Abstract
Candida albicans, an opportunistic fungal pathogen and a component of the gastrointestinal tract normal flora, is a frequent colonizer of humans. Is C. albicans capable of sensing the immune status of its host, a process we term immunosensing, and, if so, how? C. albicans causes serious disease only in immunocompromised hosts and therefore, the ability to immunosense would be advantageous to the organism. We propose a speculative model that, during colonization, C. albicans produces phenotypic variants that vary in relative concentration depending on host status. One variant is optimized for persistence as a commensal while the other variant has higher capacity to initiate pathogenic interactions. When the ratio of the two variants changes, the pathogenic potential of the population changes. The critical element of this model is that the C. albicans colonizing population is not uniform but is composed of subpopulations of phenotypic variants that are advantageous under different host conditions.
Candida albicans cells colonizing the gastrointestinal tract and causing disease in other niches are not identical
After long-term, peaceful coexistance of Candida albicans with its human host, a decline in host immunological function can lead to the development of devastating invasive candidiasis. How does this happen? Unraveling this mystery will be a crucial step in understanding the initiation and progression of disease caused by this opportunistic fungal pathogen.
C. albicans is an important cause of nosocomial infection [1, 2], most likely because hospitalized patients have multiple predisposing factors that favor candidal disease (such as immunosuppression, violation of protective barriers, and use of broad spectrum antibacterial drugs [3, 4]). However, most of the time, the interactions between C. albicans and its host are benign. Commensal colonization (see Glossary) by C. albicans is common in humans [5]; the organism typically resides in the intestinal tract, genitourinary tract or on the skin as a component of the normal flora. Colonization is established very early in life and, in one study, was observed in over 50% of one-month old infants [6]. Environmental niches are not thought to be significant in the natural history of C. albicans.
In the case of an immunosuppressed patient, C. albicans cells that were benignly colonizing can become pathogenic, escape from the intestinal tract and disseminate to distant organs via the bloodstream. Neutropenia and mucosal barrier compromise are important for C. albicans escape from the gastrointestinal (GI) tract [7]. When newborn piglets monocolonized in the gut with C. albicans are immunosuppressed, C. albicans produces invasive lesions in the gut associated lymphoid tissue, showing that C. albicans is capable of promoting its own escape from the gut of a compromised host [8].
Mechanistically, what happens when the host becomes compromised and the détente between host and colonizing C. albicans cells breaks down? It is possible that during colonization, the organism is always attempting to cause disease. In this scenario, disease is prevented by the constant vigilance of the host and the effects of competition between C. albicans and the numerous other microbial colonizers that share the resources within the host.
