Abstract
DNA methylation, histone modifications, and chromatin configuration are crucially important in the regulation of gene expression. Among these epigenetic mechanisms, silencing the expression of certain genes depending on developmental stage and tissue specificity is a key repressive system in genome programming. Polycomb (Pc) proteins play roles in gene silencing through different mechanisms. These proteins act in complexes and govern the histone methylation profiles of a large number of genes that regulate various cellular pathways. This review focuses on two main Pc complexes, Pc repressive complexes 1 and 2, and their phylogenetic relationship, structures, and function. The dynamic roles of these complexes in silencing will be discussed herein, with a focus on the recruitment of Pc complexes to target genes and the key factors involved in their recruitment.
Introduction
Several epigenetic factors are associated with gene regulatory mechanisms. Polycomb (Pc) proteins, a member of the Polycomb group (PcG) of proteins, are among the most important of these factors and are important in silencing. PcG proteins are able to silence gene expression globally, particularly during development and differentiation. During the development of an organism, particularly during mitotic cycles, several epigenetic regulatory actions occur through which genes are temporarily or permanently activated or silenced. These regulatory mechanisms, the study of which is known as epigenomics, are also maintained and reestablished during gametogenesis. The PcG proteins are biologically essential throughout development from embryogenesis to adulthood, particularly in the regulation of imprinted genes (Golbabapour et al., 2011). In this review, we refer to the assembly of PcG proteins on chromatin as a Pc complex.
Polycomb Complexes and their Biological Roles
PcG proteins form conserved regulatory structures that can suppress genes through a variety of physiological roles and types of epigenetic patterning in higher organisms (Pirrotta, 2006). Their functions mostly emerge in particular spatial patterns. Margueron and Reinberg (2011) defined the Pc complexes as a set of genes that, when mutated, cause incorrect body segmentation, similar to the initial Polycomb mutant phenotype that has been described in Drosophila. For instance, improper reactivation of pluripotent genes such as the Hox (homeobox) genes initiates carcinogenesis (for a review, see Ringrose and Paro, 2004). In Drosophila, Hox genes are active during early embryogenesis and are maintained during adult life via epigenetic mechanisms. Clustered on chromosomes (Garcia-Fernandez, 2005), the Hox gene family is important for the proper positioning of segmented structures along the body axes (Deschamps and van Nes, 2005). Crosstalk between PcG (Bantignies et al., 2011; Cao et al., 2005) and the trithorax (Petruk et al., 2001; Salvaing et al., 2006) genes was first discovered in Drosophila melanogaster. Hox genes are critical developmental regulators and are expressed in distinct regions of the anterior–posterior axis (Alexander et al., 2009). Mutations in the Hox genes have been associated with various developmental disorders, such as limb malformations (Kmita et al., 2005; Zakany and Duboule, 2007) and neural crest defects (McNulty et al., 2005). The Hox genes are also specifically expressed in the allantois of placental mammals (Scotti and Kmita, 2012; Shaut et al., 2008). Several studies have provided evidence for the roles of Hox genes in oncogenesis (for a review, see Shah and Sukumar, 2010). For instance, overexpression of certain Hox genes has been linked to oral cancer (De Souza Setubal Destro et al., 2010) and breast cancer (Hayashida et al., 2010). However, a full account of the interplay between PcG proteins and trithorax group proteins during development and carcinogenesis is beyond the scope of this article. For a better understanding of histone modifications and chromatin, a number of excellent reviews (Grimaud et al., 2006; Ringrose and Paro, 2004; Schuettengruber et al., 2007) focus on these subjects. Moreover, Pc complexes contribute crucially during embryogenesis, tissue differentiation (Pietersen and van Lohuizen, 2008; Sparmann and van Lohuizen, 2006) and tumorigenesis (Schlesinger et al., 2007; Widschwendter et al., 2007) through X-chromosome inactivation (Casanova et al., 2011; Splinter et al., 2011) and regulation of imprinted genes (Jullien et al., 2006; Makarevich et al., 2006; Schubert et al., 2006). Pc complexes are also associated with nuclear reprogramming and chromatin remodeling (for a review, see Golbabapour et al., 2011).
Polycomb Complexes in Action
Histone modifications and chromatin remodeling are two main epigenetic mechanisms in the regulation of gene expression. The nucleosome, the basic unit of chromatin, consists of a histone octamer, which contains a pair of each of the core histone proteins (H2A, H2B, H3, and H4) (Luger et al., 1997). Nucleosomes consist of 147 bp of DNA wrapped around the core histone and are attached to one another by DNA strands and the histone protein H1 (Luger et al., 1997). Histone modifications consist of any post-translational alterations, such as histone methylation and histone acetylation, imposed on histone proteins. These modifications, in turn, define the configuration of chromatin and its accessibility to transcriptional machinery (for a review, see Golbabapour et al., 2011). Methylation of H3K9 is important in constitutive heterochromatin, while that of H3K27 is a key repressive factor for the regulation of developmental genes (for a review, see Alabert and Groth, 2012).
Studies addressing the mammalian PcG family (for a review, see Jones et al., 2000) suggest a variety of tasks for Pc complexes, such as tasks related to proliferative defects in lymphoid cells (Core et al., 1997; Jacobs et al., 1999) and sex determination (Katoh-Fukui et al., 1998). These complexes contribute to silencing through histone modification. Differences in the expression of each component of Pc repressive complexes (PRCs) suggest that the composition of Pc complexes is specific to cell type and/or developmental stage (Gunster et al., 2001), which could be referred to as dynamic regulation of Pc complexes (for a review, see Lange et al., 2011). PcG proteins also contribute to the covalent post-translational modification of histones. Pc complexes repress gene expression through methylation of histone H3 (H3K27 and H3K9) (Cao and Zhang, 2004a; Lindroth et al., 2004) and ubiquitination of histone H2A (Wang et al., 2004).
Components of Polycomb Complexes
In 1985, the first Pc protein was reported by Jurgens (1985). Since then, attention has been focused on understanding its role and characteristics. Pc complexes primarily proposed as a silencing mechanism for regulating homeotic genes in Drosophila (Chan et al., 1994). Initial studies of Pc complexes showed that these complexes could limit the accessibility of homeotic genes to the transcriptional machinery through spatial modification (for a review, see Cunliffe, 2003). PcG proteins bind nucleosomes and alter the intrinsic structure of chromatin to initiate epigenetic modifications and maintain these modifications during development (for a review, see McBryant et al., 2006). These multi-protein complexes modify chromatin structure to form flexible, repressive chromatin configurations that include numerous targeted genes and maintain silencing (for a review, see Morey and Helin, 2010, and Papp and Plath, 2011). Pc response elements (PREs) are regulatory elements with which PcG complexes interact (Chan et al., 1994; Pirrotta, 1998).
