Summary
Cytosolic bacterial pathogens activate the cytosolic surveillance pathway (CSP) and induce innate immune responses, but how the host detects vacuolar pathogens like Mycobacterium tuberculosis is poorly understood. We show that M. tuberculosis also initiates the CSP upon macrophage infection via limited perforation of the phagosome membrane mediated by the ESX-1 secretion system. Although the bacterium remains within the phagosome, this permeabilization results in phagosomal and cytoplasmic mixing and allows extracellular mycobacterial DNA to access host cytosolic receptors, thus blurring the distinction between “vacuolar” and “cytosolic” pathogens. Activation of cytosolic receptors induces signaling through the STING/TBK1/IRF3 axis, resulting in IFN-β production. Surprisingly, IRF3−/− mice, which cannot respond to cytosolic DNA, are resistant to long-term M. tuberculosis infection, suggesting that the CSP promotes M. tuberculosis infection. Thus, cytosolic sensing of mycobacterial DNA plays a key role in M. tuberculosis pathogenesis and likely contributes to the high type I IFN signature in tuberculosis.
Introduction
Infection with Mycobacterium tuberculosis causes enormous worldwide morbidity and mortality, and global incidence continues to rise (Dye and Williams, 2010). A key mediator of M. tuberculosis pathogenesis is the ESX-1 specialized secretion system that modulates host-cell functions, presumably by translocating bacterial effectors into the host (Abdallah et al., 2007). Mutants lacking ESX-1 are defective for replication within macrophages and are severely attenuated in animal models of infection, but the mechanism by which this system functions to promote infection remains unclear (Guinn et al., 2004; Hsu et al., 2003; Stanley et al., 2003). A growing body of work indicates that ESX-1 contributes to M. tuberculosis virulence by modulating host innate immune responses of macrophages. For example, elicitation of type I interferon (IFN) by M. tuberculosis infection of both murine and human macrophages requires the ESX-1 secretion system (Novikov et al., 2011; Stanley et al., 2007). Although type II IFN (IFN-γ) is critical for activating host defenses, type I IFNs (IFN-α and IFN-β) can negatively regulate host resistance to M. tuberculosis in mouse models of infection (Manca et al., 2001). Bacterial pathogens that replicate in the cytoplasm of macrophages, such as Listeria monocytogenes and Franciscella tularensis, induce IFN-β transcription as part of a large transcriptional response controlled by the “cytosolic surveillance pathway” (CSP) (Henry and Monack, 2007; McCaffrey et al., 2004). CSP activation by these species occurs early after infection specifically by cytoplasmic bacteria, whereas mutants unable to breach phagosomal membranes fail to induce transcription (Henry and Monack, 2007; Leber et al., 2008). The CSP is controlled by the host transcription factor interferon regulatory factor 3 (IRF3), which is activated via phosphorylation by the TBK1 kinase. It is thought that upon phagosomal membrane rupture, bacterial products are granted cytosolic access and recognized by cytosolic receptors that lead to IRF3 activation (Vance et al., 2009). Although IRF3 activation of the CSP induces transcription of a wide range of immune response genes, including many known to be important for antiviral defense, the role of the CSP during bacterial infection is still unclear (Monroe et al., 2010).
IRF3 is activated by multiple pattern recognition receptors (PRRs), including Toll like receptors (TLRs), RIG like receptors (RLRs) and cytosolic DNA receptors (Barber, 2011). Two putative DNA receptors, DAI (Takaoka et al., 2007) and IFI204 (the mouse homolog of IFI16, (Unterholzner et al., 2010)), have been proposed to be sensors that initiate the CSP. Despite some controversies over the nature of DNA receptors, Sting is a critical adapter that clearly functions downstream of the putative sensor in the pathway, linking TBK1 and IRF3 (Ishikawa et al., 2009). Interestingly, though the host requirements for CSP activation by intracellular L. monocytogenes is identical to that of cytosolic DNA, recent work has implicated bacterial derived cyclic diadenosine monophosphate (c-di-AMP) as the relevant trigger (Woodward et al., 2010).
In contrast to cytosolic bacterial pathogens, M. tuberculosis has long thought to reside within membrane-bound phagosomes of host cells (Russell, 2011). However, one group recently reported that M. tuberculosis translocates into the host cytosol and that the ESX-1 system is required for this process (van der Wel et al., 2007). Mycobacterium marinum, an ectothermic pathogen related to M. tuberculosis that escapes from the vacuole, also requires ESX-1 secretion for cytosolic access (Smith et al., 2008). Because ESAT-6, a major secreted protein of the ESX-1 system, has membrane lytic properties at high concentrations (de Jonge et al., 2007; Hsu et al., 2003), it has been suggested that this protein is responsible for ESX-1 mediated cytosolic access.
We report that M. tuberculosis induces IFN production during macrophage infection via activation of the CSP from within the phagosome. This response requires the IFI204 DNA receptor which stimulates the STING/TBK1/IRF3 signaling axis, the same host components as required for the interferon stimulatory DNA (ISD) pathway (Ishikawa et al., 2009; Stetson et al., 2008). This is in contrast to previously published work describing an obligate role for peptidoglycan-mediated Nod signaling and IRF5 in IFN induction (Pandey et al., 2009). We provide multiple pieces of evidence that extracellular mycobacterial DNA is the critical ligand for CSP activation and that the ESX-1 system allows extracellular mycobacterial DNA access to cytoplasmic DNA receptors via limited permeabilization of the phagosomal membrane. Surprisingly, CSP activation is required for M. tuberculosis pathogenesis as IRF3−/− mice are profoundly resistant to infection.
