Abstract
The overwhelming majority of methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates exhibit a peculiar heterogeneous resistance to β-lactam antibiotics: in cultures of such strains, the majority of cells display only a low level of methicillin resistance—often close to the MIC breakpoint of susceptible strains. Yet, in the same cultures, subpopulations of bacteria exhibiting very high levels of resistance are also present with variable frequencies, which are characteristic of the particular MRSA lineage. The mechanism of heterogeneous resistance is not understood. We describe here an experimental system for exploring the mechanism of heterogeneous resistance. Copies of the resistance gene mecA cloned into a temperature-sensitive plasmid were introduced into the fully sequenced methicillin-susceptible clinical isolate S. aureus strain 476. Transductants of strain 476 expressed methicillin resistance in a heterogeneous fashion: the great majority of cells showed only low MIC (0.75 μg/ml) for the antibiotic, but a minority population of highly resistant bacteria (MIC >300 μg/ml) was also present with a frequency of ∼10−4. The genetic backgrounds of the majority and minority cells were compared by whole-genome sequencing: the only differences detectable were two point mutations in relA of the highly resistant minority population of bacteria. The relA gene codes for the synthesis of (p)ppGpp, an effector of the stringent stress response. Titration of (p)ppGpp showed increased amounts of this effector in the highly resistant cells. Involvement of (p)ppGpp synthesis genes may explain some of the perplexing aspects of β-lactam resistance in MRSA, since many environmental and genetic changes can modulate cellular levels of (p)ppGpp.
Introduction
Introduction of the semisynthetic β-lactam antibiotic methicillin into clinical practice in 1961 was followed rapidly by the appearance of methicillin-resistant Staphylococcus aureus (MRSA) isolates,4,12,26,30 which were shown to carry an extra penicillin-binding protein—PBP2A17—that was added to the four native staphylococcal PBPs—a group of membrane anchored extracytoplasmic proteins that synthesize the cell wall peptidoglycan. In contrast to the native PBPs, PBP2A was shown to have extremely low affinity to all β-lactam antibiotics and was able to continue catalyzing peptidoglycan biosynthesis even in the presence of high concentrations of antibiotics that inhibited native PBPs. The genetic determinant of PBP2A—mecA—was also identified5 as an acquired gene associated with heterologous staphylococcal chromosomal cassettes (SCCmec), which incorporate into the chromosome of MRSA strains at a unique chromosomal site.22 While the carrier cassettes—SCCs—were shown to exhibit considerable structural variation,23 the sequence of the mecA determinant is invariant in most MRSA isolates with the exception of some recently identified strains that carry a new mec gene homologue.15,28
In contrast to the rapid clarification of biochemical and genetic basis of methicillin resistance, the mechanism of the phenotype of β-lactam resistance has remained unclear. It has been known for some time that to express methicillin resistance it was necessary, but not sufficient, that the MRSA strain carried a functional copy of mecA since variations in the temperature, pH value, or salt concentration during exposure of the same MRSA strain to methicillin were shown to cause profound effects on the level of antibiotic resistance.9,18,38 Also, clinical isolates of MRSA exhibit large variations in the resistance level, which cannot be simply explained by the activity of regulatory genes (mecI/mecR or blaI/blaR) that control the transcription of the mecA determinant.
An even more complex feature of the resistant phenotype of MRSA is its heterogeneity detectable in most clinical isolates. The great majority of MRSA strains grown from single cell inocula produce cultures in which, most (over 99%) of the cells exhibit only modest or low-level resistance, sometimes approaching the breakpoint of susceptible strains. However, the same MRSA cultures also contain highly resistant cells with a frequency and degree of resistance, which are characteristic of the particular MRSA clone.18,35 Plotting the number of bacterial cells capable of growing at various concentrations of the antibiotic against the concentration of the antibiotic in the agar medium produces population analysis profiles (PAPs) the shape of which seems to represent a unique signature of the particular MRSA clone.
The purpose of studies described here was to develop a model system that would allow a better understanding of the mechanism that controls the heterogenous β-lactam-resistant phenotype in S. aureus.
