Abstract
DEAD-box helicases perform diverse cellular functions in virtually all steps of RNA metabolism from Bacteria to Humans. Although DEAD-box helicases share a highly conserved core domain, the enzymes catalyze a wide range of biochemical reactions. In addition to the well established RNA unwinding and corresponding ATPase activities, DEAD-box helicases promote duplex formation and displace proteins from RNA. They can also function as assembly platforms for larger ribonucleoprotein complexes, and as metabolite sensors. This review aims to provide a perspective on the diverse biochemical features of DEAD-box helicases and connections to structural information. We discuss these data in the context of a model that views the enzymes as integrators of RNA, nucleotide, and protein binding.
1. Introduction
DEAD-box helicases form the largest helicase family [1]. They function at many different points in RNA metabolism, and play diverse cellular roles in Bacteria, Archea, and Eukaryotes [2,3,4]. All DEAD-box helicases share a core structure that is among the most highly conserved enzyme domains [3,5]. How DEAD-box helicases accomplish diverse functions using a virtually identical core has been a central and longstanding question in RNA biology [6]. Emerging answers to this question are multifaceted and greatly affected by the cellular environment for any given DEAD-box helicase. Yet, it is clear that the molecular basis for the cellular function of each enzyme lies in its biochemical capacity.
Efforts to delineate biochemical and structural principles of DEAD-box helicase function have provided significant insight over the last years. Consistent with diverse cellular roles, the helicases have been shown to be more than duplex unwinders [3,7,8]. In addition to the well established RNA unwinding and corresponding ATPase activities, DEAD-box helicases promote duplex formation and displace proteins from RNA, but they can also function as assembly platforms for larger ribonucleoprotein complexes, and as recently reported, as sensors for metabolites [9,10,11,12,13,14].
This review aims to provide an inclusive perspective on the diverse biochemical features and their connection to structural information for DEAD-box helicases. We discuss these data in the context of a model that views the enzymes as integrators of RNA, nucleotide, and protein binding. This view has been developed through a large body of experimental and conceptual work by many groups, published over the last two decades. We believe that considering DEAD-box helicases as multi-functional modules, rather than re-purposed unwinders, is most instructive for a systematic understanding of the biochemical range of the enzymes, and thus for devising physical models of cellular functions for these essential enzymes.
2. Structural characteristics of DEAD-box helicases
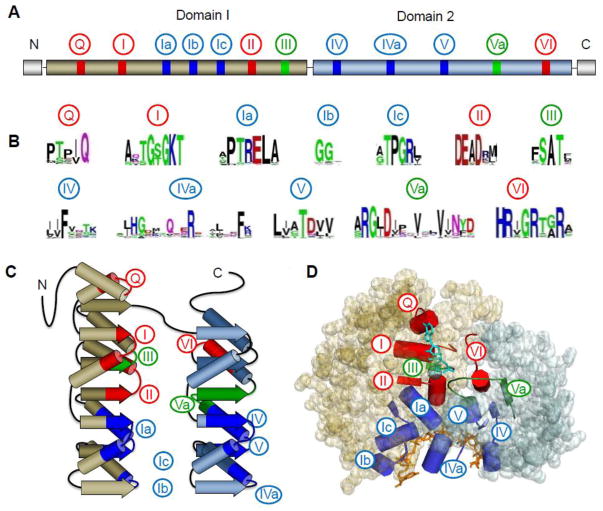
All DEAD-box helicases share the highly conserved SF2 helicase core that consists of two virtually identical domains (Fig. 1). Each of these helicase core domains resembles the recombination protein RecA [5,15]. Most DEAD-box helicases also contain C- and N-terminal extensions that are not conserved across different DEAD-box helicases [1]. Two mammalian DEAD-box helicases, DDX24 and DDX1, contain insertions in the helicase domain 1. In analogy to other SF2 helicase families, these insertions are unlikely to disrupt the RecA fold of the domain [1].
Figure 1. Architecture of the DEAD-box helicase core.
(A) Schematic of the primary structure of the DEAD-box helicase core. Domain 1 (N-terminal, tan) and 2 (C-terminal, blue) designate the two RecA-like helicase domains. Circled numbers indicate the approximate location of the characteristic sequence motifs, colored according to their primary biochemical function (red, ATP binding and hydrolysis; blue, RNA binding, green, coordination between RNA and ATP binding). (B) Sequence conservation in the characteristic sequence motifs of DEAD-box helicases. Amino acid conservation is represented by the height of letter. Coloring marks the chemical properties of a given amino acid: green and purple — polar, blue — basic, red— acidic, and black— hydrophobic (adapted from ref. [1]). (C) Schematic representation the topology of the two RecA-like helicase core domains. β-strands are indicated by arrows, α-helices by cylinders. Characteristic sequence motifs are colored as in panel A (adapted from ref [1]). (D) Position of the characteristic sequence motifs in the three-dimensional structure of the DEAD-box helicase Vasa [19]. The bound ATP analog and Mg2+ ion are colored teal; the nucleic acid is colored orange. Sequence motifs are colored as in panels A to C.
The two helicase core domains are connected by a flexible linker and can thus change their orientation to each other [16]. Opening and closing of the two domains is thought to be critical for the function of the enzymes (refs. [3,7,8], discussed in more detail below). The characteristic helicase sequence motifs are distributed over both helicase core domains [1,17] (Fig. 1). With both core domains closed (closed state), motifs involved in ATP binding and hydrolysis assemble the active ATPase site on one site of the cleft between the helicase domains (Fig. 1B,C). ATP hydrolysis occurs in the closed state [3,7,8]. However, ATP can be bound in the open state by the “partial” ATPase site on the helicase domain 1 [18].
The helicase motifs involved in RNA binding are located on the opposite site of the helicase core, perpendicular to the cleft (Fig. 1B,C). Single stranded RNA with five or more nucleotides can bind over the entire RNA binding site on both domains [19]. This arrangement explains the strong stimulation of ATP hydrolysis by RNA binding [19]. Recently, binding of duplex RNA to the RNA binding motifs in helicase domain 2 has been reported [18]. This duplex binding mode may play a critical role in the unwinding reaction (discussed below). The RNA binding regions at both helicase domains are structurally very similar to each other, and are thought to have evolved from an ancient anion-binding module [20].
The RNA binding sites in the helicase core contact only the RNA backbone [18,19,21,22,23,24,25]. This binding mode enables association with duplex RNA [26]. At the same time, preferential binding of certain RNA sequences is difficult or impossible, because no bases are contacted. Consequently, the residues in the helicase core confer no known specificity for certain RNAs [3,4]. Most full length DEAD-box helicases also show no discernible preference for particular RNA sequences or structure, even though C- or N-termini can facilitate RNA binding [27]. An exception is the bacterial DbpA and its bacterial orthologs, which bind preferentially to a ribosomal RNA hairpin, promoted by an auxiliary C-terminal domain with an RRM structure [28,29,30,31,32].