Mammalian PcG proteins have been shown to exist in several types of complexes with different components and structural configurations (Satijn et al., 1997). These complexes are composed of different nonperiodic repetitive units. Complexes of PcG proteins are mainly combinations of multi-protein structures. PRC1 consists of four core proteins: Pc [also known as Chromobox (Cbx) in mammals], polyhomeotic (Ph), posterior sex combs (Psc), and sex combs extra (Ring) (Francis et al., 2001; Saurin et al., 2001; Shao et al., 1999). PRC2 is comprised of three main, strongly conserved subcomplexes: Esc and Enhancer of Zeste [E(z)]; Suppressor of Zeste 12 (Suz12); and RbAp48/Nurf-55 (Cao et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002). There is some evidence that PRC2 is composed of more than just these four components (Cao and Zhang, 2004b; Kim et al., 2009; Li et al., 2010a; Pasini et al., 2010). Ring proteins consist of various domains such as dRing plus two additional conserved cores: Ring1A and Ring1B. Ring1 mediates the ubiquitylation of histone H2A (de Napoles et al., 2004) (for a review, see Martinez and Cavalli, 2006). PRC1 recognizes H3K27me3 to inhibit transcriptional elongation through H2A ubiquitylation (Zhou et al., 2008) and to compact the chromatin structure (Eskeland et al., 2010). PRC2 methylates H3K27 (a key chromatin mark), a main feature of chromatin silencing mediated by PcG proteins. In mammals, yin and yang 1 (Yy1) (Thomas and Seto, 1999), homologues of Drosophila Pho, contribute to histone methylation by recruiting Ez homolog 2 (Ezh2) to H3K27 (Brown et al., 1998). A schematic representation of the components of PRC1 and PRC2 is illustrated in Figure 1. The main components of PRC1 and PRC2 in Drosophila, mouse and human are summarized in Table 1. Several Pc complexes, such as PRC1 and PRC2 (for a review, see Lund and van Lohuizen, 2004), pleiohomeotic repressive complex (PhoRC), and Pc repressive deubiquitinase, are defined as PcG proteins. PcG proteins accumulate widely along chromosomes (Decamillis et al., 1992; Zink and Paro, 1989) and are recruited to PREs in association with transcription factors (for a review, see Ringrose and Paro, 2007) and DNA binding proteins such as Pho (Oktaba et al., 2008). In mammals, several transcription factors are involved in the recruitment of PcG proteins (Bracken and Helin, 2009), which has led to conflicting findings about PRE motifs, protein interaction sites that bring together different proteins in multi-protein complexes and locate them within heterochromatin (Brasher et al., 2000).
FIG. 1.

General constituents of Polycomb repression complexes (PRC1 and PRC2). PRC1 consists of Cbx, BMI1, Ring, Ph, and Pc proteins. PRC2 consists of Ezh2, Eed, Suz12, SET, and RBBP4.
Table 1.
Main Components of PRC1 and PRC2 in Drosophila, Mouse, and Human
| PcG complex | PcG subunits in Drosophila | PcG subunits in mouse | PcG subunits in human | Protein domains | Biochemical activity |
|---|---|---|---|---|---|
| PRC1 | Pc (Paro and Hogness, 1991) | Npcd, M33, Cbx4, Cbx6, Cbx7 and Cbx8 (Pearce et al., 1992; Hemenway et al., 2000; Chen and Bixby, 2005; Bernstein et al., 2006) | Cbx2, Cbx4, Cbx6, Cbx7 and Cbx8 (Levine et al., 2002; Li et al., 2010b; Vandamme et al., 2011) | Chromodomain | Binding to trimethylated H3K27 |
| PRC1 | Polyhomeotic (Decamillis et al., 1992) | Phc1, Phc2, Phc3 (Vandamme et al., 2011) | Ph1, Ph2 And Phc3 (Vandamme et al., 2011) | SAM domain and C2-C2 zinc finger | Interacting with higher-order chromatin |
| PRC1 | Posterior sex combs | Bmi1 and Mel18 | Bmi1, Mel18 and Nspc1 | Ring | Cofactor for E3 ubiquitin ligase and Sex combs extra; compacting polynucleosomes |
| PRC1 | Sex combs extra (Ring) (FlyBase-Berkeley, 2002; Fritsch et al., 2003) | Ring1A and Ring1B (National Center for Biotechnology Information and MRC Functional Genomics Unit, 2009) | Ring1 and Ring1B (Genome Sequencing Center, 2005) | Ring | E3 ubiquitin ligase for H2AK119 |
| PRC2 | Enhancer of Zeste (Jones and Gelbart, 1993; FlyBase-Berkeley, 2002) | Ezh2 and Ezh1 (Laible et al., 1999; O'Carroll et al., 2001; Margueron et al., 2008) | Ezh1 and Ezh2 (Margueron et al., 2008) | SET domain, CXC domain and homology domains I and II | Methylation of H3K9, H3K27 |
| PRC2 | Suppressor of Zeste 12 (Birve et al., 2001; Czermin et al., 2002; Tie et al., 2003) | Suz12 (Pasini et al., 2004; Shen et al., 2008) | Suz12 (Abel et al., 1996; Kirmizis et al., 2003; Cao and Zhang, 2004b) | Zn-finger and VEFS domain | Stimulating H3K27 histone methyltransferase |
| PRC2 | Extra Sex combs (Ng et al., 2000; Poux et al., 2001) | Eed (Silva et al., 2003; Montgomery et al., 2007; Shen et al., 2008) | Eed (embryonic ectoderm development) (Sewalt et al., 1998; Peytavi et al., 1999) | WD repeats | Stimulating H3K27 histone methyltransferase |
| PRC2 | Nucleosome remodeling factor 55 (Badenhorst et al., 2005) | - | Rbap48 (Rbbp4) and Rbap46 (Nicolas et al., 2000; Murzina et al., 2008) | WD repeats | Histone binding |
Chromodomains, conserved protein motifs, are shared regions of homology between two chromatin regulators, the Pc proteins and heterochromatin protein 1 (HP1) (Paro and Hogness, 1991). Bracken and Helin (2009) proposed that so-called “cell fate transcription factors” are crucial to the recruitment of PcG proteins and targeting genes. Moreover, noncoding RNAs (ncRNAs) might mediate the recruitment of PcG members through cell fate transcription factors.
Evolution of Polycomb Complexes
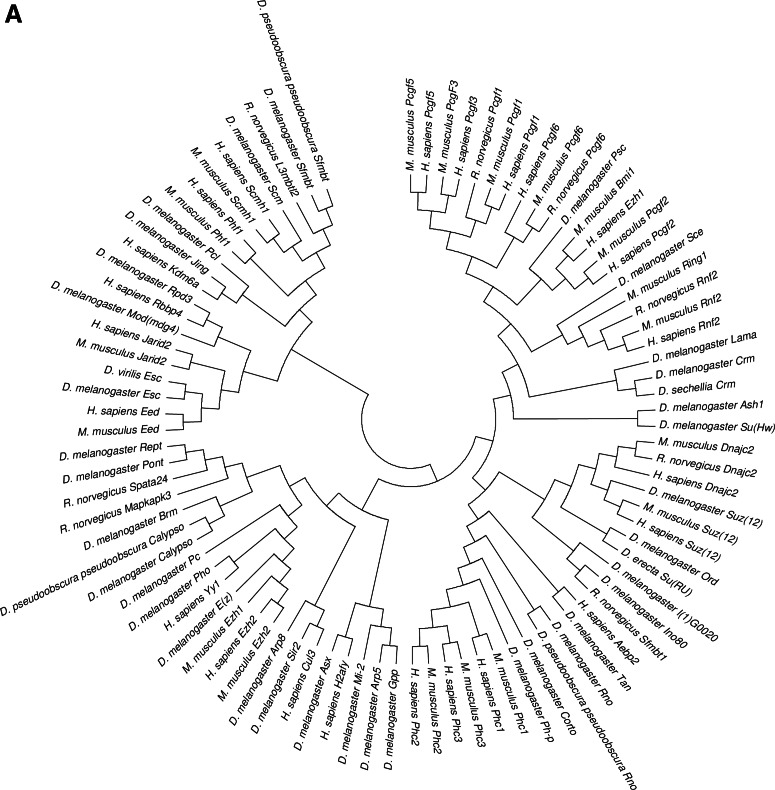
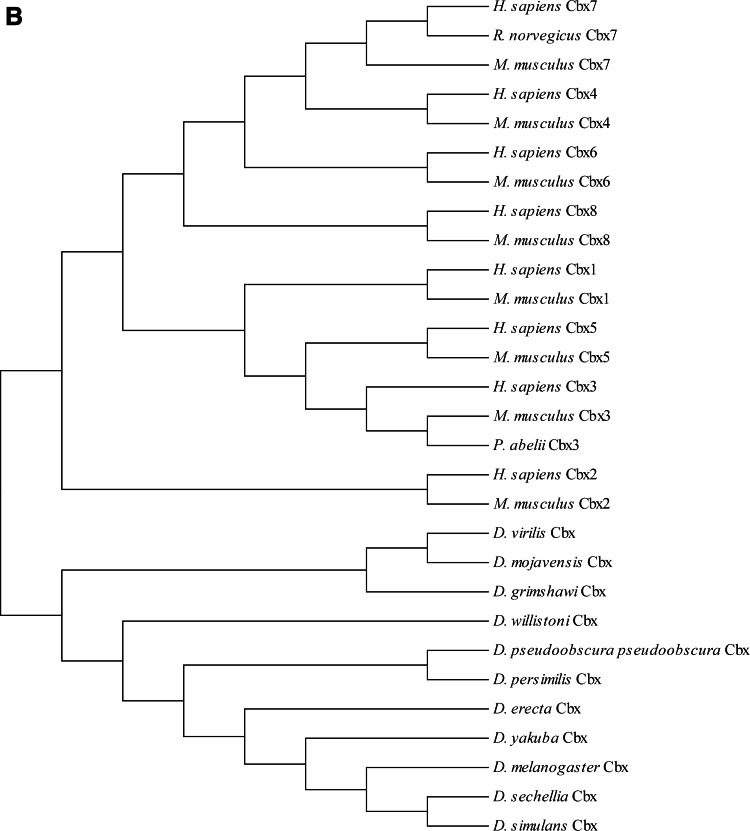
