Abstract
Fanconi anemia (FA) is a rare inherited recessive disease caused by mutations in one of fifteen genes known to encode FA pathway components. In response to DNA damage, nuclear FA proteins associate into high molecular weight complexes through a cascade of post-translational modifications and physical interactions, followed by the repair of damaged DNA. Hematopoietic cells are particularly sensitive to the loss of these interactions, and bone marrow failure occurs almost universally in FA patients. FA as a disease is further characterized by cancer susceptibility, which highlights the importance of the FA pathway in tumor suppression, and will be the focus of this review. Acute myeloid leukemia is the most common cancer type, often subsequent to bone marrow failure. However, FA patients are also at an extreme risk of squamous cell carcinoma (SCC) of the head and neck and gynecological tract, with an even greater incidence in those individuals who have received a bone marrow transplant and recovered from hematopoietic disease. FA tumor suppression in hematopoietic versus epithelial compartments could be mechanistically similar or distinct. Definition of compartment specific FA activities is now critical to assess the effects of today’s bone marrow failure treatments on tomorrow’s solid tumor development. It is our hope that current therapies can then be optimized to decrease the risk of malignant transformation in both hematopoietic and epithelial cells. Here we review our current understanding of the mechanisms of action of the Fanconi anemia pathway as it contributes to stress responses, DNA repair and squamous cell carcinoma susceptibility.
Fanconi anemia pathway mutations play key role in the development of cancer
FA is a rare, autosomal recessive, and X-linked in the case of FANCB, syndrome characterized by congenital defects, bone marrow failure (BMF), and increased susceptibility to cancers. These are predominantly acute myeloid leukemia (AML) and head and neck squamous cell carcinoma (HNSCC) [1–4]. Disease incidence is rare, estimated at 1 in 200,000 live births, with a carrier frequency of 1 in 181 [5, 6]. A diagnosis of FA is devastating with a median life expectancy of a little over 20 years [3, 6]. Symptoms frequently occur early in life and include a constellation of birth defects, hematologic abnormalities, and cellular as well as organismal hypersensitivity to agents that cause DNA interstrand crosslinks (ICLs), such as melphalan, cisplatin, and mitomycin C (MMC). Based on the variable expressivity of recognized symptoms, it is possible that the true incidence of FA is grossly underestimated. Recommendations to screen leukemia patients who recover poorly from chemotherapy, and young squamous cell carcinoma patients, particularly those who experience serious toxicity from chemotherapy and/or radiation, have been voiced [7, 8]. FA genes participate in a common pathway, which ensures genome integrity through controlling a myriad of chromatin processing and DNA damage response pathways wherein mutation of any of the FA genes individually leads to the clinical FA phenotype [9–12].
FA is described as a chromosomal instability disorder, which can affect multiple organ systems with variable severity. It is largely characterized by cellular hyper-sensitivity to DNA damaging agents that induce DNA interstrand crosslinking (ICL) [13–15], impairing DNA strand separation and unwinding, and ultimately hindering DNA replication and transcription [16, 17]. Cells cultured from individuals with FA exhibit cellular accumulation in the G2 phase of the cell cycle and pronounced chromosomal breakage when exposed to crosslinkers such as mitomycin C or diepoxybutane (DEB). In fact, the formation of radial chromosomes in FA lymphocytes treated with DEB has been utilized as a diagnostic feature of FA for years [4, 17–19]. Hypersensitivity to ICLs is a hallmark of FA cells, and the FA machinery was therefore initially regarded as a specialized ICL repair system reflected by the rare incidence of FA. However, the finding that the replication checkpoint kinase ataxia telengiectasia and Rad3-related (ATR) is required for triggering FA activation [20, 21] and that important breast cancer susceptibility genes BRCA2, PALB2 and BRIP1 are identical to FANCD1 [22], FANCN [23, 24], and FANCJ [25–27], have redefined the FA pathway as one of broad importance. Correspondingly, the FA pathway is activated beyond ICL by many forms of genotoxic stress including UV, ionizing radiation and oxidative stress, and FA deficient cells have been utilized as a general model system to study ATR signaling and BRCA functions in DNA repair [20, 28].
FA pathway activity is tightly regulated and specifically activated during the S phase of the cell cycle [29] and in response to DNA damage [15, 30]. Individuals with FA have an approximately 50-fold increased risk of developing any cancer type [2], with striking susceptibility to leukemia and squamous cell carcinomas. Carriers of FANCD1/BRCA2 and RAD51C mutations are predisposed to breast and ovarian cancer [31–33], carriers of FANCJ and FANCN mutations are predisposed to breast cancer at lower penetrance [34–37], and carriers of FANCD1/BRCA2, FANCC and other FA gene mutations are predisposed to pancreatic cancer [35, 38–41]. Importantly, defects in the FA/BRCA pathway exist in the general, non-FA population, and associated cancer susceptibility is thus not limited to the rare, inherited scenario. Tissue specific FA repression and crosslinker sensitivity have been reported. FANCD2 and FANCF deficiencies occur in ovarian tumor cells [42–44], and FANCF silencing has been detected in a significant proportion of cervical, head and neck and lung cancers [45, 46]. However, one report did not detect FANCF silencing in head and neck cancer (HNC) cells [47], and additionally, cisplatin sensitivity was not associated with the FA/BRCA pathway inactivation in HNC cell lines [48]. Similarly, Burkitt and Ljungman were unable to link cisplatin sensitivity to defective monoubiquitination of FANCD2 in four head and neck cancer cell lines. Interestingly, however, three of four cisplatin sensitive cell lines investigated were unable to induce the formation of FANCD2 nuclear foci, and both FANCD2 focus formation and cisplatin resistance could be restored through the expression of exogenous BRCA1 in these cells [49]. These findings would suggest that defects in FANCD2 nuclear focus formation, but not ubiquitination, are indicative of cisplatin sensitivity, and that BRCA1 deficiencies are responsible. Regardless, early identification of biomarkers for cancer cells, such as head and neck squamous cell carcinoma, which identify defects in the FA pathway may allow for alternative chemotherapeutic and/or irradiation options to be exploited. Because FA deficient cells are hypersensitive to DNA damage, low dose clastogenic treatments may be effective for sporadic SCCs with FA mutations. Systematic dose de-escalation studies in FA SCC models are needed to explore this possibility. The results may be particularly useful for FA patients where achieving tumor eradication while minimizing life threatening toxicity remains a difficult balancing act.
The FA pathway: multi-protein interactions coordinate DNA repair
Fifteen complementation groups and the corresponding FA genes have now been identified [14, 22–24, 26, 27, 50–64]. Their protein products function as either signal transducers and/or DNA processing factors within the larger FA-BRCA DNA damage response network as described below. Our published studies have demonstrated that multiple FA and associated genes are transcriptionally limiting and co-regulated through Rb/E2F pathways [65]. Upregulation of components of the DNA repair machinery in proliferating cells is likely to ensure maximal DNA repair capacity when the chance of replicative DNA damage and the need for DNA repair is greatest. The FA/BRCA pathway is activated during DNA replication and by DNA damage in the form of ICLs and other lesions. Protein components of the FA/BRCA pathway assemble into at least three complexes within the cell nucleus, these are the FA core complex (FACC), the ID complex composed of (mono)ubiquitinated FANCD2 and FANCI proteins, and a complex of the FANCN and BRCA2 proteins associated with homologous recombination that binds chromatin near DNA lesions downstream from the ID complex.
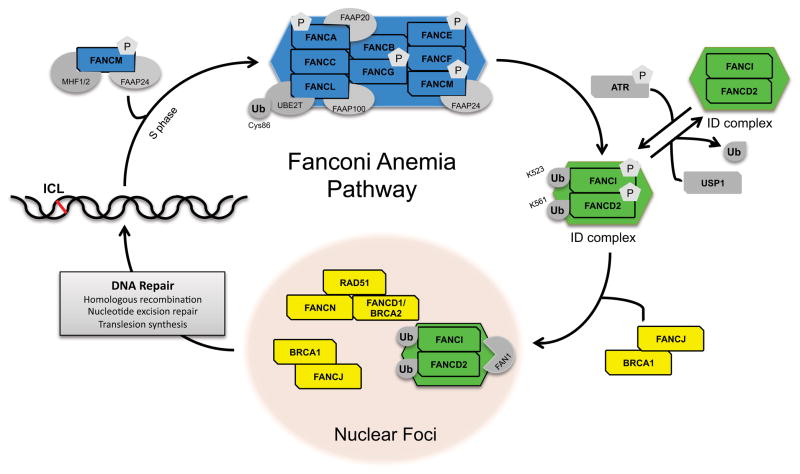
Figure 1 depicts a working model of the FA pathway: The FA core complex is composed of eight FA proteins (FANCA, B, C, E, F, G, L, and M) along with other FA-associated proteins, such as FAAP100, FAAP20 and FAAP24 [66–70]. Multiple constituents of the core complex, as well as associated components of the pathway are phosphorylated including FANCA, G, M, D1/BRCA2, D2, and I, and these phosphorylation events are important for the repair of ICLs [71–74]. The core complex forms a nuclear, high molecular weight E3 ubiquitin ligase complex, based on the sole E3 ubiquitin ligase domain of FANCL [50]. The FANCL protein contains additional domains responsible for directing substrate binding (DRWD domain) and four E2 protein interactions (RING domain) by utilizing UBE2T [75, 76]. The FANCM protein of the Fanconi anemia core complex is an important component of the pathway providing ICL resistance to cells [77]. FANCM and FAAP24, with the help of the histone-fold containing proteins MHF1 and MHF2 [78], recognize both lesions in DNA and stalled replication forks [66, 79] and subsequently generate single stranded DNA [80], which is thought to activate both ATR and its downstream target Chk1 [81].