The function of most residues involved in ATP and ssRNA binding sites is well defined, both structurally and functionally [3]. Notably, the residues in the ATP binding and hydrolysis site form a highly complex interaction network [33,34]. This interconnection complicates the introduction of mutations that prevent ATP hydrolysis without affecting ATP binding.
DEAD-box proteins are generally specific for adenosine triphosphates for driving duplex unwinding and for RNA-stimulated hydrolysis of the triphosphate [33,34]. This specificity is mostly conferred by the Q-motif, which establishes specific contacts to adenine-specific functional groups [35]. The structural basis for the communication between ATP and RNA sites during the various activities of DEAD-box proteins is not completely understood. It is clear, however, that residues in motifs III, IV and Va are critical for relaying the state of ATP binding to the RNA binding site, and vice versa [36,37,38].
In most DEAD-box helicases, C- and N-terminal domains flank the helicase core (Fig. 1A). These auxiliary domains vary in length from less than five to several hundred amino acids [1]. Auxiliary domains tend to be highly conserved among orthologs, but there is essentially no sequence conservation across all DEAD-box proteins [1]. It is therefore believed that these domains are important for the specific functions of a given DEAD-box helicase.
Structural information on auxiliary domains exists for only few DEAD-box helicases. Detected folds include RRM and oligomerization modules, and biochemical studies on these domains confirmed the functions suggested by the folds [31,39]. In Mss116p, a part of the C-terminal domain adopts an α-helical fold [23]. In several DEAD-box helicases, the sequence of C- or N-terminal extensions suggests unstructured regions [26,40]. For several enzymes, these potentially unstructured regions have been implicated in protein-protein interactions, including dimerization, but also in RNA binding [26,41].
3. DEAD-box helicases as multi-functional modules
Research from many groups over the last two decades has demonstrated that the biochemical repertoire of DEAD-box helicases extends well beyond ATP-dependent duplex unwinding. For example, DEAD-box helicases have been shown to promote duplex formation, displace proteins from RNA, function as assembly platforms for larger ribonucleoprotein complexes, and, as recently reported, sense bacterial metabolites [9,10,11,12,13,14]. These diverse functions are all accomplished with the highly conserved helicase core, suggesting a common functional theme.
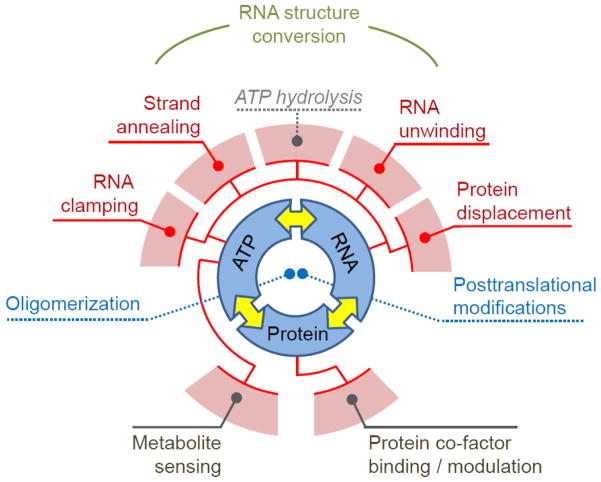
Below, we discuss the biochemical characteristics of DEAD-box proteins in the context of a model that views the enzymes as modules that integrate RNA, nucleotide, and protein binding. These three features constitute the basic level of DEAD-box helicase function (Fig. 2). Activities such as ATPase, RNA unwinding, protein displacement, or RNA clamping arise from the interaction of two or more of the basic features. For more complex functions, such as RNA structure conversion, two or more activities are utilized in a coordinated fashion (Fig. 2).
Figure 2. Biochemical features and activities of DEAD-box helicases.
The inner circle marks the basic biochemical features, RNA, nucleotide (ATP) and protein binding. The yellow arrows mark the connection between the activities. The outer circle segments (light red) designate the activities that arise from either one or the coordination of two basic biochemical features. The red lines indicate which biochemical features contribute to a given activity. RNA structure conversion is based on the coordination between two activities, strand annealing and RNA unwinding, as indicated.
This view on the biochemical functions of DEAD-box protein accommodates “exotic” roles, such as metabolite sensing and tasks that may not involve RNA [14,42], while accounting for the biochemical activities. In addition, one can readily envision the biochemical basis for enzyme regulation through posttranslational modifications or changes in oligomeric state. We believe that this inclusive and systematic view on biochemical functions of DEAD-box proteins is instructive for devising physical models for cellular roles of the enzymes. We emphasize that the discussed model is not fundamentally new, but based on work and prior insightful discussion of a large body of data by many groups.
3.1. The basic biochemical features: nucleotide, RNA, and protein binding
Like all RNA helicases, DEAD-box proteins have the ability to bind RNA, nucleotides, and other proteins (Fig. 2). Protein binding capacity has not yet been directly demonstrated for many DEAD-box helicases, but this feature is most likely shared by all family members, given their function in the context of larger protein assemblies [3,4]. The diverse biochemical activities and functions of DEAD-box helicases can be reduced to either one, or the interplay between two or all three fundamental biochemical features. We thus believe it is useful to view RNA, nucleotide, and protein binding as the basic level of DEAD-box helicase function.
RNA, nucleotide, and protein binding can affect each other (Fig. 2). It is well established that binding of ATP and ADP modulates RNA binding and that, vice versa, RNA affects nucleotide binding [43]. Protein binding has also been shown to modulate both ATP and RNA binding, and there are examples where RNA and nucleotide impact protein binding [9,25,44].
3.1.1. RNA binding and coupling to ATP binding
As briefly discussed above, RNA is bound by the helicase core of probably all DEAD-box proteins through a set of conserved residues in motifs Ia, b, c and motifs IV, IVa, V, and Va (Fig. 1). All of these residues contact the RNA backbone [18,19,21,22,23,24,25]. Crystal structures show single stranded RNA usually bound across both helicase domains, thereby promoting the closed state and thus formation of the active site for ATP hydrolysis [19,21,22,23,24,25]. Duplex RNA can be bound to the RNA binding residues at domain 2 only [18]. In addition to the contacts with the helicase core, RNA can also be bound by not universally conserved residues in the C-terminal domain. For example, in Mss116p part of the C-terminal domain extends the RNA binding site [23]. In CYT-19, unstructured regions contact RNA [26]. RNA binding through other auxiliary domains has been observed, especially through domains that contain RGG boxes [45], a motif that has been implicated in RNA binding in other, non-helicase proteins as well [46].