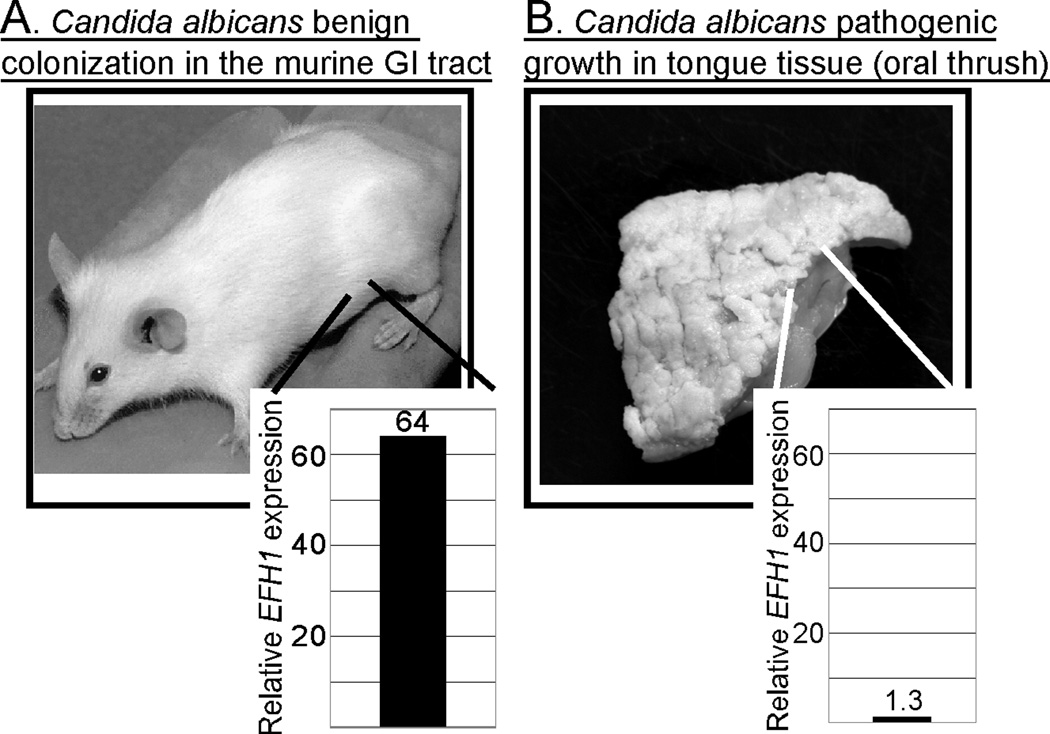
However, studies show that there are differences in gene expression between cells that are benignly colonizing the GI tract of a host and those that are actively causing disease in other niches [9–12]. For example, the EFH1 gene, encoding a transcription factor, is relatively highly expressed in cells colonizing the cecum but not in cells occupying a different niche and causing an oral infection (Figure 1) [12]. Since EFH1 is also not highly expressed during laboratory growth, the intestinal tract is the niche in which this C. albicans gene is most highly expressed. Paradoxically, however, expression of EFH1 is associated with reduced colonization of the intestinal tract. In an animal model, C. albicans cells lacking EFH1 persist in the intestinal tract at higher levels, wild-type cells expressing EFH1 at normal levels have intermediate persistence and cells that overexpress EFH1 poorly colonize the GI tract [12]. These findings show that during benign colonization C. albicans self-regulates its population size. Perhaps this regulation prevents the organisms from damaging the host or evoking a strong host response. In contrast, Efh1p is less important for pathogenesis from the bloodstream because an efh1 null mutant is indistinguishable from a wild-type strain in the intravenously inoculated mouse model of fatal systemic candidiasis [12]. These results argue that expression of EFH1 is part of a genetic program that favors commensalism. Further, organisms must then undergo a transition between the commensal state and the pathogenic state because benignly colonizing organisms in the GI tract and disease-causing organisms in different niches are not identical.
Figure 1.
Differential gene expression in C. albicans cells colonizing the gastrointestinal tract or causing oral thrush. (a) Colonization of the gastrointestinal (GI) tract without disease. Wild-type C. albicans cells established colonization of the murine GI tract as described previously [9]. RNA was extracted from colonizing C. albicans cells recovered from the murine cecum and expression of EFH1 was measured by real time quantitative reverse transcriptase PCR [12]. (b) Oral thrush in an immunosuppressed gnotobiotic newborn piglet. A germ-free piglet monocolonized in the GI tract with C. albicans and immunosuppressed developed oral thrush [8]; monocolonized piglets not treated with immunosuppressing drugs do not develop the extensive thrush shown here. (Image copyright © 2000, American Society for Microbiology. Reprinted with permission from [8]). RNA was extracted from pathogenic C. albicans cells recovered from the piglet tongue and EFH1 expression was analyzed as described above [12].
Could colonizing microorganisms detect conditions in the host in order to undergo a change in gene expression and phenotype under appropriate conditions? Here, we propose a speculative hypothesis in which C. albicans uses phenotypic variation to test host status, a process we will term ‘immunosensing’. In this article, we discuss how a complex colonizing population composed of different phenotypic variants could test host status.