Materials and Methods
Population analysis profiles
The susceptibility of S. aureus strains to oxacillin (a β-lactam antibiotic, similar to methicillin, resistant to penicillinase) was determined by PAPs. Population analysis was carried out on tryptic soy agar (TSA) plates containing increasing concentrations of oxacillin and supplemented with 20 μg/ml of chloramphenicol for 476(pmecA) and 476mut(pmecA) as described previously.10,28 Colony forming units were counted after 48 hr of incubation of the plates at 30°C.
Introduction of plasmid-borne copies of the mecA gene into the oxacillin-susceptible S. aureus strain 476
The recombinant plasmid pSPSW2C, which carries the 3,737-bp region of S. aureus COL mecA, was introduced into S. aureus strain RN4220 by electroporation, and then transduced by phage 80α into S. aureus strain 476 to yield strain 476(pmecA) as described previously.2,41
Whole-genome sequencing
Chromosomal DNA preparations isolated from the plasmid-free strains named “476mut-cured-small” and “476mut-cured-large” were sequenced and compared to the DNA sequence of strain 476. The complete genomic sequence (no contig gaps) for 476 is available at the NCBI's Website*. The 454 platform was used for the re-sequencing of the genome of strain 476—a stock of which was available in our laboratory's strain collection. The same 454 platform was also used for the sequencing of the genome of 476mut-cured-small and de novo assemblies of the reads were done. The same stock of strain 476 also served as the recipient of the pmecA in all the experiments described in this communication.
The Illumina platform was used for the sequencing of the genome of 476mut-cured-large, and a template-based assembly of the reads was done using the NCBI's complete genomic sequence for 476 as the reference. The methods for the sequencing, assemblies, and polymorphism detection were described previously.7,19,36,39
Measurement of (p)ppGpp accumulation
The level of (p)ppGpp (guanosine 5′-diphosphate, 3′-diphosphate) was determined by P13 labeling as previously described.34,37 A 3 ml culture of cells was harvested at OD 0.4 and extracted with 40 μl of 5 M formic acid by repeating freeze/thaw cycles four times, followed by incubation for 30 min on ice. Cell debris were removed by centrifugation at the maximum speed for 5 min at 4°C. The extracted samples were spotted on polyethyleneimine-cellulose F thin layer chromatography (TLC) plates pretreated with 10% sodium chloride and 100% methanol.8 The plates were developed with 1.5 M monopotassium phosphate (pH 3.5) for separation of the phosphorylated nucleotides. 32Pi-labeled nucleotides were visualized with a Typhoon9400 image scanner, and nucleotide spots were quantified by an ImageQuant software. Amounts of (p)ppGpp were described as fractions of total guanine nucleotides, including GTP and (p)ppGpp. The sample extracted from 476(pmecA) treated with 60 μg/ml of mupirocin (an agent that can induce the stringent stress response), 15 min before harvest, was used as a positive control for identification of (p)ppGpp spots on TLC plates.
Results and Discussion
Search for a simple experimental model to study the mechanism of heterogeneous resistance
While all MRSA strains carry the methicillin resistance gene mecA on a chromosomally located SCCmec cassette, experimental manipulation of these structures is difficult. Previous studies from our lab2 and other labs27 have shown that methicillin resistance can be introduced and expressed in susceptible strains of S. aureus by plasmid-borne copies of the mecA gene. It was shown that introduction of a plasmid-borne mecA into the S. aureus strain COL-S (a derivative of the highly resistant MRSA strain COL from which the SCCmec cassette was removed by precise excision), produced transductants with high and homogeneous oxacillin resistance indistinguishable from that of the original COL strain.2 Similar findings were obtained when the same plasmid was introduced into strain COL in which the resident mecA was inactivated by transposon insertion.32 Interestingly—and in contrast—introduction of the same mecA carrying plasmid into several methicillin-susceptible S. aureus (MSSA) strains produced transductants with heterogeneous methicillin resistance, similar to the phenotypes of most clinical MRSA strains, suggesting that at least some of the determinants of β-lactam resistance reside in the genetic background of the bacteria that received the plasmid-borne mecA determinant (results not shown).