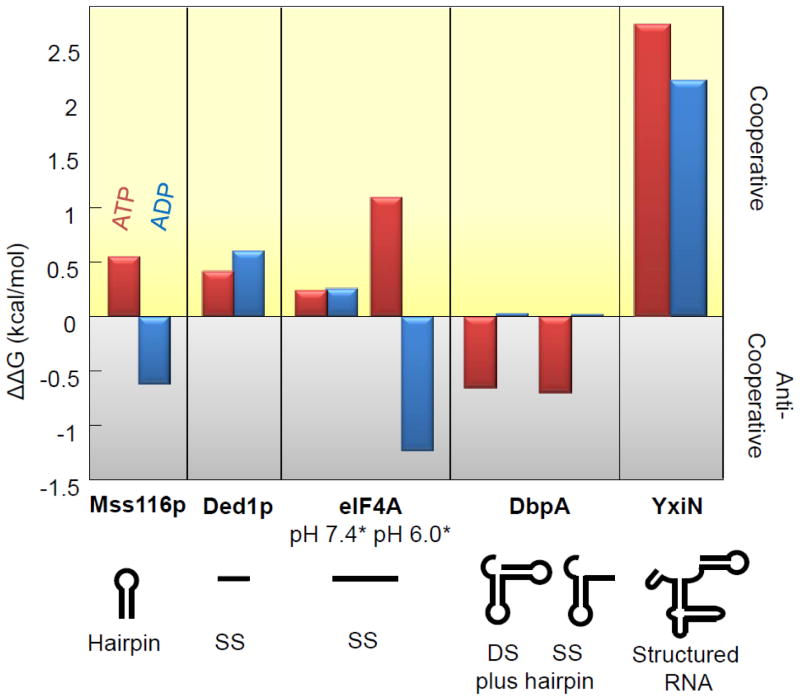
How and to which degree RNA binding is affected by nucleotides appears to vary for different DEAD-box helicases (Fig. 3). For some enzymes, RNA and ATP/ADP binding are thermodynamically coupled, i.e., ATP or ADP promote RNA binding, and vice versa [43,46,47,48]. For others, ATP/ADP and RNA binding are anti-coupled, i.e., ATP or ADP antagonize RNA binding, and vice versa (Fig. 3). At this point, it is not clear whether these differences reflect genuine functional differences between DEAD-box proteins or the use of different RNAs in the respective experiments.
Figure 3. Cooperativity between RNA and ATP or ADP binding by DEAD-box RNA helicases.
Cooperativity is expressed by changes in free binding energy [ΔΔG° (kcal/mol)] for ATP or ADP binding in the presence or absence of RNA. Values for ΔΔG° were calculated according to ΔΔG° = ΔG°(nt with RNA) − ΔG°(nt no RNA), with ΔG° = − RTlnKeq. Positive ΔΔG° values indicate cooperative binding of nucleotide and RNA, negative values indicate anti-cooperative binding. Values were calculated with published data for Mss116p, eIF4A, DbpA, YxiN [46,47,48,49,50], and for Ded1p with unpublished data (A.P. & E.J., unpublished observations). ΔΔG° = ΔG°(nt with RNA) − ΔG°(nt no RNA), with ΔG° = − RTlnKeq. RNAs used were as follows: Mss116p:28 nt RNA capable of forming a 12 bp hairpin with a 4 nt loop, Ded1p: 10 nt single stranded RNA, eIF4A: 21 nt single stranded RNA, DbpA: a 36 nt hairpin containing substrate with a single stranded region, or an additional 15 nt forming a second hairpin in place of the single stranded region, YxiN: a 154 nt structured substrate consisting of nucleotides 2481– 2634 of the B. subtilis 23S rRNA [46].
Moreover, measured affinities and thermodynamic couplings may be influenced in non-trivial ways by the experimental approach. This is because binding of ATP in the presence of RNA is likely a multi-step process, as suggested through biophysical analyses of DbpA, YxiN, and Mss116p [43,46,47,49,50]. Recent structures of Mss116p provide a possible rationalization for this observation and raise the possibility that multi-step ATP and RNA binding might be general for DEAD-box proteins [18]. The structures suggest and biochemical evidence shows ATP binding to the helicase domain 1 in the open state [18]. This state might reflect the first step of ATP binding, where effects on RNA binding are not clear. The second step of ATP binding might be represented by the well-documented ATP-bound closed state, where RNA binding is thought to be promoted [19,21,22,23,24,25]. Thus, ATP affinities and thermodynamic coupling with RNA binding that are measured by direct nucleotide association with the enzyme may differ from results obtained by approaches that require the enzyme to go through the closed state, as in ATPase or helicase reactions.
3.1.2. Nucleotide binding and metabolite sensing
Binding of ATP to DEAD-box helicases has been widely discussed in the literature, and we refer the interested reader to recent reviews on the topic [3,7,43]. Besides the obvious ability to bind ATP, some DEAD-box helicases can bind other adenine-based and related nucleotides [14,40,51]. Recent reports have even suggested functions of DEAD-box helicases based primarily on nucleotide binding, but without clear involvement of RNA [14,52].
Perhaps not unexpectedly, DEAD-box helicases bind ADP, generally with higher affinity than ATP [43,47,49,50]. The ADP-bound state has been implicated in functions for Dbp5p [25,53]. As noted above, the interplay between ADP and RNA binding may vary between different DEAD-box proteins (Fig. 3). Reported affinities raise the possibility that at ADP competes with ATP binding at certain conditions, and thus modulates DEAD-box helicase function [43,49,50]. In DbpA, the ADP-bound state is likely to be the most populated state under physiological conditions [43,49,50], and this might be true for other enzymes as well.
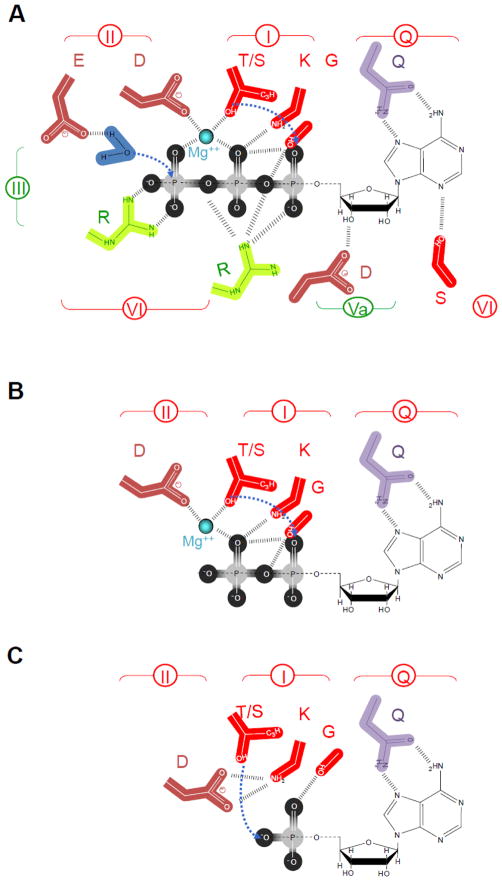
For several DEAD-box helicases, binding of AMP has been observed [40,51]. In contrast to ATP and ADP, AMP is not part of the ATP hydrolysis cycle, and AMP binding is thus not automatically expected. AMP binds exclusively to ATP binding residues in the helicase domain 1 [40,51]. The configuration of the nucleotide binding site with AMP differs in some aspects from the configuration with ATP or ADP (Fig. 4).
Figure 4. Schematic representation of nucleotide binding by DEAD-box proteins.