A hypothesis: immunosensing through population dynamics
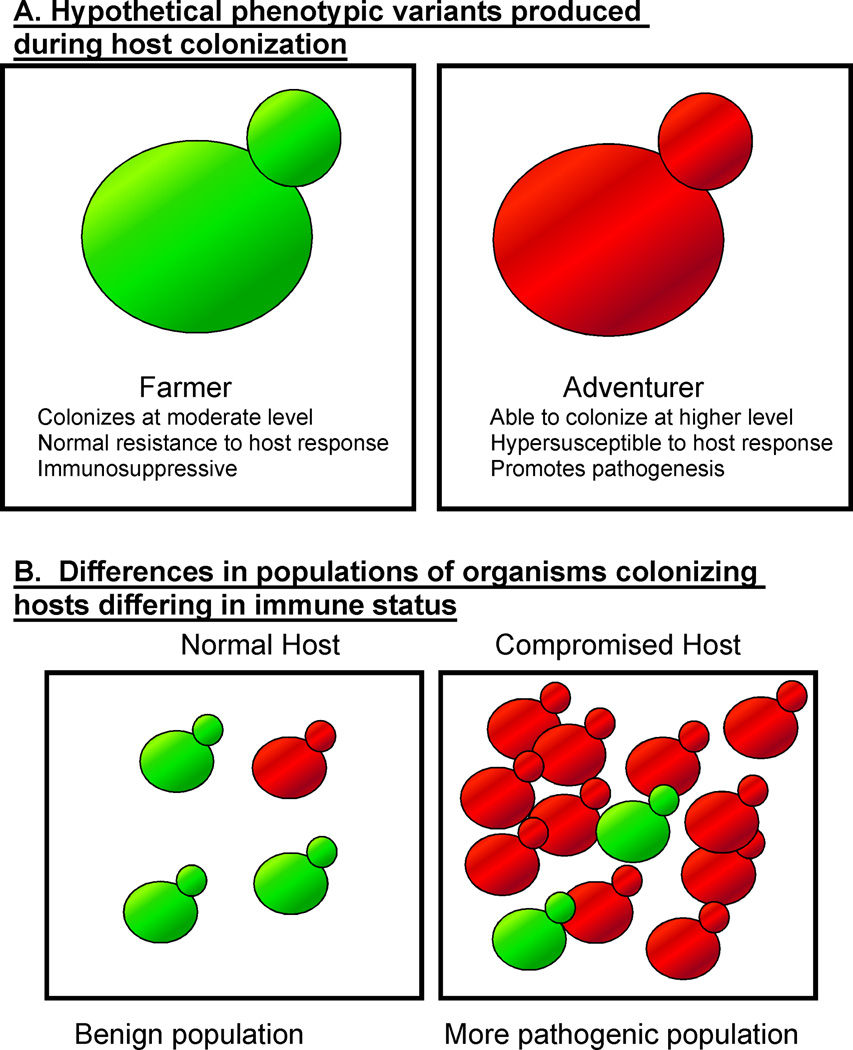
We hypothesize that during colonization, a population of C. albicans cells produces phenotypic variants differing from each other in gene expression patterns. The resulting heterogeneous population, composed of distinct subpopulations, is capable of immunosensing. For the purposes of discussion, we will term the phenotypic variants ‘farmers’ and ‘adventurers’ (Figure 2). It should be emphasized that this model is a hypothesis and cells specifically corresponding to farmers and adventurers have not been described.
Figure 2.
Hypothetical C. albicans phenotypic variants produced during colonization of a host. (a) Depicts two phenotypic variants, farmers and adventurers, that are proposed to differ in interaction with the host and susceptibility to the host response. (b) Depicts populations produced in a normal host or a compromised host.
Most of the colonizing cells are proposed to be farmers, who do not interact aggressively with the host. In fact, C. albicans farmers might pacify the host, exerting an immunosuppressive effect by promoting the expansion of regulatory T cell populations [13]. This effect could aid the survival of both farmers and adventurers.
Adventurers could interact more aggressively with the host. For example, they could colonize to higher levels, or express virulence-promoting phenotypes such as increased adhesion and increased invasiveness. In a normal host, adventurers also stimulate an anti-Candida host response. Importantly, we also propose that adventurers are more susceptible to the host response than farmers.
In a healthy host, we would envision that most of the C. albicans cells are farmers and the subpopulation of adventurers is relatively small because of their susceptibility to the host response. The rate of interconversion between farmers and adventurers could be constant or could change depending on host conditions. Interactions between farmers, adventurers, the bacterial microbiota and the host establish a relatively stable situation, where C. albicans colonizes but does not harm the host. Presumably, the ratio of farmers and adventurers in the C. albicans colonizing population is maintained at an optimal level that permits long term colonization without damage to the host.
If the host is deficient in mounting a response, due for example to neutropenia, we propose that the adventurers outcompete the farmers because they could be capable of colonizing to higher levels. As a result, adventurers could come to predominate in the population. Thus, when the host becomes compromised, the makeup of the colonizing population changes (Figure 2). The number of adventurers increases in this situation, so the chance that C. albicans cells will escape from the intestinal tract and produce disease also increases.
