Abstract
PreQ1 riboswitches regulate genes by binding the pyrrolopyrimidine intermediate preQ1 during biosynthesis of the essential tRNA base queuosine. We report the first preQ1-II riboswitch structure at 2.3 Å resolution, which uses a novel fold to achieve effector recognition at the confluence of a three-way-helical junction flanking a pseudoknotted ribosome-binding site (RBS). The results account for preQ1-II-riboswitch-mediated translational control, and expand the known repertoire of ligand binding modes utilized by regulatory RNAs.
Riboswitches are cis-acting mRNA sequences that provide an elegant solution to the problem of bacterial gene regulation. By directly sensing small molecules via an aptamer domain, riboswitches can adapt quickly to fluctuations in intracellular ligand concentration to establish a feedback loop that controls the production or import of metabolites, or the response to second messengers1. Several ligands have been identified that are recognized by multiple, structurally unrelated riboswitch classes; this group includes: SAM2, cyclic-di-GMP, and preQ13. Within the latter group, only the preQ1-II (class 2) riboswitch structure remains uncharacterized, which prompted us to elucidate this unknown fold. Comparing the modes by which such aptamers organize spatially distinct molecular determinants to recognize a common ligand provides insight into the chemical diversity attainable by regulatory RNAs4.
PreQ1 is the final metabolite on the biosynthetic pathway that produces the hypermodified nucleotide queuosine (Q) (Fig. 1a). Q is an essential modification that enhances translational fidelity5,6,7 via incorporation at the wobble position of specific anticodons in most eukaryotic and bacterial tRNAs6. Q deficiency in bacteria can lead to reduced growth fitness in the stationary phase7 and diminished virulence8. To meet regulatory needs, riboswitches responsive to preQ1 evolved in the Firmicutes, giving rise to phylogenetically distinct riboswitch classes. The preQ1-I (class 1) aptamer is distributed widely and is a compact 34 nucleotides9. The preQ1-II riboswitch is 80 nucleotides, and has been found exclusively in the Lactobacillales where it regulates expression at the translational level10. Another preQ1-II hallmark is that the complete ribosome-binding site (RBS) folds as an integral part of the aptamer in the form of an H-type pseudoknot10,11 (Fig. 1b). Biochemical analyses suggested the preQ1-II mode of ligand readout differs from preQ1-I, which recognizes the effector by canonical cis Watson-Crick base pairing10.
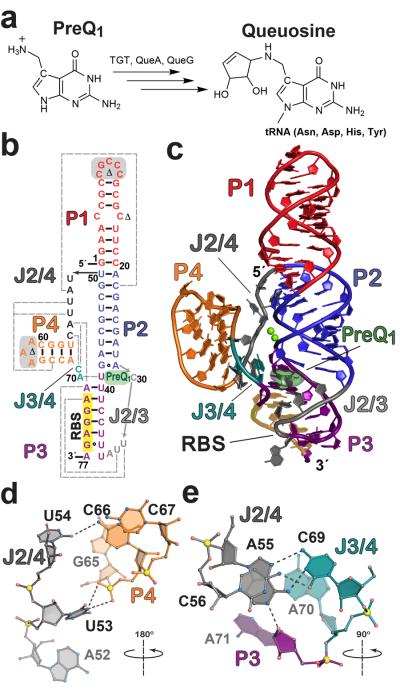
Figure 1. Queuosine biosynthesis, secondary structure, and tertiary fold of the L. rhamnosus preQ1-II riboswitch.