Oxacillin-resistant transductants of MSSA strain 476
After testing a number of methicillin-susceptible clinical isolates of S. aureus, we chose the fully sequenced MSSA strain 476 (sequence type—ST1),20 to construct a model for studying heterogeneous resistance. A heteroresistant MRSA strain—MW2—with the same sequence type 1 carrying SCCmec type IV was also sequenced recently.3
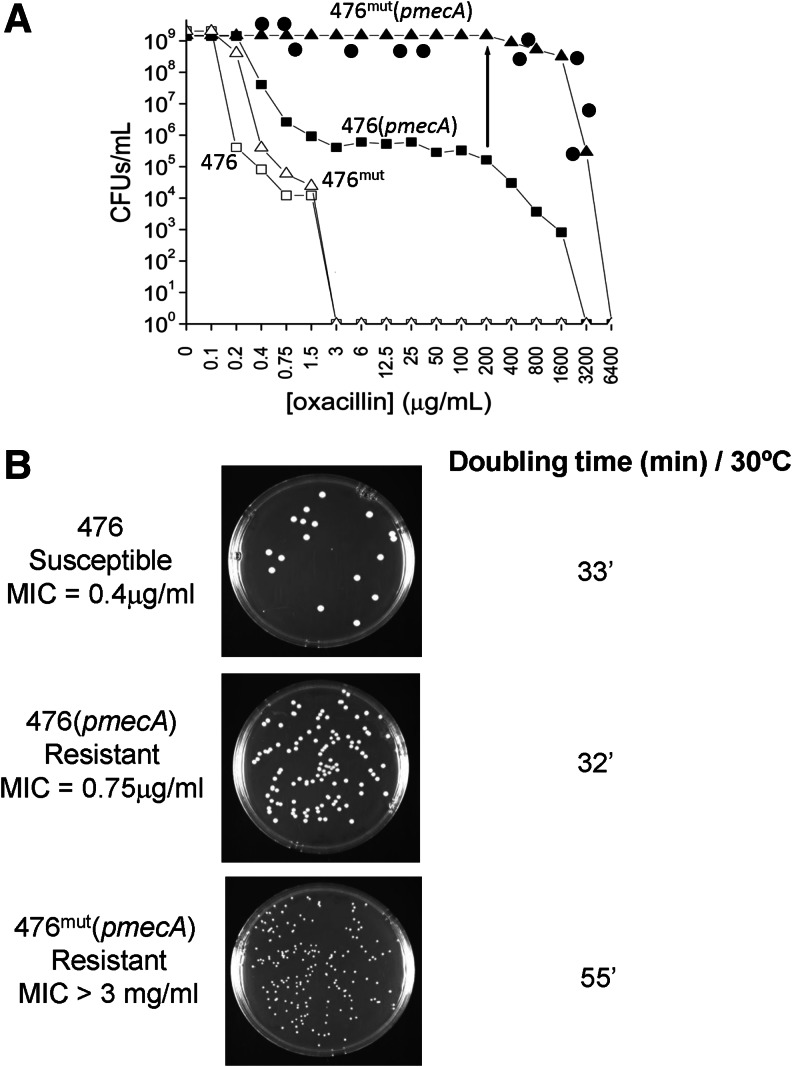
Upon transduction of the thermosensitive plasmid pmecA into strain 476, a heterogeneously resistant strain was obtained in which the majority of transductants—named 476(pmecA)—showed only low-level resistance to oxacillin (MIC 0.75 μg/ml). However, a more detailed population analysis indicated that the same culture also contained an apparently single subpopulation of a highly resistant mutant—bacteria named 476mut(pmecA)—with an approximate frequency of 10−4 and an extremely high oxacillin MIC value of >3 mg/ml (Fig. 1A). The high level resistance was specific for β-lactam antibiotics such as methicillin, oxacillin, cefuroxime, imipenem, while the bacteria remained susceptible to other antibiotics such as vancomycin, fosfomycin, rifampicin, or linezolid.
FIG. 1.