Key residues in the helicase core domain that mediate ATP binding and hydrolysis, based on the structure of several DEAD-box proteins. Sequence motifs are colored and numbered as described in Fig. 1. (A) Binding of ATP with Mg2+. (B) Binding of ADP with Mg2+. (B) Binding of AMP.
AMP binding is potentially significant because in eukaryotes the cellular AMP levels increase steeply in response to metabolic stress such as glucose deprivation in yeast, or exertion in mammalian muscle cells [54,55]. ADP and ATP concentrations vary considerably less [54,55]. Sensitivity to AMP thus raises the possibility that certain DEAD-box proteins connect RNA and energy metabolism. Unpublished results for Ded1p indeed indicate AMP affinities in the range of physiological AMP concentrations, and potent interference of AMP with RNA binding (A.P. & E.J., unpublished observations). In principle, DEAD-box helicases might thus be involved in arresting energy consuming processes in RNA metabolism in response to metabolic stresses that increase AMP levels. This is an intriguing prospect, since no other enzymes are known that link gene expression at the RNA stage to changes in AMP concentrations without detours through signaling cascades.
A perhaps even more unusual use of nucleotide binding by DEAD-box proteins has recently been reported for DDX41 (abstrakt in Drosophila) [14]. This enzyme senses the bacterial secondary messengers cyclic di-GMP (c-di-GMP) and cyclic di-AMP (c-di-AMP) [14]. In response to these metabolites, DDX41 activates a signaling cascade that ultimately induces type I interferons as part of the innate immune response to bacterial pathogens [14]. It is not known whether c-di-GMP and c-di-AMP bind like AMP to the ATP site in DDX41. However, slight re-configurations of the binding site could potentially accommodate guanosine nucleotides. Although it has not been examined whether c-di-GMP and c-di-AMP impact biochemical activities of DDX41, binding of these nucleotides, along with the AMP binding ability of several DEAD-box helicases, could suggest more unexpected cellular roles of the enzymes as metabolite sensors.
3.1.3. Protein binding
Association of other proteins with DEAD-box helicases is well recognized as key to the biological function of the enzymes [3,4]. DEAD-box helicases can bind other proteins through auxiliary domains but also through the helicase core [21,22,25,56,57,58]. A large body of experimental work and recent reviews have discussed various aspects of interactions of DEAD-box proteins with other proteins [3,4].
Comparably few studies have examined biochemical consequences of other proteins on DEAD-box helicases. It is clear that other proteins can elicit a large spectrum of effects. Those range from occlusion of RNA binding sites over inhibition or stimulation of steps in the ATP hydrolysis cycle, to increases or decreases in RNA affinity and direct modulation of the orientation of the two helicase domains to each other [25,44,58,59,60,61,62]. However, binding of other proteins to DEAD-box helicases does not always affect biochemical parameters of the enzymes [63].
How in turn DEAD-box helicases biochemically impact other proteins has not been examined in great depths. Extrapolating from investigations of other RNA helicases [64,65], one might expect considerable ability of the enzymes to impact the function of other proteins. Whether DEAD-box proteins can have functions that involve protein binding but no RNA is not clear. Several DEAD-box helicases including DDX20 (Gemin 3), p68 (DDX5) and p72 (DDX17) have been implicated in transcriptional regulation without obvious implications for simultaneous RNA binding [42,66,67]. Yet, emergence of non-coding RNA as transcriptional regulator might ultimately connect the function of these DEAD-box proteins to RNA, as suggested by a recent report [68].
3.1.4. Oligomerization and posttranslational modifications
RNA, nucleotide, and protein binding as well as their coordination likely depends on the oligomeric state of a given DEAD-box helicase, and on posttranslational modifications [3,69]. Both aspects are not well understood on a biochemical level. For several DEAD-box proteins, observed unwinding rate constants depend on the helicase concentration in a sigmoidal fashion, consistent with cooperative oligomerization [41,45,70,71]. However, we are not aware of a published, systematic study of a DEAD-box helicase oligomer. Nonetheless, oligomerization is likely for a subset of DEAD-box helicases, given that at least one DEAD-box helicase, the bacterial HerA, features a dimerization module [51], and because several other DEAD-box helicases (e.g., Xp54, p68, p72) interact with itself in immunoprecipitations, a finding consistent with oligomerization [72,73,74]. On the other hand, it has been clearly shown that several DEAD-box helicases are monomeric in biochemical studies [26,47,49].
Posttranslational modifications of DEAD-box helicases have been detected in large numbers (reviewed by Gustafson & Wessel, [69]). Significant posttranslational modification is also seen in other proteins involved in RNA metabolism [75]. In this sense, DEAD-box helicases are no exception among factors regulating the transcriptome. Phosphorylations, methylations, and sumo modifications of DEAD-box helicases are widely reported [76,77,78], but also more exotic modifications. For example, modification of eIF4A with a lipid molecule (15-deoxy-Δ-12,14-prostaglandinJ2) has been obseved [79]. While physiological significance of many modifications has been demonstrated, their biochemical consequences remain largely unclear.
3.2. RNA-related activities of DEAD-box helicases
The basic functional features of DEAD-box proteins, RNA, nucleotide and protein binding give rise to a range of biochemical activities including RNA-stimulated ATP hydrolysis, RNA duplex unwinding, protein displacement from RNA, RNA clamping, strand annealing, and RNA structure conversion (Fig. 2). Below, we discuss these activities with emphasis on how RNA, nucleotide and protein binding are coordinated to accomplish this spectrum of activities.
3.2.1. RNA-stimulated ATPase activity
DEAD-box helicases are often referred to as RNA-dependent ATPases. Virtually all DEAD-box helicases tested to date hydrolyze ATP in the presence of RNA [3,7,43]. Although RNA-dependent depletion of ATP is unlikely to be a primary cellular role for DEAD-box helicases, biochemical investigations of RNA-stimulated ATP hydrolysis have provided important insight into the function of the enzymes [7,43]. Biochemical and biophysical aspects of the ATPase reaction by DEAD-box helicases have recently been discussed by de la Cruz and co-workers in an excellent review on this topic [43].
As noted above, single stranded RNA binds across both helicase domains, thus promoting the closed state of the helicase domains and concurrent assembly of the ATPase active site [3,19]. The degree of RNA stimulation of ATP turnover ranges from less than ten-fold to orders of magnitude for different enzymes [47,48,71,80,81,82]. In our hands, careful depletion of RNA in the protein preparation reduces basal, non-RNA stimulated ATPase activities for several DEAD-box helicases to virtually undetectable levels (A.P. & E.J., unpublished observations).
Functional ATP affinities in the RNA-stimulated ATPase reaction, while expectedly dependent on the reaction conditions, are mostly in the micromolar to low millimolar range, consistent with the notion that ATP readily associates at cellular concentrations [43]. The RNA-stimulated ATPase cycle for DbpA and Mss116p have been analyzed and discussed in detail [47,49,50,80,83]. While individual rate and equilibrium constants for the reaction steps vary for each protein, common themes have emerged that also explain observations for other DEAD-box helicases [43]. These themes include one or more protein isomerization steps connected to ATP binding, traversing of tightly bound and long-lived states with both RNA and ATP, and reversibility of the chemical step [43]. Rate limiting for the overall reaction of DbpA and Mss116p are release of inorganic phosphate and either the ATP hydrolysis step or conformational changes preceding this event [47,49,50,80,83]. The biochemical parameters for the ATPase reaction depend on the particular RNA used in the reaction [43]. It is not yet clear how exactly the ATPase reaction is affected by RNA length, structure, or both.