Results
M. tuberculosis elicits the CSP via ESX-1 secretion
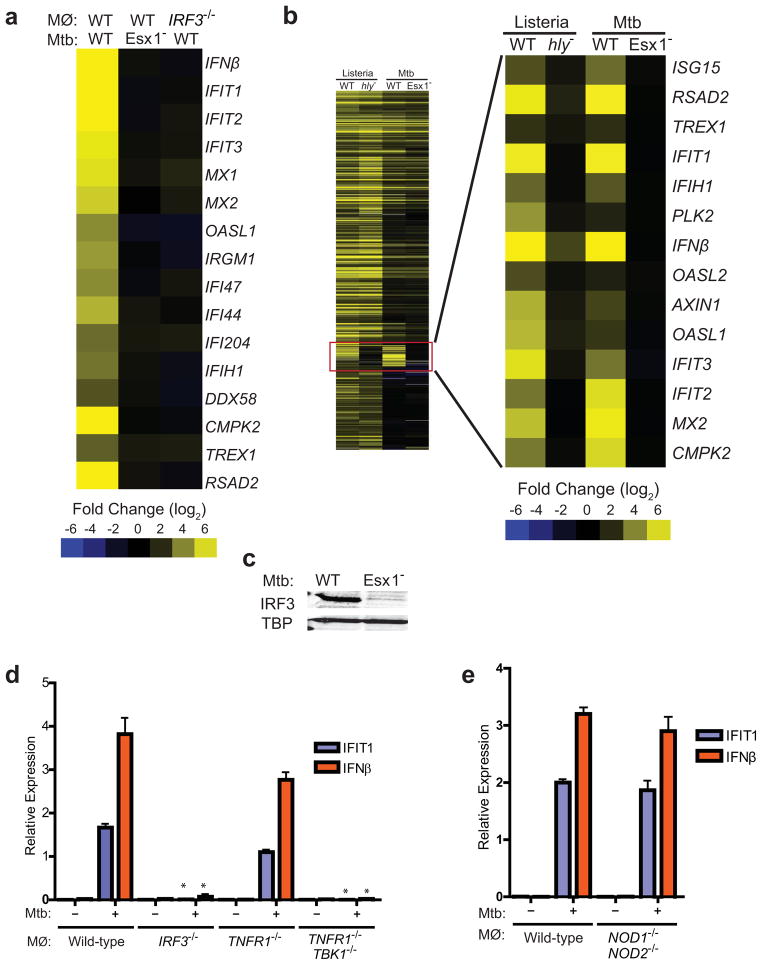
To understand how the host responds to infection with ESX-1+M. tuberculosis, we used microarrays to probe the early transcriptional response of murine bone marrow-derived macrophages (BMDMs) to infection with either wild-type or ESX-1 mutant (Tn5370::Rv3877/EccD1) M. tuberculosis cells. Overall, the transcriptional profile of macrophages in response to either strain was remarkably similar, with signatures of TLR/NFκB signaling predominating in both data sets. However, closer inspection of the data revealed that of the 861 genes significantly activated by wild-type M. tuberculosis, 162 genes were differentially expressed upon infection with ESX-1 mutant cells (Table S1). Genes in this ESX-1-dependent regulon include interferon-stimulated genes (ISGs) such as IFIT1, IFN-β, and Viperin (RSAD2), all of which are hallmarks of the cytosolic surveillance pathway (CSP) (Leber et al., 2008) (Figure 1A). RT-qPCR experiments monitoring the expression of several CSP genes over a more detailed time course demonstrated that activation was evident as early as 1h post-infection, with a peak of expression at 3h which ultimately leveled off by 5h post-infection (Figure S1A).
Figure 1. M. tuberculosis elicits the CSP via ESX-1 secretion.
(A) Microarray analysis of CSP-regulated genes in wild-type and IRF3−/− BMDMs infected with the indicated M. tuberculosis strains. Log2 fold induction versus uninfected macrophages is shown. (B) Cluster analysis of BMDM genes induced during infection with wild-type (WT) or Δhly L. monocytogenes (hly−), and with wild-type or ESX-1 mutant (Esx1−) M. tuberculosis. (C) Nuclear translocation of IRF3 in BMDMs 3 h post-infection was assessed by western blotting of nuclear fractions using IRF3 and TATA-binding protein (TBP)-specific antibodies. (D and E) IFIT1 and IFN-β mRNA levels were assessed by qRT-PCR 3 h post-infection in BMDMs (MΦ) and normalized to actin. TBK1−/− mice are viable only when TNFα signaling is abrogated, thus BMDMs from TNFR1−/− mice serve as the control for this strain. Data shown is the mean ± SD, (N=3 per group). *P<0.001 as determined by Student’s t-test comparing gene expression in each mutant macrophage and its corresponding control. See Figure S1 and Table S1 for a detailed time course analysis of CSP gene induction and microarray expression data.
Comparison of our microarray data with published bacterial response datasets deposited in the NCBI GEO database (GSE8104) revealed that the ESX-1-dependent transcriptional profile was nearly identical to the response to L. monocytogenes infection after it enters into the cytosol via the action of its pore-forming toxin, listeriolysin O (LLO) (Figure 1B, (Leber et al., 2008)). Activation of the CSP by either L. monocytogenes or cytoplasmic DNA is controlled by the host transcription factor interferon regulatory factor 3 (IRF3), which is activated via phosphorylation by the kinase TBK1 (Fitzgerald et al., 2003; Leber et al., 2008). The remarkable overlap in the transcriptional response with L. monocytogenes infection strongly suggested that M. tuberculosis also activated the IRF3 dependent cytosolic response (Leber et al., 2008). Indeed, transcriptional profiling of wild-type and IRF3−/− macrophages infected with wild-type M. tuberculosis revealed that the majority of ISGs specifically induced by ESX-1+ bacteria are also IRF3 dependent (Figure 1A, Table S1). Likewise, IRF3 was activated and translocated to the nucleus upon infection with wild-type M. tuberculosis but not with ESX-1 mutant cells (Figure 1C). Quantitative PCR analysis of both IFIT1 and IFN-β mRNA revealed that CSP activation is strictly dependent on IRF3 and ESX-1 during M. tuberculosis infection (Figure 1D, Figure S1C). BMDMs lacking TBK1 also failed to induce IFIT1 and IFN-β transcription or IRF3 nuclear translocation in response to M. tuberculosis infection, displaying defects similar to IRF3−/− cells (Stanley et al., 2007) (Figure 1D, 3E). Taken together, these results demonstrate that M. tuberculosis activates the IRF3-dependent cytosolic response in an ESX-1 dependent manner.
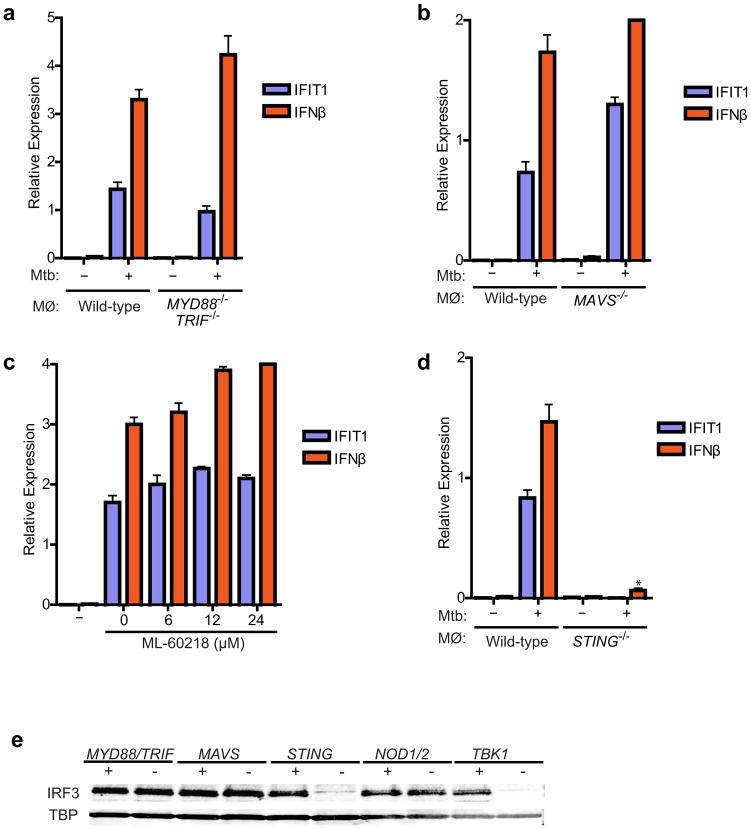
Figure 3. M. tuberculosis activates the STING/TBK1/IRF3 pathway.