During the past decade, PcG proteins have been shown to participate in a multitude of biological tasks, from stem cell regulation to differentiation, with conserved mechanisms of regulation (Muller et al., 1995). An evolutionary perspective of PcG proteins provides useful insight into their function. As highly conserved biological structures, these proteins are found in various organisms and have conserved biological activities. Figure 2 illustrates the phylogenetic tree of selected organisms (Drosophila, mouse, rat and human) and their PRC1, PRC2, and Cbx homologs (see Supplementary Fig. S1 and Supplementary Table S1 at www.liebertonline.com/omi). The mechanism underlying gene silencing by Pc complexes is conferred by their roles in the structural modification of chromatin and the post-translational modification of histones (for a review, see Margueron and Reinberg, 2011).
FIG. 2.
(A) Phylogenetic distribution of the PRC1 and PRC2 in Drosophila (40), mouse (19), rat (7), and human (22). (B) Phylogenetic distribution of the Cbx in Drosophila (11), mouse (8), rat (1), and human (8). Phylogenetic evolutionary analysis was conducted for PRC1, PRC2, and Cbx (reviewed entries in UniProtKB) using MEGA version 4 (Tamura et al., 2007).
Polycomb Complexes during Embryogenesis and Development
Developmental studies have shown that PcG proteins bind to and interact with Pc domains in more than 200 PcG target genes (Schwartz et al., 2006) and dynamically regulate their expression (Oktaba et al., 2008). For biological activity, PcG proteins need to be part of complexes consisting of core components and associated components. During development, the responses of differentiated cells to environmental signals change due to the epigenetic regulatory effects of PcG members (Paro and Hogness, 1991). As reviewed by Cunliffe (2003), Pc complexes act via a cellular memory mechanism through which cell behavior can be controlled. During the very early steps following fertilization in Drosophila, activation of homeotic genes occurs in certain segments, whereas homeotic genes are repressed in the remainder of the segments (Chan et al., 1994). The suppression of homeotic genes is maintained by a PcG-mediated mechanism that contributes to regulation in the cis-regulatory region (Felsenfeld and Groudine, 2003). During development in female mammals (presence of two X chromosomes), one X chromosome is inactivated to equalize the expression of genes to a level that is equivalent to that of males, who have only one X chromosome (for a review, see Tie et al., 2003). A small ncRNA within Xist (a gene that is generally expressed from and accumulates on the inactive X chromosome) targets PRC2 and contributes to determining chromatin configuration (Brown et al., 1992; Lucchesi et al., 2005; Zhao et al., 2008).
Polycomb Complexes in Cancer
In a variety of cancers, such as lymphoma (McCabe et al., 2012; Tonini et al., 2008), bladder cancer (Hinz et al., 2007), breast cancer and prostate cancer (Ren et al., 2012), PcG proteins repress tumor suppressor genes (Dietrich et al., 2007; Gil and Peters, 2006) through the accumulation of DNA methylation in the promoter regions of their targets, thus altering their methylation profiles (Ohm et al., 2007; Schlesinger et al., 2007). DNA methylation, the addition of a methyl group to a cytosine to form 5-methylcytosine, is a hallmark for recruiting epigenetic factors to suppress gene expression and will be discussed later in this review. Studies on abnormal silencing patterns (Ben-Porath et al., 2008; Bracken and Helin, 2009; Schlesinger et al., 2007; Widschwendter et al., 2007) in both pro- and anti-proliferative genes indicate potential cooperation between PcG proteins and DNA methylation enzymes in silencing the expression of genes (Bracken and Helin, 2009). Abnormal functions of Pc complexes appear to be the main factor involved in a variety of different types of cancerous proliferation and may be due to abnormal expression of the PcG genes themselves (mostly upregulation) (Sparmann and van Lohuizen, 2006), resulting in aberrant DNA methyltransferase silencing (Bracken and Helin, 2009). Cancers may be caused by the inactivation of tumor suppressor genes and/or reprogramming of the nucleus to pluripotent stages due to an imbalance in the dynamic chromatin configuration (Golbabapour et al., 2011; Mills, 2010). Alterations in the expression of the components of Pc complexes, along with modification of their marks, contribute to the development of a variety of cancers. For instance, during the development of cancers such as prostate cancer (Hoffmann et al, 2007; Saramaki et al., 2006), breast cancer (Yoo and Hennighausen, 2012), lymphoma (Visser et al., 2001), lung cancer (Breuer et al., 2004), and liver cancer (Cheng et al., 2011), expression of Ezh2 is increased compared with their normal counterparts. In fact, the higher the level of Ezh2 is, the poorer the cancer prognosis (for reviews, see Chang and Hung, 2012, and Simon and Lange, 2008).
Silencing Mechanisms of Polycomb Complexes
The silencing mechanism of the PcG proteins involves clonal transmission through molecular similarity between the PcG proteins and heterochromatin proteins (Paro, 1990). PcG proteins can bind to the promoter regions of genes and repress their functions (Fig. 3). However, a complete understanding of their functions in silencing genes remains to be deciphered.
FIG. 3.
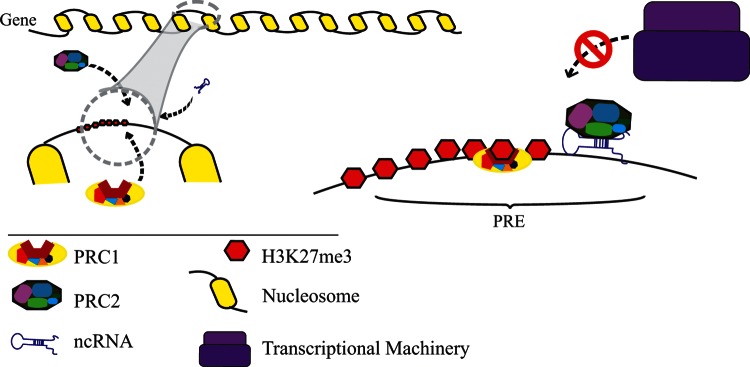
A schematic representation of recruitment of Pc complexes. Polycomb repression complexes (PRC1 and PRC2) recruit on Polycomb response elements (PRE), in association with ncRNA and other DNA-binding proteins (not drawn in the figure). PRC2 imposes high levels of H3K27me3 (as a repressive mark for chromodomains of PRC1).