Properties of Staphylococcus aureus strain 476 and its laboratory constructs carrying plasmid-borne copies of the mecA gene. The mecA gene was introduced into strain 476 by transduction using the temperature-sensitive plasmid pSTSW-2C—as described in the “Materials and Methods” section. (A) shows the oxacillin-susceptibility profile of the transductant 476(pmecA) (■) as determined by its population analysis profile (PAP). Also shown are the PAPs of the cultures of the susceptible recipient strain 476 (□); the highly resistant mutant 476mut(pmecA) (▲), which was obtained by picking a rare colony present in the culture of 476(pmecA), and the PAP of strain 476mut (Δ), which was generated from 476mut(pmecA) by deletion of the thermosensitive mecA plasmid. The PAP of 476(pmecA) was also determined by supplementing each oxacillin-containing plate with 0.03 μg/ml mupirocin (•). (B) shows colony sizes of strains 476, 476(pmecA), and 476mut(pmecA) on TSA plates incubated at 30°C for 48 hr. Also shown are growth rates (mass doubling times) of the same cultures grown in TSB at 30°C.
A single colony of 476mut(pmecA) picked from a TSA plate containing 200 μg/ml oxacillin was used as an inoculum to generate an overnight culture in tryptic soy broth (TSB). Upon plating for population analysis, this culture produced bacteria that were homogeneously and highly resistant to oxacillin (Fig. 1A).
The poorly resistant majority cells—476(pmecA)—and the highly resistant mutant— 476mut(pmecA)— selected as homogeneous cultures were next compared for a number of properties.
The two subpopulations of bacteria differed in colony size and growth rate: in contrast to 476(pmecA), the highly resistant mutant 476mut(pmecA) produced small colonies when plated on TSA (Fig. 1B) and determination of the corresponding growth rates of liquid cultures in TSB at 30°C showed mass doubling times of about 32 min for 476(pmecA) and 55 min for 476mut(pmecA) (Fig. 1B).
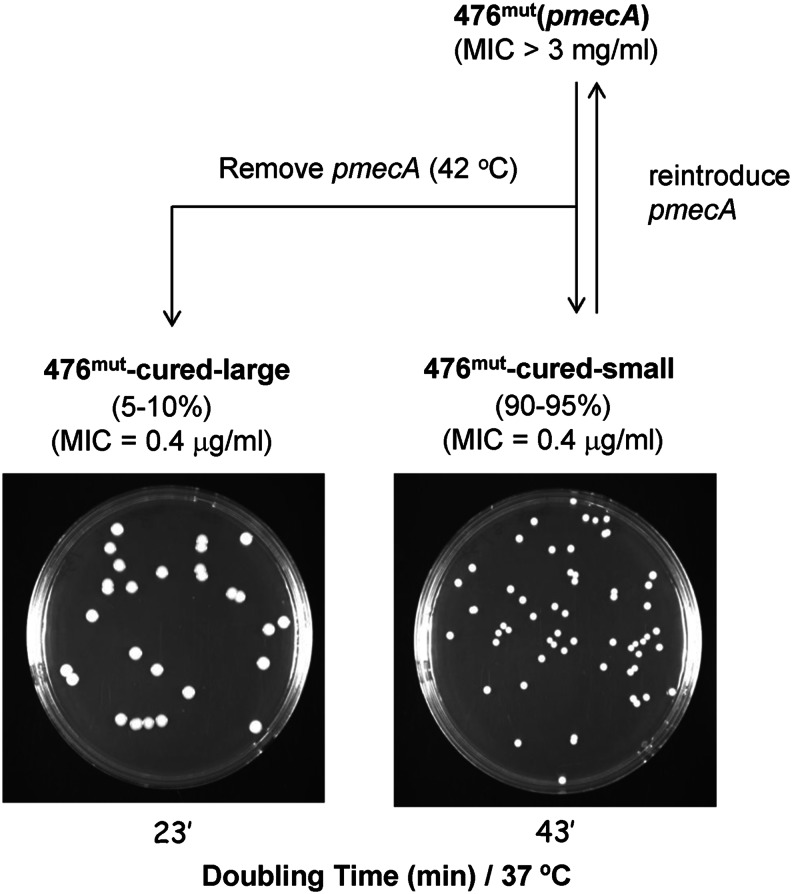
Growth of the highly resistant 476mut(pmecA) cells at 42°C, a temperature nonpermissive for plasmid replication produced bacteria that were free of the plasmid and plating of such cured (plasmid-free) bacteria on TSA produced primarily (over 95%) small colonies (named 476mut-cured-small) similar to the size of the original 476mut(pmecA). A small proportion of the cured bacteria produced somewhat larger colonies (named 476mut-cured-large) (Fig. 2). Both populations of the cured bacteria regained susceptibility to oxacillin with MIC values identical to that of the parental strain 476 (oxacillin MIC=0.4 μg/ml).
