Abstract
Background/Aims
To investigate pre-existing hepatitis B virus (HBV) quasispecies and the genotypic evolution of several variants.
Methods
From six patients with lamivudine (LAM) failure, serum samples at pretreatment, 6 months of LAM therapy, and virologic breakthrough were obtained. One hundred clones with HBV inserts in each patient were sequenced at each time point. Pretreatment serum samples were also analyzed from six patients who achieved good responses to LAM therapy.
Results
Among the six patients with LAM failure, the analysis of 100 clones from patient 1 revealed the substitutions L180M in 1% of clones and V173L in 2% of clones. Patient 2 had substitutions of L80V, W153Q, and L180M. In patient 3, mutations conferring resistance to adefovir at V84I (5%), I169L (1%), and N236H (7%) and entecavir at S202G (2%) were detected. Patient 4 had mutations at T128N (1%), I169L (1%), V173L (2%), A181V (1%), and Q215H (1%). In patient 5, M204V/I was detected in 1% and 2% of clones, respectively. L80I and V173L were also identified in patient 6. In the six patients who responded to LAM, the degree of overall quasispecies was less than those with LAM failure.
Conclusions
Various HBV quasispecies associated with drug resistance existed before treatment, and the quasispecies dynamically changed through LAM therapy.
Keywords: Quasispecies, Hepatitis B virus, Lamivudine, Entecavir, Adefovir
INTRODUCTION
Hepatitis B virus (HBV) is an enveloped DNA virus and replicates via reverse transcription of the genomic RNA template. This process includes polymerization of the minus-strand DNA, degradation of the RNA intermediate and incomplete synthesis of the plus-strand.1 However, HBV reverse transcriptase (RT) is an error-prone enzyme that lacks 3'-5' proofreading capacity.2 Hence, it is responsible for many nucleotide substitutions during replication of approximate 1011 molecules/day.
Lamivudine (LAM), the first approved oral nucleoside analogue, has an excellent safety profile and is effective in inducing viral suppression, hepatitis B e antigen (HBeAg) seroconversion, and decreasing the progression of liver fibrosis.3,4 However, it is associated with high rates of viral resistance; the prevalence of genotypic resistance after 8 years was 76%.5 The main mutation associated with LAM resistance is rtM204I/V. The rtL180M mutation is the most common compensatory mutation, contributing to increasing either replication efficiency and/or antiviral resistance.6 With various patterns of mutations leading to LAM resistance, the initial benefits provided by LAM are reduced during long-term follow-up.7 Accordingly, LAM is no longer recommended as a first-line agent for naïve patients with chronic hepatitis B (CHB).8
Entecavir (ETV), a carboxylic analogue of guanosine, was superior to LAM in HBV DNA reduction and in inducing histologic improvement in phase III trials for both HBeAg-positive and HBeAg-negative CHB.9-11 Resistance to ETV in treatment-naïve patients is known to be rare, occurring in 1.1% of patients after 4 years of therapy, and is associated with two different mutation profiles.12 Interestingly, resistance to ETV includes LAM resistance mutations, rtM250V±rtI169T+rtM204V+rtL180M and rtT184G+rtS202I+rtM204V+rtL180M.13 That is, in patients with pre-existing LAM-resistance mutations, there is a higher rate of developing ETV resistance, which was found to be 39% after 4 years.14
So far, there are not enough data on how many viral quasispecies and pre-existing drug resistance mutations exist in preantiviral therapy. Thus, the aim of this study is to investigate viral quasispecies in the polymerase gene of HBV and the presence of drug resistance mutations before and at initiation of antiviral treatment.
MATERIALS AND METHODS
1. Subjects
Six CHB patients with LAM failure were enrolled in this study. Patient sera were obtained at baseline (before treatment), 6 months after treatment and at the time of virologic breakthrough (VB) during LAM treatment. VB was defined as an increase in the serum HBV DNA levels of ≥1 log10 copies/mL during treatment in patients with initially adequate viral suppression.15 For the comparison of baseline quasispecies, we also used stored sera of six patients with HBeAg-positive CHB who showed HBeAg seroconversion and undetectable HBV DNA for more than 72 weeks during LAM therapy. Cirrhosis was clinically diagnosed with ultrasonographic evidence of surface nodularity, serum platelet count <100,000/mm3, and splenomegaly (>12 cm). Routine liver biochemical tests including aspartate aminotransferase, alanine aminotransferase, albumin, total bilirubin, prothrombin, and creatinine were performed using a sequential multiple autoanalyzer. HBeAg and antibodies to HBeAg were tested with enzyme-linked immunoassays (Dade Behring, Marburg, Germany). HBV DNA levels were quantified by polymerase chain reaction (PCR), using the COBAS TaqMan 48 analyzer (Roche Molecular Systems Inc., Branchburg, NJ, USA) with a lower detection limit of 300 copies/mL. HBV genotyping was performed using line probe genotyping assay (INNO-LiPA HBV Genotyping assay; Innogenetics, Ghent, Belgium).