Figure 1. A schematic model for the FA/BRCA pathway.
FANCM (blue) with the help of MHF1/2 (dark grey) and FAAP24 (light grey) recruit a large multi-subunit ubiquitin E3 ligase, termed the FA core complex (blue), to sites of DNA damage. The core complex then mono-ubiquitinates FANCD2 and FANCI, mono-ubiquitinated FANCD2-FANCI (green) are recruited to sites of damage by FANCJ (yellow) and BRCA1 (yellow). These FA proteins colocalize with downstream FA proteins (RAD51, FANCN, FANCD1/BRCA2) (yellow), and facilitate DNA interstrand cross-link repair.
The key upstream regulators of the FA pathway, ATR and ATM kinases, are responsible for phosphorylating several components of the FA pathway and specifically direct the cellular response to DNA damage during S-phase [15, 21]. The phosphorylation of FANCI has been identified as a potential molecular switch that turns on the FA pathway [82]. FANCI phosphorylation initiates the interactions between the FACC and the ID complex composed of FANCI and FANCD2 [82, 83]. Following phosphorylation, both components of the ID complex are monoubiquitinated by the FACC ubiquitin ligase at Lys561 of FANCD2 and Lys523 of FANCI [14, 30, 50, 59, 84].
As depicted in Figure 1, FA activation involves the assembly of high molecular weight FA complexes in the nucleus. The presence of each FA protein within the core complex is a prerequisite for proper complex assembly and subsequent DNA repair; however, several FA-interacting proteins play non-canonical roles outside of the classical pathway [85], and this explains why mutations within different FA genes yield similar, albeit not necessarily identical, clinical phenotypes. Monoubiquitination of both FANCD2 and FANCI, the components of the ID complex, are important for the downstream function of the FA pathway in DNA repair [14, 30, 59] and FANCD2 mono-ubiquitination is thus widely used as a read-out for pathway activation. It is detectable by a shift from the nonubiquitinated short form (FANCD2-S) of the protein to a slower migrating, long form (FANCD2-L) in immunoblots. FANCD2-L relocalization to chromatin is visualized using immunofluorescence (IF) by the formation of nuclear FANCD2 foci that colocalize with other nuclear DNA damage response proteins such as γ H2AX or Rad51 foci. Mutation of core complex I components (such as FANCA) is therefore reflected experimentally by the absence of FANCD2 monoubiquitination, and by the absence of FANCD2 and Rad51 foci following DNA damage induction [29, 30, 86]. This has been described for multiple cell types including keratinocytes cultured from FA patient skin biopsies, wherein retroviral FANCA complementation successfully re-instated the detection of FANCD2-L by immunoblot and IF experiments [87].
Activation of the ID complex initiates its localization to nuclear foci together with BRCA1 and Rad51, essential factors for proper homologous recombination [29, 30]. After ID complex loads onto chromatin, it binds the FAN1 protein, which is responsible for nuclease activity during the repair of damaged sites [88–91]. Although FANCI monoubiquitination is not required for proper repair of damaged DNA, it is believed that its phosphorylation may play a role in localizing the ID complex to chromatin foci [82], which are described as sites of repair because they harbor numerous repair factors, such as BRCA1, BRCA2, PCNA, or Rad51 [29, 30, 92, 93]. Subsequent DNA nuclease recruitment and activation “unhooks” the crosslink, followed by homologous recombination (HR), nucleotide excision repair (NER) on the opposing strand and translesion synthesis (TLS) to repair the gap [94].
Once DNA repair is complete, the FA pathway is turned off by de-ubiquitination of the ID complex. This inactivation of the pathway occurs when USP1, a de-ubiquitinating enzyme, associates with FANCD2 within the nuclear foci and removes the monoubiquitin moiety [95]. The absence of de-ubiquitination of FANCD2, much like the absence of ubiquitination, leads to sensitivity of the DNA to crosslinking agents, such as cisplatin or mitomycin C [96, 97]. Although the FA pathway has thus become a model system for ICL repair, it is clear that its molecular components coordinate and integrate a multitude of DNA repair machineries including HR, NER and TLS, as each of these repair mechanisms contribute to ICL repair. A recent review article by Kim and D’Andrea provides an excellent review of how the FA pathway coordinates these three critical DNA repair processes for ICL repair [98].
Clonal evolution plays a key role in progression to leukemia in the FA population
A majority of FA patients invariably experience progressive bone marrow failure (BMF), and oftentimes leukemia [5, 41, 99–101]. Marrow dysfunction occurs at approximately 7–8 years old, is associated with stem cell loss in the hematopoietic compartment, and is responsible for the majority of FA childhood mortality (for a review see [102]). The risk of BMF in FA is 90 percent by 50 years of age, although the mechanism for stem cell loss is not fully understood [3]. Rapid hematopoietic cell loss then forces compensatory chronic proliferation, which likely results in clonal evolution and leukemogenesis. Proof of concept for such clonal selection and resistance to cytokine-induced cell death has been reported [103–106]. In an environment of genomic instability, loss of function of tumor suppressors, as well as oncogenic translocations, can be acquired and selected for rapidly. Chromosomes 1, 3 and 7, for instance, were more frequently involved in FA AML cases than in de novo AML cases [7]. The specific involved clone may thus distinguish FA AML from de novo cases. The only curative therapy for hematologic FA abnormalities is a bone marrow transplant (BMT). However, genotoxic conditioning regimens and the transplant itself are a significant cause of morbidity and mortality in this patient population, which tolerates chemotherapy and radiation poorly [107].
If FA patients survive BMF through treatment, they remain susceptible to cancers, typically myeloid leukemias, and SCCs [3, 108–110]. Two research groups have shown that FA patients with biallelic mutations in BRCA2 have an exceptionally high risk of developing AML by age 5 [111, 112] and the overall prevalence of AML in FA patients is 33% by 40 years of age [3]. Many factors have led to the hypothesis that sporadic AMLs may also carry FA defects; however, only a small number of mutations have been identified. For example, FANCF silencing was demonstrated in an AML cell line [113] and hypermethylation of the promoter regions of both FANCC and FANCL have also been identified in sporadic acute leukemia [114]. Other aspects, such as the high risk of AML and myelodysplastic syndrome (MDS) in pediatric FA patients, and the similarities between chromosomal abnormalities in both FA and AML/MDS [2, 115, 116] call for further investigation into molecular link(s) between FA pathway irregularities and AML/MDS in the general population [117].
Many individuals with FA will develop AML and/or MDS if they live long enough and do not succumb to other FA related complications. It is for this reason that the FA genotype is equitably viewed as preleukemia [110]. Stabilization and prevention of pre-leukemic phenotypes in these patients is the expressed goal, but should ideally be achieved without increasing the risk of solid tumor development. However, a thorough understanding of the role of FA proteins in squamous epithelium is lacking, thus impeding the rational design of strategies that prevent both hematopoietic and epithelial transformation. Below, we highlight molecular and phenotypic aspects of FA, which might differ substantially across tissue and cancer types. If so, then care must be taken in ensuring that the prevention and treatment of leukemia in the FA patient population does not further exacerbate the development of solid tumors.
Mutations in the FA pathway lead to increased risk of squamous cell carcinoma
Results from the International FA Registry (IFAR) have revealed that FA patients are highly susceptible to non-hematologic neoplasms [3, 99]. Squamous cell carcinomas (SCCs) of the anogenital region and head and neck, the latter with an up to 1,400 fold risk over that of the normal population, are the most commonly diagnosed solid tumors in these patients. A recent report on cancer incidence in the German FA Registry also described an extreme risk of head and neck, vulvar, and esophageal SCC [115]. Cancers of the head and neck can arise throughout the mucosal linings of the entire upper aerodigestive tract, including the oral cavity, oropharynx, nasopharynx, hypopharynx and larynx. HNSCC originates from the normal mucosa [118], advances through a multistep process and evolves through a sequence of histopathologic stages [119], which is discussed in detail by Califano et al. [119, 120]. Approximately 36,500 new patients are diagnosed yearly with head and neck cancer in the United States [121]. Head and neck squamous cell carcinomas (HNSCCs) constitute the majority and are the sixth leading cancer worldwide, with prolonged tobacco and alcohol use as principal risk factors for this disease [122]. In the general population, approximately 25% of these cancers are caused by infection with human papillomavirus (HPV), particularly the HPV16 genotype [119, 123, 124]. HPV status determines significant biological differences, with HPV positive tumors exhibiting improved response to treatment and prognosis when compared to HPV negative tumors. The prevalence of HPV in FA SCCs, however, remains controversial to date [125–127].
