Abstract
Double-stranded DNA bacteriophage genomes are packaged into their icosahedral capsids at the highest densities known so far (about 50 % w:v). How the molecule is folded at such density and how its conformation changes upon ejection or packaging are fascinating questions still largely open. We review cryo-TEM analyses of DNA conformation inside partially filled capsids as a function of the physico-chemical environment (ions, osmotic pressure, temperature). We show that there exists a wide variety of DNA conformations. Strikingly, the different observed structures can be described by some of the different models proposed over the years for DNA organisation inside bacteriophage capsids: either spool-like structures with axial or concentric symmetries, or liquid crystalline structures characterised by a DNA homogeneous density. The relevance of these conformations for the understanding of DNA folding and unfolding upon ejection and packaging in vivo is discussed.
Keywords: Bacteriophage, DNA packaging, DNA ejection, Cryo-electron microscopy
Introduction
Tailed bacteriophages (Caudovirales) are remarkable supramolecular assemblies formed by a dsDNA molecule confined within an icosahedral protein capsid which diameter is in the range of the DNA persistence length. One of the apex of the icosahedron is occupied by a connector, or portal protein used as the DNA entry and exit gate, where the tail anchors. In contrast with most Eukaryotic viruses, tailed bacteriophage do not enter the cell to infect their host bacteria. After recognition and fixation upon a receptor localized on the bacterial surface, they inject their DNA content inside the host, while the phage capsid and tail remain outside, leading to the internalization of the genome. Lytic phages then use the bacterial machinery to synthesize their components and assemble inside the cell. In tailed bacteriophages (and other related eucaryotic dsDNA viruses such as herpes), a procapsid is first assembled. Then, the DNA is dragged in through the portal gate, using ATP hydrolysis as a source of energy to package the molecule against the concentration gradient, overcoming the repulsive interactions between DNA segments and its resistance to bending. The process, which is associated with a capsid conformational change, is followed by the attachment of the pre-assembled tail.
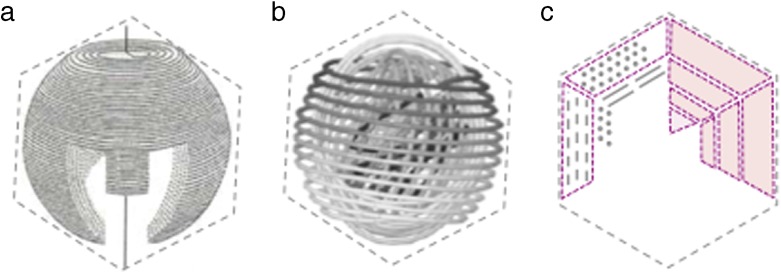
How does a tens of micrometres long, semi-flexible DNA molecule fold in the volume of an icosahedron a few tens of nanometres in diameter? The question as to how DNA is organised inside the phage capsid is a central one in the understanding of infection and packaging processes. Over the years, many models have been proposed and controversy remains. Beyond the local hexagonal packing of DNA, evidenced in the 60’s by X-Ray diffraction [1, 2] and later on by cryo-MET [3, 4], there is no definitive and complete understanding of DNA folding and conformation (see for example review in [5]). Some models are presented in Fig. 1. The inverse axial spool (Fig. 1a), proposed about 40 years ago [6, 7] is the most widely accepted, supported by the spectacular cryo-TEM images of bacteriophage T7 [8], and, more recently, by asymmetric reconstructions of phages ε15 [9], P22 [10], K1 [11] and P-SPP7 [12]. It is used as a basis by many theoretical approaches [13–16]. However, the model suffers from several limitations: (i) it seems to be restricted to phage species that possess a large inner cylindrical proteic core, and (ii) even for these species, it does only describe the path of DNA with accuracy in some very limited regions of the capsid, and is far form accounting for the complete genome folding. A variant of the axial spool model, the inverse “ball of yarn” (or concentric spool, Fig. 1b), has been predicted by recent simulations for phage lacking inner cylindrical cores [17]. As in an axial spool, DNA wounds from the periphery to the centre following successive hoops, but with spherical symmetry. In the “liquid crystalline drop” model (Fig. 1c) derived from cryo-TEM analyses [3, 18], hexagonally crystallized monodomains entirely fill the capsid volume, separated by defect walls and forming a structure related to a confined TGB (Twist Grain Boundary) liquid crystal.
Fig. 1.