In addition to other dynamics involved in their recruitment, Pc complexes bind to their targeted genes through their Pc domains and silence their expression (Bracken et al., 2006; Whitehead Institute for Biomedical Research, 2006). Several models have been proposed to describe the dynamic behavior of Pc complex binding (Beisel and Paro, 2011; Bracken and Helin, 2009; Hansen et al., 2008; Lanzuolo et al., 2011).
Packaging of DNA into chromatin structures is the main determinant of gene accessibility to the transcriptional machinery and is a regulatory mechanism of gene expression. Chromatin configuration regulates the expression of certain genes depending on cell type, which is necessary for cell differentiation and development (Golbabapour et al., 2011). In addition to the primary DNA sequence, epigenetic factors are fundamental in regulating gene expression. As such, chromatin remodeling, an epigenetic mechanism, has been considered to represent a crucial mechanism, particularly during development. This interaction between nucleic acids and proteins affects the accessibility of the DNA strand to the transcriptional machinery and, consequently, the regulation of gene expression. DNA methylation, particularly on CpG islands, defines targets for regulatory mechanisms (for a review, see Golbabapour et al., 2011). However, in different molecular physiological processes in cells, DNA methylation has been the subject of some controversy (Gilbert et al., 2007).
Chemical modifications of nucleosomal histones define the positions of nucleosomes (chromatin structure) and their accessibility to the transcriptional machinery. Different versions of PRC1 and PRC2 have been reported, and these diversities come with distinct functions (Vandamme et al., 2011). Genome-wide profiling has been widely exploited to understand PREs. A study on PRC1 and PRC2 revealed chromatin binding sites during PcG-mediated silencing (Tolhuis et al., 2006). PREs contain multiple Pc binding sites; however, no complete molecular profile of PREs has been described. This lack of understanding is due to the size of the complexes and the existence of various homologs and subunits (for a review, see Simon and Kingston, 2009). PREs contain motifs to which DNA binding proteins such as Pho (Klymenko et al., 2006), Zeste (Chen et al., 1992), and GAGA (Berger and Dubreucq, 2012) bind. Zinc-finger proteins, including Pho and Pho-like proteins, must bind to PRE to perform their biological functions (Brown et al., 2003; Fritsch et al., 1999; Mohd-Sarip et al., 2002). Upon binding to a PRE, Pho interacts with a posterior sex comb protein, SFMBT, and forms PhoRC (Klymenko et al., 2006) (for a review, see Schuettengruber and Cavalli, 2009). As mentioned previously, PcG proteins have numerous homologs and a variety of isoforms; consequently, these proteins engage in various binding patterns that, in turn, provide a diversity of complexes. Furthermore, Pc-like proteins can bind to PRC2 and alter its features and properties (O'Connell et al., 2001; Tie et al., 2003; Walker et al., 2010).
Studies addressing the PcG silencing mechanism have provided evidence for the dynamic control of gene expression (Kia et al., 2008; Kwong et al., 2008; Oktaba et al., 2008). Generally, long-term regulation of gene expression is controlled by Pc complexes and, antagonistically, by the trithorax group, through the modulation of chromatin structures and histone modifications. To exert their biochemical activities, PcG proteins must be assembled on PREs. Three main components are involved in the PcG-mediated silencing pathway: PcG proteins (Lund and van Lohuizen, 2004), DNA methylation systems (Bird, 2002), and Ezh2 (a PcG histone-lysine methyltransferase) (Vire et al., 2006). Ezh2 interacts with the other components and provides a recruitment platform for de novo methylation (Fuks et al., 2001). Generally, PRC2 inhibits transcription initiation, and PRC1 maintains the repressed status. PRC1 is able to ubiquitinate H2AK119 (Cao et al., 2005), and PRC2 can trimethylate H3K27 (Cao and Zhang, 2004a). Moreover, PRC2 exhibits catalytic activity that is involved in the methylation of H1K26 (Kuzmichev et al., 2004). Mediated by Ezh2, the addition of methyl groups to Lys27 of histone H3 by PRC2 results in silencing (Hansen et al., 2008) and is essential for the bioactivity of PRC1 (Cao et al., 2005). PcG complexes can also induce significant methylation of H3K27 (for a review, see Mueller and Verrijzer, 2009). The idea that PRC2 functions upstream of PRC1 is consistent with the fact that the Pc protein, a subcomplex, binds specifically to methylated H3K27; however, this premise remains to be proven (Margueron and Reinberg, 2011). PRC2 catalyzes the methylation of H3K27; the methylation of H3K27, particularly trimethylation, is the main hallmark of PcG-mediated silencing (Levine et al., 2004; Ringrose et al., 2003), which occurs through enzymatic activity (in association with PRC1). In addition to the nonenzymatic chromatin-compacting regulatory effect of PRC1 (Eskeland et al., 2010), this Pc complex is able to ubiquitylate histone H2A through Ring1A and Ring1B (for reviews, see Eckert et al., 2011; Richly et al., 2011; Vidal, 2009).
Furthermore, Ezh1 and Ezh2 are PRC2 subcomplexes that are able to methylate histone H3 by adding methyl groups to lysine 27 (Schuettengruber and Cavalli, 2009; Simon and Kingston, 2009). A recent review on the evolution of PRC2 discussed evidence that the primary silencing function of this multi-protein complex is accomplished through the methylation of H3K9. Specific cell lineages are needed to determine the specific functions of PRC2 (for a review, see Margueron and Reinberg, 2011). The maximum holoenzymatic activity of PRC2 is reportedly attributed to the additive contribution of each component of PRC2 (Margueron and Reinberg, 2011). Although PRC1 and PRC2 are two distinct complexes with different structures and functions (Bracken et al., 2006), they exhibit interdependence in regulating gene expression (Jorgensen et al., 2006). Methylation of H3K9 is associated with chromatin configuration and transcriptional regulation. Methylation of H3K9 is a repressive mark and a site for binding by HP1, which recognizes the methyl group and forms a protein dimer to perform its biological functions (Kwon and Workman, 2008), for example, the repression of imprinted genes (Monk et al., 2008). However, methylated H3K9 appears in the coding regions of some active genes (heterochromatin) during transcription elongation through mammalian chromatin (Vakoc et al., 2005). The specific function of H3K9 is dependent on its location in chromatin and its binding to different effector proteins (Kokura et al., 2010). The biochemical activities of the Pc complexes in Drosophila are exerted through the binding of these complexes genome-wide (Schwartz et al., 2006) and to PhoRC (Brown and Kassis, 2010; Klymenko et al., 2006). These complexes ubiquitylate histone H2A and suppress gene expression (Buchwald et al., 2006; Mueller and Verrijzer, 2009; Wang et al., 2004).
Another potential silencing mechanism is enabled by the ability of Pc complexes to bind to chromatin structures. PcG proteins are important in chromatin compaction (for a review, see Mueller and Verrijzer, 2009). To maintain their suppressive role during cell division, PcG proteins remain bound to chromatin and DNA during DNA replication (Francis et al., 2009). Bantignies and Cavalli (2011) argued that PcG proteins dynamically target chromatin in association with RNA polymerase II in a PcG-dependent manner. However, PRC1 has the ability to affect chromatin configuration independently of histone modification (Bantignies and Cavalli, 2011). In addition to post-transcriptional modifications (Berger, 2007), the dynamic structure of chromatin is mediated by two main features: chromatin remodeling and histone modifications such as acetylation, methylation, phosphorylation, ubiquitylation, deamination, ADP ribosylation, sumoylation, and proline isomerization (for a review, see Kouzarides, 2007). In addition to their interactions with DNA, chromatin modifications can affect each other. Histone modifications therefore provide spatially and temporally dependent profiles (known as the “histone code”) to mediate gene regulation and accessibility to the transcriptional machinery (Guil and Esteller, 2009; Turner, 2002). Certain types of DNA sequences, such as CpG islands, promoters, and repetitive elements, along with epigenetic factors, are the main features in determining chromatin configuration.