Because only a small number of persons exposed to tobacco, alcohol or HPV ultimately develop cancer, it is believed that an individual must also have an inherent predisposition, which collaborates with these genotoxic exposures for carcinogenesis to occur [128]. Along with the previously mentioned environmental factors, HNSCC has also been linked to FA, and patients with this BMF syndrome are susceptible to chromosomal instability and the development of SCC [3, 126, 129]. Cloos et al. have proposed that there are different levels of DNA safeguarding capability within the general population which play a role in the development of cancer [130]. Unfortunately, overall treatment outcomes for HNC have not improved in decades and conventional clastogenic therapies have substantial side effects on normal physiological functions such as swallowing, speech and physical appearance. These responses to therapy, together with tumor development, are also predicted to be modified by genetic predispositions, and appear to be greatly amplified in FA patients [131]. Therefore, while FA tumors are predicted to be exquisitely sensitive to conventional chemotherapy and radiation, the FA individual’s global hyper-sensitivity to DNA damage limits the effectiveness of such therapies, and particularly radiotherapy, dramatically [131–134].
Array based comparative genome hybridization of 21 sporadic oral squamous cell carcinomas revealed de-regulation of a number of FA and FA-associated genes, including BRCA1, BRCA2 (FANCD1), FANCG and FANCD2 [135]. Two recent publications in Science utilized whole-exome sequencing approaches in order to screen for genetic mutations in primary head and neck cancers and prior to drug treatment [136, 137]. Stransky et al. identified numerous mutations listed in Table 1 in either FA genes themselves, or more generally in a select subset of genes associated with DNA repair [136]. The list of genes shown in Table 1 represents new analyses of the data previously published by Stransky et al., which reveal that 38 out of the 74 (51%) sequenced tumors harbored one or more somatic mutations in the indicated gene subset. More strikingly, a great majority of these tumors (18/38 tumors; 47%) harbored multiple mutations ranging from 2–7. Among these mutated genes, BRCA2, FANCM (5 mutations reported), ATR, UBE4A (4 mutations reported), BRCA1, USP43 and USP44 (3 mutations reported) are most frequent. These recent findings indicate that a subset of primary, therapy-naïve HNSCCs harbor mutations in important DNA repair pathways including FA. Such mutations may contribute functionally to increased tumor growth and/or may modify sensitivity to conventional drug therapies. Of note, mutations in FA-related genes were also identified by Agrawal et al. [137], albeit at a lesser frequency. This might be due to a distinct clinical cohort, sample size and/or technical differences.
Table 1. Somatic mutations of HNSCC patient tumors in a select subset of FA- and DNA-repair associated genes.
[136] Each letter in parentheses represent each individual patient. Mutations were identified by exome sequencing of tumors isolated from sporadic head and neck squamous cell carcinoma patients [136].
| FA-Associated Genes | Description | Mutation Types | Amino Acid Change |
|---|---|---|---|
| ATM | ataxia telangiectasia mutated isoform 1 | Missense | p.I2899M |
| Missense | p.S974F (a) | ||
|
| |||
| ATR | ataxia telangiectasia and Rad3 related protein | Nonsense | p.W1784* (b) |
| Missense | p.S1701F (b) | ||
| Missense | p.E1840Q (c) | ||
| Missense | p.A248S (c) | ||
|
| |||
| BRCA1 | breast cancer 1, early onset isoform 2 | Missense | p.E554G |
| Missense | p.R1670K (d) | ||
| Missense | p.V627I (d) | ||
|
| |||
| BRCA2 | breast cancer 2, early onset | Nonsense | p.Y2884* |
| Missense | p.A1411T | ||
| Missense | p.E97V (e) | ||
| Splice_Site | (f) | ||
| Missense | p.K1530E | ||
|
| |||
| BLM | Bloom syndrome protein | Missense | p.D294N (g) |
|
| |||
| ERCC5 | XPG-complementing protein | Missense | p.Q1002R (a) |
|
| |||
| ERCC6 | excision repair cross-complementing rodent | Missense | p.R928T (h) |
|
| |||
| ERCC8 | excision repair cross-complementing rodent | Missense | p.D371H (f) |
|
| |||
| FANCC | Fanconi anemia, complementation group C | Missense | p.T319R (b) |
|
| |||
| FANCI | Fanconi anemia, complementation group I isoform | Missense | p.C558F |
|
| |||
| FANCM | Fanconi anemia, complementation group M | Missense | p.I633M (i) |
| Missense | p.V1951E (a) | ||
| Missense | p.H2016D (j) | ||
| Missense | p.S981L (k) | ||
| Nonsense | p.Q1701* | ||
|
| |||
| RAD50 | RAD50 homolog isoform 1 | Missense | p.E1106Q (e) |
|
| |||
| RAD51AP1 | RAD51 associated protein 1 isoform a | Missense | p.S263C |
|
| |||
| RAD51AP2 | RAD51 associated protein 2 | Nonsense | p.Q5* (d) |
| Missense | p.S355R | ||
|
| |||
| RAD51C | RAD51 homolog C isoform 1 | Missense | p.M118I (i) |
| Missense | p.Q340K (m) | ||
|
| |||
| RAD51L1 | RAD51-like 1 isoform 2 | Missense | p.E167K (g) |
|
| |||
| RAD52 | RAD52 homolog | Missense | p.G59R (n) |
|
| |||
| RAD54B | RAD54 homolog B | Missense | p.L428V (i) |
|
| |||
| RAD9B | RAD9 homolog B | Frame_Shift | p.V412fs (h) |
|
| |||
| RADIL | Rap GTPase interactor | Missense | p.P249R (o) |
|
| |||
| REV1 | REV1-like isoform 1 | Missense | p.R874W (p) |
| RMI1 | RMI1, RecQ mediated genome instability 1, | Missense | p.E617D (m) |
|
| |||
| UBE2E2 | ubiquitin-conjugating enzyme E2E 2 | Missense | p.S46F (h) |
| Missense | p.D22H (g) | ||
|
| |||
| UBE2I | ubiquitin-conjugating enzyme E2I | Missense | p.W103C (i) |
|
| |||
| UBE2J2 | ubiquitin conjugating enzyme E2, J2 isoform 1 | Missense | p.I113F (j) |
|
| |||
| UBE2NL | ubiquitin-conjugating enzyme E2N-like | Nonsense | p.K95* (r) |
|
| |||
| UBE2Q1 | ubiquitin-conjugating enzyme E2Q | Missense | p.M206I (k) |
|
| |||
| UBE3A | ubiquitin protein ligase E3A isoform 2 | Missense | p.K489E (a) |
| Missense | p.S166R | ||
|
| |||
| UBE4A | ubiquitination factor E4A | Missense | p.E616K |
| Missense | p.E822D (i) | ||
| Missense | p.E827K (i) | ||
| Missense | p.L605V | ||
|
| |||
| UBE4B | ubiquitination factor E4B isoform 1 | Nonsense | p.E114* (o) |
|
| |||
| USP12 | ubiquitin thiolesterase 12 | Missense | p.A341E (j) |
|
| |||
| USP13 | ubiquitin thiolesterase 13 | Missense | p.A561T (s) |
| Missense | p.P729L (d) | ||
|
| |||
| USP19 | ubiquitin thioesterase 19 | Nonsense | p.Y941* |
|
| |||
| USP40 | ubiquitin thioesterase 40 | Nonsense | p.G641* (o) |
|
| |||
| USP44 | ubiquitin thiolesterase 44 | Missense | p.E226Q (n) |
| Missense | p.E167Q (n) | ||
| Missense | p.S64T (n) | ||
|
| |||
| USP7 | ubiquitin specific peptidase 7 | Missense | p.M1070V |
|
| |||
| USP8 | ubiquitin specific peptidase 8 | Missense | p.D359H (f) |
| Missense | p.V736I | ||
|
| |||
| USP29 | ubiquitin specific peptidase 29 | Missense | p.T584N (r) |
|
| |||
| USP36 | ubiquitin specific peptidase 36 | Missense | p.I191M (k) |
|
| |||
| USP45 | ubiquitin specific peptidase 45 | Missense | p.K180R (t) |
|
| |||
| USP17L2 | deubiquitinating enzyme 3 | Missense | p.Q430E (k) |
|
| |||
| USP4 | ubiquitin specific protease 4 isoform a | Missense | p.R179P (f) |
| Missense | p.R40Q (f) | ||
|
| |||
| USP24 | ubiquitin specific protease 24 | Missense | p.Y343H (c) |
|
| |||
| USP26 | ubiquitin-specific protease 26 | Missense | p.K734N |
| Missense | p.V906L (p) | ||
|
| |||
| USP28 | ubiquitin specific protease 28 | Missense | p.E371Q (k) |
|
| |||
| USP35 | ubiquitin specific protease 35 | Missense | p.E55K (k) |
|
| |||
| USP43 | ubiquitin specific protease 43 | Missense | p.S455F |
| Missense | p.R206Q (k) | ||
| Missense | p.E834V (t) | ||
|
| |||
| USP47 | ubiquitin specific protease 47 | Missense | p.I1263V |
|
| |||
| USP51 | ubiquitin specific protease 51 | Missense | p.F624L (j) |
|
| |||
| USP9X | ubiquitin specific protease 9, X-linked isoform | Missense | p.P1105L (s) |
| Missense | p.W512L (p) | ||
|
| |||
| USP9Y | ubiquitin specific protease 9, Y-linked | Missense | p.I226F |
|
| |||
| WDR48 | WD repeat domain 48 | Splice_Site | (i) |
As stated previously, individuals with FA have an astonishing probability of one in three for developing solid tumors, most commonly HNSCC, by age 48 [2, 99]. The cumulative risk of developing HNSCC is more than 4% per year, an estimated 700-fold greater risk than in non-FA individuals. In patients that also received bone marrow transplants, this risk increases further by four-fold [138], likely due to deficiencies in DNA damage repair, and subsequent rises in genomic instability, in epithelial cells of the head and neck region. BMT conditioning regimens are presumed to lead to an increased risk of epithelial cancers through genome instability and clonal selection in epithelial FA compartments. A uniquely high susceptibility to head and neck cancers, however, remains unexplained. Possible factors involved might be extensive tissue destruction and regeneration following the preparative regimen, or a role for oncogenic pathogens including, but not limited, to HPV. FA can affect various organs and individuals generally present with complications such as growth retardation, congenital malformations, learning disability, hyper-pigmentation and an elevated risk of secondary malignancies including HNSCC at a early age [139].