(a) Queuosine biosynthesis from preQ1 with known enzymes shown. Although animals must obtain Q from dietary sources or gut flora, bacteria can produce it by de novo synthesis (reviewed in 21, 22). TGT, tRNA:guanine transglycosylase; QueA, epoxyqueuosine synthase; and QueG, oQ (epoxyqueuosine) reductase (b) Secondary structure of the wild type L. rhamnosus preQ1-II riboswitch used in this investigation based on the crystal structure. PreQ1 is dark green; various pairing regions, P, are color coded with long-range interactions indicated by dashed gray lines; junctions are labeled J. Sites modified for crystallization are highlighted in gray or marked with a Δ. See Supplementary Fig. 1 for the modified construct (MC) used in crystallization and isothermal titration calorimetry (ITC); numbering is based on the MC 77-mer sequence. The consensus RBS sequence 5′-AGGAG-3′ is highlighted in yellow. (c) Cartoon depiction of the preQ1-bound crystal structure. Coloring is the same as b with the preQ1 effector depicted as a semitransparent surface model. The RBS is labeled and highlighted in yellow. (d) Hydrogen-bond tertiary interactions (dashed lines) between P4 and J2/4 that stabilize the core fold; the view is rotated ~180° about the axis shown, relative to the orientation in c. (e) Tertiary interactions that knit together J2/4, J3/4, and A71 of the three-way helical junction; the view is rotated ~90° about the indicated axis relative to c.
To elucidate the basis for ligand recognition and gain insight into how effector binding is communicated to the RBS, we identified and investigated a preQ1-II riboswitch from Lactobacillus rhamnosus (Fig. 1b, Online Methods, Supplementary Results). Binding of preQ1 to the wild type L. rhamnosus preQ1-II riboswitch sequence produced a KD of 17.9 ± 0.6 nM (Supplementary Table 1 and Supplementary Fig. 2a), which is comparable to the 100 nM affinity reported for the preQ1-II riboswitch from Streptococcus pneumonia10. To grow diffraction-quality crystals, we made modifications to the wild type sequence to produce a modified construct (MC) (Fig. 1b and Supplementary Fig. 1a). These changes had no tangible effects in terms of preQ1 affinity (Supplementary Table 1 and Supplementary Fig. 2b).
We then determined the crystal structure of the MC L. rhamnosus preQ1-II riboswitch in the ligand-bound state by Cs+ SAD phasing, followed by refinement to 2.3 Å resolution (Supplementary Table 2 and Supplementary Fig. 3). The “J”-shaped tertiary fold has dimensions of 77 Å × 43 Å × 27 Å and exhibits only subtle differences from the consensus secondary structure in J3/4 and P310 (Figs. 1b,c), thus making it a likely representative of the preQ1-II riboswitch fold. P1, P2, and P3 form the longest coaxial helical stack with P1 most distal from the ligand-binding site (Fig. 1c), consistent with its dispensability in preQ1 binding10. The most prominent structural feature is an H-type pseudoknot wherein loop J2/3, which flanks P2, engages in base pairing with the RBS to form stem P3. The latter stem is preceded by J2/4, P4, and J3/4, which serves as “loop 3” in the classical H-type fold (Supplementary Fig. 1c,d). Despite topological similarities, the preQ1-II architecture differs entirely from that of preQ1-I aptamers12–14.
The preQ1-II ligand-binding pocket resides at a three-way junction comprising P2, P3, and P4 (Fig. 1c). This topological transition is facilitated by interactions from junction bases and Mg2+ ions. N3 of U53 in J2/4 forms a bifurcated hydrogen bond with the non-bridging oxygens of core base C66 in P4 (Fig. 1d); similarly, the O2 group of U54 interacts with the C66 N4 amine. The nearby base C56 of J2/4 contributes its N4 to hydrogen bond with O2 of C69 in J3/4, while C69 and A55 form a trans Watson-Crick/Hoogsteen pair, and N1 of A55 interacts with O2′ of A71 in P3 (Fig. 1e). Two Mg2+ ions form contacts between the phosphate backbones of P2, P3, and J2/4 (Fig. 1c and Supplementary Fig. 4), knitting together irregular topological features that compose the nearby ligand-binding site.