Recruitment of Polycomb Complexes
PcG proteins cannot specifically bind to DNA sequences. Studies on the associations of the transcriptional factors that recruit these proteins have revealed that multiple transcriptional factors are involved in specific binding to target genes (Bracken and Helin, 2009). During development in Drosophila, a number of transcription factors recruit PcG proteins to PREs (Ringrose and Paro, 2007), which are a combination of several binding sites (elements) (Bracken and Helin, 2009). Promoters in mammals mainly contain either low or high GC dinucleotides, which are classified as low- and high-CpG content promoters, respectively. These classes exhibit different patterns of histone modification and roles in regulation (Broad Institute of Harvard and MIT, 2007; Weber et al., 2007). CG-rich regions are important in PRC2 recruitment, and CpG islands are required for initial localization (Mendenhall et al., 2010) (for a review, see Margueron and Reinberg, 2011). PRC1 and PRC2 mostly target high-CpG content promoters (Boyer et al., 2006; Ku et al., 2008) (for a review, see Zhou et al., 2011). DNA-binding factors and CpG islands are two main features involved in the recruitment of Pc complexes to chromatin, particularly on PREs (Gal-Yam et al., 2008; Meissner et al., 2008; Mendenhall et al., 2010). Nucleosomal array analyses have shown that Pc components are able to remodel chromatin structure and compact chromatin independently of histone modifications (Eskeland et al., 2010; Francis et al., 2004; Margueron et al., 2008). PRC1 inhibits transcription at the promoter region of the targeted gene through chromatin remodeling (Lavigne et al., 2004), although this conclusion is controversial (Eskeland et al., 2010). A genome-wide study comparing active and inactive PcG-targeted regions revealed different levels of PRC1 recruitment. Coincidently, active regions lacking E(z) and Pc exhibit significant trimethylation at H3K9 and H3K27 (Breiling et al., 2004). Trimethylation usually occurs around the promoter regions of active genes, suppressing their expression (Bernstein et al., 2005; Kim et al., 2005). Studies of histone methylation in humans have demonstrated that monomethylation is a hallmark of transcriptional activation, while trimethylation is a hallmark of gene repression (Barski et al., 2007).
Polycomb Complexes and ncRNAs
ncRNAs are involved in the repressive function of silencing complexes and their interactions with nucleosomes. Many ncRNAs have been identified that interact directly with Pc complexes (Kanhere et al., 2010). For instance, HOTAIR and Xist RepA, two ncRNAs, interact with PRC2 to impose a silencing status on their respective genes (Rinn et al., 2007; Zhao et al., 2008). A microarray study of immunoprecipitated PcG proteins demonstrated that ncRNAs associate with PRC2 (Khalil et al., 2009). The SET domain is a sequence motif that catalyzes lysine methylation on histones. PcG proteins generally contain SET domains and Cbx, are mainly present in histone methyltransferases, such as Ezh2, and are involved in protein–genome interactions (Krajewski et al., 2005).
Studies have revealed a link between ncRNAs (which act as recruiters of PcG proteins) and Pc components such as PRC2 (Khalil et al., 2009). ncRNAs are important in the recruitment of PRC1 complexes and interact with Cbx (Yap et al., 2010). Critical in the recruitment of PcG proteins and targeting genes, long ncRNAs are able to recruit PRC1 and PRC2 complexes (for a review, see Bracken and Helin, 2009). Long ncRNAs (such as HOTAIR, KCNq1OT1, and REPA) recruit PcG proteins to chromatin (Bracken and Helin, 2009; Khalil et al., 2009; Zhao et al., 2008).
In addition to the transcriptional roles of long ncRNAs, the mechanism underlying their recruitment is based on their ability to bind specifically to the promoter regions of target genes (Fig. 3) and SET domains/Cbx (for a review, see Bracken and Helin, 2009). Long ncRNAs, such as HOTAIR (Rinn et al., 2007; Tsai et al., 2010), KCNq1OT1 (Kotake et al., 2011; Mohammad et al., 2010) and REPA (Zhao et al., 2008), mediate the methylation of H3K27 by PRC2. In humans, many long ncRNAs interact with PRC2 complexes and are implicated in regulatory functions in PcG-mediated silencing in trans (Khalil et al., 2009). Most ncRNAs exert their activity in cis (such as KCNq1OT1), although some act in trans (such as HOTAIR5). In a proposed model, long ncRNAs bind specifically to the promoters of their target genomic sequences and recruit Pc complexes (SET domains/Cbx) (Mercer et al., 2009; O'Meara and Simon, 2012; Yang et al., 2011).
ncRNAs mediate the relocation of target genes based on the interaction between nuclear subcompartments and nonhistone protein methylation to regulate gene expression (Yang et al., 2011). Yang et al. (2011) concluded that dimethylation of PRC2 on specific residues localized the targeted gene transcriptional machinery through NEAT2, an ncRNA related to interchromatin granules.
Argonaute proteins are approximately 100 kDa (Ender and Meister, 2010) and consist of two principal domains: a Piwi-Argonaute-Zwille domain and a Piwi domain (for reviews, see Ender and Meister, 2010; Parker and Barford, 2006 ). These proteins act as the molecular scaffolds in RNA silencing mechanisms and are necessary for the complementary binding of small ncRNAs to their targets (Parker and Barford, 2006). The silencing mechanism of Argonaute proteins is enabled by small ncRNAs that guide these proteins to their target sites (Takeda et al., 2008).
To exert their biological activity, particularly for guiding small RNAs in gene silencing activity, small ncRNAs are incorporated into Argonaute protein-containing complexes (Ender and Meister, 2010). These complexes contain specific domains that bind to small ncRNAs. Based on phylogenetic analyses, Argonaute proteins are classified into the Ago subfamily, which mostly associates with miRNAs and siRNAs, and the Piwi subfamily, which mainly associates with the germline and piRNAs (Ender and Meister, 2010). The biogenesis and germline functions of the Piwi subfamily are generally enabled by piRNAs (Girard et al., 2006; Houwing et al., 2007). Interactions between the Argonaute proteins and guide molecules are mainly mediated through interactions between the sugar–phosphate backbone of the nucleic acids and these proteins (Wang et al., 2008). The complementary levels of small ncRNAs and their RNA targets represent the main regulatory feature involved in Ago-mediated silencing mechanisms. Gene silencing by small ncRNAs that contain mismatches in their midregion occurs at the level of translation, particularly during its early steps (Ender and Meister, 2010). However, the expression levels and roles of the ncRNAs during development are different. Argonaute proteins exert their silencing role in the cytoplasm at the translational level and in the nucleus at the transcriptional level (for a review, see Guang et al., 2008). Despite recent progress in understanding and modeling the biological functions of ncRNAs in PcG-mediated silencing pathways (for a review, see Beisel and Paro, 2011), our understanding of epigenetics is still in its infancy, and further studies are required to thoroughly understand and grasp epigenetic silencing mechanisms.