In contrast to leukemogenesis, the development of solid tumors in individuals with FA, particularly SCCs of the head and neck (HN) and anogenital tract, is understudied at a molecular level and thus poorly understood. Such tumors occur at an early age in the FA population and with striking aggressiveness. Surgical treatment remains the mainstay, but relapse-free, two-year survival rates are below 50%. Chemotherapy and radiation treatments are associated with high mortality and morbidity due to the patients’ global hypersensitivity to DNA damage. A recent report by Birkeland et al highlights a particularly high degree of complications from radiotherapy, including mucositis and pancytopenia, and poor overall survival [131]. Conventional clastogenic treatments are problematic, and new therapies for the clinical management of FA associated SCC are thus urgently needed. Early intervention is the best line of defense, and will be key for increasing the chance of complete surgical resection and survival in this patient population. However, the absence of relevant biomarkers impedes attempts for early detection of tumor cells in individuals with FA. It is important to note that keratinocytes are the cell type of origin for basal cell carcinomas and squamous cell carcinomas; therefore, FA functions in SCC need to be studied in this same cell type.
Sporadic malignancies with inactivated FA pathway components and associated proteins have been identified [140]. Factors such as hypermethylation, loss of function mutations in FA genes, and increased expression of proteins that influence FA protein functionality and binding interactions have been proposed as mechanisms that disrupt the FA pathway [46, 141, 142]. Transcriptional BRCA1 repression in the absence of genetic mutations has been reported for sporadic tumors and linked to poor prognosis in patients with breast cancer [143, 144]. Transcriptional repression of one or several FA genes by an unknown mechanism was also reported for sporadic head and neck SCCs [145], oral cancers from particularly young patients [146], and ovarian cancers [42]. Finally, disruption of transcriptional FA co-regulation may also occur through epigenetic mechanisms, as has been shown with the hypermethylation of FANCF and at a lower frequency for BRCA1 and BRCA2 in cervical, oral, ovarian and lung cancers [45, 46, 147]. DNA methylation profiling of laryngeal SCC cell lines and primary laryngeal carcinomas revealed very high frequencies of hypomethylation for FANCA, and hypomethylation for BRCA1 and hypermethylation for BRCA2 [148]. Together with a recent report that identified a polymorphism in FANCA associated with cervical cancer progression [149], it is likely that insights into the role of FA in SCC in the rare FA patient population will advance our understanding of mechanisms and consequences of FA inactivation in a subset of SCC in the general population.
Oncogenicity: a selection driven or active process in the FA population?
The onset of cancers, such as leukemia and squamous cell carcinoma (SCC), takes place at an unusually early age in persons with FA when compared to control populations. The observed accelerated transformation of FA hematopoietic cells to leukemia, and that of keratinocytes to SCC, may occur through similar or distinct molecular mechanisms. Recent unpublished data generated in our lab indicate that at the level of gene regulation, several pathways specific to epithelial (rather than hematopoietic) cell growth and development are affected by FA loss in vitro. However, a small number of genes that are regulated in FA-deficient epithelial cells have also previously been shown to be similarly modulated in hematopoietic cells. We thus hypothesize that FA loss regulates some cellular pathways universally, and others in a tissue specific manner. Distinguishing between these possibilities will be of great interest and importance to the FA scientific community. A number of compelling hypotheses have been put forth, with an excellent recent review describing the general importance of “fields” in HNSCC. Field cancerization involves the existence of geographic areas within the mucosa from which tumors tend to arise [119, 150, 151]. Cells within the fields tend to share a specific set of chromosomal translocations and thus an increased risk for transformation. Leemans et al. supported a patch-field-tumor-metastasis progression model for sporadic HNSCCs; given the tendency for chromosomal instability in FA, it is conceivable that individuals with FA might harbor a greater number of fields for greater tumor initiation. This hypothesis remains to be tested, and it is equally possible that FA loss stimulates tumor growth and progression subsequent to or in addition to initiation [119].
Published work suggests that the development of leukemia in an individual with FA is likely, at least in part, a result of clonal evolution. FA hematopoietic stem cells (HSCs), unlike normal stem cells, are hypersensitive to apoptosis-inducing cytokines such as tumor necrosis factor-alpha (TNF-alpha). Exposure to these cytokines reduces the number of FA HSCs by increasing the rate of apoptosis in these cells. These events lead to rapid bone marrow failure with decreased production of all differentiated cell types. FA HSCs adapt to this hostile environment through the development of mutations, which confer resistance to TNF-alpha but retain the characteristic sensitivity to crosslinkers. This evolutionary event was demonstrated via long term, low dose exposure of FANCC murine stem cells to TNF-alpha, followed by transplantation into congenic, lethally irradiated mice. Mice transplanted with TNF-alpha resistant Fancc−/−, but not Fancc-complemented cells developed leukemia. Based on the data, Li et al. proposed that selection for TNF-alpha resistant cells in an environment of genome instability allows for the rapid evolution of pre-leukemic clones [152]. In a recent review, Meyer et al. thus stress the importance of reducing the selection pressure in FA patients in order to decrease the transformation of FA hematopoietic cells towards leukemia [153].
Unlike the strong selection of FA normal to pre-leukemic and leukemic cells, the transformation of FA to squamous cell carcinoma may be a more active process. FA SCC patients have a high propensity for developing carcinomas in the oral cavity. Over 60% of FA patient tumors occur in this anatomical region, frequently in the absence of common risk factors like smoking and alcohol use [129]. The elevated incidence of head and neck and anogenital SCC in FA patients, as well as the unusual location of the tumors, has led some to hypothesize that the onset of HNSCC in FA patients may be the result of a “double-hit”, meaning that the inherent genomic instability may cooperate with an unknown environmental factor. It is equally possible that genetic instability is particularly transforming in these anatomical areas and specific cells therein.
In recent years, human papillomavirus (HPV) infection has been recognized as a risk factor in the development of HNSCC in the general population [154–157]. Interestingly, the frequency of HPV-positive HNSCC is increasing in the United States, while the frequency of HPV-negative HNSCC is declining [158]. In the general population, such tumors often share high risk HPV and expression of the viral E6/E7 oncogenes [159, 160]. The role of HPV in FA HNSCC carcinogenesis remains unclear. Dutch samples did not harbor HPV [127], but work by Kutler et al suggested HPV is present in a majority of primary tumors in the United States [126]. Furthermore, a recent report from Brazil demonstrated increased prevalence of HPV in oral rinses from individuals with FA without lesions when compared to controls, supporting the argument that HPV infection might precede tumor development [125].
Over 30 HPV types cause lesions of the anogenital tract and are subgrouped into high and low risk HPVs depending upon their association with malignant versus benign tumors. By far the most commonly detected anogenital HPV type is HPV16, followed by HPV18 and HPV31 [161]. Their oncogenic potential is reflected in vitro by the ability of high risk, but not low risk, HPV genomic DNA to immortalize primary human keratinocytes [162–165]. When grown under differentiating conditions such as in organotypic epithelial rafts, high risk HPV E6/E7 expressing keratinocytes display abnormalities reminiscent of precancerous lesions [166].