PreQ1 was well defined in electron density maps (Fig. 2a) and no artifacts were introduced near the ligand-binding site from crystal contacts (Supplementary Fig. 5). Effector specificity is based primarily on base stacking and hydrogen bonding. In the preQ1-II riboswitch, the ligand stacks between a “floor” A71•U31 pair (Fig. 2a) and a “ceiling” G42•A29 pair (Fig. 2b). Collectively, 297 of 333 Å2 of the preQ1 molecule is inaccessible to solvent when bound to the preQ1-II riboswitch, which is slightly less than the 330 of 337 Å2 sequestered by the preQ1-I translational riboswitch12. Hydrogen-bond readout of preQ1 occurs at its Watson-Crick face, which engages in a trans-Watson-Crick/Watson-Crick base pair with conserved base C30 donated from J2/3, and a nearby water that mediates a hydrogen bond between O6 of preQ1 and O2′ of C30 (Fig. 2a). This latter interaction accounts for the ability of the preQ1-II riboswitch to accommodate the N6 group of 2,6-diaminopurine10. Additional hydrogen bonds are contributed by conserved base U41, where its Watson-Crick face recognizes the N2-N3-N9 edge of preQ1 (Fig. 2a). Under physiological conditions15 and the crystallization pH, the preQ1 methylamine is positively charged, allowing it to engage in a salt-bridge with the pro-RP non-bridging oxygen of A71, with an additional hydrogen bond to O2 of U31 (Fig. 2a).
Figure 2. Architecture of the effector binding site and tertiary interactions involved in preQ1-II riboswitch ligand recognition.
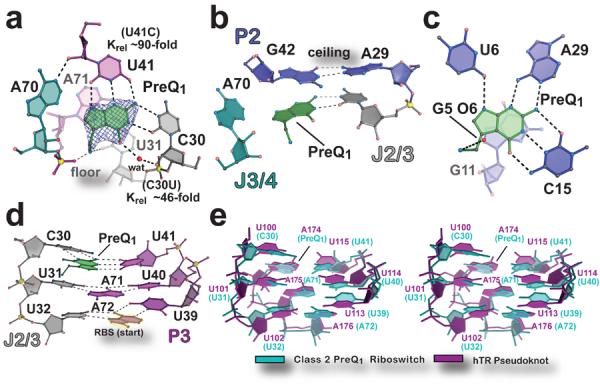
(a) View of the preQ1 ligand-binding site. The final refined ligand is covered by an unbiased Fo-Fc omit electron density map, contoured at the 3.0 σ level, that was calculated prior to inclusion of preQ1 in the model. The “floor” of the binding pocket is formed by a Hoogsteen base pair between A71•U31. Krel (KD mutant / KD wild type) of binding site mutants, determined by ITC, is shown next to the respective base. (b) The “ceiling” of the preQ1 binding pocket is formed by a cis-Watson-Crick/Watson-Crick base pair between G42 and A29. (c) The preQ1-I translational riboswitch in complex with preQ1 (PDB ID 3Q50)12. (d) Major-groove base triples that stack on the RBS to facilitate formation of the P3 pseudoknot. (e) Stereo view of an all-atom superposition between the eight nucleotides of the preQ1-II riboswitch base triples in d and equivalent base triples from the hTR pseudoknot (PDB ID 1YMO)19. The average rmsd was 1.46 Å (excluding hTR A174, which spatially overlaps preQ1).
To probe our structural observations, we constructed C30U and U41C mutants of the preQ1-II riboswitch, and measured their respective preQ1-binding affinities (Supplementary Table 1 and Supplementary Fig. 2c,d). Our results revealed that C30U binds preQ1 with a KD of 0.81 ± 0.12 μM, representing a 46-fold loss in relative affinity, which is comparable to the ~100-fold affinity loss reported for the C41U mutant of the S. pneumonia preQ1-II riboswitch10. The U41C mutant binds preQ1 with a KD of 1.60 ± 0.02 μM, representing an ~90-fold loss in relative binding. The respective ΔΔG values for C30U and U41C were 2.2 and 2.6 kcal mol−1, which is energetically consistent with the loss of two or three hydrogen bonds to preQ1. The overall affinity-loss corroborates structural observations, and suggests that disrupting one or two hydrogen bonds can abrogate an additional interaction within the mutated base (Fig. 2a).