2. Cloning and sequencing
DNA extraction was carried out with QIAamp DNA Mini Kit (Qiagen Inc., Hilden, Germany) following the manufacturer's instructions. The primers were designed to the RT region of the HBV polymerase gene: forward 5'-GGTCACCATATTCTTGGGAA-3', reverse 5'-GTGGGGGTTGCGTCAGCAAA-3'. PCR reactions were performed in 50 µL reaction volumes. The reaction contained 10 mL of DNA extract, 1 µL of each primer (10 µmoL), 0.2 µL of Taq polymerase high fidelity (5 U/µL), 5 µL of 10×Taq buffer, 2 µL of dNTP mixture, and 36.8 µL H2O. The PCR programs used were as follows: denaturing at 94℃ for 5 minutes, followed by 40 cycles of denaturing at 94℃ 40 seconds, annealing at 52℃ for 1 minute and elongation at 72℃ for 1 minute and then a final extension at 72℃ for 10 minutes.
PCR amplified HBV DNA was cloned into pGEM T Easy Vector (Promega Co., Madison, WI, USA) according to the manufacturer's instructions. After transformation, 100 clones with the HBV insert were picked up at each time point. The colonies were grown overnight at 37℃ in 3 mL Luria-Bertani broth mixed with ampicillin (100 µg/mL). Recombinant plasmid DNA was purified using the Wizard Plus SV Minipreps DNA Purification System (Promega Co.) following the manufacturer's instructions. Purified plasmids were electrophoresed after digestion with restriction enzymes Xba I and Pst I (New England BioLabs, Beverly, MA, USA) and sequenced using primers, SP6 (5'-ATT TAG GTG ACA CTA TAG-3') or T7 (5'-TAA TAC GAC TCA CTA TAG GG-3'). The reference gene of HBV genotype C was NCBI GenBank No. AF286594, which was considered the wild type.
RESULTS
1. Clinical characteristics and clone sequencing of HBV
All six patients with LAM failure had chronic infection by HBV of genotype C. There were four males and two females with a mean age of 54±12.5 years. Three of the patients had clinical evidence of cirrhosis. The mean duration of LAM therapy was 30.5±10.7 months, and the mean time to VB was 27.8±11.3 months. ETV 1 mg had been administered for 2nd-line therapy in patient 1 and 4, and adefovir (ADV) 10 mg was started in the remaining patients (Table 1). Six patients (three males, mean age of 46±8.6 years) who responded to LAM therapy also had genotype C, and HBeAg seroconversion occurred at a mean of 22.5±5.6 months.
Table 1.
Characteristics of the Patients
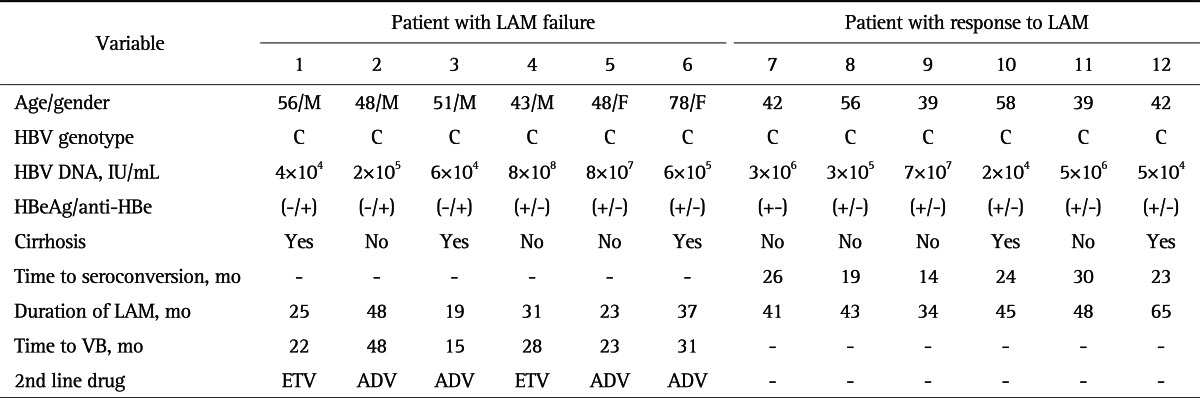
LAM, lamivudine; M, male; F, female; HBV, hepatitis B virus; HBeAg, hepatitis B e antigen; anti-HBe, antibodies to HBeAg; VB, virologic breakthrough; ETV, entecavir 1 mg; ADV, adefovir.