HPV-driven carcinogenesis is initiated and maintained by the oncogenic actions of two proteins, E6 and E7, which bind and inhibit p53 and retinoblastoma (Rb) family members, and many other cellular proteins, respectively. The inactivation of these key players stimulates survival and proliferation in infected cells [161, 167, 168]. E7 is considered the predominant oncogene as it can immortalize keratinocytes in the absence of a cooperating oncogene, and stimulates head and neck as well as cervical cancer development in transgenic mouse models [169]. E7 drives differentiating keratinocytes into a proliferative state, and is required for viral genome maintenance and amplification [170, 171].
In human cancer cell lines derived from sporadic tumors, E7 protein levels are controlled by ubiquitin mediated proteasome degradation. Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells, and several candidate cellular regulators have already been described in vitro [172–175].
Signaling pathways that modify E7 abundance, however, and their effects on the viral life cycle and SCC development are poorly understood. Since p53 and Rb inactivation through mutation is also almost universally observed during the development of HPV-negative solid tumors in humans [176], HPV mediated cellular immortalization and transformation has been viewed as a model system for both HPV-related and -unrelated SCC. Previous findings have demonstrated that FA loss stimulates, and that FA correction inhibits, cellular proliferation and hyperplasia in differentiated epidermis expressing either the HPV18 E6/E7 oncogenes or full-length HPV16 viral genomes. Importantly, proliferative differences were not observed under standard culture conditions, indicating that differentiation-specific cell cycle checkpoints are controlled by functional FA pathways [87]. Recent evidence from our laboratory suggests specific molecular links between the FA pathway and HPV E7: First, acute FANCA or FANCD2 knockdown in E7-transduced human keratinocytes dramatically increased E7 protein but not message levels. This upregulation is likely due to protein stabilization, and may be biologically important since exogenous E7 expression stimulated HPV-induced hyperplasia in FA proficient epidermis. Second, FA knockdown increased cellular proliferation and viral genome copy number in HPV16 and HPV31 positive organotypic epithelial rafts. Hyperplasia was specific to HPV positive keratinocyte at least in one particular isogenic cell system, and was not observed in the corresponding immortalized HPV negative cell line. Based on these data, the overarching hypothesis is that FA loss cooperates with HPV oncogene activities to stimulate SCC phenotypes in human epidermis, and that this process involves, at least in part, increased E7 oncoprotein abundance [87, 177]. Whether the same cellular proteins, which mediate FA-controlled E7 regulation in the HPV positive environment, also stimulate tumor phenotypes in the HPV negative tumor environment under certain circumstances will require elucidation of the underlying molecular mechanism(s). This is particularly important in view of the fact that the etiology of FA HNSCCs remains unclear.
Disease biomarker discovery for FA-associated HNSCC
Individuals with sporadic HNSCC frequently present at advanced disease stages, which is a direct result of inferior diagnostic approaches, and necessitates aggressive chemo- and/or radiation therapy [178]. The downside of this scenario in FA patients diagnosed with HNSCC is that these therapeutic methods typically lead to extreme toxicity and sometimes death [129, 132, 133]. To add to the complexity of the situation, in over 20% of FA patients diagnosed with a solid tumor, a diagnosis of FA was not made until after the cancer was detected, and thus treatment toxicity is not easily predictable [99]. Therefore, there is an urgent need for improved, early diagnosis of HNSCC in this patient population, but also for a diagnosis of FA prior to the development and treatment of cancer. Although HNSCC solid tumors in FA patients may be treatable in the future with the advancement of molecularly targeted therapies, cancer prevention in this patient population should be the primary goal.
The predominant research focus to identify markers of sporadic HNSCCs has been largely restricted to global genome and proteome analyses [179]. Zimmermann et al. discuss genomic analysis with regards to biomarkers for oral cancers in great detail, and propose that transcriptomics will prove to be a beneficial method for biomarker identification [180]. A specific type of genomic analysis, metagenomics, has emerged in the study of oral diseases [181]. This technique isolates nucleic acids and performs unprejudiced DNA sequencing to categorize viruses and microorganisms present in a particular sample. This approach was utilized to characterize the normal flora present in the oral cavity of healthy persons and the data obtained revealed a connection between oral health and specific types of bacteria present within the oral cavity [182].
Burbelo et al. describe multiple diagnostic tools including proteomics, and emphasize the promise and potential of this technique to diagnose head and neck cancers [183]. Mass spectrometry has previously been used to study changes in salivary levels of numerous proteins such as M2BP, MRP14, and CD59 [184], and these potential markers could be validated by immunoassays [184]. With a focus on detecting head and neck cancer at an early stage in the disease process using a semi-invasive collection and analysis method, Remmerbach et al. advanced the field further by introducing proteome/peptidome analysis of brush biopsies from OSCC patients and matched controls. Indeed, Matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS) analysis could differentiate OSCC patients from healthy controls with high specificity and sensitivity. However, further optimization of this method is needed to establish its practicality in detecting early stages of OSCC in a clinical setting by taking into account numerous factors including time and cost [185]. The above-mentioned “omics” techniques have identified several potential biomarkers of head and neck cancer. Although these techniques are highly powerful methods, it is important to note that the combined information from these approaches still predict only a portion of the complexity of OSCCs, and additional techniques used in conjunction with the aforementioned approaches will likely be beneficial.
In addition to genomics and proteomics, the area of cancer research could benefit from the coupling of these practices with additional “omic” biomarker discovery approaches. The results obtained from these “omic” methods may not fully correlate to one another in the sense that the regulation of certain genes does not always result in abundance in the corresponding proteins. This is not surprising, since the production of a protein may occur some time after its gene is transcriptionally activated. This caveat placed on the field of transcriptomics/genomics may lead researchers to place more emphasis on the protein expression levels within a given system. Proteins provide an excellent reflection of the biological processes that take place at a defined time in a certain system, and metabonomics is the next most accurate technology for observing the intermediates and final products relating to both gene transcription and protein regulation.
Metabonomics was first defined as “the quantitative measurements of the multiparametric metabolic response of a living system to pathophysiological stimuli or genetic modifications” [186]. The technique has been minimally utilized thus far in cancer biology; however, it has shown to be a potentially useful tool in the area of biomarker identification in disease [187–189]. Metabonomics is the study of an organism’s metabolome, which makes this a powerful “omics” technique because the metabolome is downstream of both the genome and proteome and is complementary to other “omic” methods [186, 190]. Metabonomics has been employed in drug surveillance as well as disease diagnosis, identification of changes in metabolites associated with cell apoptosis, cancer cell growth, proliferation rates and metabolic effects related to the Warburg hypothesis of modified energy production [190–193]. The main goal of metabonomics research in HNSCC should be to understand normal metabolic function in epidermal compartments and to define how changes in this metabolic fingerprint can function as early diagnostic tools [194, 195]. Defining changes in the metabolome related to disease onset and progression in the FA HNSCC will provide significant information about regulatory pathways in cancer in general, and should drive the identification of novel cancer biomarkers and therapeutic targets in FA.
Acknowledgments
Research on Fanconi anemia in the Wells laboratory was supported by a Public Health Service Grant CA102357, and by a grant from the Fanconi Anemia Research Fund. We thank Elizabeth Hoskins and Drs. Paul Andreassen, Kasiani Myers, Timothy Chlon and Lisa Privette Vinnedge for critical comments on this report.