Some models of DNA organisation in the phage capsid. a Inverse axial spool, reproduced from [2]. b Inverse concentric spool (or ball of yarn), reproduced from [41]. c Liquid crystalline drop. Monodomains (in purple) of DNA segments hexagonally crystallised entirely fill the capsid and are separated by defect walls
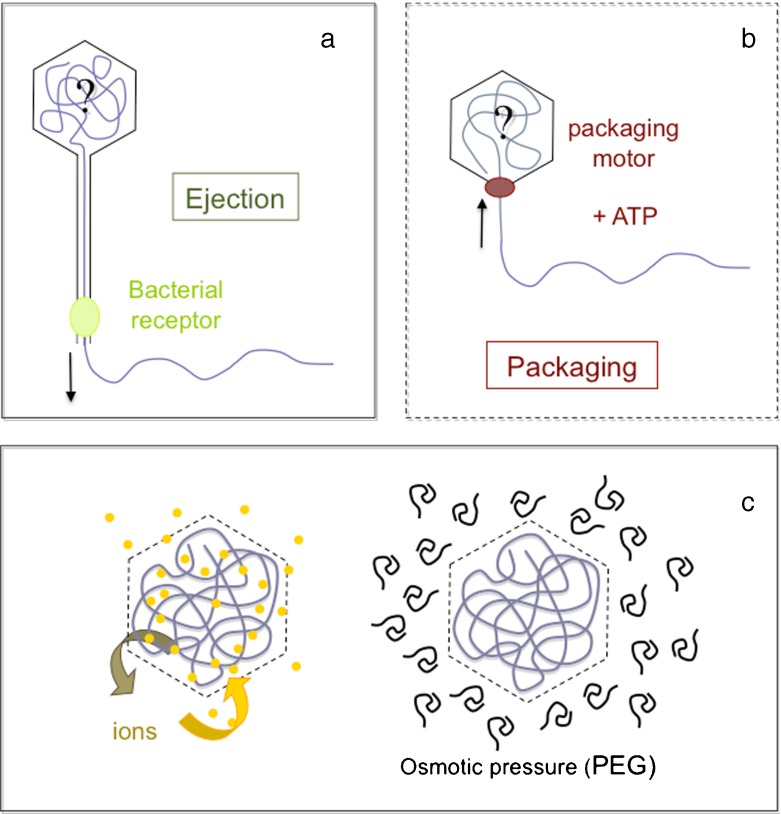
How does the DNA conformation evolve during the ejection and packaging of the molecule? What do we know about the interactions (DNA-DNA or DNA-capsid interactions) involved in these processes? During the last ten years the mechanisms underlying the ejection as well as the packaging processes have been extensively investigated, both experimentally and theoretically. Novel in vitro experimental approaches have become possible, thanks to the DNA packaging and ejection kits now available for some phage species (Fig. 2a, b). DNA packaging in ϕ29, T4 and λ can be monitored in vitro in the presence of ATP [19–21]. In contrast, DNA ejection is in vitro a passive process, triggered by the interaction of the phage with its bacterial protein receptor. Receptors of phages λ, T5 and SPP1 are now identified and purified [22–24]. This has lead to new wealth of information regarding the forces and energies involved in these processes ([19, 25], see reviews in [26–28]). The capsid of bacteriophages λ, T5 and SPP1 is semi-permeable, permeable to water and small ions, impermeable to proteins and polymers (Fig. 2c). In vitro experiments are thus prone to the exploration of wide variety of chemical environments: the nature and concentrations of ionic species inside the capsid can be tuned nearly at will, as well as the osmotic pressure applied outside [25, 29–32]. Combined to a cryo-EM approach, ejection (or packaging) experiments in vitro provide direct insights in the DNA conformational changes. In this article we focus on the multiplicity of DNA conformations that can be found inside phage capsids according to the length of the DNA molecule and the physico-chemical environment: temperature, presence or absence of DNA-condensing multivalent cations, or osmolytes such as Poly Ethylene Glycol (PEG). We will discuss the relevance of this polymorphism and its possible relationships with the packaging and ejection processes in vivo.
Fig. 2.
aIn vitro, DNA ejection is a passive process triggered upon interaction of the phage tail and its purified receptor (in acid green). b DNA is packed in vitro into the purified procapsid by the ATP-driven portal motor (in red). c The capsid of many bacteriophages is permeable to water and ions, impermeable to crowding polymers such as PEG, which allows us to tune at will the osmotic pressure and the ionic environment of the molecule
Results: DNA organisation in partially filled capsids
The length of the DNA molecule encapsidated can be varied upon partial ejection. In is λ and SPP1 the extent of the ejection can be tuned precisely applying a controlled osmotic pressure leading to populations of phages containing a given DNA length [24, 25]. In T5, the ejection is a discrete multisteps process interrupted by a series of pauses [33, 34]. This character provides a unique opportunity to dispose of a polydisperse population of phages, permanently (under osmotic pressure [35]) or transiently (in the absence of pressure [18]) arrested at one or another of these pauses, and containing from a few kbp to tens of kbp.
DNA organisation in the presence of monovalent and divalent counterions
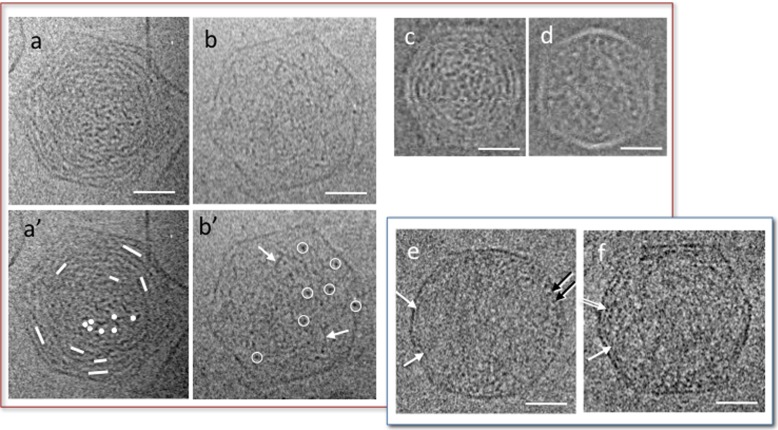
We have triggered DNA ejection from bacteriophage T5 in the presence of its receptor FhuA at 37 °C, and analysed the DNA conformation in partially filled capsid quenched at low temperature for cryo-TEM observation. In the presence of mono- and di-valent cations (here 10 mM Tris-Cl, 100 mM NaCl, 1 mM MgCl2, 1 mM CaCl2), the DNA organisation inside the capsids varies with its length [18]. Some of these conformations are shown in Fig. 3a, b. Above 380 mg/ml, the DNA is hexagonally packed (not illustrated, see [18]). At lower filling, a cholesteric organisation forms. It can be recognised in Fig. 3a, a′ with its characteristic rotation of the molecular orientations between the centre and the periphery of the capsid (underlined in Fig. 3a′). This organisation is observed between 380 and 160 mg/ml (i.e. for encapsidated lengthes Lin comprised between 70 and 30 % L0, L0 being the total genome length in the full-filled capsid). Figure 3b, b′ shows an example of isotropic (disordered) conformation, observed for DNA concentration below 160 mg/ml (Lin < 30 % L0). Portions of the chain are seen randomly in all possible orientations over the whole projected volume, from perpendicular (dot-like, white circles in Fig. 3b′) to oblique and parallel (threads-like, black arrows). In all cases, and at all stages of the ejection, the DNA takes up all the available volume and fills homogeneously the capsid [18]. There is neither preferential crowding, nor exclusion at the periphery. Interhelix spacing, that can be measured locally on cryo-TEM micrographs of hexagonal and cholesteric organisations when the molecule is favourably oriented, is roughly constant within a given capsid, and varies between capsids with the DNA concentration.