Ligand recognition by the preQ1-II riboswitch does not involve standard cis Watson-Crick readout and thus appears unique compared to the preQ1-I and purine-sensing riboswitches (Fig 2a,c and Supplementary Fig. 6). However, extension of this comparison to riboswitches that bind purine nucleosides or purine-like effectors revealed canonical and non-canonical base recognition (Supplementary Fig. 7a–f). In this respect, ligand recognition by preQ1-II is more similar to riboswitches that recognize second messengers or enzyme cofactors, as observed for the c-di-GMP-I and -II; SAM-I, -II, and –III2; and THF riboswitches3 (Supplementary Fig. 7b–h). In particular, the C30 and U41 interactions with preQ1 are reminiscent of one of the two modes of folinic acid recognition by the THF riboswitch, which uses C53 and U25 of the aptamer (Supplementary Fig. 7g,h) – spatially equivalent to C30 and U41, respectively, of the preQ1-II riboswitch – to form a trans-Watson-Crick/Watson-Crick pair with the effector's pterin moiety16,17. Such comparisons of ligand binding demonstrate that non-canonical pairing is feasible even for riboswitches that bind small purine-like nucleobase effectors – such as preQ1 – but non-Watson-Crick binding is more prevalent than canonical modes of ligand recognition.
Another outstanding question is how ligand binding by the preQ1-II riboswitch effects gene regulation. Our structure confirms that access to the RBS is obstructed by formation of P3, which produces the predicted H-type pseudoknot10 (Fig 1c and Supplementary Fig. 3e). This feature prevents recognition of the mRNA by the ribosome's anti-RBS, effectively blocking translation. Importantly, the preQ1-II ligand-binding site is located at the intersection of the P2-P3-P4 helical junction, and establishes a unique folding environment that places preQ1 in close proximity to the RBS. The associated pseudoknot is buttressed by three tiers of stacked base triples that are 97% conserved in nucleotide identity, and comprise C30•preQ1•U41, U31•A71-U40 and U32•A72-U39 (Fig. 2d), which harbors base A72 from the 5′-end of the RBS (Figs. 1b and 2d). The spatial location of preQ1 in the preQ1-II riboswitch exhibits striking similarities to other RNAs harboring stacked, major-groove U•A-U base triples including the SAM-II riboswitch18 and human telomerase RNA (hTR)19 (Fig. 2e and Supplementary Fig. 8). Significantly, the U100C mutation in one of the hTR triples resulted in a significant destabilization of the tertiary structure19. By analogy, effector binding to the preQ1-II riboswitch completes a C30• preQ1•U41 base triple that is expected to enhance fold stability, supporting RBS sequestration in favor of a gene “off” state. This model is supported by in-line probing of the L. rhamnosus preQ1-II riboswitch, wherein diminution of backbone flexibility was observed for: (i) the three major-groove base-triples, (ii) P4, and (iii) bases of the anti-RBS, but an increase in flexibility at A64 within the P4 loop when preQ1 levels increased (Supplementary Figure 9). The resulting KD of 0.3 μM for preQ1 was similar to the KD of 0.10 μM reported for a comparably sized S. pneumonia preQ1-II riboswitch analyzed under similar conditions10. (See Supplementary Table 1 and Supplementary Fig. 2e for ITC under in-line probing controls). The proposed mechanism is likely applicable to other preQ1-II riboswitches.
Our analysis of the L. rhamnosus preQ1-II riboswitch reveals a novel fold and mode of effector recognition that governs RBS sequestration. Although the mode of ligand recognition differs from a prior model10, our structure accounts for the observed binding preferences for various preQ1 analogues. This work enhances our understanding of the diverse ligand-recognition mechanisms that have evolved for riboswitch-mediated gene regulation. In this respect, the preQ1-II riboswitch is notable because of its prominence in Streptococcus pathogens and its responsiveness to a ligand that is foreign to the mammalian metabolome – factors that form a basis for antimicrobial targeting20.
