used to label SNPs based on their degree of linkage disequilibrium with rs509208. As displayed, rs509208 is 450 kb upstream of BCHE (blue arrows) and is approximately 1 Mb away from the next closest genes (orange arrows). The association plot was generated using the Locus Zoom software (http://csg.sph.umich.edu/locuszoom).
Supplementary Table 1: Peak association signals (p < 5 × 10−6) from unique genes or intergenic loci in the GWAS of global cortical Aβ load