(A, B) IFIT1 and IFN-β mRNA levels were assessed by qRT-PCR in BMDMs (MΦ) as in Figure 1. Data shown is the mean ± SD, (N≥3 per group). (C) Wild-type C57L/B6 BMDMs were pre-treated for two-hours with the RNA polymerase III inhibitor ML-60218 and infected with wild-type M. tuberculosis for 3 hours prior to RNA extraction. Data shown is the mean ± SD, (N=3 per group). (D) IFIT1 and IFN-β mRNA levels were assessed 3 h post-infection of STING−/− macrophages wild-type M. tuberculosis. Data shown is the mean ± SD, (N=3 per group). *P<0.001 as determined by Student’s t-test. (E) Nuclear translocation of IRF3 in BMDMs was determined as described in Figure 1C.
These results, as well as other published work (Leber et al., 2008; Stockinger et al., 2004), are inconsistent with a recent report that type I IFN induction requires MDP-initiated Nod signaling and IRF5 (Pandey et al., 2009). This was surprising as we demonstrated previously that Rip2, a component absolutely required for Nod signaling, is dispensable for activation of IFN-β transcription during M. tuberculosis infection of macrophages (Stanley et al., 2007). To independently test the contribution of the putative Nod/IRF5 pathway in CSP activation, we infected macrophages from NOD1−/−NOD2 −/− double knockout mice with M. tuberculosis. These macrophages generated robust IRF3 nuclear translocation and produced amounts of IFN-β mRNA that were indistinguishable from wild-type cells (Figures 1E and 3E). Similarly, macrophages from IRF5−/− mice were also able to activate IFN-β mRNA transcription to nearly wild-type levels. Although there was a 2-fold decrease in mRNA levels compared to various IRF knockout strains, this was in sharp contrast to IRF3−/− macrophages that are completely blocked for IFN-β induction (Figure S1B). While we cannot completely account for the discrepancies between the two studies at present, the recent finding that many of the phenotypes associated with the IRF5−/− knockout mouse, including Type I IFN induction, are due to a spontaneous mutation in the DOCK2 gene suggests that a reevaluation of the role of IRF5 in IFN activation is warranted. Despite these discrepancies, our results support the model that, like with L. monocytogenes infection (Leber et al., 2008), initial CSP activation by M. tuberculosis critically requires the TBK1/IRF3 pathway.
M. tuberculosis permeabilizes the phagosomal membrane early after infection
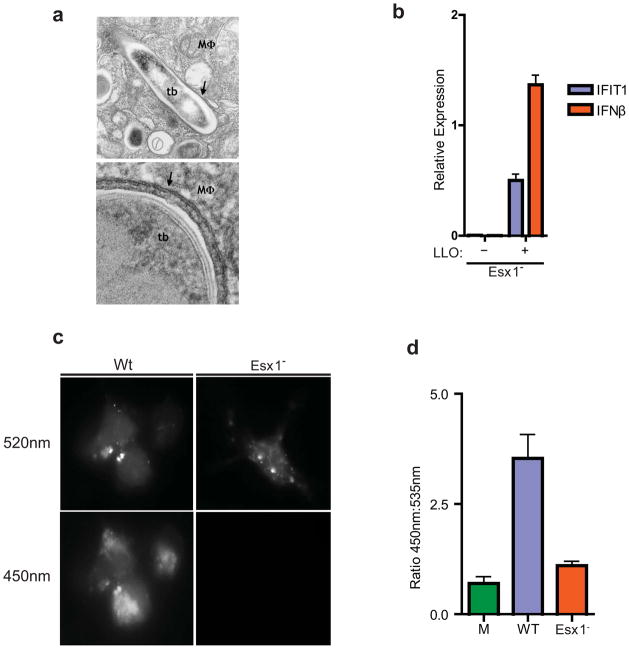
The concordance between CSP activation by L. monocytogenes, a cytosolic pathogen, and M. tuberculosis, a vacuolar pathogen, was unexpected given the seemingly different pathogenic strategies of these two microbes (Russell, 2011). However, ESX-1 has been reported to allow M. tuberculosis to rupture the phagosomal membrane (van der Wel et al., 2007). Importantly, van der Wel et al. observed M. tuberculosis in the cytosol only after several days of infection, whereas we detected robust CSP activation as early as three hours post-infection. Electron microscopy studies confirmed that all bacteria at this early time point were clearly encircled by phagosomal membranes (Figure 2A). Thus, it is likely that early CSP activation represents limited, ESX-1 mediated perforation of the phagosomal membrane early after infection rather than wholesale membrane dissolution.
Figure 2. M. tuberculosis permeabilizes the phagosomal membrane early after infection.
(A) Electron microscopy of C57/B6 BMDMs infected with wild-type M. tuberculosis 3 h post-infection. Macrophage cytosol (MΦ), M. tuberculosis cell (tb) and intact phagosomal membrane (arrows) are indicated. (B) IFIT1 and IFN-β mRNA levels were assessed by qRT-PCR (actin normalized) in BMDMs infected with ΔesxAM. tuberculosis cells containing an LLO expression plasmid or empty vector. Data shown is the mean ± SD, (N=3 per group). *P<0.001 as determined by Student’s t-test comparing gene expression in each mutant macrophage and its corresponding control. (C) Macrophages were preloaded with CCF4-am substrate and infected with wild-type or ΔesxAM. tuberculosis strains, both of which over-expressed the blaC gene. 3 h post infection, cells were excited with a 405nm laser, and percent CCF4 cleavage was measured as the ratio of 450nm:535nm emission by fluorescence microscopy of infected cells (D). Data shown is the mean ± SD, (N=3 per group).
To begin to test if the requirement for ESX-1 in CSP activation was due to membrane permeabilization, we infected macrophages with ESX-1 mutants expressing an auto-activated form of LLO from L. monocytogenes (Singh et al., 2008). As shown in Figure 2B, LLO expression restored CSP activation to ESX-1 mutant cells, demonstrating that membrane permeabilization, and not substrates secreted by ESX-1 per se, is sufficient for CSP activation. To test if ESX-1 functions to permeabilize the phagosomal membrane during infection, we employed a fluorescent β-lactamase assay previously used to measure vacuolar rupture of phagosomes (Ray et al., 2010). This provides a direct way to detect mixing of phagosomal contents with the cytosol during the course of infection through the use of CCF4, a membrane impermeable FRET probe. CCF4 contains a coumarin and a fluorescein molecule connected by a β-lactam substrate (Ray et al., 2010). After loading the cytosol with CCF4, excitation of coumarin leads to efficient FRET and emission of green (520nm) light by fluorescein, as there is no β-lactamase present in eukaryotic cells. If the substrate is cleaved by a β-lactamase, however, FRET is inhibited and coumarin emits blue fluorescence (450nm) upon excitation. Infection of CCF4-loaded BMDMs with wild-type M. tuberculosis over-expressing the secreted mycobacterial β-lactamase BlaC led to blue fluorescence by 3 h post infection, indicating mixing of phagosomal contents with the cytosol (Figure 2C). Importantly, ESX-1 mutant bacteria over-expressing BlaC led to no cleavage of the CCF4 substrate. Taken together, these results demonstrate that the ESX-1 secretion system leads to rapid permeabilization of the phagosomal membrane upon macrophage infection. Thus, ESX-1-mediated activation of the CSP by M. tuberculosis infection requires phagosomal membrane permeabilization, but not its complete dissolution.