ATPase measurements have also been instructive in delineating biochemical effects of protein co-factors on DEAD-box helicases. For example, the RNA export factor Gle1p promotes ADP release from Dbp5p, thereby accelerating ATP turnover [25]. For the DEAD-box helicase eIF4A, the eukaryotic translation initiation factor 4G promotes RNA binding and also phosphate release, thus stimulating ATP hydrolysis by eIF4A at several steps [25,44,59]. In contrast, the protein dimer MAGOH-Y14 inhibits phosphate release in the DEAD-box helicase eIF4A-III, which allows the enzyme to remain stably bound to the RNA for prolonged periods of time [84].
3.2.2. RNA Unwinding
ATP-driven RNA unwinding requires sophisticated coordination between ATP and RNA binding and associated conformational changes of the enzyme [7]. At least a subset of DEAD-box proteins likely unwinds RNA duplexes in the cell, and most, if not all DEAD-box helicases unwind RNA duplexes in vitro, if appropriate substrates are used [85,86,87]. Mutations in DEAD-box helicases that diminish duplex unwinding often impair cellular functions of the enzymes [19,25,34,36,71,88,89,90]. However, this correlation does not automatically imply that a given helicase unwinds RNA duplexes in the cell. Rather, the correlation indicates that both, cellular function and duplex unwinding rely on similar residues to coordinate RNA with ATP binding and hydrolysis [3]. For this reason, ATP-driven duplex unwinding with defined model substrates is an attractive proxy to biochemically assess the coupling of ATP binding and hydrolysis to RNA remodeling for a given enzyme [7,85,91].
As for all helicases, RNA duplex unwinding by DEAD-box proteins is a multi-step process that requires ATP [7,8]. However, DEAD-box helicases employ an unwinding mechanism that differs in key aspects from the translocation-based strand separation seen for many DNA and some viral RNA helicases [41,70,92,93,94,95,96]. Instead of moving along one of the RNA strands, DEAD-box helicases load directly to the duplex and then pry the strands apart in an ATP-dependent fashion (Fig. 5). This unwinding mode is termed local strand separation, since the enzymes separate only several (estimated 5 to 7) basepairs per ATP-dependent unwinding event [41]. The remainder of the duplex dissociates non-enzymatically [41,70,92,97,98,99]. Accordingly, DEAD-box helicases effectively unwind duplexes with up to ~10 to 12 bp [41,92,97,99]. Unwinding of longer or more stable duplexes becomes dramatically slower, and virtually no unwinding is seen for duplexes with more than two helical turns [45].
Figure 5. Duplex unwinding by local strand separation.
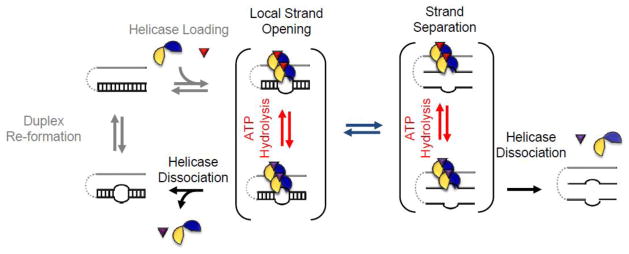
RNA strands are represented by black lines. Single-stranded region is shown in gray. The dotted line indicates requirement for proximity, but not physical connection to the duplex. The DEAD-box helicase is depicted by yellow and blue semi-circles representing the helicase N- and C-terminal domains respectively. More than 2 helicase protomers may participate in the unwinding for a subset of DEAD-box helicases, but as mentioned, DEAD-box helicases can also function as monomers. ATP is shown as red triangle. ATP hydrolysis and subsequent formation of ADP (purple triangle) and free phosphate is required for release of products and recycling of the enzyme. Dissociation of the helicase prior to complete strand separation results in an RNA species with a partially opened helix which will rapidly return to a duplex structure (adapted from refs. [7,91]).
Many, but not all DEAD-box helicases use single stranded or structured RNA adjacent to the duplex to facilitate loading of the enzyme [96,99,100]. The polarity of the single stranded regions is not critical for unwinding, as long as the loading region is proximal to the duplex [70]. Loading can be facilitated by auxiliary domains on the enzyme, through oligomerization, and via protein-cofactors [29,41,44]. Recent reports show that eIF4G, which facilitates loading of the DEAD-box helicase eIF4A to duplexes, also induces preferences for 5′ vs. 3′ single stranded loading regions [101].
DEAD-box helicases use only one ATP per unwinding event, regardless of the duplex length [97,102]. The actual strand separation for several DEAD-box helicases requires only ATP binding; hydrolysis is necessary for recycling of the enzymes [97,102]. Other DEAD-box helicases appear to couple strand separation to the hydrolysis step, as judged from kinetic measurements [47]. At the same time, structures for several DEAD-box helicases show virtually identical conformations at the pre-ATP hydrolysis step and the transition state [19,23,25,84]. We therefore believe that strand separation occurs as soon as ATP is productively bound by the enzyme, regardless of whether or not it is hydrolyzed [3]. Enzymes with a fast hydrolysis step couple strand separation to the hydrolysis event, those with slower hydrolysis steps couple strand separation to ATP binding. Coupling of strand separation to ATP binding also explains why unwinding of very short duplexes is connected to the hydrolysis of less than one ATP per unwinding event [97].
Because DEAD-box helicases open only several basepairs in an ATP-dependent manner, there is a certain probability that the enzyme dissociates from the RNA before complete non-enzymatic strand separation of the remaining basepairs (Fig. 5). The probability of such futile ATP turnovers increases sharply with length and stability of the duplex [92,97,102]. Unwinding of longer duplexes is thus connected to the turnover of multiple, sometimes more than a dozen ATPs [97].
Recent structural data for Mss116p, together with previous results now suggest a model for critical structural changes of DEAD-box helicases during duplex unwinding (Fig. 6) [18]. Without ligands, the two helicase domains are in the open conformation [15,25,46]. Here, ATP initially binds to domain 1, and duplex RNA to domain 2 [18]. This structure rationalizes the direct action of the enzyme on the duplex. The helicase-RNA-ATP complex then transits into the closed conformation seen with the ATP ground state analog ADP-BeFx, where both, ATP and single stranded RNA bind across both helicase domains [23,25]. The transition between the open and closed structure is incompatible with duplex RNA binding [18,19,23]. RNA contacts formed by motifs 1a, 1b, 1c and by a conserved region past motif 2 region are sterically incompatible with duplex RNA [18,19,23]. As a result, unwinding likely occurs during the formation of the closed complex required for ATP hydrolysis. The second RNA strand, which is initially contacted by only very few amino acids in domain 2, is expected to be readily released after duplex opening.