FIG. 2.
Schematic representation of methods used to generate various derivatives of the transductants of strain 476. 476mut cells free of the pmecA plasmid were produced by incubation of 476mut(pmecA) at 42°C, resulting in 95% small colonies (named 476mut-cured-small) and 5% large colonies (named 476mut-cured-large). Reintroduction of the pmecA plasmid into 476mut-cured-small cells produced bacteria with homogenous and high-level resistance to oxacillin indistinguishable from that of the original 476mut(pmecA) transductants.
Reintroduction of the pmecA plasmid into the small-size cells in the second round of transduction produced a homogenous culture of highly oxacillin-resistant bacteria indistinguishable from the original 476mut(pmecA) transductants—both in the antibiotic resistance level and in their small colony size.
These observations clearly indicate that important determinant(s) that define the level of oxacillin resistance (MIC value) in strain 476mut(pmecA) must reside in the genetic background of the bacteria.
Identification of the mutations in the highly resistant bacteria
To identify the mutation(s) responsible for the high oxacillin resistance of 476mut(pmecA), whole-genomic sequencing was done using the Genome Sequencer FLX from Roche 454.31 The only mutations found in the highly resistant mutant were two single-nucleotide substitutions that occur in the relA gene on the chromosomea (Fig. 3). PCR sequencing confirmed that these mutations were not sequencing or assembly errors.
FIG. 3.
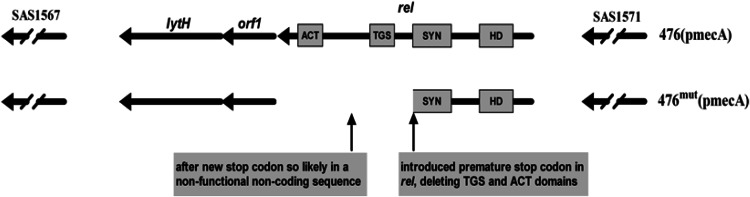
Structure of the relA gene in Staphylococcus aureus strains 476(pmecA) and location of the two-point mutations in the relA gene of 476mut(pmecA). In the 736 aa-long wild-type RelA protein, the Pfam search found an aa 57–156 match hydrolase domain (HD; E-score=10−17); an aa 247–357 match synthetase domain (SYN; E-score=10−51); and an aa 402–461 and 661–727 match, respectively, TGS and ACT (respective E-scores=10−26 and 10−7). In the 2,211-bp-long relA gene, the actual base changes are g→a at the 1,052nd base introducing a new premature stop codon and g→c at the 1,546th base. The former substitution caused the mutant RelA protein lack of TGS and ACT domains.
The relA gene has been partially characterized experimentally in a study unrelated to antibiotic resistance16 and is involved in the production of the alarmone (p)ppGpp. When faced with nutrient starvation, virtually all bacterial species synthesize high levels of (p)ppGpp,25 with S. aureus being no exception.8 The effector (p)ppGpp then goes on to initiate a complex cellular program called the stringent response that modulates the expression of many genes and slows down growth.25
To better understand the effects of the mutations in relA, a Pfam search13 was done that predicted that the wild-type Rel protein consists of at least four domains (Fig. 3). Listed in order from the amino- to carboxy-terminus, the domains are as follows: hydrolysis domain (HD), responsible for hydrolyzing/degrading (p)ppGpp; synthetase (SYN), responsible for synthesizing (p)ppGpp; and TGS and ACT, whose functions are poorly understood.