M. tuberculosis activates the STING/TBK1/IRF3 pathway
TBK1 is activated by a variety of pattern recognition receptors, including Toll-like receptors (TLR), cytosolic RNA receptors, and cytosolic DNA receptors (Nakhaei et al., 2009). To determine which pathway is activated by M. tuberculosis, we monitored CSP activation in a variety of mouse knockout cells. Macrophages deficient either for TLR signaling (MYD88−/− TRIF−/−) or RNA sensing (MAVS−/−) were indistinguishable from wild-type cells, producing similar amounts of IFIT1 and IFN-β transcription and IRF3 nuclear translocation in response to M. tuberculosis infection (Figures 3A, 3B and 3E). Furthermore, chemical inhibition of the RNA-Polymerase III/Rig-I pathway (Chiu et al., 2009) had no effect on M. tuberculosis activation of CSP transcripts in BMDMs (Figure 3C).
Sting is an adapter protein specifically required for both DNA and cyclic di-nucleotide mediated activation of IRF3 and IFN-β transcription (Burdette et al., 2011; Ishikawa et al., 2009). To test if M. tuberculosis activates the CSP via Sting, we infected STING−/− BMDMs and monitored CSP activation. Surprisingly, macrophages lacking Sting were unable to activate IRF3 translocation and CSP transcription upon infection (Figure 3D, 3E). Additionally, in wild-type macrophages, Sting translocated from the ER to cytosolic puncta in an ESX-1 dependent manner (data not shown). Thus, Sting is an essential component required for M. tuberculosis-activation of IRF3.
We next determined the bacterial ligand necessary for CSP activation via the STING/TBK1/IRF3 pathway during M. tuberculosis infection. L. monocytogenes produces c-di-AMP that elicits the CSP (Woodward et al., 2010), and Sting has recently been found to be a direct sensor of cyclic di-nucleotides (Burdette et al., 2011; McWhirter et al., 2009). c-di-GMP is unlikely to be the trigger of the CSP as the Erdman strain used in our studies contains a genomic deletion, common in several TB clinical isolates, that removes the only identified di-guanylate cyclase, MT1397 (Tsolaki et al., 2004). In addition, infection with the CDC1551 strain of M. tuberculosis that contains MT1397 elicited similar levels of IFN-β transcription as the Erdman strain, and over-expression of MT1397 in the Erdman strain had no effect on IFN-β induction (Figure S2A). Likewise, M. tuberculosis infection of macrophages expressing the c-di-GMP phosphodiesterase, RocR, had no effect on IFN-β production (data not shown) (Burdette et al., 2011; McWhirter et al., 2009). Furthermore, infection of BMDMs with strains of Erdman either lacking or overexpressing the sole di-adenylate cyclase gene in M. tuberculosis, disA (Rv3586), led to equivalent levels of IFN-β production (Figures S2B, S2C, and S2D). Taken together, our data suggests that mycobacterial derived c-di-GMP and c-di-AMP are not responsible for M. tuberculosis-mediated CSP activation.
The cytosolic DNA sensor IFI204/IFI16 contributes to CSP activation
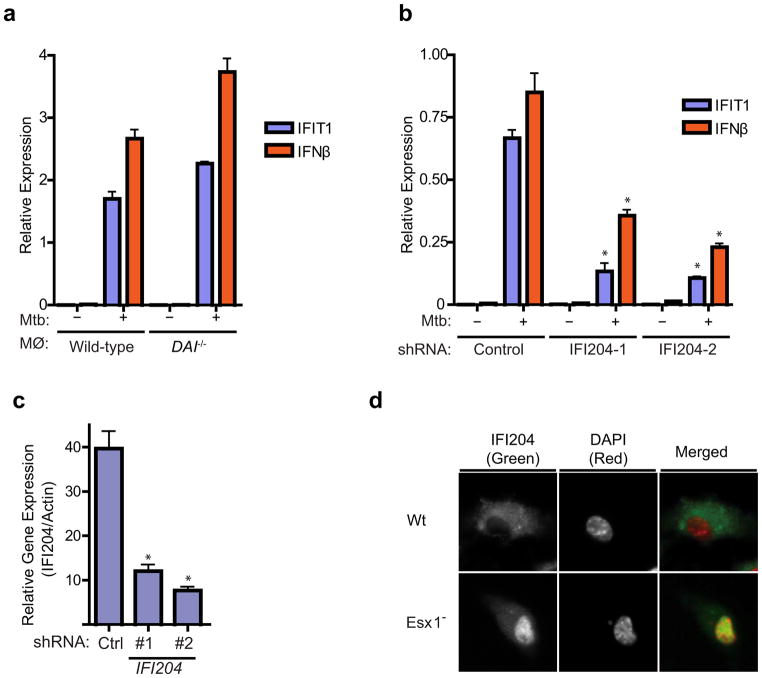
We next explored the possibility that M. tuberculosis DNA is the trigger for activation of Sting and the CSP response. We first tested the role of two reported cytosolic DNA sensors, DAI (ZBP1) and IFI16/IFI204, in M. tuberculosis induction of the CSP (Takaoka et al., 2007; Unterholzner et al., 2010). Although BMDMs from DAI−/− mice responded normally to both DNA transfection and M. tuberculosis infection (Figure 4A), knockdown of IFI204 expression (the mouse homolog of human IFI16) in immortalized BMDMs significantly reduced IFIT1 and IFN-β induction upon M. tuberculosis infection (Figures 4B and 4C), suggesting that IFI16/IFI204 and AIM-2 like receptors (ALRs), may serve as the predominant DNA sensors in BMDMs (Unterholzner et al., 2010). In addition, wild-type M. tuberculosis induced cytosolic translocation of IFI204, consistent with its relocalization upon DNA transfection and IFN stimulation (Unterholzner et al., 2010) (Figure 4D). Taken together, these results demonstrate that the IFI204 cytosolic DNA sensor contributes to the ESX-1-specific response of macrophages.
Figure 4. The cytosolic DNA sensor IFI204/IFI16 contributes to CSP activation.
(A) IFIT1 and IFN-β mRNA levels were assessed by qRT-PCR in BMDMs (MΦ) from wild-type and DAI knockout mice infected with wild-type M. tuberculosis for 3 h. Data shown is the mean ± SD, (N=3 per group). (B) IFIT1 and IFN-β mRNA levels were assessed by qRT-PCR after M. tuberculosis infection of immortalized BMDMs (iBMDMs) transduced with lentiviral constructs expressing one of two different shRNAs targeting IFI204 (IFI204–1 and IFI204–2) or a scrambled shRNA control. Data shown is the mean ± SD, (N=3 per group). *P<0.005 by Student’s T-test. (C) IFI204 mRNA levels were assessed by qRT-PCR in the transduced iBMDMs described in (B). *P<0.005 by Student’s T-test. (D) Wild-type C57L/B6 BMDMswere infected with either wild-type or ΔesxAM. tuberculosis for 3 hours and cellular localization of IFI204 as assessed via immunofluorescence. See Table S2 for shRNA sequences and primers used for this study.