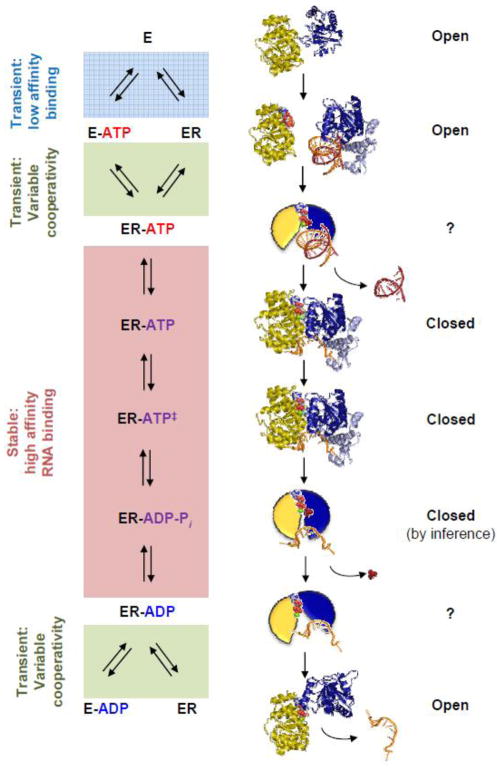
Figure 6. Structural basis for duplex unwinding by DEAD-box proteins.
The left panel shows the scheme for the RNA-dependent ATP hydrolysis cycle. Relative RNA affinities for each stage of the cycle is indicated on the left of the scheme. The structures show each stage for which structural information is available, as discussed in the text. For stages without direct structural information, cartoons represent speculative structural arrangements. Structures depict in yellow the N-terminal domain and in blue the C-terminal domain. The following structures are shown: eIF4A without nucleotide or RNA (1FUU), N-terminal domain of DDX20 with ADPNP (3B7G), C-terminal domain of Mss116p with C-terminal extension (CTE, light blue) and duplex RNA (red and orange (4DB2), Mss116p with CTE (light blue), ADP-BeF3, Mg2+and single stranded RNA (orange) (3I61), Mss116p with CTE (light blue), ADP-AlF4, Mg2+, and single stranded RNA (3I62), and UAP56, ADP, and Mg2+ (1XTJ). On the right, the orientation of the helicase domains relative to each other is indicated for each stage. “?” indicates insufficient evidence for domain orientation.
Additional contacts with the single stranded RNA after initial strand separation provide further energy for domain closure and the unwinding reaction [18, 23]. The second RNA strand, which is initially contacted by very few amino acids in domain 2 [18], is expected to be readily released after duplex opening. Initial opening of the duplex might actively be promoted by the enzyme, or passively by capturing single stranded intermediates arising by breathing of the duplex [18].
The model accounts for the isomerization of the enzyme between ATP and RNA unwinding, and explains why ATP binding can lead to strand separation in some enzymes. The structure in the transition state for ATP hydrolysis, approximated with the non-hydrolyzable analog ADP-AlFx, is virtually indistinguishable from the structure in the ATP ground state [23,84]. This observation may also rationalize why duplex unwinding is coupled in some enzymes to ATP binding and in others to hydrolysis [3,43]. There are not yet structures for the enzyme bound to ADP, inorganic phosphate and ssRNA only, and none for the enzyme bound to ADP and RNA. The latter structure might illuminate why ADP appears to antagonize RNA binding in some enzymes, but to promote RNA binding in others (Fig. 3). With only ADP bound, the helicase adopts the open conformation again [46,103]. Despite the missing intermediate structures, the model explains key biochemical observations of duplex unwinding, and is thus likely to be generally applicable to DEAD-box helicases [18].
3.2.3. RNA clamping
RNA clamping refers to the ability of a DEAD-box helicase to remain bound to RNA for prolonged periods of time [9,84]. RNA clamping was first observed for eIF4A-III, which functions as assembly platform for the multi-component exon junction complex (EJC, refs. [9,104]). On a phenomenological level, a function as immobile RNA clamp seemed antithetical to “highly dynamic” remodeling enzymes. However, careful structural, mechanistic and molecular biological studies on eIF4A-III and EJC components revealed that the tight RNA binding of eIF4A-III is accomplished by other EJC factors that prevent the release of the ATP hydrolysis products from eIF4A-III [9,21,22,84]. The helicase thus remains trapped in a tightly RNA-bound state [84]. This notion is consistent with kinetic data for the DEAD-box proteins DbpA and Mss116p that predict these states to be tightly bound and long-lived [43,47,49,50].
To which extent DEAD-box helicases have general inherent capacity to form long-lived, tightly bound RNA states has not been analyzed, even though RNA clamping is often invoked for enzymes for which it has not been experimentally shown. There is indirect evidence for Ded1p and Mss116p for long-lived states where RNA is tightly bound in vitro [102], but to date, direct and systematic analyses of RNA clamping has only been reported for eIF4A-III [84]. Given that eIF4A-III, a protein without significant C- and N-terminal domains, functions as RNA clamp, it is likely that this function does not require specific auxillary domains.
3.2.4. Protein Displacement
Remodeling of RNA-protein complexes (RNPs) is thought to be a key function of DEAD-box and other helicases [91]. Molecular biological evidence for several DEAD-box helicases points to functions in the displacement of other proteins [86,105,106,107]. However, in none of these cases is it clear whether the protein displacement occurs through direct action of the helicase, or indirectly through RNA remodeling either in proximity or at a distance from the displaced proteins. Mechanistic analysis of displacement of biologically relevant proteins by DEAD-box proteins has not yet been reported, in large part owing to poorly defined physiological environments for most of the enzymes.
Protein displacement has been directly demonstrated on model substrates for Dbp5p and Ded1p [10,53,108]. Ded1p removes proteins from single stranded RNA in an ATP-dependent fashion, provided the proteins have a small footprint [10,108]. This observation may reflect the inability Ded1p to move along RNA in a substantial manner, given that a translocating RNA helicase can remove proteins with larger footprints [10].
Notably, Ded1p is unable to remove a protein (U1A) bound to duplex RNA [108]. This observation is consistent with unwinding by local strand separation, which does not involve movement of the enzyme on the duplex [41]. The inability of Ded1p to remove U1A from a duplex substrate may be noteworthy for discussions of protein removal by DEAD-box helicases. Extrapolating from this observation makes it seem possible that other proteins bound to duplex RNA could simply prevent unwinding by the DEAD-box helicase. Such proteins could thus control the unwinding activity without requiring direct contacts between the protein and the helicase [106].