Conclusions
Throughout the course of this review, we have discussed the structure and recruitment mechanisms of Pc complexes and their connection to ncRNAs. The phylogenetic distributions of PRC1, PRC2, and Cbx in Drosophila, mouse, rat, and human were also briefly presented. PcG proteins and their regulatory roles in epigenetics, particularly during development, were discussed. Multiple epigenetic pathways govern cell fates and developmental states. Thus, homeostasis, or cellular balance, is the outcome of epigenetic regulation with proper timing. Cancer generally results from an imbalance among epigenetic mechanisms, such as those mediated by PcG proteins and trithorax group proteins that maintain homeostasis in the activity of certain genes. For instance, improper reactivation of pluripotent genes such as the Hox genes initiates carcinogenesis (for a review, see Ringrose and Paro, 2004). In Drosophila, Hox genes are active during early embryogenesis and maintained during adult life by epigenetic mechanisms.
In the last decade, several researchers have focused on understanding the epigenetic mechanisms involved in modifying chromatin. Among these factors, Pc complexes impact the structural modification of chromatin and maintain the silenced state with respect to the expression of certain genes. In fact, PcG proteins cooperate with specific domains of chromatin to silence the expression of genes. Polycomb proteins form at least two distinct complexes, PRC1 and PRC2. The components of Pc complexes, which are known to regulate homeotic genes, vary in different stages of development to control hundreds of other genes in mammals and insects. These complexes are also important in a variety of different cancers. The dynamic nature of Pc complexes increases their flexibility and permits their accurate interaction with environmental signals to regulate the expression of genes as global epigenetic repressors. In cancerous diseases, epigenetic studies have shown that these complexes have roles in transcriptional misregulation, particularly of tumor suppressor genes, leading to unscheduled activation or repression of undesired pathways and thereby enhancing cancerous proliferation. Pc complexes govern the methylation marks on histone H3 through their chromodomains, establishing the chromatin configuration and the spatial distribution of genes within the nucleus. Consequently, Pc complexes control the level of expression of a gene. Numerous studies have emphasized the importance of ncRNAs in epigenetic regulation mediated by PcG proteins. However, our knowledge of Pc complexes and their roles and mechanisms does not provide insight into epigenetic networks, nuclear reprogramming or diseases.
Supplementary Material
Acknowledgments
Preparation of this review was supported by University of Malaya Grants: UM/MOHE High Impact Research Grant No. F000009-21001, PG016-2012B and FP041/2010A.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Abel KJ. Brody LC. Valdes JM, et al. Characterization of Ezh1, a human homolog of Drosophila enhancer of zeste near BRCA1. Genomics. 1996;37:161–171. doi: 10.1006/geno.1996.0537. [DOI] [PubMed] [Google Scholar]
- Alabert C. Groth A. Chromatin replication and epigenome maintenance. Nature Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- Alexander T. Nolte C. Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Ann Rev Cell Develop Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- Badenhorst P. Xiao H. Cherbas L, et al. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Devel. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F. Cavalli G. Polycomb group proteins: Repression in 3D. Trends Genet. 2011;27:454–464. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Bantignies F. Roure V. Comet I, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Barski A. Cuddapah S. Cui KR, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beisel C. Paro R. Silencing chromatin: Comparing modes and mechanisms. Nature Rev Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I. Thomson MW. Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N. Dubreucq B. Evolution goes GAGA: GAGA binding proteins across kingdoms. Biochim Biophys Acta (BBA)-Gene Reg Mech. 2012;1819:863–868. doi: 10.1016/j.bbagrm.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bernstein BE. Kamal M. Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein E. Duncan EM. Masui O. Gil J. Heard E. Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Devel. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Birve A. Sengupta AK. Beuchle D, et al. Su(z)12, a novel Drosophila polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128:3371–3379. doi: 10.1242/dev.128.17.3371. [DOI] [PubMed] [Google Scholar]
- Boyer LA. Plath K. Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP. Dietrich N. Pasini D. Hansen KH. Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Devel. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP. Helin K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nature Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Brasher SV. Smith BO. Fogh RH, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A. O'neill LP. D'eliseo D. Turner BM. Orlando V. Epigenome changes in active and inactive polycomb-group-controlled regions. EMBO Rep. 2004;5:976–982. doi: 10.1038/sj.embor.7400260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer RH. Snijders PJ. Smit EF, et al. Increased expression of the Ezh2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia. 2004;6:736–743. doi: 10.1593/neo.04160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad Institute of Harvard and MIT. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ. Hendrich BD. Rupert JL, et al. The human xist gene. Analysis of a 17 kb inactive x-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brown JL. Fritsch C. Mueller J. Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- Brown JL. Kassis JA. Spps, a Drosophila Sp1/KLF family member, binds to PREs and is required for PRE activity late in development. Development. 2010;137:2597–2602. doi: 10.1242/dev.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL. Mucci D. Whiteley M. Dirksen ML. Kassis JA. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Buchwald G. Van Der Stoop P. Weichenrieder O. Perrakis A. Van Lohuizen M. Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R. Tsukada Y. Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cao R. Wang LJ. Wang HB, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R. Zhang Y. The functions of E(Z)/Ezh2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Devel. 2004a;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Cao R. Zhang Y. Suz12 is required for both the histone methyltransferase activity and the silencing function of the EED–Ezh2 complex. Mol Cell. 2004b;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Casanova M. Preissner T. Cerase A, et al. Polycomblike 2 facilitates the recruitment of PRC2 polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development. 2011;138:1471–1482. doi: 10.1242/dev.053652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS. Rastelli L. Pirrotta V. A polycomb response element in the ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ. Hung MC. The role of Ezh2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. Bixby JL. Neuronal pentraxin with chromo domain (NPCD) is a novel class of protein expressed in multiple neuronal domains. J Comp Neurol. 2005;481:391–402. doi: 10.1002/cne.20391. [DOI] [PubMed] [Google Scholar]
- Chen JD. Chan CS. Pirrotta V. Conserved DNA binding and self-association domains of the Drosophila zeste protein. Mol Cell Biol. 1992;12:598–608. doi: 10.1128/mcb.12.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AS. Lau SS. Chen Y, et al. Ezh2-mediated concordant repression of Wnt antagonists promotes beta-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71:4028–4039. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- Core N. Bel S. Gaunt SJ, et al. Altered cellular proliferation and mesoderm patterning in polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- Cunliffe VT. Memory by modification: The influence of chromatin structure on gene expression during vertebrate development. Gene. 2003;305:141–150. doi: 10.1016/s0378-1119(03)00386-x. [DOI] [PubMed] [Google Scholar]
- Czermin B. Melfi R. Mccabe D. Seitz V. Imhof A. Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- De Napoles M. Mermoud JE. Wakao R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Devel Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- De Souza Setubal Destro MF. Bitu CC. Zecchin KG, et al. Overexpression of HoxB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Intl J Oncol. 2010;36:141–149. [PubMed] [Google Scholar]
- Decamillis M. Cheng NS. Pierre D. Brock HW. The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with polycomb. Genes Devel. 1992;6:223–232. doi: 10.1101/gad.6.2.223. [DOI] [PubMed] [Google Scholar]
- Deschamps J. Van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- Dietrich N. Bracken AP. Trinh E, et al. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007;26:1637–1648. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL. Adhikary G. Rorke EA. Chew YC. Balasubramanian S. Polycomb group proteins are key regulators of keratinocyte function. J Invest Dermatol. 2011;131:295–301. doi: 10.1038/jid.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C. Meister G. Argonaute proteins at a glance. J Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- Eskeland R. Leeb M. Grimes GR, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Flybase-Berkeley. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 2002 doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ. Follmer NE. Simon MD. Aghia G. Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ. Kingston RE. Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Francis NJ. Saurin AJ. Shao ZH. Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Fritsch C. Beuchle D. Muller J. Molecular and genetic analysis of the polycomb group gene Sex combs extra/Ring in Drosophila. Mech Devel. 2003;120:949–954. doi: 10.1016/s0925-4773(03)00083-2. [DOI] [PubMed] [Google Scholar]