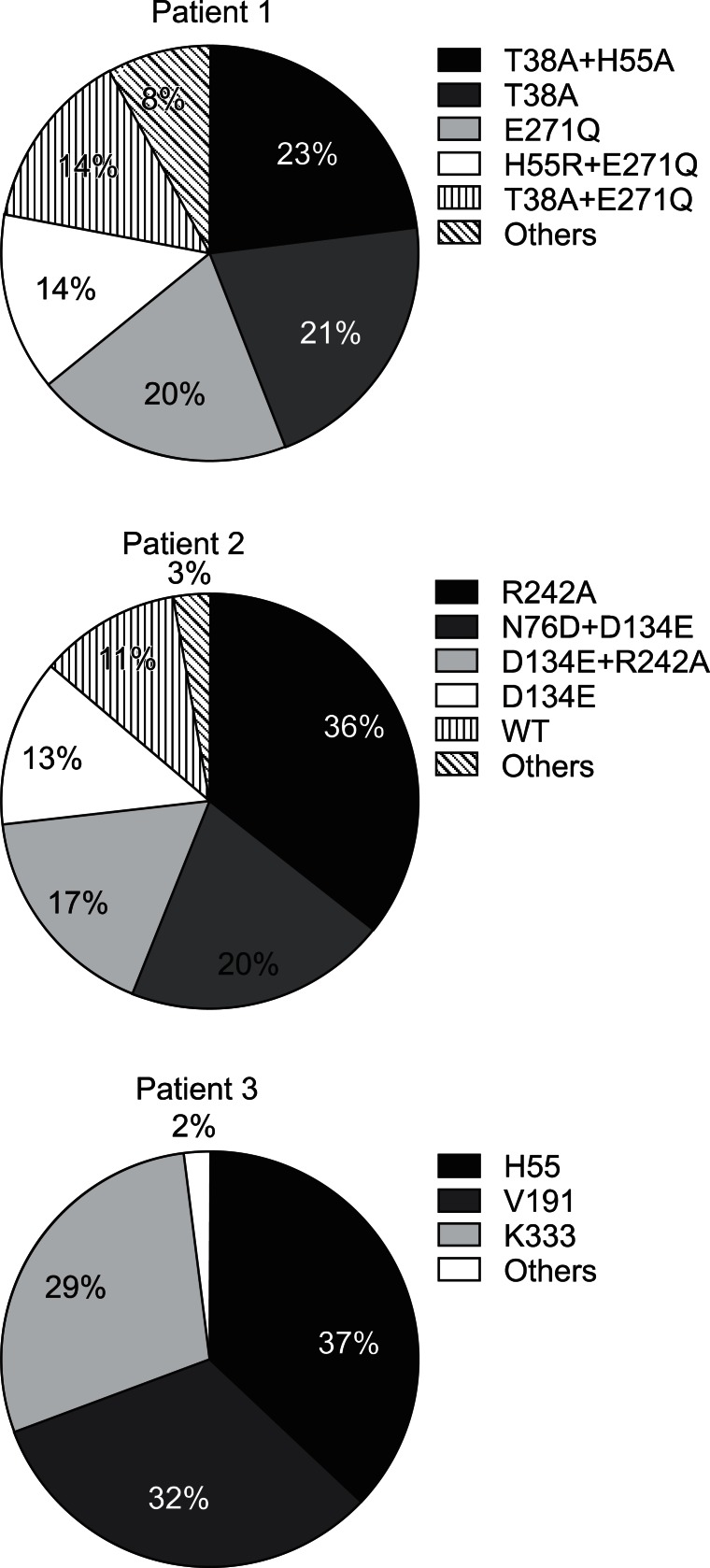
In order to determine the composition and changes of HBV quasispecies before and during LAM treatment in the patients with LAM failure, clone sequencing was performed at the baseline (prior to LAM therapy), at 6 months, and at VB. Fig. 1 shows examples of the viral quasispecies in HBV P genes of patient 1 to 3 before treatment. In patient 1, 23 of 100 clones (23%) contained T38A+H55A, which was followed by T38A (21%), and E271Q (20%). The predominant HBV quasispecies in patient 2 was R242A (36%), and the remaining were N76D+D134E (20%), D134E+R242A (17%), and D134E (13%).