Fig. 3.
Conformations observed by cryo-TEM of partially filled capsid in the presence of mono- and di-valent ions and quenched at room temperature (a–d) and at 4 °C (e, f). Cholesteric (a, c) and isotropic (b, d) organisations observed in bacteriophage T5 (a, b) and T3 (c, d, reproduced from [37]) in Tris 10 mM, NaCl 100 mM, MgCl2 1 mM, CaCl2 1 mM (T5) and Tris 10 mM, NaCl 200 mM, MgCl2 1 mM (T3). In the cholesteric conformations the orientation of the DNA segments (underlined by dots and lines in a′) rotates between the central region and the periphery, in a double twist organisation. In the isotropic conformation, DNA is randomly oriented. DNA segments locally perpendicular to the observation plane are circled in white (b′). Some of the DNA segments locally oblique or parallel to the observation plane are pointed by white arrows. e, f Partially filled capsids of T5 in the same ionic conditions (Tris 10 mM, NaCl 100 mM, MgCl2 1 mM, CaCl2 1 mM), quenched after 15 min stabilisation at 4 °C. DNA segments locally visible in transverse view (dots, black arrows) or oblique or longitudinal views (white arrow) accumulate at the periphery, forming a monolayer in some regions. A double layer is also visible locally (double arrow). Scale bars = 20 nm
The same behaviour is observed under similar ionic conditions, in partially filled capsids of other phages ϕ29 [36] and T3 [37] that are markedly smaller than T5. In particular, partially filled T3 capsids show a homogeneous distribution of the DNA, with cholesteric and isotropic configurations, found in the same concentration ranges (here 285 and 110 mg/ml respectively) (Fig. 3c, d).
This series of phases also forms in bulk solutions of short DNA fragments [38, 39] and the critical concentrations at phase transitions are the same in both cases, suggesting that, under these physico-chemical conditions, electrostatic repulsions dominate the contribution of the bending energy and govern the genome organisation in the capsid, even in capsids of small radii with high bending constraints such as T3.
A comparable series of structural and energetic stages was reported in numerical simulations of DNA in bacteriophage capsids [40], with an isotropic to liquid crystalline (nematic) transition, but with significant differences: the transition is predicted at 80 % filled capsids (in contrast to about 30 % here). In addition, the simulations show a heterogeneous DNA distribution, with a higher density at the periphery. The observation of an homogeneous distribution within the capsid whatever the encapsidated length is indeed in contrast with most theoretical [14–16] and numerical approaches [40–42] that predict higher DNA concentration in the outer region of the capsid. The importance of this heterogeneity at low DNA contents depends on the balance of the relative contributions of the electrostatic energy and bending energy. Our observations show that the contribution of the elastic energy is probably overestimated in the models.
Screening DNA-DNA repulsions at high ionic strength should in principle modify this balance, then possibly leading to an increase of the DNA density at the capsid periphery. We did not analyse so far the effect of increasing the ionic strength on the observed conformations. Yet, an X-ray scattering analysis in a λ mutant containing 78 % of the wt genome has shown a decrease of the interhelix spacing with the increase of the added salt concentration (either mono or di-valent from a few mM to molar concentration), probably indicative of a concentration of the DNA molecule at the periphery of the capsid [43].
Changes in the temperature can also be expected to modify the balance between the electrostatic and elastic energies. Increase of the DNA persistence length was reported with the decrease of temperature [44, 45]. The ejection process itself is very sensitive to temperature [46]. After partial ejection at 37 °C, T5 capsids were transferred and stabilised at 4 °C before freezing for cryo-TEM observation. Most images show that the DNA distribution is not homogeneous throughout the volume of the capsid. There is a DNA accumulation at the periphery. A monolayer, and in some cases a second innermore layer, is (are) locally pressed against the capsid wall (Fig. 3e, f, arrows). Although we do not dispose of 3-dimensional reconstructions of the path of the molecule, the absence of dotted patterns in the central region suggests that the inner part of the capsid contains little or even no DNA. In contrast with capsid quenched at room temperature, DNA therefore does not distribute over all the capsid volume, but crowds at its periphery and would wound into concentric layers taking an inverse ball of yarn conformation (concentric spool). Nonetheless, in these samples, about 10 % of the phages show a DNA homogeneous distribution with conformations similar to those observed at room temperature. It is therefore likely that our experimental conditions here correspond to a region of transition between the two conformations.