We propose that adventurers are more pathogenic than farmers. Therefore, it could be advantageous to limit their numbers during colonization of a healthy host. The proposed mechanism ensures that large numbers of adventurers only accumulate in a compromised host.
Our speculative model thus proposes that in response to changes in the host, an overall change in the phenotype of the colonizing C. albicans population occurs. In this way, the organisms respond to host status, maintaining colonization when the host is healthy but invading tissue and causing disease when the host is compromised. The ability to produce a complex, heterogeneous colonizing population composed of phenotypic variants with different properties is proposed to be crucial for optimal host interaction.
Phenotypic variation is well known in C. albicans
The phenotypic variation discussed above is hypothetical but there are several known examples of phenotypic variation in C. albicans. A well-studied case involves two cell types, termed ‘white’ and ‘opaque,’ that exhibit cooperative behavior. The existence of white cells, opaque cells and switching between them was first observed in a particular clinically isolated strain of C. albicans [14]. It was later appreciated that switching between white and opaque cells occurred in cells that were homozygous at the mating type locus, MTL [15, 16]. Further, opaque cells are the mating-competent form of C. albicans [15–18]. White and opaque cells differ in the expression of hundreds of genes [19] showing that their phenotypes are distinct in many ways.
In a mixed population, white and opaque cells cooperate to enhance mating [20]. Through release of pheromone, opaque cells signal white cells. As a result, the white cells aggregate, adhere to a substratum and form a biofilm [21]. Within the biofilm, pheromone gradients produced by opaque cells are stabilized, promoting mating. Thus, through pheromone secretion, a small number of opaque cells influence the behavior of a large number of white cells and the white cells create an environment that is conducive for mating. This example shows that interaction between phenotypic variants in a mixed population can enhance a biological activity.
Other examples of phenotypic variation in C. albicans involve either cellular or colonial morphology. For example, C. albicans can grow in either a yeast form or as filamentous hyphae [22]. The transcription profiles of the two forms are quite different, with large numbers of genes that are differentially regulated during laboratory growth [23–25]. Variation in cellular morphology at the population level produces differences in colony morphology [26]. Switching between colony morphologies is regulated and is derepressed in the absence of the histone deacetylase Sir2 [27].
These examples show that phenotypic variation in C. albicans is not unusual. During murine intestinal colonization, both yeast and hyphal forms are produced [12, 28]. Thus, at least one type of phenotypic variation occurs during colonization. Further, the cooperation between white and opaque cells shows that a mixed population need not be simply the sum of its parts; rather, a mixed population might have increased capabilities that neither single population possesses.
For other microorganisms, phenotypic variation contributes to colonization and persistence
The notion that a colonizing population of organisms can be composed of distinct subpopulations of phenotypic variants that promote colonization, virulence and persistence during stress is well established for several microorganisms. Organisms that persist in a host for long periods of time and cause chronic infections often exhibit antigenic variation [29], i.e. variation in expression of surface proteins that are antigenic. Antigenic variation is proposed to contribute to colonization or infection in several ways [30]. Antigenic variation enhances persistence by allowing organisms to evade the host immune response. An infection that lasts longer could prolong the period of high infectivity, enhancing transmission of the organism. Antigenic variation can also allow an organism to infect a previously infected host. In addition, different variants of an antigenic protein might differ in activity, altering the course of the disease. All of these effects can be beneficial for the infecting organism.
Antigenic variation has been described in bacterial, fungal and protozoan species [30]. For example, in Borrelia species, antigenic variation contributes to persistence in a host during relapsing fever or Lyme disease [31–33]. In Plasmodium spp., the parasites that cause malaria, the products of the var genes are proteins that are expressed on the surface of mammalian red blood cells infected with the organism. Different var genes encode proteins that differ in antigenicity and ligand binding specificity [34, 35]. Through differential expression and recombinational generation of new sequences [36, 37], the parasites produce extensive antigenic diversity, allowing them to evade the host immune response.