Fig. 1.
Viral quasispecies in the hepatitis B virus (HBV) P genes of three patients. The identified number of quasispecies of each patient were 58, 31, and 81, respectively. The frequency of wild-type HBV was 11% in patient 2. However, there was no wild-type virus in patients 1 and 3. WT, wild-type (reference sequence): NCBI GeneBank no. AF286594 (HBV Pol_genotype C).
In patient 3, there were a total of 81 quasispecies with various amino acid substitutions in H55, V191, and K333, which were almost equally distributed in 100 clones. The frequency of wild type HBV was 11% in patient 2. However, there was no wild type virus in patient 1 and 3.
2. Pre-existing antiviral drug-resistant mutations
Pretreatment viral quasispecies was compared among patients with LAM failure and those who responded to LAM therapy. Among six patients with LAM failure, cloning and sequencing data on the RT region of HBV revealed that LAM resistance mutation, L180M or M204V/I substitutions, pre-existed in patient 1, 2, 5, and 6, albeit a small percentage (1% to 2%). Moreover, 1% of the clones in patient 1 and 4 harbored ADV resistance mutations, A181V. Whereas, only M204V/I substitution was identified in 1% of the clones in one patient who responded to LAM therapy. Neither L180M nor A181V mutation was detected in the six responders (Table 2).
Table 2.
Frequency of Pre-Existing Drug-Resistant Mutations According to Response to Lamivudine (LAM)
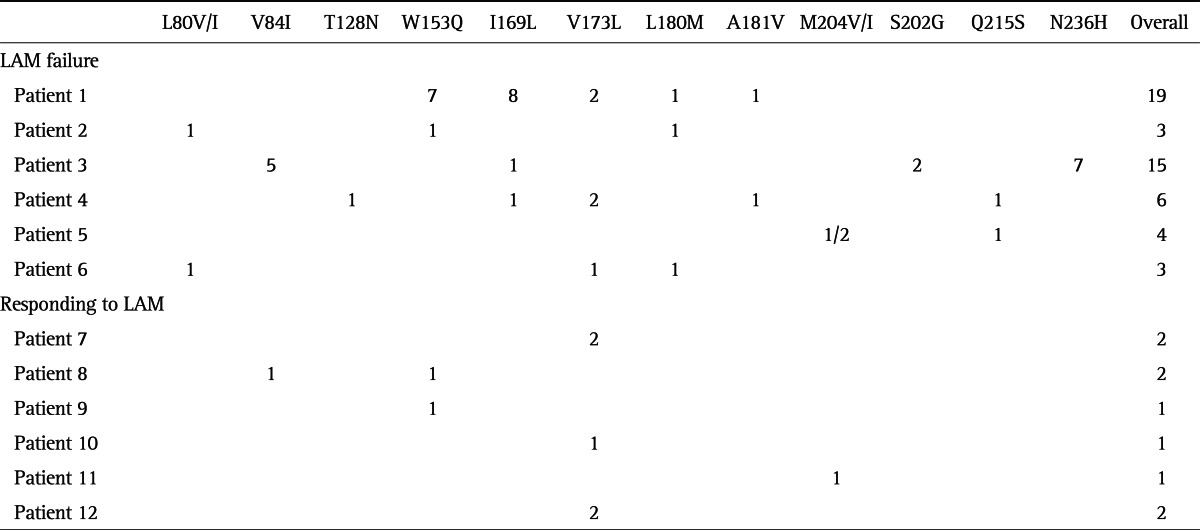
Data are presented as percentage.
3. Evolution of drug-resistant mutations
Table 3 shows the dynamic changes and proportions of quasispecies in HBV P gene of six patients with LAM failure during treatment. At baseline, some colonies contained LAM or ETV resistant mutations such as L80V/I, L180M, M204V/I, A181V, and N236H. In patient 1, 3, and 4, I169L substitution was also detected. Interestingly, the colonies of patient 3 contained S202G, which is known as ETV signature resistance, in conjunction with I169L. At 6 months of LAM therapy, quasispecies variants bearing LAM resistance mutations, M204V/I+L180M, appeared in all six patients. In patient 3 and 4, ADV related changes, N236H and A181T, became undetectable at this time point. In patient 1, ADV-associated variant N236H newly emerged in conjunction with M204I substitution. While ETV-related mutations became undetectable in patient 1, 3, and 4, I169L+L180M+M204V was detected in patient 6. At VB, ADV resistance mutations were not detected in any patient.