DNA organisation in the presence of multivalent cations
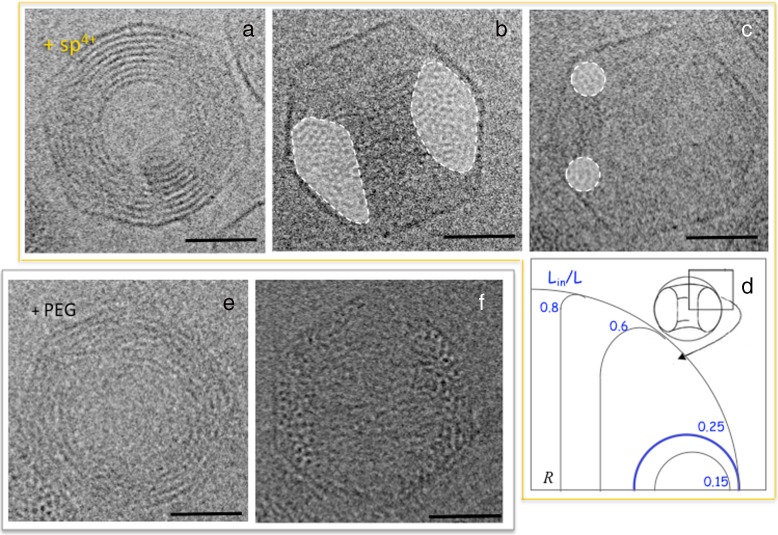
In the presence of multivalent cations such as spermine 4 + , spermidine 3 + or Cobalt hexamine 3 + , DNA molecules in solution collapse into toroids [47–49]. The same phenomenon can occur inside bacteriophage capsids. After partial ejection triggered in the presence of mono and divalent ions only (as above), we added spermine at a DNA condensing concentration known to induce a hexagonal packing when DNA is condensed in solution (here 5 mM, see [50, 51]). Spermine diffuses through the capsid wall inducing attractive DNA-DNA interactions. The molecule collapses into a toroid which is confined inside the capsid, as illustrated in Fig. 4a and b which show two spermine-induced toroids observed in top and side views respectively. The fine DNA structure within these confined toroids is analysed in detail elsewhere [52]. Briefly, DNA packs with a hexagonal order with a lattice parameter aH that depends on the ionic conditions (relative amounts of spermine and mono + di-valent cations). Note that the distance aH is always larger than aH0, the distance in the initial full capsid. Complex phenomena superimpose to the hexagonal order. The competition with the chirality of the molecule, at work in all dense states of DNA [53], results here in the rotation of the hexagonal lattice along the toroid circumference. In combination with correlations of neighbouring segments of double helix and the strong curvature due to the confinement, the phenomenon results in periodic variations of the DNA helical pitch around the toroid [52].
Fig. 4.
Toroidal conformations observed by cryo-TEM in partially filled capsid after condensation of the DNA segments remaining in the capsid with spermine 4+ (a–c), or PEG (e, f, reproduced from [32]). Toroids are observed in top (a, e) and side views (b, c, f). On side views, the surface of the cross section is shaded in white. d Theoretical prediction of the shape of toroids in the absence of DNA-capsid interactions for different ratio of encapsidated DNA length (Lin) to the full genome length (L0), redrawn from [15]. Scale bars = 25 nm
We analysed confined toroids formed by DNA molecules of various lengths, from 3 to 55 kbp. Due to the confinement, not only the size but also the shape of the toroid varies with this length. The main characteristics of the shapes of the toroids are revealed by the observation of side views (Fig. 4b, c). For toroids formed by DNA molecules smaller than 28.5 kbp (i.e. Lin/L0 ≤ 0.25, with L0 being the total genome length), the cross section can be approximated to a circle, the toroid then corresponding to a true torus (Fig. 4c). Within this range of DNA lengths, the radii of the toroids are smaller than that of the capsid container Rc. For the smallest molecules observed here (about 3000 kbp), it can be as small as Rc/2 [52]. For longer molecules (Lin/L0 > 0.25), the cross section elongates in the perpendicular plane (Fig. 4b), with a geometry that approaches that of a cylindrical spool.
A theoretical approach of the shape of a DNA toroidal condensate inside the phage capsid predicts a similar phenomenon with a torus to spool transition at Lin = 0.25L0, in the absence of interactions (either repulsive or attractive) between the DNA and the capsid wall, the only constraint being the confinement [15]. Figure 4d shows the theoretical cross sections predicted for different filling ratio. Our experimental data are in good agreement regarding altogether the overall shape of the condensate and the threshold for the transition.
In addition, we observed that the position and orientation of the toroid inside the capsid also influences its shape, due to the anchoring on the triangular faces of the icosahedron (not illustrated). This effect should be analysed more precisely.
DNA organisation under osmotic stress
DNA condensation into toroids can be obtained not only in the presence of polyvalent cations as above, but also under osmotic stress with crowding agents such as Poly Ethylene Glycol (PEG) [54, 55]. Adding 15 % PEG6000 to the external medium after partial DNA ejection results, as the addition of spermine does, into the collapse of the encapsidated DNA into a toroid. The PEG does not permeate the capsid (which allows only water and ion exchanges) but creates a pressure gradient across the capsid wall, which would origin toroid formation. The mechanisms that govern this phenomenon remain to be clarified. Whatsoever, the local order within the toroids is also hexagonal. The shape of the toroids is markedly different from those obtained in the presence of spermine, as can been seen on side view images (Fig. 4f). They present a non-convex cross section with a crescent shape. This unique shape would result from attractive interactions between the capsid proteic surface and the DNA molecules [32].