Extracellular M. tuberculosis DNA triggers the CSP
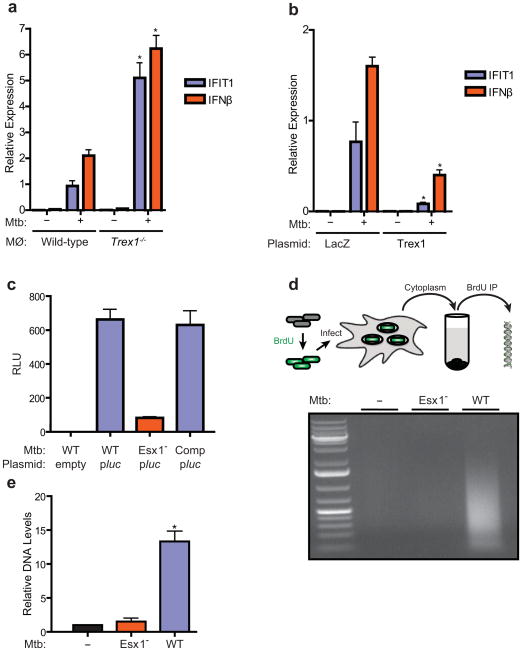
To further explore the link between cytosolic DNA responses and M. tuberculosis infection, we determined whether cytosolic DNAses could modulate CSP activation during M. tuberculosis infection. Trex1 is a cytosolic DNase that negatively regulates the ISD pathway by decreasing the half-life of cytoplasmic DNA, and mice lacking TREX1 display enhanced IFN-β production in response to cytosolic DNA but not other inducers such as RNA (Stetson et al., 2008; Yan et al., 2010). Importantly, infection of TREX1−/− knockout macrophages with wild-type M. tuberculosis led to over three-fold increased induction of IFN-β and IFIT1 as compared to control cells (Figure 5A). Conversely, over-expression of TREX1 significantly reduced IFIT1 and IFN-β transcription during M. tuberculosis infection (Figure 5B). This data strongly suggests that extracellular M. tuberculosis DNA (eDNA) exposed to the macrophage cytoplasm is the trigger for IRF3 activation.
Figure 5. Extracellular M. tuberculosis DNA triggers the CSP.
(A) IFIT1 and IFN-β mRNA levels were assessed by qRT-PCR in infected BMDMs (MΦ) as described in Figure 1. (B) RAW264.7 cells were stably transfected with a plasmid over-expressing either Trex1 or LacZ and infected with wild-type M. tuberculosis for 4 h. IFIT1 and IFN-β gene expression was analyzed by qRT-PCR. (C) Luminescence readings from BMDMs infected with either wild-type (Wt), ESX-1 mutant (Esx1−), or complemented ESX-1 mutant (Comp) M. tuberculosis carrying either empty vector or a plasmid encoding a secreted form of Metridia luciferase under the control of the eukaryotic-specific CMV promoter (pMAN4). Luminescence readings were taken at 16 h post infection. (D) M. tuberculosis cells were labeled with BrdU prior to macrophage infection. After 3 h, macrophage cytosolic fractions were isolated and BrdU-labeled bacterial DNA was recovered by immunoprecipitation using anti-BrdU antibodies, and separated by agarose gel electrophoresis. (E) Lysates from (D) were used as templates for qPCR quantification of M. tuberculosis DNA. Data shown for (A) (B) and (D) is the mean ± SD, (N=3 per group). *P<0.01 by Student’s T-test, comparing mutant and overexpression strains to their corresponding controls. Also see Figure S2 for analysis of M. tuberculosis c-di-nucleotide mutants in CSP activation and Table S3 for M. tuberculosis strains used in this study.
Although it was curious that DNA would be a ligand during bacterial infection, we reasoned that eDNA, perhaps released as a result of bacterial lysis, may be the ligand that triggers the CSP. To test this idea, we first asked whether M. tuberculosis was capable of transferring DNA into infected macrophages. We created an M. tuberculosis strain carrying an episomal plasmid encoding a luciferase reporter gene under the control of eukaryotic expression signals, including the CMV promoter (Sauer et al., 2010). The enzyme also contains an N-terminal signal sequence that directs its secretion from eukaryotic cells. As expected, this bacterial strain produced no luminescence signal when grown in culture, though directly transfecting the plasmid into human and mouse cell lines resulted in robust luminescence activity in cell supernatants (not shown). Infection of macrophages with the luciferase reporter strain gave rise to significant luminescence signal during infection (Figure 5C). Importantly, infection with ESX-1 mutant bacteria carrying the same reporter construct failed to elicit any luminescence signal. These results strongly suggest that mycobacterial DNA is released into the macrophage cytosol during infection in an ESX-1 dependent manner.
To directly detect M. tuberculosis eDNA within the macrophage cytosol, we infected macrophages with BrdU-labeled M. tuberculosis, gently lysed the cells, and then fractionated via centrifugation to obtain a cleared cytosolic extract (Figure 5D). Immunoprecipitation using anti-BrdU antibodies, followed by agarose gel electrophoresis, revealed a significant enrichment of M. tuberculosis DNA from macrophages infected with wild-type versus ESX-1 mutant bacteria (Figure 5D). Quantification of M. tuberculosis DNA from the immunoprecipitate using qPCR primers for the sigF gene showed greater than 10-fold enrichment of DNA recovered from wild-type infected cells over mock or ESX-1 mutant infected cells (Figure 5E). These results demonstrate that the ESX-1 system exposes M. tuberculosis eDNA to the host cytosol, which then triggers the activation of the cytosolic surveillance pathway via activation of the STING/TBK1/IRF3 axis.
Cytosolic signaling is required for virulence of M. tuberculosis
A growing body of work indicates that ESX-1 contributes to M. tuberculosis virulence by modulating host innate immune responses of macrophages (Stanley et al., 2007). Thus, activation of the CSP by M. tuberculosis may be an important virulence mechanism. To test the functional role of the CSP during M. tuberculosis infection, we performed low-dose aerosol infections of IRF3−/− mice and congenic C57BL/6 controls. Surprisingly, while wild-type mice began to succumb at approximately 120 days post infection, all IRF3−/− mice survived more than one year, suggesting that activation of IRF3 by M. tuberculosis is deleterious to the host (Figure 6A). Enumeration of colony forming units (CFUs) from lungs and spleens of infected mice revealed that though the kinetics of bacterial replication during the acute phase of infection was identical in IRF3−/− and wild-type mice, M. tuberculosis viability declined continuously during the chronic stage of infection in IRF3−/− mice (Figure 6B, 6C). Cytokine analysis of mouse serum demonstrated decreased levels of IFN-β protein in IRF3−/− mice relative to wild-type mice, demonstrating that IRF3 controls M. tuberculosis induction of type I IFNs and ISGs during infection (Figure 6D). In addition, serum from infected IRF3−/− mice had reduced levels of the chemokines RANTES and MCP-1, and increased levels of IL-12p70 and IL-1β (Figure 6D). Collectively, this data demonstrates that IRF3 activation plays a critical role to promote M. tuberculosis virulence, likely driven by DNA mediated activation of the CSP, and the induction of cytokines including type I IFN.