Of the two single-nucleotide substitutions in the highly resistant mutant, one introduced a premature stop codon in relA that deleted the TGS and ACT domains. The other mutation occurs after the new stop codon in what is likely to be a noncoding nonfunctional sequence. That only the TGS and ACT domains were deleted may be significant as a larger disruption of relA was proposed to be lethal.16
Studies of orthologs of relA in other bacterial species offer some insight into the functions of the TGS and ACT domains. Blasts33 of sequences in GenBank6 indicate that relA has a very close ortholog relMsm in Mycobacterium smegmatis MC2 155.b The RelMsm protein is known to have the same domain organization as depicted in Fig. 3, and in an in vitro system, deletion of the TGS and ACT domains has been found to increase in the net rate of (p)ppGpp synthesis by this protein.24 An increased cellular level of (p)ppGpp in the highly resistant mutant would be consistent with the mutant's observed reduced growth rate.
A partial revertant with reduced resistance carries an extra mutation in a gene relQ that codes for another (p)ppGpp synthetase
Removal of the pmecA plasmid from strain 476mut(pmecA) by growing bacteria at 42°C produced antibiotic-susceptible cells named 476mut-cured-small most of which (95%) have retained the small colony size characteristic of 476mut(pmecA). Nevertheless, a fraction of the cured cells (about 5%) produced larger colonies called 476mut-cured-large.
Reintroduction of the pmecA plasmid into the majority of the cured cells, which had the small colony size (476mut-cured-small) produced transductants indistinguishable from 476mut(pmecA) both in the resistance level and colony size. Reintroduction of the plasmid-borne mecA into the rare larger colonies (476mut-cured-large) also produced homogeneous oxacillin-resistant cultures, but the actual oxacillin MIC value was somewhat less (about 800 μg/ml) compared with the majority 476mut(pmecA) cells (MIC >3 mg/ml).
PCR sequencing of the relA gene of 476mut-large and 476mut-small showed that both strains carried the same two-nucleotide substitutions in relA (Fig. 3). To identify the presumed additional mutation in 476mut-cured-large, whole-genome sequencing was done using a shortread technology. The only mutation found between the highly resistant mutant and the partial revertant was a single-nucleotide substitution in the latter strain in the gene relQ, which is separated from relA by 728,007 bases on the chromosome. PCR sequencing confirmed that the base change in relQ was not a sequencing or assembly error.
The relQ gene has not been characterized experimentally, but its function can be inferred using standard bioinformatics tools. A Pfam search13 found that the wild-type RelQ protein consists of a (p)ppGpp synthetase domain SYN.c The base change in the partial revertant converted a valine to a glycine in the middle of SYN, possibly affecting (p)ppGpp synthesis.d Relative to the highly resistant mutant, a decrease in the cellular (p)ppGpp level in the partial revertant would be consistent with the strain's observed increased growth rate and reduced antibiotic resistance.
Interestingly, blasts33 of sequences in GenBank6 reveal that the two genes relA and relQ (and another gene relP also in S. aureus 476) have very close orthologs (relASmu, relQSmu, and relPSmu respectively), in another gram-positive bacterium Streptococcus mutans UA159.1,e Very recently, these three genes in S. mutans were all shown experimentally to code for (p)ppGpp synthetases.29
That the extra gene found to be mutated in the partial revertant, is relQ, is highly statistically significant. There are roughly 2,800,000 bp and 2,500 predicted genes in the S. aureus genome, so it is exceedingly unlikely that it is a mere coincidence that the one extra gene found to be mutated in the partial revertant also codes for a (p)ppGpp synthetase.