Discussion
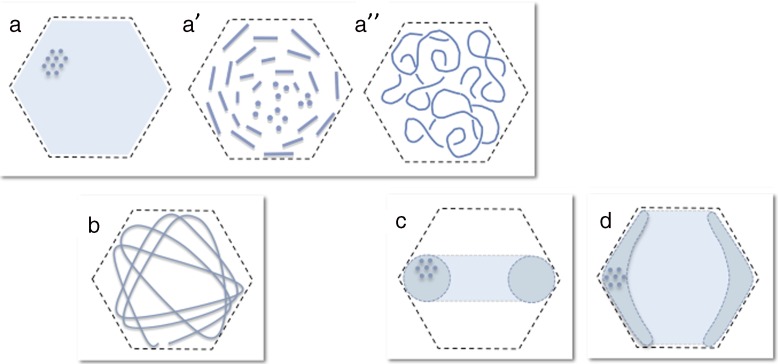
We have shown that, in partially filled bacteriophage capsids, DNA can adopt a multiplicity of conformations depending on both the length of the encapsidated portion of the genome, and the physico-chemical environment (ionic species, temperature, osmotic pressure), while these conditions have no effect on full-filled capsid (unpublished results, see also [43]). This polymorphism is sketched on Fig. 5. It is important to note that these conformations are probably not the only possible ones. Other may occur under other—yet unexplored—conditions. Both the nature and concentrations of ionic species should be explored thoroughly, as well as a wide range of temperature and osmotic pressure. For example, the organisation and density of DNA condensates obtained in the presence of polycations depends on the composition of the specimen (i.e. nature and relative amounts mono-, di- and multi-valent ions [50, 51, 56]). Thus, it could well be that toroids with different local packing (cholesteric-like order for example rather than hexagonal) could also exist.
Fig. 5.
Polymorphism of DNA conformation in partially filled capsids. a–a″ Capsids homogeneously filled with DNA with a series of liquid crystalline organisations (hexagonal (a) and cholesteric (a′) at high and intermediate filling) and an isotropic confined coil at low filling (a″). b Centro symmetric inverse ball-of-yarn conformation with accumulation at the periphery. c Confined toroidal globule. d Confined toroidal globule with capsid-DNA attractive interactions. a–a″ No symmetry. b Centro-symmetric conformation. c, d Axial symmetry
The role of the nature and concentration of divalent cations could also provide new insights. Some experimental and numerical data suggest that, in situation of confinement, Mg2 + could, at certain concentration, induce DNA condensation [57, 58], possibly leading to novel conformations.
The role of the size of the capsid should also be addressed more specifically, although the available data suggest that (at least within the 50–80 nm range that can be considered so far) it does not play a dominant role. Cholesteric and isotropic conformations occur in T3 as well as in T5 (Fig. 3). Toroids form in the presence of spermine or PEG in partially filled capsids of both T5 and λ [59, 60]. Nonetheless, the structure of the toroids formed in the smallest capsids remains to be analysed in details. The capsid size could possibly play a role in many features of importance such as the shape of the toroids, the defects of the hexagonal packing [18], or the variations of the DNA helical pitch observed in confined toroids [52]. It can also be expected to shift the phase transitions between different states, especially between random coil and concentric inverse ball conformations for example (Fig. 5a′, b). In addition, other factors, able to introduce or relax geometrical constraints, are also probably involved here such as the presence of proteic core inside the capsid in some species, or the shape of the capsid—isometric or oblate [17].
Can we learn about the DNA organisation in full-filled capsids looking at partially filled capsids? It is striking to notice that the different structural models presented in Fig. 1 are directly related to the different conformations described here and sketched in Fig. 5. The series of liquid crystalline organisations (Fig. 5a–a″) point to the liquid crystalline drop model with its homogeneous DNA density (Fig. 1c). The conformational changes observed with the decrease of temperature result in a concentric spool-like structure (Figs. 2b and 5b). Lastly the condensation of the molecule either in the presence of polycations or under pressure induces the collapse of the molecule into toroidal structures (Fig. 5c, c′), closely related to the axial spool model. Numerical and theoretical approaches do indeed predict a variety of conformations according to the environment of the molecule [15, 40, 42, 58]. However, while it is clear in all these approaches that attractive interactions lead to toroid or spool-like configurations (in qualitative agreement with the observations), none of them can account for the homogeneous filling found at room temperature with repulsive DNA-DNA interactions (see for example [41, 42]). We have hypothesized that the bending energy is overestimated in theoretical and numerical studies. The persistence length of the DNA is generally taken as 50 nm [61], while it has been measured as 35 nm in the presence of divalent cations (present in all phage buffers) [62]. In addition, at very small scale (5 to 15 nm), it has been shown that the DNA does not behave like a Worm Like Chain, and shows an increased flexibility [63, 64]. The underlying mechanisms are still unknown but could involve local opening of the double helix with the formations of single strand “bubbles” and/or kinks [63]. This would introduce discontinuities that are not taken into account by theoretical and numerical models. Supercoiling of the molecule could also play a role. Whatsoever, the diversity of the models happens to reflect the diversity of the conformations observed experimentally. The different models proposed for DNA organisation may therefore not be exclusive, but valid under certain physico-chemical (ionic environment, osmotic pressure, temperature) and geometrical (absence or presence of large inner proteic core, size and shape of the capsid) conditions. Note that other models such as the folded and twisted toroids proposed for elongated capsids (although not recalled here) may also be relevant.
Models of DNA organisation in bacteriophage capsid are not only structural but also mechanistic and should account for packaging and ejection processes. The polymorphism of partially filed capsids shows that the way the DNA folds and unfolds is strongly dependent on the environment of the molecule. This is to say that the two processes are not symmetrical in vivo. Indeed, in situ, the two processes occur in capsid immersed into very different media (extracellular for the ejection, intracellular for the packaging) that differ in terms of osmotic pressure and ionic environment. Intermediate stages are therefore very unlikely to be the same. Although the ionic and osmotic pressure and ionic conditions are not precisely known in vivo, it could well be that some of the conditions explored here may be relevant for the understanding of the two processes. The few atmosphere osmotic pressure of bacterial cytoplasm for example could favour DNA condensation at the capsid periphery as here in the presence of PEG (Fig. 5d). During ejection, the molecule would rather fill homogeneously the capsid and follow the series of phase transitions described above (Fig. 5a–a″). Note that other factors may also play a crucial role in DNA conformational changes, and in particular the very different kinetics of the two processes.