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alter BP. Fanconi’s anemia and malignancies. Am J Hematol. 1996;53:99–110. doi: 10.1002/(SICI)1096-8652(199610)53:2<99::AID-AJH7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003;101:822–826. doi: 10.1182/blood-2002-05-1498. [DOI] [PubMed] [Google Scholar]
- 3.Kutler DI, Singh B, Satagopan J, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 4.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A. 2011;155A:1877–1883. doi: 10.1002/ajmg.a.34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rochowski A, Olson SB, Alonzo TA, et al. Patients with Fanconi anemia and AML have different cytogenetic clones than de novo cases of AML. Pediatr Blood Cancer. 2012 doi: 10.1002/pbc.24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rochowski A, Rosenberg PS, Alonzo TA, et al. Estimation of the prevalence of Fanconi anemia among patients with de novo acute myelogenous leukemia who have poor recovery from chemotherapy. Leuk Res. 2012;36:29–31. doi: 10.1016/j.leukres.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montes de Oca R, Andreassen PR, Margossian SP, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 10.Zhang F, Fan Q, Ren K, et al. FANCJ/BRIP1 recruitment and regulation of FANCD2 in DNA damage responses. Chromosoma. 2010;119:637–649. doi: 10.1007/s00412-010-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song IY, Barkley LR, Day TA, et al. A novel role for Fanconi anemia (FA) pathway effector protein FANCD2 in cell cycle progression of untransformed primary human cells. Cell Cycle. 2010;9:2375–2388. doi: 10.4161/cc.9.12.11900. [DOI] [PubMed] [Google Scholar]
- 12.Sato K, Ishiai M, Toda K, et al. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 2012;31:3524–3536. doi: 10.1038/emboj.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garner E, Smogorzewska A. Ubiquitylation and the Fanconi anemia pathway. FEBS Lett. 2011;585:2853–2860. doi: 10.1016/j.febslet.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smogorzewska A, Matsuoka S, Vinciguerra P, et al. Identification of the FANCI Protein, a Monoubiquitinated FANCD2 Paralog Required for DNA Repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi T, Garcia-Higuera I, Xu B, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki MS. Is Fanconi’s anaemia defective in a process essential to the repair of DNA cross links? Nature. 1975;257:501–503. doi: 10.1038/257501a0. [DOI] [PubMed] [Google Scholar]
- 17.Auerbach AD. A test for Fanconi’s anemia. Blood. 1988;72:366–367. [PubMed] [Google Scholar]
- 18.Sasaki MS, Tonomura A. A high susceptibility of Fanconi’s anemia to chromosome breakage by DNA cross-linking agents. Cancer Res. 1973;33:1829–1836. [PubMed] [Google Scholar]
- 19.Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21:731–733. [PubMed] [Google Scholar]
- 20.Wang W. A major switch for the Fanconi anemia DNA damage-response pathway. Nat Struct Mol Biol. 2008;15:1128–1130. doi: 10.1038/nsmb1108-1128. [DOI] [PubMed] [Google Scholar]
- 21.Andreassen PR, D’Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 23.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 24.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2006 doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 25.Litman R, Peng M, Jin Z, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Levitus M, Waisfisz Q, Godthelp BC, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 27.Levran O, Attwooll C, Henry RT, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 28.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi T, Garcia-Higuera I, Andreassen PR, et al. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 31.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meindl A, Ditsch N, Kast K, et al. Hereditary breast and ovarian cancer: new genes, new treatments, new concepts. Dtsch Arztebl Int. 2011;108:323–330. doi: 10.3238/arztebl.2011.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 34.Cantor SB, Guillemette S. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. 2011;7:253–261. doi: 10.2217/fon.10.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70:7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campeau PM, Foulkes WD, Tischkowitz MD. Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet. 2008;124:31–42. doi: 10.1007/s00439-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 37.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nature Genetics. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 38.van der Heijden MS, Brody JR, Gallmeier E, et al. Functional defects in the fanconi anemia pathway in pancreatic cancer cells. Am J Pathol. 2004;165:651–657. doi: 10.1016/S0002-9440(10)63329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couch FJ, Johnson MR, Rabe K, et al. Germ line Fanconi anemia complementation group C mutations and pancreatic cancer. Cancer Res. 2005;65:383–386. [PubMed] [Google Scholar]
- 40.van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–2588. [PubMed] [Google Scholar]
- 41.Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25:5875–5884. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- 42.Pejovic T, Yates JE, Liu HY, et al. Cytogenetic instability in ovarian epithelial cells from women at risk of ovarian cancer. Cancer Res. 2006;66:9017–9025. doi: 10.1158/0008-5472.CAN-06-0222. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Li M, Lu S, et al. Promoter hypermethylation of FANCF plays an important role in the occurrence of ovarian cancer through disrupting Fanconi anemia-BRCA pathway. Cancer Biol Ther. 2006;5:256–260. doi: 10.4161/cbt.5.3.2380. [DOI] [PubMed] [Google Scholar]
- 45.Narayan G, Arias-Pulido H, Nandula SV, et al. Promoter hypermethylation of FANCF: disruption of Fanconi Anemia-BRCA pathway in cervical cancer. Cancer Res. 2004;64:2994–2997. doi: 10.1158/0008-5472.can-04-0245. [DOI] [PubMed] [Google Scholar]
- 46.Marsit CJ, Liu M, Nelson HH, et al. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23:1000–1004. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- 47.Ameziane N, Chen F, Leemans CR, et al. No evidence for FANCF gene silencing in head-and-neck squamous cell carcinomas. Cell Oncol. 2009;31:53–56. doi: 10.3233/CLO-2009-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snyder ER, Ricker JL, Chen Z, Waes CV. Variation in cisplatinum sensitivity is not associated with Fanconi Anemia/BRCA pathway inactivation in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2007;245:75–80. doi: 10.1016/j.canlet.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 49.Burkitt K, Ljungman M. Compromised Fanconi anemia response due to BRCA1 deficiency in cisplatin-sensitive head and neck cancer cell lines. Cancer Lett. 2007;253:131–137. doi: 10.1016/j.canlet.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 50.Meetei AR, de Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 51.Meetei AR, Levitus M, Xue Y, et al. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 52.Meetei AR, Medhurst AL, Ling C, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Winter JP, Leveille F, van Berkel CG, et al. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am J Hum Genet. 2000;67:1306–1308. doi: 10.1016/s0002-9297(07)62959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Winter JP, Waisfisz Q, Rooimans MA, et al. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 55.Lo Ten Foe JR, Rooimans MA, Bosnoyan-Collins L, et al. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 56.Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. 1992;358:434. doi: 10.1038/358434a0. [DOI] [PubMed] [Google Scholar]
- 57.Timmers C, Taniguchi T, Hejna J, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 58.de Winter JP, Rooimans MA, van Der Weel L, et al. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat Genet. 2000;24:15–16. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- 59.Sims AE, Spiteri E, Sims RJ, 3rd, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 60.Positional cloning of the Fanconi anaemia group A gene. Nat Genet. 1996;14:324–328. doi: 10.1038/ng1196-324. [DOI] [PubMed] [Google Scholar]
- 61.Dorsman JC, Levitus M, Rockx D, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 63.Stoepker C, Hain K, Schuster B, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 64.Kim Y, Lach FP, Desetty R, et al. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoskins EE, Gunawardena RW, Habash KB, et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008;27:4798–4808. doi: 10.1038/onc.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ciccia A, Ling C, Coulthard R, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Ling C, Ishiai M, Ali AM, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. Embo J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ali AM, Pradhan A, Singh TR, et al. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood. 2012;119:3285–3294. doi: 10.1182/blood-2011-10-385963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leung JW, Wang Y, Fong KW, et al. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci U S A. 2012;109:4491–4496. doi: 10.1073/pnas.1118720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collins NB, Wilson JB, Bush T, et al. ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood. 2009;113:2181–2190. doi: 10.1182/blood-2008-05-154294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiao F, Mi J, Wilson JB, et al. Phosphorylation of fanconi anemia (FA) complementation group G protein, FANCG, at serine 7 is important for function of the FA pathway. J Biol Chem. 2004;279:46035–46045. doi: 10.1074/jbc.M408323200. [DOI] [PubMed] [Google Scholar]