Table 3.
Evolution of Antiviral Drug-Resistant Mutations
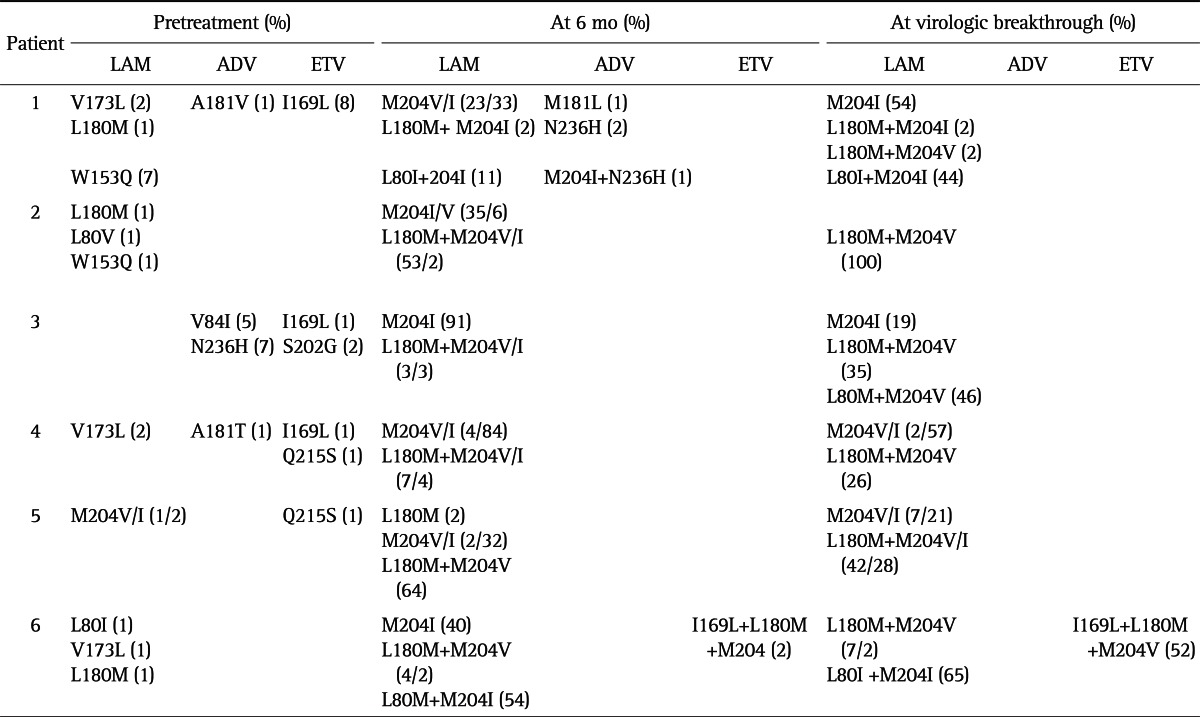
LAM, lamivudine; AVD, adefovir; ETV, entecavir.
4. Concomitant overlapping S gene changes
All patients showed at least one kind of concomitant envelope gene change, which overlaps the polymerase gene. In patient 1, changes of the P gene, rtW153Q, rtI169L, rtV173L, rtA181V, and rtM204V, corresponded to change of the S gene, rtG145R, rtF161H, rtE164D, and rtI195M, respectively (Table 4). In patient 2, rtW153Q and rtW196L change, a termination codon caused by rtM204, was detected. The ADV resistance mutation, rtA181V resulted in a concomitant change of rtL173F in patient 4.
Table 4.