The role of stress response in the expression of β-lactam resistance
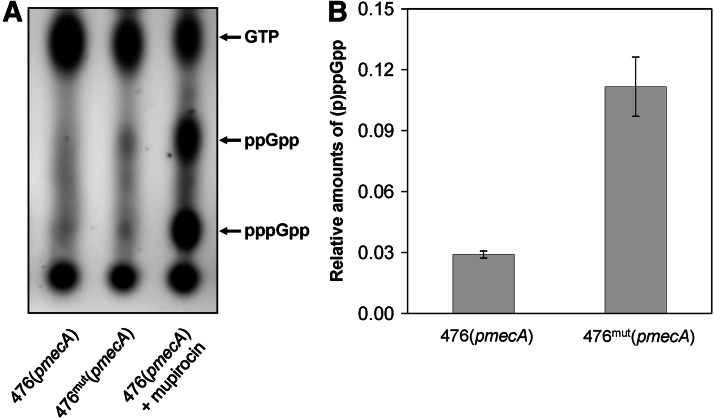
The findings described in this communication suggest that expression of high-level resistance in our model system was connected to mutations in the relA and/or relQ genes, which should cause production of increased amounts of (p)ppGpp in the resistant bacteria. To test this, we determined the cellular amounts of this effector. Figure 4 shows that the model strain 476mut(pmecA) indeed had larger relative amounts of (p)ppGpp—approximately fourfold—as compared to strain 476(pmecA). The fractions of (p)ppGpp in strains 476(pmecA) and 476mut(pmecA) were 0.029 and 0.113, respectively. Increased relative level of (p)ppGpp in 476mut(pmecA) is similar to that in the small colony variant of MRSA JKD6210, which carries a point mutation in the relA gene causing reduction of the RelA hydrolase activity and constitutive activation of the stringent response.14
FIG. 4.
Oxacillin resistance and cellular amounts of (p)ppGpp. (A) Cell extracts were obtained from identical amounts of strains 476(pmecA) and 476mut(pmecA). The relative amounts of (p)ppGpp were determined by thin layer chromatography followed by autoradiography of extracts from 32P-labeled bacteria. The sample from 476(pmecA)+mupirocin was used as a control for the (p)ppGpp spots on TLC plates. (B) Relative amounts of (p)ppGpp are expressed as a fraction of the total 32P-labeled guanine phosphate precursor pool.
Involvement of a stress response in the β-lactam-resistant phenotype of MRSA has been proposed earlier.11 A direct effect of the stringent stress response on the β-lactam resistance level is shown in Fig. 1A. Plating strain 476(pmecA) on oxacillin-containing agar plates (see curve with solid square symbols) produced a heterogeneous phenotype in which, the overwhelming majority of bacteria expressed only very low levels of antibiotic resistance (MIC around 0.75 μg/ml). When the same population analysis was repeated using agar plates that also contained a sub-MIC concentration of mupirocin (see curve with solid circle symbols in Fig. 1A), culture strain 476(pmecA) was able to express homogenous and high-level resistance indistinguishable from the resistance level obtained in strain 476mut pmecA, that is, the strain that carried the relA mutation.
Of the roughly 2,500 predicted genes in the S. aureus genome, the only two genes found to be mutated in the highly resistant mutant and its partial revertant both code for (p)ppGpp synthetases. Involvement of (p)ppGpp may explain many of the peculiar properties of β-lactam resistance in MRSA, since variables such as shifts in temperature, salt concentration, and pH, which were shown to impact on β-lactam resistance in MRSA, are also known to affect cellular (p)ppGpp levels in several bacterial species.21,40 Any one of the many mutations in any one of the many genes can also be expected to modulate (p)ppGpp levels, including genes important for the uptake and utilization of nutrients.25
Footnotes
The gene relA has the identifier SAS1570 in the annotation for the 476 genomic sequence produced by the Sanger Institute.
The two proteins were found to be excellent best blastp reciprocal matches (E-scores<10−160, with 41% aa identity extending over their entire lengths).
In the 211 aa long wildtype relQ protein, aa 43-162 match SYN (E-score=10−43).
In the 636 bp long relQ gene, the actual base change is t→g at the 299th translated base.
The gene relP has the identifier SAS2394 in the annotation for the 476 genomic sequence. The rel and relstrep proteins were found to be excellent best blastp reciprocal matches (E-scores=0, with 50% identity over their entire lengths). Ditto for the relQ and relQstrep proteins (E-scores<10−68, 58% aa identity over entire lengths). Ditto for the relP and relPstrep proteins (E-scores<10−27, 39% aa identity over entire lengths).