3.2.5. Strand Annealing
A number of DEAD-box helicases promote the formation of duplexes [13,45,71,109]. For Ded1p and Mss116p, strand annealing activity has been shown to be not simply the reverse of duplex unwinding [45,71]. Strand annealing does not require ATP or ADP in most DEAD-box helicases that have been tested for this activity, but ATP is required for strand annealing in other family members [45,109]. Notwithstanding, ATP and ADP modulate annealing activity in Ded1p [45]. DDX21 catalyzes the intramolecular variant of strand annealing - RNA folding [110]. The newly discovered ability of DEAD-box helicases to bind duplex RNA through domain 2 provides a straightforward rationale for the annealing activity [18]. In several DEAD-box proteins, however, auxiliary domains contribute to the capacity to promote duplex formation [45,110].
Strand annealing is also facilitated by numerous DNA and other RNA helicases [91,111]. In most cases the physiological role of the activity is not clear. Many RNA binding proteins display a certain capacity to promote duplex formation, perhaps simply by affecting the electrostatic environment of the nucleic acid. However, the DEAD-box helicases Ded1p and Mss116p are among the strongest strand annealers known to date, accelerating duplex formation by orders of magnitude over the uncatalyzed reaction [45,71,91]. It has been speculated that the ability to promote duplex formation might contribute to the function of the enzymes as RNA chaperones [7,71].
3.2.6. RNA structure conversion
The activities of DEAD-box helicases outlined above can, in principle, work in a coordinated fashion to accomplish more complex biochemical functions. For example, a combination of protein displacement and RNA unwinding would enable RNP remodeling, although this activity has not yet been reported for DEAD-box proteins. However, it has also been proposed that the DEAD-box helicase UAP56 promotes protein deposition on RNA [112]. This reaction could potentially resemble those catalyzed by ATP-dependent clamp loader proteins, which also belong to a helicase superfamily [113]. Direct biochemical evidence for such activities is not available. However, biochemical evidence for promotion of protein binding to RNA by the DEAD-box helicase DDX1 has recently been reported [114].
While the molecular basis for these functions is not yet clear, another “complex” activity, the conversion of one RNA structure into another, has been directly demonstrated for DEAD-box helicases [115,116]. For Ded1p, RNA structure conversion relies on both RNA unwinding and strand annealing activities [116]. The ability of DEAD-box helicases to facilitate RNA structure conversion is thought to be of potential significance for the function of the enzymes as chaperones [7,71]. Coupling of ATP binding and hydrolysis to structural changes in RNA allows DEAD-box helicases to populate RNA structures that are thermodynamically less stable than competing structures [115,116], thus potentially increasing the functional range of RNAs or RNPs [117].
4. Perspective
Research over the past two decades has provided a firm perspective on the biochemical range of DEAD-box helicases and on structure-function relations in the enzymes. However, several significant questions still await answers. First, the structural basis for communication between ATP and RNA binding sites in the DEAD-box core remains to be defined during the various activities of the enzymes. Second, impact of RNA length and structure on sub-steps of ATPase and helicase reactions will have to be delineated, and, along these lines, potential roles for oligomerization of DEAD-box helicases. Third, the impact of posttranslational modifications on the biochemical features of the enzymes will need to be analyzed. Yet, the perhaps biggest challenge lies in defining cellular RNA substrates, co-factors, and the physiological reaction steps catalyzed by of DEAD-box helicases, such that it becomes possible to analyze cellular reactions by these enzymes on a biochemical and quantitative level.
Highlights.
DEAD-box helicases perform diverse cellular functions in RNA metabolism.
DEAD-box helicases share a highly conserved core domain.
We discuss DEAD-box proteins as integrators of RNA, nucleotide, and protein binding.
Acknowledgments
We apologize to all colleagues whose research could not be discussed here because of space limitations. Research on DEAD-box helicases in the author’s laboratory is supported by the NIH (GM 067700 to E.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fairman-Williams ME, Guenther U-P, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 4.Linder P. The Dynamic Life with DEAD-box RNA Helicases. In: Jankowsky E, editor. RNA Helicases: RSC Biomolecular Sciences. Vol. 19. 2010. pp. 32–60. [Google Scholar]
- 5.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen T. RNA 1997–2007: a remarkable decade of discovery. Mol Cell. 2007;14:715–720. doi: 10.1016/j.molcel.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:12370–11250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 9.Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 10.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 11.Grifo JA, Abramson RD, Satler CA, Merrick WC. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 12.Ray BK, Lawson TG, Kramer JC, Cladaras MH, Grifo JA, Abramson RD, Merrick WC, Thach RE. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 13.Rössler OG, Straka A, Stahl H. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 2001;29:2088–2096. doi: 10.1093/nar/29.10.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nature Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreou AZ, Klostermeier D. Conformational changes of DEAD-box helicases monitored by single molecule fluorescence resonance energy transfer. Meth Enzymol. 2012;511:75–109. doi: 10.1016/B978-0-12-396546-2.00004-8. [DOI] [PubMed] [Google Scholar]
- 17.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 18.Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490:121–125. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 20.Milner-White EJ, Pietras Z, Luisi BF. An ancient anion-binding structural module in RNA and DNA helicases. Proteins. 2012;78:1900–1908. doi: 10.1002/prot.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CLP, Pedersen JS, Séraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 22.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins R, Karlberg T, Lehtiö L, Schütz P, van den Berg S, Dahlgren LG, Hammarström M, Weigelt J, Schüler H. The DEXD/H-box RNA helicase DDX19 is regulated by an alpha-helical switch. J Mol Biol. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci USA. 2011;108:12254–12259. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J Mol Biol. 2002;32:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 30.Nicol SM, Fuller-Pace FV. The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc Natl Acad Sci USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006;12:959–967. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strohmeier J, Hertel I, Diederichsen U, Rudolph MG, Klostermeier D. Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop. Biol Chem. 2011;392:357–369. doi: 10.1515/BC.2011.034. [DOI] [PubMed] [Google Scholar]
- 34.Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 36.Banroques J, Doère M, Dreyfus M, Linder P, Tanner NK. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J Mol Biol. 2010;396:949–966. doi: 10.1016/j.jmb.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 37.Banroques J, Cordin O, Doère M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol. 2008;28:3359–3371. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karow AR, Klostermeier D. A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res. 2009;37:4464–4471. doi: 10.1093/nar/gkp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klostermeier D, Rudolph MG. A novel dimerization motif in the C-terminal domain of the Thermus thermophilus DEAD box helicase Hera confers substantial flexibility. Nucleic Acids Res. 2009;37:421–430. doi: 10.1093/nar/gkn947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, Flores A, Karlsson Hedestam GB, Schiavone LH. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol. 2007;372:150–159. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 41.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 42.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henn A, Bradley MJ, De La Cruz EM. ATP utilization and RNA conformational rearrangement by DEAD-box proteins. Annu Rev Biophys. 2012;41:247–267. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers J, GW, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 45.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 46.Theissen B, Karow AR, Köhler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci USA. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao W, Coman MM, Ding S, Henn A, Middleton ER, Bradley MJ, Rhoades E, Hackney DD, Pyle AM, De La Cruz EM. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J Mol Biol. 2011;409:399–414. doi: 10.1016/j.jmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorsch JR, Herschlag D The DEAD box protein eIF4A1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 49.Henn A, Cao W, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henn A, Cao W, Licciardello N, Heitkamp SE, Hackney DD, De La Cruz EM. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci U S A. 2010;107:4046–4050. doi: 10.1073/pnas.0913081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. Crystal structure and nucleotide binding of the Thermus thermophilus RNA helicase Hera N-terminal domain. J Mol Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Lin C, Liu Z-R. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 53.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 54.Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 55.Walther T, Novo M, Rossger K, Letisse F, Loret MO, Portais JC, Francois JM. Control of ATP homeostasis during the respiro-fermentative transition in yeast. Mol Syst Biol. 2010;6:344. doi: 10.1038/msb.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 57.Nielsen KH, Behrens MA, He Y, Oliveira CL, Jensen LS, Hoffmann SV, Pedersen JS, Andersen GR. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res. 2011;39:2678–2689. doi: 10.1093/nar/gkq1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011;39:2260–2270. doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rozovsky N, Butterworth AC, Moore MJ. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008;14:2136–2148. doi: 10.1261/rna.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alcázar-Román AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 62.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 63.Shen J, Zhang L, Zhao R. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem. 2007;282:22544–22550. doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- 64.Jia H, Wang X, Anderson JT, Jankowsky E. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc Natl Acad Sci USA. 2012;109:7292–7297. doi: 10.1073/pnas.1201085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E. The RNA Helicase Mtr4p Modulates Polyadenylation in the TRAMP Complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuller-Pace FV, Jacobs AM, Nicol SM. Modulation of transcriptional activity of the DEAD-box family of RNA helicases, p68 (Ddx5) and DP103 (Ddx20), by SUMO modification. Biochem Soc Trans. 2007;35:1427–1429. doi: 10.1042/BST0351427. [DOI] [PubMed] [Google Scholar]
- 67.Yan X, Mouillet JF, Ou Q, Sadovsky Y. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol Cell Biol. 2003;23:414–423. doi: 10.1128/MCB.23.1.414-423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cloutier SC, Ma WK, Nguyen LT, Tran EJ. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J Biol Chem. 2012;287:26155–26166. doi: 10.1074/jbc.M112.383075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gustafson EA, Wessel GM. DEAD-box helicases: posttranslational regulation and fuction. Biochem Biophys Res Comm. 2010;395:1–6. doi: 10.1016/j.bbrc.2010.02.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 71.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM Involvement of DEAD-box Proteins in Group I and Group II Intron Splicing. . Biochemical Characterization of Mss116p, ATP Hydrolysis-dependent and -independent Mechanisms, and General RNA Chaperone Activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ernoult-Lange M, Baconnais S, Harper M, Minshall N, Souquere S, Boudier T, Benard M, Andrey P, Pierron G, Kress M, Standart N, le Cam E, Weil D. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA. 2012;18:1702–1715. doi: 10.1261/rna.034314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ogilvie VC, Wilson BJ, Nicol SM, Morrice NA, Saunders LR, Barber GN, Fuller-Pace FV. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 2003;31:1470–1480. doi: 10.1093/nar/gkg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blackwell E, Ceman S. Arginine methylation of RNA-binding proteins regulates cell function and differentiation. Mol Reprod Dev. 2012;79:163–175. doi: 10.1002/mrd.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathew R, Hartmuth K, Möhlmann S, Urlaub H, Ficner R, Lührmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nature Struct Mol Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 78.Jacobs AM, Nicol SM, Hislop RG, Jaffray EG, Hay RT, Fuller-Pace FV. SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene. 2007;24:543–553. doi: 10.1038/sj.onc.1210387. [DOI] [PubMed] [Google Scholar]
- 79.Kim WJ, Kim JH, Jang SK. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 2007;26:5020–5032. doi: 10.1038/sj.emboj.7601920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talavera MA, De La Cruz EM. Equilibrium and kinetic analysis of nucleotide binding to the DEAD-box RNA helicase DbpA. Biochemistry. 2005;44:959–970. doi: 10.1021/bi048253i. [DOI] [PubMed] [Google Scholar]
- 81.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 82.Garcia I, Uhlenbeck OC. Differential RNA-dependent ATPase activities of four rRNA processing yeast DEAD-box proteins. Biochemistry. 2008;47:12562–12573. doi: 10.1021/bi8016119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Talavera MA, Matthews EE, Eliason WK, Sagi I, Wang J, Henn A, De La Cruz EM. Hydrodynamic characterization of the DEAD-box RNA helicase DbpA. J Mol Biol. 2006;355:697–707. doi: 10.1016/j.jmb.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 84.Nielsen KH, Chamieh H, Andersen CBF, Fredslund F, Hamborg K, Le Hir H, Andersen GR. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jankowsky E, Putnam A. Duplex unwinding with DEAD-box proteins. Meth Mol Biol. 2010;587:245–264. doi: 10.1007/978-1-60327-355-8_18. [DOI] [PubMed] [Google Scholar]
- 86.Chen JY, Stands L, Staley JP, Jackups J, RR, Latus LJ, Chang TH. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 87.Kos M, Tollervey D. The Putative RNA Helicase Dbp4p Is Required for Release of the U14 snoRNA from Preribosomes in Saccharomyces cerevisiae. Mol Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 88.Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 91.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 93.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 94.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 96.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rogers J, GW, Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 99.Rogers J, GW, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 100.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J Biol Chem. 2012;287:20301–20312. doi: 10.1074/jbc.M112.347278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi H, Cordin O, Minder CM, Linder P, Xu R-M. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci U S A. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shibuya T, Tange TØ, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nature Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 105.Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perriman R, Barta I, Voeltz GK, Abelson J, Ares MJ. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci USA. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bowers HA, Maroney PA, Fairman ME, Kastner B, Lührmann R, Nilsen TW, Jankowsky E. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chamot D, Colvin KR, Kujat-Choy SL, Owttrim GW. RNA structural rearrangement via unwinding and annealing by the cyanobacterial RNA helicase, CrhR. J Biol Chem. 2005;280:2036–2044. doi: 10.1074/jbc.M409700200. [DOI] [PubMed] [Google Scholar]
- 110.Valdez BC. Structural domains involved in the RNA folding activity of RNA helicase II/Gu protein. Eur J Biochem. 2000;267:6395–6402. doi: 10.1046/j.1432-1327.2000.01727.x. [DOI] [PubMed] [Google Scholar]
- 111.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nature Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 112.Taniguchi I, Ohno M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol Cell Biol. 2008;28:601–608. doi: 10.1128/MCB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:23–30. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 114.Robertson-Anderson RM, Wang J, Edgcomb SP, Carmel AB, Williamson JR, Millar DP. Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J Mol Biol. 2011;410:959–971. doi: 10.1016/j.jmb.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang Q, Fairman ME, Jankowsky E. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J Mol Biol. 2007;368:1087–1100. doi: 10.1016/j.jmb.2007.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chu VB, Herschlag D. Unwinding RNA’s secrets: advances in the biology, physics, and modeling of complex RNAs. Curr Opin Struct Biol. 2008;18:305–314. doi: 10.1016/j.sbi.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]