Reciprocal Changes of P and S Genes at Pretreatment
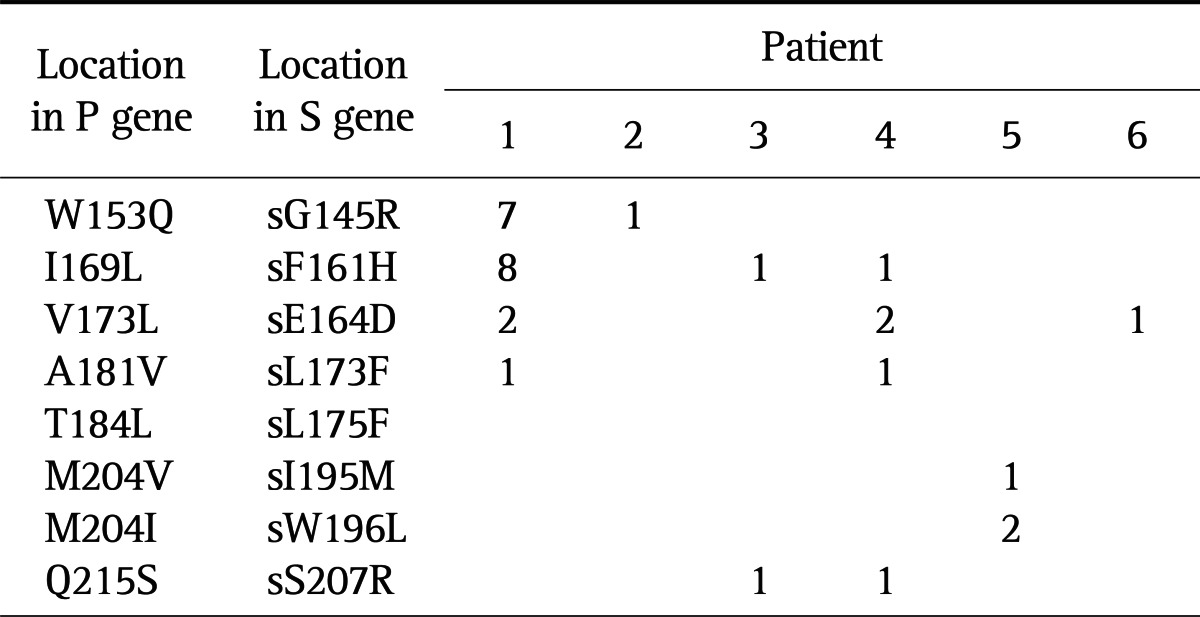
Data are presented as percentage.
DISCUSSION
In the present study, the HBV quasispecies of polymerase RT domain in six patients with LAM resistance were examined before antiviral therapy, at 6 months of LAM therapy, and at VB. In addition, pretreatment sera were also analyzed for the viral quasispecies in six patients who had a good response to LAM therapy. The finding that more diverse HBV quasispecies was identified in LAM-failure patients than in LAM-responding patients and that the frequency of pre-existing M204V/I substitution was not different between two groups suggests that degree of overall viral quasispecies rather than M204V/I substitition might predict response to LAM. Sequence analysis of 100 HBV clones isolated from sera of each patient with LAM failure at each time point revealed the evolution of the viral quasispecies. Although LAM is no longer recommend as a first-line therapy for CHB due to high incidence of drug resistance,15 many patients especially in Asian countries have been exposed to this drug. Hence, analysis of substitutions in the sequence of the polymerase gene of this error-prone HBV would be required to select the rescue agent for patients with LAM resistance.16 Furthermore, it was reported that substitutions at rtI184, rtS202, and/or rtM250 were already detected in LAM-refractory subjects at baseline ETV treatment.10 As seen in Fig. 1, many viral quasispecies of HBV polymerase gene existed even before LAM therapy. Pre-existing LAM-resistance mutation, rtM204V/I, was found in 3% (3/100) of clones in patient 5, whereas rtL180M mutation was detected in 1% (1/100) of clones in patient 1 and 2. LAM compensatory mutations, rtL80V/I and/or rtV173L, were seen in patient 1, 2, 4, and 6. Our result is in agreement with a previous report that some viral populations harbor YMDD mutants in LAM-untreated asymptomatic hepatitis B carriers.17 Emergence of ETV resistance seems to be infrequent, in particular in treatment-naïve patients. The importance of pre-existing ETV resistant HBV variants was highlighted in a previous study, in which three out of 111 (2.7%) patients had substitutions in the HBV polymerase gene corresponding to variants associated with ETV resistance (rtS202S/I) prior to LAM treatment.18 In another large-scale study, three out of 255 (1.2%) treatment-naïve patients had the rtM250L/V mutation typical of ETV resistance.19 In our sequencing data, rtI169L mutation was present in 8%, 1%, and 1% of 100 colonies in patient 1, 3, and 4, as well as rtA181V/T ADV resistance mutations in patient 1 and 4. The significance and clinical relevance of these pre-existing ETV or ADV resistance variants of minor fractions remains to be elucidated by a large number of patients and long term follow-up. Nevertheless, the selective pressure of nucleos(t)ide analogue therapy will enable the pre-existing mutants to generate a progeny virus that is more fit and can spread more rapidly in the liver.