- 73.Ho GP, Margossian S, Taniguchi T, D’Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26:7005–7015. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 75.Machida YJ, Machida Y, Chen Y, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 76.Cole AR, Lewis LPC, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nature Structural & Molecular Biology. 2010;17:294–U254. doi: 10.1038/nsmb.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh TR, Bakker ST, Agarwal S, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114:174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh TR, Saro D, Ali AM, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim JM, Kee Y, Gurtan A, D’Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gari KDC, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hovest MG, Krieg T, Herrmann G. Differential roles for Chk1 and FANCD2 in ATR-mediated signalling for psoralen photoactivation-induced senescence. Exp Dermatol. 2011;20:883–889. doi: 10.1111/j.1600-0625.2011.01365.x. [DOI] [PubMed] [Google Scholar]
- 82.Ishiai M, Kitao H, Smogorzewska A, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takata M, Ishiai M, Kitao H. The Fanconi anemia pathway: insights from somatic cell genetics using DT40 cell line. Mutat Res. 2009;668:92–102. doi: 10.1016/j.mrfmmm.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 84.Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol. 2010;17:294–298. doi: 10.1038/nsmb.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaddar T, Carreau M. Fanconi anemia proteins and their interacting partners: a molecular puzzle. Anemia. 2012;2012:425814. doi: 10.1155/2012/425814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang YG, Herceg Z, Nakanishi K, et al. The Fanconi anemia group A protein modulates homologous repair of DNA double-strand breaks in mammalian cells. Carcinogenesis. 2005;26:1731–1740. doi: 10.1093/carcin/bgi134. [DOI] [PubMed] [Google Scholar]
- 87.Hoskins EE, Morris TA, Higginbotham JM, et al. Fanconi anemia deficiency stimulates HPV-associated hyperplastic growth in organotypic epithelial raft culture. Oncogene. 2009;28:674–685. doi: 10.1038/onc.2008.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.MacKay C, Declais AC, Lundin C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smogorzewska A, Desetty R, Saito TT, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Donnell L, Durocher D. DNA repair has a new FAN1 club. Mol Cell. 2010;39:167–169. doi: 10.1016/j.molcel.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 91.Liu T, Ghosal G, Yuan J, et al. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 92.Wang X, Andreassen PR, D’Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Howlett NG, Taniguchi T, Durkin SG, et al. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 94.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nijman SM, Huang TT, Dirac AM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 96.Oestergaard VH, Langevin F, Kuiken HJ, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim JM, Parmar K, Huang M, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alter BP. Cancer in Fanconi anemia, 1927–2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 100.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 101.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 102.MacMillan ML, Hughes MR, Agarwal S, Daley GQ. Cellular therapy for fanconi anemia: the past, present, and future. Biol Blood Marrow Transplant. 17:S109–114. doi: 10.1016/j.bbmt.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 103.Schultz JC, Shahidi NT. Tumor necrosis factor-alpha overproduction in Fanconi’s anemia. Am J Hematol. 1993;42:196–201. doi: 10.1002/ajh.2830420211. [DOI] [PubMed] [Google Scholar]
- 104.Rosselli F, Sanceau J, Gluckman E, et al. Abnormal lymphokine production: a novel feature of the genetic disease Fanconi anemia. II. In vitro and in vivo spontaneous overproduction of tumor necrosis factor alpha. Blood. 1994;83:1216–1225. [PubMed] [Google Scholar]
- 105.Dufour C, Corcione A, Svahn J, et al. TNF-alpha and IFN-gamma are overexpressed in the bone marrow of Fanconi anemia patients and TNF-alpha suppresses erythropoiesis in vitro. Blood. 2003;102:2053–2059. doi: 10.1182/blood-2003-01-0114. [DOI] [PubMed] [Google Scholar]
- 106.Lensch MW, Rathbun RK, Olson SB, et al. Selective pressure as an essential force in molecular evolution of myeloid leukemic clones: a view from the window of Fanconi anemia. Leukemia. 1999;13:1784–1789. doi: 10.1038/sj.leu.2401586. [DOI] [PubMed] [Google Scholar]
- 107.Myers KC, Davies SM. Hematopoietic stem cell transplantation for bone marrow failure syndromes in children. Biol Blood Marrow Transplant. 2009;15:279–292. doi: 10.1016/j.bbmt.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 108.Alter BP. Bone marrow failure syndromes in children. Pediatr Clin North Am. 2002;49:973–988. doi: 10.1016/s0031-3955(02)00031-7. [DOI] [PubMed] [Google Scholar]
- 109.Tischkowitz M, Dokal I. Fanconi anaemia and leukaemia - clinical and molecular aspects. Br J Haematol. 2004;126:176–191. doi: 10.1111/j.1365-2141.2004.05023.x. [DOI] [PubMed] [Google Scholar]
- 110.Butturini A, Gale RP, Verlander PC, et al. Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study. Blood. 1994;84:1650–1655. [PubMed] [Google Scholar]
- 111.Wagner JE, Tolar J, Levran O, et al. Germline mutations in BRCA2: shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood. 2004;103:3226–3229. doi: 10.1182/blood-2003-09-3138. [DOI] [PubMed] [Google Scholar]
- 112.Hirsch B, Shimamura A, Moreau L, et al. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103:2554–2559. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- 113.Tischkowitz M, Ameziane N, Waisfisz Q, et al. Bi-allelic silencing of the Fanconi anaemia gene FANCF in acute myeloid leukaemia. Br J Haematol. 2003;123:469–471. doi: 10.1046/j.1365-2141.2003.04640.x. [DOI] [PubMed] [Google Scholar]
- 114.Hess CJ, Ameziane N, Schuurhuis GJ, et al. Hypermethylation of the FANCC and FANCL promoter regions in sporadic acute leukaemia. Cell Oncol. 2008;30:299–306. doi: 10.3233/CLO-2008-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rosenberg PS, Alter BP, Ebell W. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica. 2008;93:511–517. doi: 10.3324/haematol.12234. [DOI] [PubMed] [Google Scholar]
- 116.Mehta PA, Harris RE, Davies SM, et al. Numerical chromosomal changes and risk of development of myelodysplastic syndrome--acute myeloid leukemia in patients with Fanconi anemia. Cancer Genet Cytogenet. 2010;203:180–186. doi: 10.1016/j.cancergencyto.2010.07.127. [DOI] [PubMed] [Google Scholar]
- 117.D’Andrea AD. Targeting DNA repair pathways in AML. Best Pract Res Clin Haematol. 2010;23:469–473. doi: 10.1016/j.beha.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 118.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 119.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 120.Califano J, van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 121.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 122.Baron AE, Franceschi S, Barra S, et al. A comparison of the joint effects of alcohol and smoking on the risk of cancer across sites in the upper aerodigestive tract. Cancer Epidemiol Biomarkers Prev. 1993;2:519–523. [PubMed] [Google Scholar]
- 123.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 124.Field JK. Oncogenes and tumour-suppressor genes in squamous cell carcinoma of the head and neck. Eur J Cancer B Oral Oncol. 1992;28B:67–76. doi: 10.1016/0964-1955(92)90016-t. [DOI] [PubMed] [Google Scholar]
- 125.de Araujo MR, Rubira-Bullen IR, Santos CF, et al. High prevalence of oral human papillomavirus infection in Fanconi’s anemia patients. Oral Dis. 2011;17:572–576. doi: 10.1111/j.1601-0825.2011.01803.x. [DOI] [PubMed] [Google Scholar]
- 126.Kutler DI, Wreesmann VB, Goberdhan A, et al. Human papillomavirus DNA and p53 polymorphisms in squamous cell carcinomas from Fanconi anemia patients. J Natl Cancer Inst. 2003;95:1718–1721. doi: 10.1093/jnci/djg091. [DOI] [PubMed] [Google Scholar]
- 127.van Zeeburg HJ, Snijders PJ, Wu T, et al. Clinical and molecular characteristics of squamous cell carcinomas from Fanconi anemia patients. J Natl Cancer Inst. 2008;100:1649–1653. doi: 10.1093/jnci/djn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li FP, Montesano R. Interactions of cancer susceptibility genes and environmental carcinogens. American Association for Cancer Research (AACR)-International Agency for Research on Cancer (IARC) Joint Conference. Cancer Res. 1994;54:4243–4247. [PubMed] [Google Scholar]
- 129.Kutler DI, Auerbach AD, Satagopan J, et al. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2003;129:106–112. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- 130.Cloos J, Spitz MR, Schantz SP, et al. Genetic susceptibility to head and neck squamous cell carcinoma. J Natl Cancer Inst. 1996;88:530–535. doi: 10.1093/jnci/88.8.530. [DOI] [PubMed] [Google Scholar]
- 131.Birkeland AC, Auerbach AD, Sanborn E, et al. Postoperative clinical radiosensitivity in patients with fanconi anemia and head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2011;137:930–934. doi: 10.1001/archoto.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alter BP. Radiosensitivity in Fanconi’s anemia patients. Radiother Oncol. 2002;62:345–347. doi: 10.1016/s0167-8140(01)00474-1. [DOI] [PubMed] [Google Scholar]
- 133.Bremer M, Schindler D, Gross M, et al. Fanconi’s anemia and clinical radiosensitivity report on two adult patients with locally advanced solid tumors treated by radiotherapy. Strahlenther Onkol. 2003;179:748–753. doi: 10.1007/s00066-003-1099-8. [DOI] [PubMed] [Google Scholar]
- 134.Spanier G, Pohl F, Giese T, et al. Fatal course of tonsillar squamous cell carcinoma associated with Fanconi anaemia: A mini review. J Craniomaxillofac Surg. 2011 doi: 10.1016/j.jcms.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 135.Sparano A, Quesnelle KM, Kumar MS, et al. Genome-wide profiling of oral squamous cell carcinoma by array-based comparative genomic hybridization. Laryngoscope. 2006;116:735–741. doi: 10.1097/01.mlg.0000205141.54471.7f. [DOI] [PubMed] [Google Scholar]
- 136.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rosenberg PS, Socie G, Alter BP, Gluckman E. Risk of head and neck squamous cell cancer and death in patients with Fanconi anemia who did and did not receive transplants. Blood. 2005;105:67–73. doi: 10.1182/blood-2004-04-1652. [DOI] [PubMed] [Google Scholar]
- 139.Kaplan MJ, Sabio H, Wanebo HJ, Cantrell RW. Squamous cell carcinoma in the immunosuppressed patient: Fanconi’s anemia. Laryngoscope. 1985;95:771–775. [PubMed] [Google Scholar]