Figure 6. Cytosolic signaling is required for virulence of M. tuberculosis.
(A) Kaplan-Meier survival analysis of IRF3−/− and C57BL/6 control mice infected with 102 CFU wild-type M. tuberculosis via the aerosol route (n = 10 per group). IRF3−/− mice showed significantly improved survival compared to wild-type (WT) mice as calculated by log-rank test (**P < 0.001). (B, C) Bacterial burdens as measured by colony forming units (CFU) in the lungs (B) and spleen (C) of IRF3−/− and wild-type mice infected as described in (A) (n = 5 per time point) at the indicated time points. Data shown are the mean ± SD. *P < 0.02 and **P <0.005 comparing C57BL/6 and IRF3−/− mice by Kruskal Wallis. (D) IFN-β, Rantes, MCP1, IL-12 and IL-1β levels were measured by ELISA in serum from IRF3−/− and C57BL/6 control mice 21 days post infection (n = 4 per group). Data shown is the mean ± SD. *P < 0.005 comparing C57BL/6 and IRF3−/− mice as determined by Student’s T-test.
Discussion
We have identified a mechanism by which virulent M. tuberculosis, a vacuolar pathogen, elicits the same innate immune transcriptional response induced by cytoplasmic pathogens. The requirement for the mycobacterial ESX-1 secretion system to elicit interferon stimulatory genes from macrophages has been well documented (Novikov et al., 2011; Schnappinger et al., 2003; Stanley et al., 2007), but how this occurred was unknown. By showing that host molecules required for sensing of DNA in the cytoplasm (IFI204, Sting, and Trex1) are also involved in the response to ESX-1+ M. tuberculosis, as well as identifying mycobacterial DNA specifically in the cytoplasm in an ESX-1 specific fashion, we conclude that M. tuberculosis triggers the CSP via exposure of extracellular mycobacterial DNA to the cytosol of macrophages. Importantly, this response is independent of TLR/Nod signaling and distinct from cytoplasmic RNA sensing molecules. Recognition of DNA by IFI204 leads to phosphorylation of IRF3 by TBK1 and, ultimately, the induction of a defined set of approximately 160 genes. Importantly, mice lacking IRF3 are more resistant to M. tuberculosis infection. While this is consistent with a role of type I IFN playing a negative role in host resistance, the phenotype of the IRF3−/− mouse is likely due to many factors besides type I IFN, as discussed below. Moreover, sensing of M. tuberculosisDNA likely takes place during human tuberculosis, as ESX-1 mediated activation of the CSP is operative in human macrophages and is likely responsible for the robust type I IFN signature associated specifically with active disease (Berry et al., 2010; Novikov et al., 2011).
Although Type I IFN is absolutely critical for resistance to viruses, there is growing literature about the role of IFN-α/β in bacterial infections (Barber, 2011; Monroe et al., 2010). Because type I IFN inhibits IL-1β, a cytokine that promotes M. tuberculosis clearance, it suggests that differential cytokine responses mediated by IFN-α/β contribute to the phenotype of the IRF3−/− mouse (Novikov et al., 2011). It is important to note that while IFN-α/β likely plays a role in this phenotype, the IRF3−/− mouse is much more resistant to M. tuberculosis infection than the IFNAR−/− mouse that is only deficient for type I IFN signaling (Stanley et al., 2007). These results indicate that other immune modulators regulated by the CSP contribute to pro-M. tuberculosis inflammatory immune responses.
Given the evidence that ESAT-6 perturbs membranes (de Jonge et al., 2007; Hsu et al., 2003), we propose that M. tuberculosis utilizes the ESX-1 system to deliver ESAT-6 to create conduits in phagosomal membranes, allowing bacterial eDNA to contact cytosolic receptors and activate the CSP. Since activation occurs prior to overt lysis of the phagosomal membrane (van der Wel et al., 2007), our results suggest that ESX-1 mediates limited perforation of the lipid bilayer early after infection. Because virulence-associated protein secretion systems activate cytosolic responses in other pathogens (Monroe et al., 2010), we envision that the primary role of ESAT-6-mediated membrane damage is to provide access of secreted virulence factors to enter the host cytosol, and eDNA exposure to cytosolic receptors is an indirect consequence of membrane perforation. Furthermore, the ability of M. tuberculosis to perforate the membrane without its dissolution is likely under a delicate balance of factors, and fine control of ESX-1 secretion may serve to limit membrane perforation and cytosolic signaling (Ohol et al., 2010).
Identification of mycobacterial DNA as the ligand raises the question of how eDNA is liberated and exposed during M. tuberculosis infection. Although it could be released from bacteria that have lysed after phagocytosis, the fact that CSP activation occurs within hours after infection of naïve macrophages, conditions in which bacterial viability is very high, argues against this idea. Alternatively, eDNA may be naturally occurring on the surface of the bacterium during normal growth, and thus pre-existing prior to infection. Indeed, a strong body of literature supports the notion that eDNA can play important roles in normal bacterial physiology, most notably biofilm formation (Whitchurch et al., 2002). Furthermore, eDNA has been identified within outer membrane vesicles secreted by various gram-negative and gram-positive bacterial species (Ellis and Kuehn, 2010) and has recently been found to be encapsulated within mycobacterial membrane vesicles (Prados-Rosales et al., 2011) (personal communication). Thus, we favor a model in which the mycobacterial eDNA sensed by the macrophage also plays a fundamental role in mycobacterial cell biology that is distinct from CSP triggering, perhaps to promote biofilm formation.
Although our data support the involvement of eDNA as the ligand that triggers IFN-β, the work of Pandey et al. suggests that peptidoglycan fragments detected by cytosolic Nod receptors is responsible for initiating the response (Pandey et al., 2009). While we cannot account for this discrepancy, our previous work showed that the effect of Nod activation of NFκB on IFN-β induction was apparent only in cells in which TLR signaling was inhibited by chronic stimulation, suggesting that it plays only a minor role in the initiation of the CSP (Leber et al., 2008). While Nod2 can contribute partially to IFN-β production by modulating NFκB activity in response to M. tuberculosis and L. monocytogenes infection, Nod signaling alone does not appear to affect IRF3 activation (Balachandran and Beg, 2011; Leber et al., 2008). Moreover, the finding that the course of infection in NOD2−/− mice is similar to that in wild-type mice demonstrates that this signaling pathway plays only a minor role in M. tuberculosis control (Divangahi et al., 2008). Overall, our data supports the conclusion that extracellular DNA is the incipient ligand that is recognized and activates the CSP in vivo.