As expected, while rtL180M and rtM204V/I variants were present as a single mutation prior to LAM treatment, the rtL180M+rtM204V/I combination mutations became predominant at 6 months and at VB. Interestingly, the rtV173A/L mutation that had been described in previous reports20,21 was detected at baseline, but not thereafter in patient 1, 4, and 6. Regarding HBV quasispecies associated with ADV resistance, the rtV84I/S, rt N236H, rtA181T, and rtQ215H mutations, were detected at baseline and at 6 months of LAM therapy in patient 1, 3, 4, and 6. However, no colony harboring such variants was observed at VB. In addition to classical ADV resistance mutations such as rtN236S or rtA181T, a few other related-variants including rtV84I/S, rtL84S, and rtQ215H were identified before VB in patient 1, 3, 4, and 6. It is known that, for nucleos(t)de-naive individuals treated with ETV, LAM resistant quasispecies are necessary prior to emergence of ETV resistance variants. This, so-called two-hit model was suggested for the development of ETV resistance in that LAM resistance mutations were selected initially and one or two additional ETV resistance mutations were acquired in the same virus afterwards.12,20,22 Guo et al.23 cloned and sequenced the HBV RT of a patient who had been treated with ETV 0.5 mg as a first line therapy developing ETV resistance. The LAM resistance quasispecies (rtM204V±rt180M) emerged as early as 48 weeks, and ETV resistant quasispecies (rtM204±rtL180M plus S202G) were found after 112 weeks.23 Unlike the previous study, rtI169S/L and/or rtS202G mutations were detected in the absence of LAM resistance variants at pretreatment, even though those variants disappeared during LAM therapy until VB. This finding suggests that some viral population might harbor ETV resistance mutations, without preceding LAM resistance variants in treatment-naïve patients who receive ETV 0.5 mg. Although we demonstrated viral diversity even at pretreatment by cloning and sequencing, it needs to be kept in mind that most of viral quasispecies shown in Fig. 1 have no clinical implication because we did not perform any susceptibility test for those variants.
The polymerase gene overlaps the envelope gene and thus mutations selected during antiviral resistance can cause concomitant changes to the overlapping reading frame with the major resistance mutations for LAM, ADV, and ETV.16 We found that HBsAg changes corresponding to resistance mutations existed even at pretreatment. For instance, a change at sI195M, sW196L associated with rtM204V/I was detected in one and two of 100 colonies in patient 1 and 5, respectively. The mutation at sF161H, which is located adjacent to the region that was defined as the "a" determinant, in association with rtI169L mutation was observed in patient 1, 3, and 4. The clinical implication of these reciprocal changes of the P and S genes is that they might affect diagnostic assays, vaccine escape, replication fitness, and pathogenicity of HBV.
In conclusion, we found that various HBV quasispecies associated with drug resistance existed as well as S gene variants before treatment and the quasispecies dynamically changed along with LAM therapy.
ACKNOWLEDGEMENTS
This work was supported by a faculty grant of Department of Internal Medicine, Yonsei University College of Medicine for 2007, and in part by a grant of the Korea Healthcare Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (no. A102065).