This highlights the need to be able to follow DNA packaging and ejection in situ. Structural approaches of bacteriophage ejection in situ have already been attempted by cryo-TEM of entire bacteria trapped within a film [65–67]. However, little attention has been paid to the DNA conformation, which observation is hampered by the thickness of the specimens. Cryo-TEM of vitreous sections, which has already proved powerful for the analysis of liquid crystalline organisations in situ as well as in vitro [53, 68, 69], appears here as a possible alternative approach.
All the conditions explored here, although probably relevant, do not reproduce exactly the environment of DNA in interaction with its bacterial host. In addition, this environment is very likely to vary, not only between species, but also between individuals. For example, concentrations gradients inside the bacterial cytoplasm could result in local changes in the DNA folding. In addition, we have already shown that under certain conditions, different conformation can coexist within a specimen (see above at 4 °C). It is therefore possible that there also exists a polymorphism within a phage population in vivo, which could even lead, in some cases to different types of full-filled capsids, as suggested by the multiplicity of configurations observed after DNA release from phage l [70]. The comparison of systematic in vitro experiments performed in well-defined conditions, with in situ analyses could help to clarify the mechanisms involved in both packaging and ejection, and possibly provide an answer to the question as to how DNA is folded in the phage capsid.
Materials & methods
Biochemical purification T5 st(0) bacteriophages were produced in E. coli, purified, and concentrated in 10 mM Tris-Cl, pH 7.6, 100 mM NaCl, 1 mM CaCl2 and 1 mM MgCl2. The receptor FhuA was purified as described in [23]. T5 and FhuA were kindly provided by M. de Frutos, M. Renouard and P. Boulanger (IBBMC, Orsay, France).
Obtention of a population of phages containing various amounts of DNA Partially filled capsids were obtained in two different ways: either by quenching the specimen before the entire DNA molecule is ejected, or by applying an external osmotic pressure to stop the ejection at some intermediate stages, as described in [18, 32] respectively. Briefly, the phage solution was mixed with an excess of FhuA, in the absence or presence of an osmolyte (PEG MW 6000), and transferred to 37 °C to trigger the ejection. DNAse I (Sigma) was present in the solution to digest the ejected DNA.
Change of the physico-chemical environment and obtention of multiple DNA conformations After partial ejection at 37 °C, the specimens were either immediately frozen at room temperature for cryo-TEM observation, or transferred and let to stabilise for 15 min at 4 °C before freezing. To condense DNA remaining in the capsid, two different protocols were used: i) the addition of spermine at a final concentration of 5 mM (followed, when needed, by dilution of the sample to decrease the PEG concentration below 1 % while keeping constant the ionic conditions), ii) DNA ejection in the presence of 15 % PEG 6000 followed by direct freezing of the sample in the PEG solution. The methods are extensively described in [18, 32, 35, 52].
Cryo-electron microscopy 3 μl of the phage solutions were deposited on an EM grid covered with a holey carbon film (Quantifoil R2/2) previously treated by plasma glow discharge. The grid was then blotted to remove the excess of material and quenched by immersion in liquid ethane cooled down by liquid nitrogen. All the process was carried out in a home-made plunging device to control the temperature and hygrometry, to prevent any change of the ionic concentrations of the sample. The grids were transferred to a JEOL 2010 LaB6 or a JEOL 2010-FEG TEM operated at 200 kV. Images were recorded on Kodak S0163 films at a direct magnification of ×50000 under low dose conditions of imaging (10–20 e − /Å2). The defocus was set at 850 nm to optimize the imaging of the DNA lattice spacing in the capsids.
Acknowledgements
The research on T5 was carried out with F. Livolant at the Laboratoire de Physique des Solides (Orsay). We thank G. Pehau Arnaudet (Institut Pasteur, Paris) for his support on the federative FEG-TEM. We thank M. de Frutos, P. Boulanger and M. Renouard (Institut de Biochimie et Biophysique Moléculaire et Cellulaire, Orsay) for purification of T5 and FhuA. I also thank M. de Frutos for discussions and critical reading of the manuscript.