Online Methods
Identification of the L. rhamnosus riboswitch
Previously our work on the L. casei preQ1-II riboswitch – identified by Breaker and co-workers10 – resulted in crystals with X-ray diffraction limited to 5.6 Å resolution23. In an effort to improve crystal quality, we conducted BLASTn refseq RNA searches24 starting with the L. casei riboswitch sequence. Our goal was to identify closely related sequences, especially those with shorter joining regions that might be more amenable to high-resolution structural analysis. This approach led to the identification of a putative preQ1-II riboswitch sequence in the 5′ UTR of a COG4708 gene from L. rhamnosus, which is characteristic of other preQ1-II riboswitches10. The COG4708 gene is hypothesized to produce a protein that transports Q precursors into the cell. Despite genomic synteny and similar probiotic properties, L. casei and L. rhamnosus are classified as distinct species25. Unlike L. casei, L. rhamnosus has also been associated with endocarditis and is not considered strictly beneficial26. Significantly, two differences were apparent in the respective riboswitch sequences between positions 21 and 77 – a region documented previously as important for preQ1-binding function10. Variations included a deletion of L. casei position 34 (ΔC34) in J2/3 of L. rhamnosus, and a C53U change in J2/4. Using ITC and in-line probing approaches (described below), we found the L. rhamnosus sequence is responsive to preQ1, and crystallization trials led to well diffracting crystals. At present, the basis for differences in crystal diffraction resulting from the use of the respective sequences is unknown.
RNA production and isothermal titration calorimetry (ITC)
L. rhamnosus preQ1-II riboswitch sequences (Fig. 1b and Supplementary Fig. 1a,b) and mutants thereof were prepared by in vitro transcription and purified by denaturing PAGE as described23. PreQ1 was prepared by organic synthesis27 (LeadGen Labs, LLC). ITC measurements were conducted using a VP-ITC calorimeter (MicroCal, Inc) at 20 °C or 25 °C. Lyophilized RNA was resuspended in 0.010 M Na-HEPES pH 7.0 or HEPPS pH 8.3 and 0.10 M NaCl or 0.10 M KCl. For MgCl2-free conditions 0.5 mM EDTA pH 8.0 was included in lieu of multivalent ions. The RNA was heated to 65 °C for 5 min, then MgCl2 was added to a final concentration of 6.0 mM, or 20 mM, or 1.0 mM of Co(NH3)6Cl3 was added, followed by slow cooling to 24 °C; MgCl2 and Co(NH3)6Cl3 were omitted completely for folding conditions containing EDTA. The RNA was dialyzed against 4 L of 0.10 M NaCl or 0.10 M KCl, 6.0 mM, 20.0 mM MgCl2, 1 mM Co(NH3)6Cl3 or 0.5 mM EDTA, and 0.050 M Na-HEPES pH 7.0 or 0.050 M Na-HEPPS pH 8.3 overnight at 4 °C, then diluted with dialysis buffer to: 3.3 μM for the wild type riboswitch, 13.8 μM for the C30U mutant, 23.3 – 26.2 μM for the U41C mutant, 1.5 – 5.7 μM for wild type with 0.5 mM EDTA, and 3.6 μM for wild type with Co(NH3)6Cl3. PreQ1 was dissolved in dialysis buffer to a concentration 10-fold higher than the RNA. Measurements were carried out by titrating preQ1 into the riboswitch located in the sample cell (cell volume is ~1.7 mL) using 28 or 29 injections of 10 μL each, except for the first injection of 3 μL, with 120 or 240 s intervals between injections; the reference power was 15 μcal s−1. The thermograms were analyzed with Origin 7.0 (MicroCal) using a 1:1 binding model. Experiments were performed in triplicate for wild type and MC variant L. rhamnosus sequences with MgCl2, and in duplicate for mutant riboswitches and for the wild type in all other conditions (Supplementary Table 1); representative titrations and curve fits are shown in Supplementary Fig. 2.