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Hollinger FB. Hepatitis B virus genetic diversity and its impact on diagnostic assays. J Viral Hepat. 2007;14(Suppl 1):11–15. doi: 10.1111/j.1365-2893.2007.00910.x. [DOI] [PubMed] [Google Scholar]
- 2.Cane PA, Mutimer D, Ratcliffe D, et al. Analysis of hepatitis B virus quasispecies changes during emergence and reversion of lamivudine resistance in liver transplantation. Antivir Ther. 1999;4:7–14. [PubMed] [Google Scholar]
- 3.Lai CL, Chien RN, Leung NW, et al. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 4.Dienstag JL, Schiff ER, Wright TL, et al. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341:1256–1263. doi: 10.1056/NEJM199910213411702. [DOI] [PubMed] [Google Scholar]
- 5.Yuen MF, Seto WK, Chow DH, et al. Long-term lamivudine therapy reduces the risk of long-term complications of chronic hepatitis B infection even in patients without advanced disease. Antivir Ther. 2007;12:1295–1303. [PubMed] [Google Scholar]
- 6.Xiong X, Yang H, Westland CE, Zou R, Gibbs CS. In vitro evaluation of hepatitis B virus polymerase mutations associated with famciclovir resistance. Hepatology. 2000;31:219–224. doi: 10.1002/hep.510310132. [DOI] [PubMed] [Google Scholar]
- 7.Dienstag JL, Cianciara J, Karayalcin S, et al. Durability of serologic response after lamivudine treatment of chronic hepatitis B. Hepatology. 2003;37:748–755. doi: 10.1053/jhep.2003.50117. [DOI] [PubMed] [Google Scholar]
- 8.European Association For The Study Of The Liver. EASL Clinical Practice Guidelines: management of chronic hepatitis B. J Hepatol. 2009;50:227–242. doi: 10.1016/j.jhep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Lai CL, Shouval D, Lok AS, et al. Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N Engl J Med. 2006;354:1011–1020. doi: 10.1056/NEJMoa051287. [DOI] [PubMed] [Google Scholar]
- 10.Chang TT, Gish RG, de Man R, et al. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N Engl J Med. 2006;354:1001–1010. doi: 10.1056/NEJMoa051285. [DOI] [PubMed] [Google Scholar]
- 11.Lai CL, Rosmawati M, Lao J, et al. Entecavir is superior to lamivudine in reducing hepatitis B virus DNA in patients with chronic hepatitis B infection. Gastroenterology. 2002;123:1831–1838. doi: 10.1053/gast.2002.37058. [DOI] [PubMed] [Google Scholar]
- 12.Colonno RJ, Rose R, Baldick CJ, et al. Entecavir resistance is rare in nucleoside naive patients with hepatitis B. Hepatology. 2006;44:1656–1665. doi: 10.1002/hep.21422. [DOI] [PubMed] [Google Scholar]
- 13.Bartholomeusz A, Locarnini SA. Antiviral drug resistance: clinical consequences and molecular aspects. Semin Liver Dis. 2006;26:162–170. doi: 10.1055/s-2006-939758. [DOI] [PubMed] [Google Scholar]
- 14.Tenney DJ, Rose RE, Baldick CJ, et al. Two-year assessment of entecavir resistance in Lamivudine-refractory hepatitis B virus patients reveals different clinical outcomes depending on the resistance substitutions present. Antimicrob Agents Chemother. 2007;51:902–911. doi: 10.1128/AAC.00833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507–539. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 16.Bartholomeusz A, Locarnini S. Hepatitis B virus mutations associated with antiviral therapy. J Med Virol. 2006;78(Suppl 1):S52–S55. doi: 10.1002/jmv.20608. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi S, Ide T, Sata M. Detection of YMDD motif mutations in some lamivudine-untreated asymptomatic hepatitis B virus carriers. J Hepatol. 2001;34:584–586. doi: 10.1016/s0168-8278(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 18.Jardi R, Rodriguez-Frias F, Schaper M, et al. Hepatitis B virus polymerase variants associated with entecavir drug resistance in treatment-naive patients. J Viral Hepat. 2007;14:835–840. doi: 10.1111/j.1365-2893.2007.00877.x. [DOI] [PubMed] [Google Scholar]
- 19.Mirandola S, Campagnolo D, Bortoletto G, Franceschini L, Marcolongo M, Alberti A. Large-scale survey of naturally occurring HBV polymerase mutations associated with anti-HBV drug resistance in untreated patients with chronic hepatitis B. J Viral Hepat. 2011;18:e212–e216. doi: 10.1111/j.1365-2893.2011.01435.x. [DOI] [PubMed] [Google Scholar]
- 20.Yim HJ, Hussain M, Liu Y, Wong SN, Fung SK, Lok AS. Evolution of multi-drug resistant hepatitis B virus during sequential therapy. Hepatology. 2006;44:703–712. doi: 10.1002/hep.21290. [DOI] [PubMed] [Google Scholar]
- 21.Moriconi F, Colombatto P, Coco B, et al. Emergence of hepatitis B virus quasispecies with lower susceptibility to nucleos(t)ide analogues during lamivudine treatment. J Antimicrob Chemother. 2007;60:341–349. doi: 10.1093/jac/dkm187. [DOI] [PubMed] [Google Scholar]
- 22.Tenney DJ, Levine SM, Rose RE, et al. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498–3507. doi: 10.1128/AAC.48.9.3498-3507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo JJ, Li QL, Shi XF, et al. Dynamics of hepatitis B virus resistance to entecavir in a nucleoside/nucleotide-naive patient. Antiviral Res. 2009;81:180–183. doi: 10.1016/j.antiviral.2008.09.004. [DOI] [PubMed] [Google Scholar]