- Fritsch C. Brown JL. Kassis JA. Muller J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- Fuks F. Burgers WA. Godin N. Kasai M. Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN. Egger G. Iniguez L, et al. Frequent switching of polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez J. Hox, ParaHox, ProtoHox: Facts and guesses. Heredity (Edinb) 2005;94:145–152. doi: 10.1038/sj.hdy.6800621. [DOI] [PubMed] [Google Scholar]
- Genome Sequencing Center. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005;434:724–731. doi: 10.1038/nature03466. [DOI] [PubMed] [Google Scholar]
- Gil J. Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Nature Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Gilbert N. Thomson I. Boyle S. Allan J. Ramsahoye B. Bickmore WA. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J Cell Biol. 2007;177:401–411. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A. Sachidanandam R. Hannon GJ. Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Golbabapour S. Abdulla MA. Hajrezaei M. A concise review on epigenetic regulation: Insight into molecular mechanisms. Intl J Mol Sci. 2011;12:8661–8694. doi: 10.3390/ijms12128661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C. Negre N. Cavalli G. From genetics to epigenetics: The tale of polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- Guang S. Bochner AF. Pavelec DM, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S. Esteller M. DNA methylomes, histone codes and miRNAs: Tying it all together. Intl J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Gunster MJ. Raaphorst FM. Hamer KM, et al. Differential expression of human polycomb group proteins in various tissues and cell types. J Cell Biochem. 2001;81:129–143. doi: 10.1002/jcb.1093. [DOI] [PubMed] [Google Scholar]
- Hansen KH. Bracken AP. Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nature Cell Biology. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Hayashida T. Takahashi F. Chiba N, et al. HoxB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci USA. 2010;107:1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway CS. Halligan BW. Gould GC. Levy LS. Identification and analysis of a third mouse Polycomb gene, MPc3. Gene. 2000;242:31–40. doi: 10.1016/s0378-1119(99)00540-5. [DOI] [PubMed] [Google Scholar]
- Hinz S. Kempkensteffen C. Weikert S, et al. Ezh2 polycomb transcriptional repressor expression correlates with methylation of the APAF-1 gene in superficial transitional cell carcinoma of the bladder. Tumour Biology. 2007;28:151–157. doi: 10.1159/000103380. [DOI] [PubMed] [Google Scholar]
- Hoffmann MJ. Engers R. Florl AR. Otte AP. Muller M. Schulz WA. Expression changes in Ezh2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer. Cancer Biol Ther. 2007;6:1403–1412. doi: 10.4161/cbt.6.9.4542. [DOI] [PubMed] [Google Scholar]
- Houwing S. Kamminga LM. Berezikov E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ. Kieboom K. Marino S. Depinho RA. Van Lohuizen M. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Jones DO. Cowell IG. Singh PB. Mammalian chromodomain proteins: Their role in genome organisation and expression. Bioessays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Jones RS. Gelbart WM. The Drosophila polycomb-group gene enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF. Giadrossi S. Casanova M, et al. Stem cells primed for action. Polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle. 2006;5:1411–1414. doi: 10.4161/cc.5.13.2927. [DOI] [PubMed] [Google Scholar]
- Jullien PE. Katz A. Oliva M. Ohad N. Berger F. Polycomb group complexes self-regulate imprinting of the polycomb group gene MEDEA in Arabidopsis. Curr Biol. 2006;16:486–492. doi: 10.1016/j.cub.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Jurgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985;316:153–155. [Google Scholar]
- Kanhere A. Viiri K. Araujo CC, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh-Fukui Y. Tsuchiya R. Shiroishi T, et al. Male-to-female sex reversal in M33 mutant mice. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- Khalil AM. Guttman M. Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia SK. Gorski MM. Giannakopoulos S. Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. Kang K. Kim J. AEBP2 as a potential targeting protein for polycomb repression complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH. Barrera LO. Zheng M, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A. Bartley SM. Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther. 2003;2:113–121. [PubMed] [Google Scholar]
- Klymenko T. Papp B. Fischle W, et al. A polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Devel. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M. Tarchini B. Zakany J. Logan M. Tabin CJ. Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435:1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- Kokura K. Sun L. Bedford MT. Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010;29:3673–3687. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y. Nakagawa T. Kitagawa K, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krajewski WA. Nakamura T. Mazo A. Canaani E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25:1891–1899. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M. Koche RP. Rheinbay E, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A. Jenuwein T. Tempst P. Reinberg D. Different Ezh2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A. Nishioka K. Erdjument-Bromage H. Tempst P. Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Devel. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SH. Workman JL. The heterochromatin protein 1 (HP1) family: Put away a bias toward HP1. Mol Cells. 2008;26:217–227. [PubMed] [Google Scholar]
- Kwong C. Adryan B. Bell I, et al. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 2008;10:1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G. Haynes AR. Lebersorger A, et al. The murine polycomb-group genes Ezh1 and Ezh2 map close to Hox gene clusters on mouse chromosomes 11 and 6. Mammal Genome. 1999;10:311–314. doi: 10.1007/s003359900993. [DOI] [PubMed] [Google Scholar]
- Lange M. Demajo S. Jain P. Di Croce L. Combinatorial assembly and function of chromatin regulatory complexes. Epigenomics. 2011;3:567–580. doi: 10.2217/epi.11.83. [DOI] [PubMed] [Google Scholar]
- Lanzuolo C. Lo Sardo F. Diamantini A. Orlando V. PcG complexes set the stage for epigenetic inheritance of gene silencing in early S phase before replication. PLoS Genetics. 2011;10:1002370. doi: 10.1371/journal.pgen.1002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne M. Francis NJ. King IFG. Kingston RE. Propagation of silencing: Recruitment and repression of naive chromatin - In trans by polycomb repressed chromatin. Mol Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- Levine SS. King IFG. Kingston RE. Division of labor in polycomb group repression. Trends Biocheml Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Levine SS. Weiss A. Erdjument-Bromage H. Shao Z. Tempst P. Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. Margueron R. Ku M. Chambon P. Bernstein BE. Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Devel. 2010a;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Wang X. Lu Z, et al. Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. Plos One. 2010b;10:0013732. doi: 10.1371/journal.pone.0013732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM. Shultis D. Jasencakova Z, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with chromomethylase 3. EMBO J. 2004;23:4146–4155. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC. Kelly WG. Parming B. Chromatin remodeling in dosage compensation. Ann Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Luger K. Mader AW. Richmond RK. Sargent DF. Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lund AH. Van Lohuizen M. Polycomb complexes and silencing mechansims. Curr Opin Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Makarevich G. Leroy O. Akinci U, et al. Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7:947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R. Li GH. Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R. Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AM. Cavalli G. The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle. 2006;5:1189–1197. doi: 10.4161/cc.5.11.2781. [DOI] [PubMed] [Google Scholar]
- Mcbryant SJ. Adams VH. Hansen JC. Chromatin architectural proteins. Chromosome Res. 2006;14:39–51. doi: 10.1007/s10577-006-1025-x. [DOI] [PubMed] [Google Scholar]
- McCabe MT. Graves AP. Ganji G, et al. Mutation of A677 in histone methyltransferase Ezh2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci USA. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty CL. Peres JN. Bardine N. Van Den Akker WM. Durston AJ. Knockdown of the complete Hox paralogous group 1 leads to dramatic hindbrain and neural crest defects. Development. 2005;132:2861–2871. doi: 10.1242/dev.01872. [DOI] [PubMed] [Google Scholar]
- Meissner A. Mikkelsen TS. Gu HC, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM. Koche RP. Truong T, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010 doi: 10.1371/1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR. Dinger ME. Mattick JS. Long non-coding RNAs: Insights into functions. Nature Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mills AA. Throwing the cancer switch: Reciprocal roles of polycomb and trithorax proteins. Nature Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F. Mondal T. Guseva N. Pandey GK. Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A. Venturini F. Chalkley GE. Verrijzer CP. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D. Wagschal A. Arnaud P, et al. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res. 2008;18:1270–1281. doi: 10.1101/gr.077115.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ND. Yee D. Montgomery SA. Magnuson T. Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J Mol Biol. 2007;374:1145–1157. doi: 10.1016/j.jmb.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L. Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Mueller J. Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genetics Devel. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Muller J. Gaunt S. Lawrence PA. Function of the polycomb protein is conserved in mice and flies. Development. 1995;121:2847–2852. doi: 10.1242/dev.121.9.2847. [DOI] [PubMed] [Google Scholar]