Riboswitch crystallization and X-ray data collection
A solution of 0.25 mM RNA (prepared as described above for ITC) in 0.01 M Na-cacodylate pH 7.0 was heated to 65 °C for 3 min. MgCl2 was added to a final concentration of 6 mM and preQ1 – synthesized as described12 – was added to a final concentration of 0.5 mM followed by heating to 65 °C for 5 min. The solution was slow cooled to 24 °C. Crystallization was by the hanging-drop vapor diffusion method. A volume of 1.7 μl of pre-folded RNA was mixed 1:1 with well solution comprising: 14.4–14.8% (w/v) poly(ethylene) glycol 6000 (PEG6K), 0.14–0.16 M MgOAc2, 0.05 M Na-cacodylate pH 6.0, 1 mM spermine, and 0.15 M CsCl. Crystals appeared at 20 °C within 2 weeks. Crystals grew as rectangular plates of size 0.20 mm × 0.05 mm × 0.02 mm. Cryoprotection was by a 30 s transfer into synthetic mother liquor comprising: 17.2–17.8% (w/v) PEG6K, 168–192 mM MgOAc2, 0.18 M CsCl, 0.06 M Na-cacodylate pH 6.0, and 1.2 mM spermine supplemented with 20% (v/v) 2-methyl-2,4-pentanediol and 20% (v/v) ethanol. The sample was vitrified by plunging in N2(l). X-ray diffraction intensities were recorded at Stanford Synchrotron Radiation Lightsource (SSRL, Menlo Park CA) beamline 7–1 at −173 °C and reduced with the HKL2000 software package28.
Phase determination, structural refinement, and analysis
Experimental phases were obtained by single-wavelength anomalous diffraction (SAD). A single crystal was used to obtain an 18-fold redundant, 2.6 Å resolution X-ray data set that was recorded at a wavelength of 1.7 Å to optimize the Cs+ anomalous signal (Supplementary Table 2). Phenix AutoSol29 located 14 site-bound Cs+ atoms, which were used for initial phase calculations in combination with density modification (Supplementary Fig. 3). The figure of merit before density modification in Resolve (as implemented in Phenix) was 0.41. Initial model building was performed by autobuilding in Phenix, followed by iterative rounds of building in Coot30 and refinement in Phenix. The trace and solvent content were consistent with a single molecule in the asymmetric unit. A 2.3 Å resolution native dataset was collected at SSRL on a second crystal at a wavelength of 1.127 Å, which was used to extend the resolution of the SAD model. At a late stage, preQ1 was modeled into reduced bias 2Fo-Fc and Fo-Fc maps (e.g. Fig. 2a) using starting coordinates derived from the small-molecule crystal structure31. Cs+ ions were included in the refined model based on their anomalous diffraction signal (Supplementary Fig. 3c) and coordination geometry. Mg2+ ions were modeled based on their electron density and octahedral coordination geometry32. The final Rwork/Rfree values were 19.3/24.4% with reasonable geometry (Supplementary Table 2). Nearly the entire 77-nucleotide riboswitch was resolved in electron density maps including the ligand-binding pocket and RBS (Fig. 2a and Supplementary Fig. 3). The area of preQ1 that is inaccessible to solvent was calculated using a water probe with a radius of 1.4 Å using UCSF Chimera33. The averaged kicked map in Supplementary Fig. 3d was generated in Phenix.
In-line probing of the L. rhamnosis preQ1-II riboswitch
The in-line probing reaction was carried out on an extended preQ1-II sequence (Supplementary Fig. 1b) essentially as described34. RNA was prepared by in vitro transcription (as described above for ITC), dephosphorylated with alkaline phosphatase, and radiolabeled with [γ32P]ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs), which was then PAGE purified. RNA was folded by heating in 0.05 M Tris-HCl (pH 8.3 at 25 °C) to 65 °C, snap cooling on ice, then adding KCl to 0.10 M, MgCl2 to 0.020 M, and preQ1 at one of the concentrations described (Supplementary Fig. 9). In-line probing reactions each contained 1.0 × 106 CPM and were incubated at 25 °C for ~40 h. Reaction products were separated by denaturing 7.5% PAGE, the gel was dried, and then exposed to a phosphor storage screen for ~40 h. Imaging was carried out using a GE Storm 860, and gel quantification was conducted by use of SAFA with data normalized against invariant nucleotides selected by SAFA35; the nucleotides chosen as invariant bases were: 34, 40, 50, 53 and 54. The fraction of RNA cleaved was determined by setting the maximum amount cleaved to 1 and the minimum to 0 for all concentrations tested at a given nucleotide position, as described34. Binding curves were generated in PRISM 6 by fitting the corrected intensities measured to a dose response curve, with the apparent KD equal to a level of 0.5 cleaved.