Since bacterial infection correlates with type I IFN production, it is tempting to speculate that CSP induction is a “strategy” adopted by M. tuberculosis to promote infection, and that the ability of the bacteria to expose eDNA arose for the sole purpose of triggering this response (Novikov et al., 2011). The phenotype of the IRF3−/− mouse is certainly consistent with this notion. However, if eDNA provides a fitness advantage in vivo, an alternative view is that eDNA exposure to the cytosol is an inevitable consequence of membrane perforation that allows virulence proteins a passageway into the cytosol. In this way, while CSP activation initiates profound inflammatory responses, M. tuberculosis may have evolved to require these changes in host tissues to activate virulence mechanisms or produce an environment conducive to growth. Consistent with this idea, recent studies have shown that Salmonella promotes inflammatory immune responses which, in turn, enhances persistent infection (Arpaia et al., 2011; Winter et al., 2010). Thus, M. tuberculosis may have evolved a dependency that requires the effects of robust innate immune signaling triggered by DNA. In either case, it is likely that excessive host immunopathology triggered by CSP activation contributes to the susceptibility of wild-type mice to M. tuberculosis (Philips and Ernst, 2012).
Although CSP activation via detection of nucleic acids is a key mechanism by which cells sense intracellular pathogens, cytosolic DNA sensing likely initiates other innate immune responses in addition to the transcriptional changes described here. For example, Franciscella tularensis, a cytosolic pathogen, activates the DNA receptor AIM2, leading to Caspase1 inflammasome activation (Fernandes-Alnemri et al., 2010). Likewise, links between nucleic acid sensing and autophagy, an important mediator of bacterial clearance, are also beginning to emerge (Saitoh et al., 2009). Hence, eDNA generation may be common to a wide variety of bacterial species, and its exposure to the cytosol during infection allows host cells to elicit a multifactorial innate response. Furthermore, the placement of DNA receptors in the cytosol allows host cells to specifically elicit these responses only in response to bacteria that access the cytosol (pathogens) verses those that do not (non-pathogens) (Vance et al., 2009).
Experimental Procedures
Electron Microscopy
BMDMs were seeded onto Aclar disks in DME medium within 12-well tissue culture dishes and grown for 48 h prior to infection. Monolayers were infected with M. tuberculosis at an MOI of 10 for 4 h and adherent cells were fixed in 3% glutaraldehyde in 0.1M Sodium cacodylate for 2 h. Fixed cells were rinsed in sodium cacodylate buffer, stained with uranyl acetate, embedded and sectioned.
Mice and macrophages
IRF3−/− mice were a gift from T. Taniguchi (University of Tokyo, Tokyo, Japan). Wild-type C57BL/6 mice were purchased from Jackson laboratories. BMDMs were obtained from the following mouse strains: MYD88−/−/TRIF−/− (Barbalat et al., 2009), TREX1−/− (Stetson et al., 2008), STING−/− (Ishikawa et al., 2009), TNFR1−/− & TBK1−/−/TNFR1−/− (Ishii et al., 2008), IFNAR1−/−, IRF1−/−, IRF3−/−, IRF5−/−, and IRF7−/− (Takaoka et al., 2007). RAW264.7 cells were obtained from ATCC. C57BL/6 immortalized BMDMs (iBMDMs) were a gift from R. Vance (UC Berkeley).
Cell culture
RAW264.7 cells were cultured in DMEM-H21 containing 10%FBS. Immortalized BMDMs (iBMDMs) were cultured in RPMI-1640 containing 10% FBS. BMDMs were obtained from mouse femurs as previously described (Leber et al., 2008) and cultured in DMEM H-21 supplemented with 10% MCSF derived from 3T3-MCSF cells.
Bacterial strains
Unless noted, the Erdman strain of M. tuberculosis was used as the wild-type strain. The CDC1551 M. tuberculosis strain was obtained from the Colorado State TBVTRM contract. The M. tuberculosis Erdman mutants ΔesxA and Tn5370::Rv3877/EccD1 were previously described (Stanley et al., 2003). The M. tuberculosisRv3676 deletion mutant (ΔdisA) was constructed by replacing the entire Rv3676 open reading frame with the hygromycin resistance gene by homologous recombination. The deletion plasmid was delivered into M. tuberculosis using specialized phage transduction as described previously (Ohol et al., 2010). Deletion of Rv3676 was confirmed by Southern blotting using the DIG nucleic acid detection kit (Roche). All strains were cultured in 7H9 medium supplemented with 10% OADC, 0.5% Glycerol, and 0.05% Tween-80.
Macrophage infection
Macrophages were infected as previously described (Ohol et al., 2010), with some modifications. Briefly, M. tuberculosis cultures were washed twice with PBS, gently sonicated to disperse clumps, and resuspended in DMEM supplemented with 10% horse serum. Media was removed from cells, monolayers overlaid with the bacterial suspension, and centrifuged for 10 min at 1,000 RPM. Cells were washed twice in PBS and returned to macrophage media.
Cellular fractionation and western blotting
Cytosolic and nuclear protein fractions were obtained from M. tuberculosis infected macrophages using the Qproteome Nuclear Protein Kit (Qiagen) according to manufacturer’s instructions. Micro BCA protein kit (Pierce) was used to measure protein levels and equal amounts of protein were run on 4–20% Tris-HCL Criterion gels (Biorad). Proteins were transferred onto nitrocellulose membranes and incubated simultaneously with anti-IRF3 (Invitrogen) and anti-TBP (Abcam) antibodies. Western blots were analyzed using an Odyssey Imager (Licor) according to manufacturer’s instructions. Western blot figures are a representative of at least three independent experiments.
RNA isolation and qPCR
RNA was isolated and purified from macrophages using the Trizol micro-midi RNA isolation kit (Invitrogen) per manufacturer’s instructions. For qPCR analysis 1μg of RNA was reverse transcribed using the VILO cDNA synthesis kit (Invitrogen) and qPCR analysis was performed in triplicate, as previously described (Ohol et al., 2010) using gene specific primers. Data presented is a representative of at least three independent experiments.
Microarrays
Microarray analysis was performed using total RNA isolated from BMDMs infected with M. tuberculosis. Total RNA was amplified and hybridized to MEEBO mouse microarrays as previously described (Leber et al., 2008). Acuity software (Molecular probes) was used for microarray data analysis (Leber et al., 2008). Statistically significant differences in gene expression were identified using the Statistical Analysis of Microarray (SAM) software tool version 3.1, using a false discovery rate (FDR) of less than 5%. Microarray data shown are from two independent experiments. Datasets can be found in the NCBI Geo Database (GSE36686).
Mouse infection
IRF3−/− and wild-type C57BL/6 mice were infected with M. tuberculosis (Erdman) via low-dose aerosol infection as previously described (Ohol et al., 2010). Lungs and spleens were harvested, homogenized, and plated on 7H10 agar plates. Serum from infected mice was harvested via cardiac puncture. For survival experiments, infected mice were sacrificed when they had lost 15% of their maximal body weight.