- Muller J. Hart CM. Francis NJ, et al. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Murzina NV. Pei XY. Zhang W, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information and Mrc Functional Genomics Unit. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009 doi: 10.1371/1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. Hart CM. Morgan K. Simon JA. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E. Morales V. Magnaghi-Jaulin L. Harel-Bellan A. Richard-Foy H. Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- O'Carroll D. Erhardt S. Pagani M. Barton SC. Surani MA. Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell S. Wang LJ. Robert S. Jones CA. Saint R. Jones RS. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J Biol Chem. 2001;276:43065–43073. doi: 10.1074/jbc.M104294200. [DOI] [PubMed] [Google Scholar]
- O'Meara M. Simon J. Inner workings and regulatory inputs that control polycomb repressive complex 2. Chromosoma. 121:221–234. doi: 10.1007/s00412-012-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE. McGarvey KM. Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K. Gutierrez L. Gagneur J, et al. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Devel Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Papp B. Plath K. Reprogramming to pluripotency: Stepwise resetting of the epigenetic landscape. Cell Res. 2011;21:486–501. doi: 10.1038/cr.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS. Barford D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem Sci. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- Paro R. Hogness DS. The polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D. Bracken AP. Jensen MR. Lazzerini Denchi E. Helin K. Suz12 is essential for mouse development and for Ezh2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D. Cloos PaC. Walfridsson J, et al. JARID2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Pearce JJ. Singh PB. Gaunt SJ. The mouse has a polycomb-like chromobox gene. Development. 1992;114:921–929. doi: 10.1242/dev.114.4.921. [DOI] [PubMed] [Google Scholar]
- Petruk S. Sedkov Y. Smith S, et al. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- Peytavi R. Hong SS. Gay B, et al. HEED, the product of the human homolog of the murine eed gene, binds to the matrix protein of HIV-1. J Biol Chem. 1999;274:1635–1645. doi: 10.1074/jbc.274.3.1635. [DOI] [PubMed] [Google Scholar]
- Pietersen AM. Van Lohuizen M. Stem cell regulation by polycomb repressors: Postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Polycomb silencing mechanisms and genomic programming. Berger S.L.N.O.H.B. Histone Code and Beyond: New Approaches to Cancer Therapy. 2006:97–113. doi: 10.1007/3-540-37633-x_6. [DOI] [PubMed] [Google Scholar]
- Poux S. Melfi R. Pirrotta V. Establishment of polycomb silencing requires a transient interaction between PC and ESC. Genes Devel. 2001;15:2509–2514. doi: 10.1101/gad.208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. Baritaki S. Marathe H, et al. Polycomb protein Ezh2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72:3091–3104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- Richly H. Aloia L. Di Croce L. Roles of the polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011 doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L. Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Ann Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Ringrose L. Paro R. Polycomb/trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Ringrose L. Rehmsmeier M. Dura JM. Paro R. Genome-wide prediction of polycomb/trithorax response elements in Drosophila melanogaster. Devel Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- Rinn JL. Kertesz M. Wang JK, et al. Functional demarcation of active and silent chromatin domains in human Hox loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvaing J. Decoville M. Mouchel-Vielh E, et al. Corto and DSP1 interact and bind to a maintenance element of the Scr Hox gene: Understanding the role of enhancers of trithorax and polycomb. BMC Biol. 2006;4:9. doi: 10.1186/1741-7007-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramaki OR. Tammela TL. Martikainen PM. Vessella RL. Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (Ezh2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- Satijn DP. Gunster MJ. Van Der Vlag J, et al. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ. Shao ZH. Erdjument-Bromage H. Tempst P. Kingston RE. A Drosophila polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y. Straussman R. Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature Genetics. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Schubert D. Primavesi L. Bishopp A, et al. Silencing by plant polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B. Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B. Chourrout D. Vervoort M. Leblanc B. Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB. Kahn TG. Nix DA, et al. Genome-wide analysis of polycomb targets in Drosophila melanogaster. Nature Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Schwartz YB. Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Scotti M. Kmita M. Recruitment of 5' HoxA genes in the allantois is essential for proper extra-embryonic function in placental mammals. Development. 2012;139:731–739. doi: 10.1242/dev.075408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt RG. Van Der Vlag J. Gunster MJ, et al. Characterization of interactions between the mammalian polycomb-group proteins Enx1/Ezh2 and EED suggests the existence of different mammalian polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. Sukumar S. The Hox genes and their roles in oncogenesis. Nature Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- Shao Z. Raible F. Mollaaghababa R, et al. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shaut CA. Keene DR. Sorensen LK. Li DY. Stadler HS. HoxA13 Is essential for placental vascular patterning and labyrinth endothelial specification. PLoS Genet. 2008 doi: 10.1371/1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X. Liu Y. Hsu YJ, et al. Ezh1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J. Mak W. Zvetkova I, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Devel Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Simon JA. Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nature Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simon JA. Lange CA. Roles of the Ezh2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Sparmann A. Van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Splinter E. De Wit E. Nora EP, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Devel. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A. Iwasaki S. Watanabe T. Utsumi M. Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evolut. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thomas MJ. Seto E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Tie F. Prasad-Sinha J. Birve A. Rasmuson-Lestander A. Harte PJ. A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol Cell Biol. 2003;23:3352–3362. doi: 10.1128/MCB.23.9.3352-3362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B. Muijrers I. De Wit E, et al. Genome-wide profiling of PRC1 and PRC2 polycomb chromatin binding in Drosophila melanogaster. Nature Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Tonini T. D'andrilli G. Fucito A. Gaspa L. Bagella L. Importance of Ezh2 polycomb protein in tumorigenesis process interfering with the pathway of growth suppressive key elements. J Cell Physiol. 2008;214:295–300. doi: 10.1002/jcp.21241. [DOI] [PubMed] [Google Scholar]
- Tsai MC. Manor O. Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Vakoc CR. Mandat SA. Olenchock BA. Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Vandamme J. Volkel P. Rosnoblet C. Le Faou P. Angrand PO. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol Cell Proteom. 2011;10(M110):002642. doi: 10.1074/mcp.M110.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Intl J Devel Biol. 2009;53:355–370. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- Vire E. Brenner C. Deplus R, et al. The polycomb group protein Ezh2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Visser HPJ. Gunster MJ. Kluin-Nelemans HC, et al. The polycomb group protein Ezh2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol. 2001;112:950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- Walker E. Chang WY. Hunkapiller J, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB. Wang LJ. Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang Y. Juranek S. Li H. Sheng G. Tuschl T. Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–U972. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M. Hellmann I. Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Whitehead Institute for Biomedical Research. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M. Fiegl H. Egle D, et al. Epigenetic stem cell signature in cancer. Nature Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Yang LQ. Lin CR. Liu W, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL. Li S. Muñoz-Cabello AM, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo KH. Hennighausen L. Ezh2 methyltransferase and H3K27 methylation in breast cancer. Intl J Biol Sci. 2012;8:59–65. doi: 10.7150/ijbs.8.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J. Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Devel. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Zhao J. Sun BK. Erwin JA. Song J-J. Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW. Goren A. Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- Zhou W. Zhu P. Wang J, et al. Histione H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink B. Paro R. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature. 1989;337:468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.