References
- 1.North ACT, Rich A. X-ray diffraction studies of bacterial viruses. Nature. 1961;191:1242–1245. doi: 10.1038/1911242a0. [DOI] [PubMed] [Google Scholar]
- 2.Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 3.Lepault J, Dubochet J, Baschong W, Kellenberger E. Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. EMBO J. 1987;6:1507–1512. doi: 10.1002/j.1460-2075.1987.tb02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, L.W., Thomas, J.A.: Condensed genome structure. In: Rosmann, M.G., Rao, V.B. (eds.) Viral Molecular Machines, Advances in Exeprimental Medecine and Biology 726. Springer (2012) [DOI] [PMC free article] [PubMed]
- 6.Richards KE, Williams RC, Calendar R. Mode of DNA packing within bacteriophage heads. J. Mol. Biol. 1973;78:255–259. doi: 10.1016/0022-2836(73)90114-9. [DOI] [PubMed] [Google Scholar]
- 7.Earnshaw WC, Casjens SR. DNA packaging by the double-standed DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 8.Cerritelli ME, Cheng NQ, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Ecapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/S0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 9.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–216. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Mühlenhoff, Garady-Schahn R, Scholl D, Molineux IJ. The structure of bacteriophages K1E and K1-5 expleain processive degradation of polysaccharide capsules and evolution of new host specificities. J. Mol. Biol. 2007;371:836–849. doi: 10.1016/j.jmb.2007.05.083. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, Chiu W. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat. Struct. Mol. Biol. 2010;17:830–837. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odijk T. Hexagonally packed DNA within bacteriopahge T7 stabilised by curvature stress. Biophys. J. 1998;75:1223–1227. doi: 10.1016/S0006-3495(98)74041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odijk T, Slok F. Nonuniform Donnan equilibrium within bacteriophage packed with DNA. J. Phys. Chem. B. 2003;107:8074–8077. doi: 10.1021/jp0224822. [DOI] [Google Scholar]
- 15.Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purohit PK, Kondev J, Phillips R. Mechanics of DNA packaging in viruses. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrov AS, Boz MB, Harvey SC. The conformation of double-stranded DNA inside bacteriophages depends on capsid size and shape. J. Struct. Biol. 2007;160:241–248. doi: 10.1016/j.jsb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leforestier A, Livolant F. The bacteriophage genome undergoes a succession of intracapsid phase transitions upon DNA ejection. J. Mol. Biol. 2010;396:384–395. doi: 10.1016/j.jmb.2009.11.047. [DOI] [PubMed] [Google Scholar]
- 19.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 20.Gaussier H, Yang Q, Catalano CE. Building a virus from scratch assembly of an infectious virus using purified components in a rigourously defined biochemical assay. J. Mol. Biol. 2006;357:1154–1166. doi: 10.1016/j.jmb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Kondabagil KR, Zhang ZH, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J. Mol. Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 22.Roa M. Receptor-triggered ejection of DNA and protein in phage lambda. FEMS Microbiol. Lett. 1981;11:257–252. doi: 10.1111/j.1574-6968.1981.tb06976.x. [DOI] [Google Scholar]
- 23.Boulanger P, Le Maire M, Bonhivers M, Dubois S, Desmadril M, Letellier L. Purification and structural and functional characterization of FhuA, a transporter of the Escherichia coli outer membrane. Biochemistry. 1996;35:14216–14224. doi: 10.1021/bi9608673. [DOI] [PubMed] [Google Scholar]
- 24.São-José C, de Frutos M, Raspaud E, Santos MA, Tavares P. Pressure built by DNA packing inside virions: enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J. Mol. Biol. 2007;374:346–355. doi: 10.1016/j.jmb.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 25.Evilevitch A, Lavelle C, Knobler C, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA from phage. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knobler CM, Gelbart WM. Physical chemistry of DNA viruses. Ann. Rev. Phys. Chem. 2009;60:367–383. doi: 10.1146/annurev.physchem.59.032607.093728. [DOI] [PubMed] [Google Scholar]
- 27.Gelbart WM, Knobler CM. The physics of phages. Phys. Today. 2008;61:42–44. doi: 10.1063/1.2835152. [DOI] [Google Scholar]
- 28.Smith DE. Single molecule studies of viral DNA packaging. Curr. Opin. Virol. 2011;1:134–141. doi: 10.1016/j.coviro.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu D, Van Valen D, Hu Q, Phillips R. Ion-dependent dynamics of DNA ejections for bacteriophage lambda. Biophys. J. 2010;99:1101–1119. doi: 10.1016/j.bpj.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evilevitch A, Fang LT, Yoffe AM, Castelnovo M, Rau DC, Parsegian VA, Gelbart WM, Knobler CM. Effects of salt concentrations and bending energy on the extent of ejection of phage genomes. Biophys. J. 2008;94:1110–1120. doi: 10.1529/biophysj.107.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bactériophage phi29. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leforestier A, Siber A, Livolant F, Podgornik R. Protein-DNA interactions determine the shapes of DNA toroids condensed in virus capsids. Biophys. J. 2011;100:2209–2216. doi: 10.1016/j.bpj.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangenot S, Hochrein M, Rädler J, Letellier L. Real-time imaging of DNA éjection from single phage particles. Curr. Biol. 2005;15:430–435. doi: 10.1016/j.cub.2004.12.080. [DOI] [PubMed] [Google Scholar]
- 34.Chiaruttini N, de Frutos M, Augarde E, Boulanger P, Letellier L, Viasno V. Is the in vitro ejection of bacteriophage dna quasistatic? A bulk to single virus study. Biophys. J. 2010;99:447–455. doi: 10.1016/j.bpj.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leforestier A, Brasiles S, de Frutos M, Raspaud E, Letellier L, Tavares P, Livolant F. Bacteriophage T5 DNA ejection under pressure. J. Mol. Biol. 2008;384:730–739. doi: 10.1016/j.jmb.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 36.Comolli LR, Spakowitz AJ, Siegerist CE, Jardine PJ, Grimes S, Anderson DL, Bustamante C, Downing KH. Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 37.Fang PA, Wright ET, Weintraub ST, Hakala K, Wu W, Serwer P, Jiang W. Visualization of bacteriophage T3 capsids with DNA incompletely packaged in vivo. J. Mol. Biol. 2008;384:1384–1399. doi: 10.1016/j.jmb.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durand D, Doucet J, Livolant F. A study of the structure of highly concentrated phases of DNA by X-ray diffraction. J. Phys. II. 1992;2:1769–1783. doi: 10.1051/jp2:1992233. [DOI] [Google Scholar]