Trex1 Overexpression
TREX1 was cloned into the pTracer CMV/BSD vector (Invitrogen) using cDNA generated from RAW264.7 macrophage total RNA. Plasmids were electroporated into RAW264.7 cells using the Amaxa nucleofector kit IV. Twenty-four hours post electroporation, cells were cultured in media containing blasticidin (5ug/mL) for 5 days, followed by FACs sorting for GFP positive cells.
Cytokine measurements
IFN-β levels in mouse serum was measured using the Verikine mouse Interferon-β ELISA kit (PBL). IL-12p70, MCP-1, RANTES, and IL-1β serum levels were measured using SearchLight Multiplex ELISA Array (Pierce).
Luminescence Assay
Bacterial DNA release was measured as previously described (Sauer et al., 2010), with the following modifications. The CMV promoter, the Metridia secreted luciferase, and SV40 early polyA of the pMet plasmid (Clonetech) were subcloned into the mycobacterial plasmid pMV261-Zeo (PMAN-1). M. tuberculosis carrying the luciferase reporter construct was used to infect IFNAR1−/− macrophages and cell supernatants were removed 16 h post infection and analyzed for luciferase activity using the Ready-to-Glow assay kit (Clonetech).
Knockdown cells
The mouse TRC1 lentiviral library (Moffat et al., 2006) was obtained from Sigma and used to knockdown IFI204 mRNA in immortalized bone marrow derived cells according to manufacturer’s protocol.
CCF4-am Beta-lactamase Assay
Prior to infection, macrophages were pre-loaded with CCF4-am, using the Live-BLAzer FRET B/G load kit (Invitrogen) per manufacturer’s instructions. After loading, macrophages were than infected with either wild-type M. tuberculosis or ΔesxAM. tuberculosis strains over-expressing the mycobacterial beta-lactamase gene, BlaC. CCF4 cleavage in the cytosol was measured via excitation of cells at 405nm and measuring the emission at 535nm(intact) and 450nm(cleaved).
BrdU Assay
M. tuberculosis strains were grown in 5μM BrdU (Sigma) for 3–4 days in the dark. BrdU labeled cells were used to infect 30 million macrophages at an MOI of 10. 3 h post infection, cells were washed once in K buffer (20 mM HEPES, pH 7.6, 150 mM KCl, 5 mM MgCl2 + protease inhibitors) and then lysed in K buffer containing .0125% digitonin for 10 min on ice. Lysates were centrifuged at 1,500×g for 5 min at 4°C, supernatants transferred to a fresh tube and then centrifuged at 15,000×g for 5 min at 4°C. The resulting supernatants were than filtered through a 0.45 micron filter, boiled for 2 mins and then applied to Protein-G Dynal beads conjugated with anti-BrdU antibody (Sigma). Beads were incubated overnight, washed twice in PBS .01%Tween-20 and the bound BrdU labeled DNA was then released via boiling of beads followed by phenol-chloroform extraction.
Supplementary Material
Highlights.
M. tuberculosis permeabilizes phagosomes and activates cytosolic signaling pathways
Host cytoplasmic DNA receptors sense M. tuberculosis extracellular DNA
DNA recognition activates the Sting/TBK1/IRF3 pathway
Cytosolic sensing promotes M. tuberculosis infection
Acknowledgments
We thank T. Taniguchi, R. Vance, G. Barber, G. Barton, D. Stetson, S. Akira and members of their labs for reagents and macrophages. We are grateful to Charlie Kim for assistance with mice, J. MacGurn for EM images, L. Kohler for generating M. tuberculosis KO strains, A. Casadevall for sharing unpublished results, and Lynn Connolly for critical reading of the manuscript. This work was supported by NIH grant P01 AI063302 (D.A.P. and J.S.C.) and NIH training grant T32 AI060537 (P.S.M.). D. A. Portnoy has a consulting relationship with and a financial interest in Aduro BioTech.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. Type VII secretion--mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, Peterson SN, Monack DM, Barton GM. TLR signaling is required for Salmonella typhimurium virulence. Cell. 2011;144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Beg AA. Defining emerging roles for NF-kappaB in antivirus responses: revisiting the interferon-beta enhanceosome paradigm. PLoS Pathog. 2011;7:e1002165. doi: 10.1371/journal.ppat.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, Honore N, Marchal G, Jiskoot W, England P, et al. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol. 2007 doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divangahi M, Mostowy S, Coulombe F, Kozak R, Guillot L, Veyrier F, Kobayashi KS, Flavell RA, Gros P, Behr MA. NOD2-deficient mice have impaired resistance to Mycobacterium tuberculosis infection through defective innate and adaptive immunity. J Immunol. 2008;181:7157–7165. doi: 10.4049/jimmunol.181.10.7157. [DOI] [PubMed] [Google Scholar]
- Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Monack DM. Activation of the inflammasome upon Francisella tularensis infection: interplay of innate immune pathways and virulence factors. Cell Microbiol. 2007;9:2543–2551. doi: 10.1111/j.1462-5822.2007.01022.x. [DOI] [PubMed] [Google Scholar]
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, 3rd, Freedman VH, Kaplan G. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc Natl Acad Sci U S A. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey RL, Fawcett P, O’Riordan M, Lee KD, Havell EA, Brown PO, Portnoy DA. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci U S A. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Induction of type I interferons by bacteria. Cell Microbiol. 2010;12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Novikov A, Cardone M, Thompson R, Shenderov K, Kirschman KD, Mayer-Barber KD, Myers TG, Rabin RL, Trinchieri G, Sher A, et al. Mycobacterium tuberculosis Triggers Host Type I IFN Signaling To Regulate IL-1β Production in Human Macrophages. J Immunol. 2011 doi: 10.4049/jimmunol.1100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. Mycobacterium tuberculosis MycP1 Protease Plays a Dual Role in Regulation of ESX-1 Secretion and Virulence. Cell Host Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, Fitzgerald KA, Sassetti CM, Kelliher MA. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annual review of pathology. 2012;7:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest. 2011;121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Bobard A, Danckaert A, Paz-Haftel I, Clair C, Ehsani S, Tang C, Sansonetti P, Tran GV, Enninga J. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell Microbiol. 2010;12:545–556. doi: 10.1111/j.1462-5822.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev. 2011;240:252–268. doi: 10.1111/j.1600-065X.2010.00984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Jamieson A, Cresswell P. GILT is a critical host factor for Listeria monocytogenes infection. Nature. 2008;455:1244–1247. doi: 10.1038/nature07344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Manoranjan J, Pan M, Bohsali A, Xu JJ, Liu J, McDonald KL, Szyk A, LaRonde-LeBlanc N, Gao LY. Evidence for Pore Formation in Host Cell Membranes by ESX-1-Secreted ESAT-6 and Its Role in Mycobacterium marinum Escape from the Vacuole. Infection and Immunity. 2008;76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Tsolaki AG, Hirsh AE, DeRiemer K, Enciso JA, Wong MZ, Hannan M, Goguet de la Salmoniere YO, Aman K, Kato-Maeda M, Small PM. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A. 2004;101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.