An invasive subpopulation that promotes intestinal colonization by Salmonella Typhimurium illustrates how one phenotypic variant can influence colonization by other cells in the population [38, 39]. By invading intestinal tissue, the invasive cells provoke the host inflammatory response. Inflammation promotes luminal colonization by S. Typhimurium possibly because S. Typhimurium possesses multiple mechanisms for resisting the toxic inflammatory response while competing commensal organisms do not. Although the invasive subpopulation is killed when the bacteria invade intestinal tissue, their activity ensures that conditions that facilitate colonization by the bulk of the bacterial population are established.
’Persisters,’ dormant cells that are highly resistant to antimicrobial compounds, represent another example of phenotypic variants that form a subpopulation of cells within a population [40, 41]. Treatment of a population with an antimicrobial drug kills the vast majority of the cells. However, the persisters survive due to their high drug resistance, enabling the population to recover when the drug is removed. Whether persisters promote host colonization by allowing colonizing populations to resist host-produced antimicrobial compounds or contribute to recurrence of infection following antibiotic treatment remain open questions.
In addition, organisms adapt to the host environment upon introduction into a host. For example, Vibrio cholerae organisms shed by an infected host suffering from cholera differ phenotypically from V. cholerae organisms that are present in the environment [42]. Therefore, within a host, the organisms undergo a transition from one phenotypic state to another. At certain times or in certain locations within the host, the V. cholerae population will likely be composed of mixtures of cells in different phenotypic states. Whether the mixed population exhibits properties distinct from those of the individual subpopulations is not known.
As these examples show, populations of microorganisms are often composed of distinct subpopulations. Such complex populations promote colonization, persistence and transmission.
Concluding remarks
Many different organisms generate phenotypic variants during growth within a host. The production of variants affects colonization or pathogenesis in several important ways, for example, allowing immune evasion and long term persistence in the host. For C. albicans, we propose that production of a particular type of phenotypic variant allows the organism to monitor the immune status of its host. This immunosensing function represents a new proposed function for phenotypic variation. Future experimental tests of this model will include identification of phenotypic variants produced during colonization by C. albicans and analysis of the contribution of variants to pathogenicity (Box 1). A prediction of this model is that mutants that are unable to generate the appropriate phenotypic variants will be attenuated in producing endogenous candidiasis.
Box 1. Outstanding questions.
Can a subpopulation of cells corresponding to adventurers be identified and isolated? The speculation that subpopulations exist would be supported by the demonstration that such cells occur during intestinal colonization.
How do adventurers contribute to pathogenesis in animal models of candidiasis? The speculation that farmers and adventurers differ in pathogenic potential could be established experimentally.
In addition to the properties described here, in what other ways do adventurers and farmers differ from each other? Transcription profiling could demonstrate the full spectrum of activities that are co-regulated with the farmer or adventurer phenotype.
How commonly in nature do heterogeneous colonizing populations promote colonization and host exploitation? The strategy of producing subpopulations that differ in their behavior in the host might be shared by many microorganisms.
Immunosensing through phenotypic variation might not be restricted to fungi. The ability of bacteria to sense the host’s immune response and respond to the host by undergoing phenotypic change has been recently discussed [43]. Therefore, the strategy of using a complex colonizing population to monitor and respond to the host could represent a general microbial mechanism for optimizing host-microbe interactions.
Acknowledgements
We are particularly grateful to Drs. Ralph Isberg and Stanley Falkow for careful review of the manuscript and discussion of the ideas. We also thank Drs. Joan Mecsas, Linden Hu, and Linc Sonenshein for helpful discussion. Our research is supported by grant AI087194 from the National Institute of Allergy and Infectious Diseases (to C.A.K.). J.P. was supported by training grant T32 AI07422 from the National Institutes of Health.
Glossary
- Colonize
to establish a population in a particular niche, e.g. in a host.
- Commensal
an organism that benefits from an interaction with another organism without causing damage to the second organism.
- Disease
an alteration of the body that results in the inability of the body or a part of the body to perform its normal function, e.g. due to the effects of a microorganism.
- Endogenous candidiasis
disease caused by Candida organisms that are already colonizing a host.
- Immunosensing
the process by which colonizing microorganisms detect a change in the immune status of the host.
- Opportunistic pathogen
a microorganism that can be pathogenic if the host is compromised but is not harmful to a healthy host.