- 140.Lyakhovich A, Surralles J. Disruption of the Fanconi anemia/BRCA pathway in sporadic cancer. Cancer Lett. 2006;232:99–106. doi: 10.1016/j.canlet.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 141.Valeri A, Martinez S, Casado JA, Bueren JA. Fanconi anaemia: from a monogenic disease to sporadic cancer. Clin Transl Oncol. 2011;13:215–221. doi: 10.1007/s12094-011-0645-6. [DOI] [PubMed] [Google Scholar]
- 142.Wei M, Xu J, Dignam J, et al. Estrogen receptor alpha, BRCA1, and FANCF promoter methylation occur in distinct subsets of sporadic breast cancers. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9766-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Russell PA, Pharoah PD, De Foy K, et al. Frequent loss of BRCA1 mRNA and protein expression in sporadic ovarian cancers. Int J Cancer. 2000;87:317–321. doi: 10.1002/1097-0215(20000801)87:3<317::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 144.Thompson ME, Jensen RA, Obermiller PS, et al. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9:444–450. doi: 10.1038/ng0495-444. [DOI] [PubMed] [Google Scholar]
- 145.Wreesmann VB, Estilo C, Eisele DW, et al. Downregulation of Fanconi anemia genes in sporadic head and neck squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec. 2007;69:218–225. doi: 10.1159/000101542. [DOI] [PubMed] [Google Scholar]
- 146.Tremblay S, Pintor Dos Reis P, Bradley G, et al. Young patients with oral squamous cell carcinoma: study of the involvement of GSTP1 and deregulation of the Fanconi anemia genes. Arch Otolaryngol Head Neck Surg. 2006;132:958–966. doi: 10.1001/archotol.132.9.958. [DOI] [PubMed] [Google Scholar]
- 147.Dhillon VS, Shahid M, Husain SA. CpG methylation of the FHIT, FANCF, cyclin-D2, BRCA2 and RUNX3 genes in Granulosa cell tumors (GCTs) of ovarian origin. Mol Cancer. 2004;3:33. doi: 10.1186/1476-4598-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Szaumkessel M, Richter J, Giefing M, et al. Pyrosequencing-based DNA methylation profiling of Fanconi anemia/BRCA pathway genes in laryngeal squamous cell carcinoma. Int J Oncol. 2011;39:505–514. doi: 10.3892/ijo.2011.1039. [DOI] [PubMed] [Google Scholar]
- 149.Wang SS, Bratti MC, Rodriguez AC, et al. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. J Infect Dis. 2009;199:20–30. doi: 10.1086/595563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Braakhuis BJ, Bloemena E, Leemans CR, Brakenhoff RH. Molecular analysis of surgical margins in head and neck cancer: more than a marginal issue. Oral Oncol. 2010;46:485–491. doi: 10.1016/j.oraloncology.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 151.Braakhuis BJ, Tabor MP, Kummer JA, et al. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 152.Li J, Sejas DP, Zhang XL, et al. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. Journal of Clinical Investigation. 2007;117:3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Meyer S, Neitzel H, Tonnies H. Chromosomal aberrations associated with clonal evolution and leukemic transformation in fanconi anemia: clinical and biological implications. Anemia. 2012;2012:349837. doi: 10.1155/2012/349837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 155.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 156.Schlecht NF, Burk RD, Adrien L, et al. Gene expression profiles in HPV-infected head and neck cancer. J Pathol. 2007;213:283–293. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- 157.Adelstein DJ, Ridge JA, Gillison ML, et al. Head and neck squamous cell cancer and the human papillomavirus: summary of a National Cancer Institute State of the Science Meeting, November 9–10, 2008, Washington, D. C Head Neck. 2009;31:1393–1422. doi: 10.1002/hed.21269. [DOI] [PubMed] [Google Scholar]
- 158.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gillison ML, Shah KV. Chapter 9: Role of mucosal human papillomavirus in nongenital cancers. J Natl Cancer Inst Monogr. 2003:57–65. doi: 10.1093/oxfordjournals.jncimonographs.a003484. [DOI] [PubMed] [Google Scholar]
- 160.Herrero R. Chapter 7: Human papillomavirus and cancer of the upper aerodigestive tract. J Natl Cancer Inst Monogr. 2003:47–51. doi: 10.1093/oxfordjournals.jncimonographs.a003482. [DOI] [PubMed] [Google Scholar]
- 161.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 162.Münger K, Phelps WC, Bubb V, et al. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hawley-Nelson P, Vousden KH, Hubbert NL, et al. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pecoraro G, Morgan D, Defendi V. Differential effects of human papillomavirus type 6, 16 and 18 DNAs on immortalization and transformation of human cervical epithelial cells. Proc Natl Acad Sci USA. 1989 doi: 10.1073/pnas.86.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Woodworth CD, Doninger J, DiPaolo JA. Immortalization of human foreskin keratinocytes by various human papillomavirus DNAs corresponds to their association with cervical carcinoma. J Virol. 1989;63:159–164. doi: 10.1128/jvi.63.1.159-164.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McCance DJ, Kopan R, Fuchs E, Laimins LA. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc Natl Acad Sci USA. 1988;85:7169–7173. doi: 10.1073/pnas.85.19.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Syrjanen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol. 2005;32 (Suppl 1):S59–66. doi: 10.1016/j.jcv.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 168.Snijders PJ, Cromme FV, van den Brule AJ, et al. Prevalence and expression of human papillomavirus in tonsillar carcinomas, indicating a possible viral etiology. Int J Cancer. 1992;51:845–850. doi: 10.1002/ijc.2910510602. [DOI] [PubMed] [Google Scholar]
- 169.Munger K, Baldwin A, Edwards KM, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.DiMaio D, Liao JB. Human papillomaviruses and cervical cancer. Adv Virus Res. 2006;66:125–159. doi: 10.1016/S0065-3527(06)66003-X. [DOI] [PubMed] [Google Scholar]
- 172.Klingelhutz AJ, Qian Q, Phillips SL, et al. Amplification of the chromosome 20q region is associated with expression of HPV-16 E7 in human airway and anogenital epithelial cells. Virology. 2005;340:237–244. doi: 10.1016/j.virol.2005.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang J, Sampath A, Raychaudhuri P, Bagchi S. Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells. Oncogene. 2001;20:4740–4749. doi: 10.1038/sj.onc.1204655. [DOI] [PubMed] [Google Scholar]
- 174.Reinstein E, Scheffner M, Oren M, et al. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene. 2000;19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- 175.Lin CH, Chang HS, Yu WC. USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. J Biol Chem. 2008;283:15681–15688. doi: 10.1074/jbc.M708278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hickman ES, Moroni MC, Helin K. The role of p53 and pRB in apoptosis and cancer. Curr Opin Genet Dev. 2002;12:60–66. doi: 10.1016/s0959-437x(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 177.Hoskins EE, Morreale RJ, Werner SP, et al. The Fanconi Anemia Pathway Limits Human Papillomavirus Replication. J Virol. 2012 doi: 10.1128/JVI.00408-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wadsworth JT, Somers KD, Cazares LH, et al. Serum protein profiles to identify head and neck cancer. Clin Cancer Res. 2004;10:1625–1632. doi: 10.1158/1078-0432.ccr-0297-3. [DOI] [PubMed] [Google Scholar]
- 179.Nagaraj NS. Evolving ‘omics’ technologies for diagnostics of head and neck cancer. Brief Funct Genomic Proteomic. 2009;8:49–59. doi: 10.1093/bfgp/elp004. [DOI] [PubMed] [Google Scholar]
- 180.Zimmermann BG, Park NJ, Wong DT. Genomic targets in saliva. Ann N Y Acad Sci. 2007;1098:184–191. doi: 10.1196/annals.1384.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Willner D, Furlan M, Schmieder R, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci U S A. 2011;108 (Suppl 1):4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Crielaard W, Zaura E, Schuller AA, et al. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Burbelo PD, Bayat A, Lebovitz EE, Iadarola MJ. New technologies for studying the complexity of oral diseases. Oral Dis. 2012;18:121–126. doi: 10.1111/j.1601-0825.2011.01863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hu S, Arellano M, Boontheung P, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Remmerbach TW, Maurer K, Janke S, et al. Oral brush biopsy analysis by matrix assisted laser desorption/ionisation-time of flight mass spectrometry profiling--a pilot study. Oral Oncol. 2011;47:278–281. doi: 10.1016/j.oraloncology.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 186.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 187.Fan TW, Lane AN, Higashi RM. The promise of metabolomics in cancer molecular therapeutics. Curr Opin Mol Ther. 2004;6:584–592. [PubMed] [Google Scholar]
- 188.Giskeodegard GF, Grinde MT, Sitter B, et al. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J Proteome Res. 2010;9:972–979. doi: 10.1021/pr9008783. [DOI] [PubMed] [Google Scholar]
- 189.Yang Y, Li C, Nie X, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6:2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 190.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 191.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 192.Zhang J, Bowers J, Liu L, et al. Esophageal cancer metabolite biomarkers detected by LC-MS and NMR methods. PLoS One. 2012;7:e30181. doi: 10.1371/journal.pone.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Napoli C, Sperandio N, Lawlor RT, et al. Urine metabolic signature of pancreatic ductal adenocarcinoma by (1)h nuclear magnetic resonance: identification, mapping, and evolution. J Proteome Res. 2012;11:1274–1283. doi: 10.1021/pr200960u. [DOI] [PubMed] [Google Scholar]
- 194.Bu Q, Huang Y, Yan G, et al. Metabolomics: A Revolution for Novel Cancer Marker Identification. Comb Chem High Throughput Screen. 2012 doi: 10.2174/138620712799218563. [DOI] [PubMed] [Google Scholar]
- 195.Ma Y, Zhang P, Yang Y, et al. Metabolomics in the fields of oncology: a review of recent research. Mol Biol Rep. 2012 doi: 10.1007/s11033-012-1584-1. [DOI] [PubMed] [Google Scholar]