Accession codes
Protein Data Bank: Coordinates and structure factors have been deposited under accession code 4jf2.
Supplementary Material
Acknowledgments
We thank J. Jenkins, D. Turner and C. Kielkopf for suggestions, and V. Bandarian for preQ0. J.A.L. was funded by NIH T32 GM068411 and a Hooker fellowship. This research was funded by NIH grants RR026501 and GM063162 to J.E.W. Portions of this research were conducted at SSRL, which is funded in by the DOE and NIH grants GM103393 and RR001209.
Footnotes
Author Contributions M.S. identified the L. rhamnosus riboswitch, produced RNA, and conducted in-line probing; J.K. and J.A.L grew crystals; J.A.L. conducted ITC, interpreted in-line probing, and solved the structure. J.E.W. supervised the project. J.A.L. and J.E.W. prepared the manuscript.
Competing Financial Interests The authors declare no competing financial interests.
Additional Information Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Bastet L, Dube A, Masse E, Lafontaine DA. Mol Microbiol. 2011;80:1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x. [DOI] [PubMed] [Google Scholar]
- 2.Batey RT. Wiley Interdiscip Rev RNA. 2011;2:299–311. doi: 10.1002/wrna.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batey RT. Q Rev Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breaker RR. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz M, Kubli E. Nature. 1981;294:188–190. doi: 10.1038/294188a0. [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama S, et al. Nature. 1979;282:107–109. doi: 10.1038/282107a0. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi S, Nishimura Y, Hirota Y, Nishimura S. J Biol Chem. 1982;257:6544–6550. [PubMed] [Google Scholar]
- 8.Durand JM, et al. J Bacteriol. 1994;176:4627–4634. doi: 10.1128/jb.176.15.4627-4634.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth A, et al. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 10.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. RNA. 2008;14:685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg Z, et al. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins JL, Krucinska J, McCarty RM, Bandarian V, Wedekind JE. J Biol Chem. 2011;286:24626–24637. doi: 10.1074/jbc.M111.230375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein DJ, Edwards TE, Ferre-D'Amare AR. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang M, Peterson R, Feigon J. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Hoops GC, Park J, Garcia GA, Townsend LB. J Heterocyclic Chem. 1996;33:767–781. [Google Scholar]
- 16.Trausch JJ, Ceres P, Reyes FE, Batey RT. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Ishibe-Murakami S, Patel DJ, Serganov A. Proc Natl Acad Sci U S A. 2011;108:14801–14806. doi: 10.1073/pnas.1111701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 19.Theimer CA, Blois CA, Feigon J. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Deigan KE, Ferre-D'Amare AR. Acc Chem Res. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarty RM, Bandarian V. Bioorg Chem. 2012;43:15–25. doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grosjean H, de Crécy-Lagard V, Björk GR. Trends Biochem Sci. 2004;29:519–522. doi: 10.1016/j.tibs.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Lippa GM, et al. Methods Mol Biol. 2012;848:159–184. doi: 10.1007/978-1-61779-545-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Morita H, et al. J Bacteriol. 2009;191:7630–7631. doi: 10.1128/JB.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avlami A, Kordossis T, Vrizidis N, Sipsas NV. J Infect. 2001;42:283–285. doi: 10.1053/jinf.2001.0793. [DOI] [PubMed] [Google Scholar]
- 27.Akimoto H, Imamiya E, Hitaka T, Nomura H, Nishimura S. J Chem Soc Perk T 1. 1988:1637–1644. [Google Scholar]
- 28.Otwinowski Z, Minor W. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klepper F, Polborn K, Carell T. Helv Chim Acta. 2005;88:2610–2616. [Google Scholar]
- 32.Wedekind JE. Metal Ions Life Sci. 2011;9:299–345. [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Soukup GA, Breaker RR. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.