- Pathogenic
able to cause disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Gudlaugsson O, et al. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis. 2003;37:1172–1177. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ampel NM. Emerging disease issues and fungal pathogens associated with HIV infection. Emerg Infect Dis. 1996;2:109–116. doi: 10.3201/eid0202.960205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderone RA. Introduction and Historical Perspectives. In: Calderone RA, editor. Candida and Candidiasis. ASM Press; 2002. pp. 3–13. [Google Scholar]
- 5.Odds FC. Candida infections: an overview. Crit Rev Microbiol. 1987;15:1–5. doi: 10.3109/10408418709104444. [DOI] [PubMed] [Google Scholar]
- 6.Russell C, Lay KM. Natural history of Candida species and yeasts in the oral cavities of infants. Arch Oral Biol. 1973;18:957–962. doi: 10.1016/0003-9969(73)90176-3. [DOI] [PubMed] [Google Scholar]
- 7.Koh AY, et al. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008;4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrutis KA, et al. Intestinal lesions associated with disseminated candidiasis in an experimental animal model. J Clin Microbiol. 2000;38:2317–2323. doi: 10.1128/jcm.38.6.2317-2323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbach A, et al. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell. 2010;9:1075–1086. doi: 10.1128/EC.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thewes S, et al. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol. 2007;63:1606–1628. doi: 10.1111/j.1365-2958.2007.05614.x. [DOI] [PubMed] [Google Scholar]
- 11.Zakikhany K, et al. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]
- 12.White SJ, et al. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Luca A, et al. Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J Immunol. 2007;179:5999–6008. doi: 10.4049/jimmunol.179.9.5999. [DOI] [PubMed] [Google Scholar]
- 14.Slutsky B, et al. "White-opaque transition": a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart SR, et al. In Candida albicans white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 17.Hull CM, et al. Evidence for mating of the "asexual" yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 18.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 19.Lan CY, et al. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci U S A. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- 21.Daniels KJ, et al. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 2006;25:2240–2252. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudbery P, et al. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Goyard S, et al. The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albicans. Mol Biol Cell. 2008;19:2251–2266. doi: 10.1091/mbc.E07-09-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nantel A, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slutsky B, et al. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Martin J, et al. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinneberg KM, et al. Effect of INT1 gene on Candida albicans murine intestinal colonization. J Surg Res. 1999;87:245–251. doi: 10.1006/jsre.1999.5755. [DOI] [PubMed] [Google Scholar]
- 29.Palmer GH, et al. 'Nothing is permanent but change'- antigenic variation in persistent bacterial pathogens. Cell Microbiol. 2009;11:1697–1705. doi: 10.1111/j.1462-5822.2009.01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deitsch KW, et al. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank SA, Barbour AG. Within-host dynamics of antigenic variation. Infect Genet Evol. 2006;6:141–146. doi: 10.1016/j.meegid.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Barbour AG, et al. Pathogen escape from host immunity by a genome program for antigenic variation. Proc Natl Acad Sci U S A. 2006;103:18290–18295. doi: 10.1073/pnas.0605302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bankhead T, Chaconas G. The role of VlsE antigenic variation in the Lyme disease spirochete: persistence through a mechanism that differs from other pathogens. Mol Microbiol. 2007;65:1547–1558. doi: 10.1111/j.1365-2958.2007.05895.x. [DOI] [PubMed] [Google Scholar]
- 34.Beeson JG, Brown GV. Pathogenesis of Plasmodium falciparum malaria: the roles of parasite adhesion and antigenic variation. Cell Mol Life Sci. 2002;59:258–271. doi: 10.1007/s00018-002-8421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handunnetti SM, et al. Antigenic variation of cloned Plasmodium fragile in its natural host Macaca sinica Sequential appearance of successive variant antigenic types. J Exp Med. 1987;165:1269–1283. doi: 10.1084/jem.165.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scherf A, et al. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 37.Frank M, et al. Frequent recombination events generate diversity within the multi-copy variant antigen gene families of Plasmodium falciparum. Int J Parasitol. 2008;38:1099–1109. doi: 10.1016/j.ijpara.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ackermann M, et al. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454:987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- 39.Stecher B, et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 42.Merrell DS, et al. Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade WF, O'Toole GA. Antibodies and immune effectors: shaping Gram-negative bacterial phenotypes. Trends Microbiol. 2010;18:234–239. doi: 10.1016/j.tim.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]