- 39.Livolant F, Leforestier A. Condensed phases of DNA: structure and phase transitions. Prog. Polym. Sci. 1996;21:1115–1164. doi: 10.1016/S0079-6700(96)00016-0. [DOI] [Google Scholar]
- 40.Petrov AS, Harvey SC. Structural and thermodynamic principles of viral packaging. Structure. 2007;15:21–27. doi: 10.1016/j.str.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Petrov AS, Harvey SC. Packaging double-helical DNA into viral capsids: structures, forces, and energetics. Biophys. J. 2008;95:497–502. doi: 10.1529/biophysj.108.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys. J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu X, Rau DC, Parsegian VA, Fang LT, Knobler CM, Gelbart WM. Salt-dependent DNA-DNA spacings in intact bacteriophage λ reflect relative importance of DNA self-repulsion and bending energies. Phys. Rev. Lett. 2011;106:28102. doi: 10.1103/PhysRevLett.106.028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geggier S, Kotlyar A, Vologodskii A. Temperature dependence of DNA persistence length. Nucleic Acid. Res. 2011;39:1419–1426. doi: 10.1093/nar/gkq932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forties R, Bundschuh R, Poirier MG. The flexibility of locally melted DNA. Nucleic Acid. Res. 2009;37:4580–4586. doi: 10.1093/nar/gkp442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Frutos M, Letellier L, Raspaud E. DNA ejection from bacteriophage T5: analysis of the kinetics and energetics. Biophys. J. 2005;88:1364–1370. doi: 10.1529/biophysj.104.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gosule LC, Schellman JA. Compact form of DNA induced by spermidine. Nature. 1976;259:333–335. doi: 10.1038/259333a0. [DOI] [PubMed] [Google Scholar]
- 48.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 49.Widom J, Baldwin RL. Cation-induced toroidal condensation of DNA studies with Co3+(NH3)6. J. Mol. Biol. 1980;144:431–453. doi: 10.1016/0022-2836(80)90330-7. [DOI] [PubMed] [Google Scholar]
- 50.Raspaud E, Olvera de la Cruz M, Sikorav JL, Livolant F. Precipitation of DNA by polyamines: a polyelectrolyte behavior. Biophys. J. 1998;74:381–393. doi: 10.1016/S0006-3495(98)77795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raspaud E, Durand D, Livolant F. Interhelical spacing in liquid crystalline spermine and spermidine-DNA precipitates. Biophys. J. 2005;88:392–403. doi: 10.1529/biophysj.104.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leforestier A, Livolant F. Structure of toroidal DNA collapsed inside the phage capsid. Proc. Natl. Acad. Sci. 2009;106:9157–9162. doi: 10.1073/pnas.0901240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leforestier A, Bertin A, Dubochet J, Richter K, Sartori Blanc N, Livolant F. Expression of chirality in columnar hexagonal phases or DNA and nucleosomes. C.R.A.S Chimie. 2008;11:229–244. doi: 10.1016/j.crci.2007.09.008. [DOI] [Google Scholar]
- 54.Evdokimov YuM, Platonov AL, Tikhonenko AS, Varshavsky YaM. A compact form of double stranded DNA in solution. FEBS Lett. 1972;23:180–184. doi: 10.1016/0014-5793(72)80335-1. [DOI] [PubMed] [Google Scholar]
- 55.Lerman LS. A transition to a compact form of DNA in polymer solutions. PNAS. 1971;68:1886–1890. doi: 10.1073/pnas.68.8.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pelta J, Livolant F, Sikorav JL. DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem. 1996;271:5656–5662. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 57.Lee S, Tran CV, Nguyen TT. Inhibition of DNA ejection from bacteriophage by Mg(+2) counterions. J. Chem. Phys. 2011;134:125104. doi: 10.1063/1.3569133. [DOI] [PubMed] [Google Scholar]
- 58.Li ZD, Wu JZ, Wang ZG. Osmotic pressure and packaging structure of caged DNA. Biophys. J. 2007;94:737–746. doi: 10.1529/biophysj.107.112508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeembaeva M, Castelnovo M, Larsson F, Evilevitch A. Osmotic pressure: resisting of promoting DNA ejection from phage λ. J. Mol. Biol. 2008;381:310–323. doi: 10.1016/j.jmb.2008.05.081. [DOI] [PubMed] [Google Scholar]
- 60.Evilevitch A. Effects of condensing agent and nuclease on the extent of ejection from phage lambda. J. Phys. Chem. B. 2006;110(44):22261–22265. doi: 10.1021/jp060573j. [DOI] [PubMed] [Google Scholar]
- 61.Hagerman PJ. Flexibility of DNA. Ann. Rev. Biophys. Biophys. Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 62.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 2000;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiggins PA, van der Heijden T, Moreno-Herrero F, Spakowitz A, Phillips R, Widom J, Dekker C, Nelson PC. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat. Nanotechnol. 2006;1:137–141. doi: 10.1038/nnano.2006.63. [DOI] [PubMed] [Google Scholar]
- 64.Yuan CL, Chen HM, Xion WL, Archer LA. DNA bending stiffness on small length scales. Phys. Rev. Lett. 2008;100:18102. doi: 10.1103/PhysRevLett.100.018102. [DOI] [PubMed] [Google Scholar]
- 65.Guerrero-Ferreira RC, Wright ER. Cryo-electron tomography of bacterial viruses. Virology. 2013;435:179–186. doi: 10.1016/j.virol.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, Chen CY, Shiomi D, Niki H, Margolin W. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology. 2011;417:304–311. doi: 10.1016/j.virol.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gautier A, Salamin L, Tosi-Couture E, Mc Dowall A, Dubochet J. Electron microscopy of the chromosome of dinoflagellates in situ: confirmation of Bouligand’s liquid crystal hypothesis. J. Ultrastr. Mol. Struct. Res. 1986;97:10–30. doi: 10.1016/S0889-1605(86)80003-9. [DOI] [Google Scholar]
- 69.Sartori Blanc N, Senn A, Leforestier A, Livolant F, Dubochet J. DNA in Human and Stallion spermatozoa forms local hexagonal packing with twist and many defects. J. Struct. Biol. 2001;134:76–81. doi: 10.1006/jsbi.2001.4365. [DOI] [PubMed] [Google Scholar]
- 70.Virrankoski-Castrodeza VN, Parish JH. Evidence for supercoiling in the DNA of bacteriophages heads. Arch. Microbiol. 1980;126:277–283. doi: 10.1007/BF00409932. [DOI] [PubMed] [Google Scholar]