Acknowledgments
We thank Shang Wei Wu, Yanjiao Zhou, and Aude Antignac for assistance in setting up the experimental model described in this communication. This work was supported by a grant from the U.S. Public Health Service 2 RO1 AI457838-13 and grant # UL1 TR000043-07S1 from the National Center for Advancing Translational Sciences (NCATS, National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) Program awarded to A.T., and M.M. was a recipient of the Rockefeller University Henry and Marie Josee Kravis Postdoctoral Fellowship.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ajdic D., et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antignac A. Tomasz A. Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob. Agents Chemother. 2009;53:435–441. doi: 10.1128/AAC.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba T., et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 4.Bancroft E.A. Antimicrobial resistance: it's not just for hospitals. JAMA. 2007;298:1803–1804. doi: 10.1001/jama.298.15.1803. [DOI] [PubMed] [Google Scholar]
- 5.Beck W.D. Berger-Bachi B. Kayser F.H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J. Bacteriol. 1986;165:373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson D.A., et al. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Cassels R. Oliva B. Knowles D. Occurrence of the regulatory nucleotide ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 1995;177:5161–5165. doi: 10.1128/jb.177.17.5161-5165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers H.F. Hackbarth C.J. Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1987;31:1982–1988. doi: 10.1128/aac.31.12.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lencastre H., et al. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 1991;35:632–639. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Lencastre H., et al. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 1999;5:163–175. doi: 10.1089/mdr.1999.5.163. [DOI] [PubMed] [Google Scholar]
- 12.de Lencastre H. Oliveira D. Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 2007;10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn R.D., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao W., et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Álvarez L., et al. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentry D., et al. The rel gene is essential for in vitro growth of Staphylococcus aureus. J. Bacteriol. 2000;182:4995–4997. doi: 10.1128/jb.182.17.4995-4997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman B.J. Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman B.J. Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 1986;29:85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillier L.W., et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 20.Holden M.T., et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U S A. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikehara K., et al. Accumulation of relA gene-independent ppGpp in Bacillus subtilis vegetative cells upon temperature shift-down. J. Biochem. 1984;95:895–897. doi: 10.1093/oxfordjournals.jbchem.a134684. [DOI] [PubMed] [Google Scholar]
- 22.Ito T. Katayama Y. Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.IWG-SCC. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain V., et al. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 2006;15:1449–1464. doi: 10.1110/ps.062117006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain V. Kumar M. Chatterji D. ppGpp: stringent response and survival. J. Microbiol. 2006;44:1–10. [PubMed] [Google Scholar]
- 26.Jevons M.P. Celbenin-resistant staphylococci. Br. Med. J. 1961;1:124–125. [Google Scholar]
- 27.Katayama Y. Zhang H.Z. Chambers H.F. PBP 2a mutations producing very high-level resistance to β-lactams. Antimicrob. Agents Chemother. 2004;48:453–459. doi: 10.1128/AAC.48.2.453-459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim C., et al. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J. Biol. Chem. 2012;287:36854–36863. doi: 10.1074/jbc.M112.395962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemos J.A., et al. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 2007;65:1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- 30.Lowy F.D. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 2003;111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margulies M., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews P. Tomasz A. Insertional inactivation of the mec gene in a transposon mutant of a methicillin-resistant clinical isolate of Staphylococcus aureus. Antimicrob. Agents Chemother. 1990;34:1777–1779. doi: 10.1128/aac.34.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGinnis S. Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mechold U. Malke H. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J. Bacteriol. 1997;179:2658–2667. doi: 10.1128/jb.179.8.2658-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami K. Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 1989;171:874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poly F., et al. Genome sequence of a clinical isolate of Campylobacter jejuni from Thailand. Infect. Immun. 2007;75 doi: 10.1128/IAI.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raskin D.M. Judson N. Mekalanos J.J. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. U S A. 2007;104:4636–4641. doi: 10.1073/pnas.0611650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sieradzki K. Chung M. Tomasz A. Role of a sodium-dependent symporter homologue in the thermosensitivity of beta-lactam antibiotic resistance and cell wall composition in Staphylococcus aureus. Antimicrob. Agents Chemother. 2008;52:505–512. doi: 10.1128/AAC.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velicer G.J., et al. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. U S A. 2006;103:8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells D.H. Gaynor E.C. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J. Bacteriol. 2006;188:3726–3729. doi: 10.1128/JB.188.10.3726-3729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S.W. de Lencastre H. Tomasz A. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 2001;183:2417–2424. doi: 10.1128/JB.183.8.2417-2424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]