Abstract
There are two important problems in the assembly of small, icosahedral RNA viruses. First, how does the capsid protein select the viral RNA for packaging, when there are so many other candidate RNA molecules available? Second, what is the mechanism of assembly? With regard to the first question, there are a number of cases where a particular RNA sequence or structure—often one or more stem-loops—either promotes assembly or is required for assembly, but there are others where specific packaging signals are apparently not required. With regard to the assembly pathway, in those cases where stem-loops are involved, the first step is generally believed to be binding of the capsid proteins to these “fingers” of the RNA secondary structure. In the mature virus, the core of the RNA would then occupy the center of the viral particle, and the stem-loops would reach outward, towards the capsid, like stalagmites reaching up from the floor of a grotto towards the ceiling. Those viruses whose assembly does not depend on protein binding to stem-loops could have a different structure, with the core of the RNA lying just under the capsid, and the fingers reaching down into the interior of the virus, like stalactites. We review the literature on these alternative structures, focusing on RNA selectivity and the assembly mechanism, and we propose experiments aimed at determining, in a given virus, which of the two structures actually occurs.
Keywords: RNA virus, RNA secondary structure, Virus assembly, Icosahedral viruses
Introduction
Viruses were among the first biological objects to be investigated by physicists, beginning with the founding of the “Phage School” by Max Delbrück shortly after World War II. Viruses often combine great biological complexity with apparent physical simplicity, and the main goal of that early work was the determination of the principles of viral structure. In recent years, this work has expanded into investigations of the mechanisms of viral assembly [1–3]. Such understanding should offer novel opportunities for pharmacological intervention, and it should facilitate the design of a wide range of useful nanoparticles. As a consequence, these processes are currently under intense experimental and computational investigation.
Small, icosahedral RNA viruses are among the most popular objects of current studies of structure and assembly. These viruses contain a single-stranded RNA (ssRNA) genome surrounded by a protein shell (the capsid). Some important human pathogens are small ssRNA viruses (e.g., hepatitis E, rubella, rhinovirus). Assembly of ssRNA viruses is spontaneous, at least in the cells that they infect (some viruses can be assembled in vitro, but others cannot).
At present, we do not understand how the formation of the mature virus depends on the secondary and tertiary structure of the RNA molecule that is packaged. We do not know to what extent these structures vary from particle to particle in an ensemble of mature viruses, we do not know whether these structures change (whether the RNA refolds) as the capsid proteins bind to the RNA, and we do not know what pathway(s) the RNA follows in achieving its final packaged structure.
For several ssRNA viruses, studies using X-ray crystallography have revealed the capsid structure at atomic resolution. This work takes advantage of the icosahedral symmetry of the virus because the presence of multiple copies of identical proteins in identical conformations enhances the signal-to-noise ratio. Examples of particular interest to us in this work include satellite tobacco mosaic virus (STMV) [4, 5], Pariacoto virus (PaV) [6], and MS2 [7]. For STMV and PaV, the crystal structures revealed some of the RNA density, but the icosahedral averaging precluded identification of the sequences associated with the density. The MS2 crystal structure did not reveal any information on the structure of the RNA, but the crystal structure was later determined for a virus-like particle obtained by soaking an RNA stem-loop into previously crystallized MS2 capsids [8].
The crystallographic studies have been supplemented by single-particle reconstructions using cryo-electron microscopy (cryo-EM) [6, 9, 10]. Some of these studies use the native virus, while others target virus-like particles obtained by assembling the capsid proteins around RNA molecules other than those of the native genomes.
The combination of X-ray crystallography and cryo-EM has provided glimpses of the RNA organization inside these viruses. Unfortunately, the RNA genome does not possess a highly symmetric sequence, so the icosahedral averaging blurs all features of the RNA structure except those that do have approximate icosahedral symmetry. The genomes of STMV and PaV both contain multiple double-helical regions that vary in sequence and in structural details, but that are visible in the structural studies. We have developed detailed all-atom models for the genomic RNA of these two viruses, which, when added to the previously determined capsid structures, provide the first complete models for any viruses.
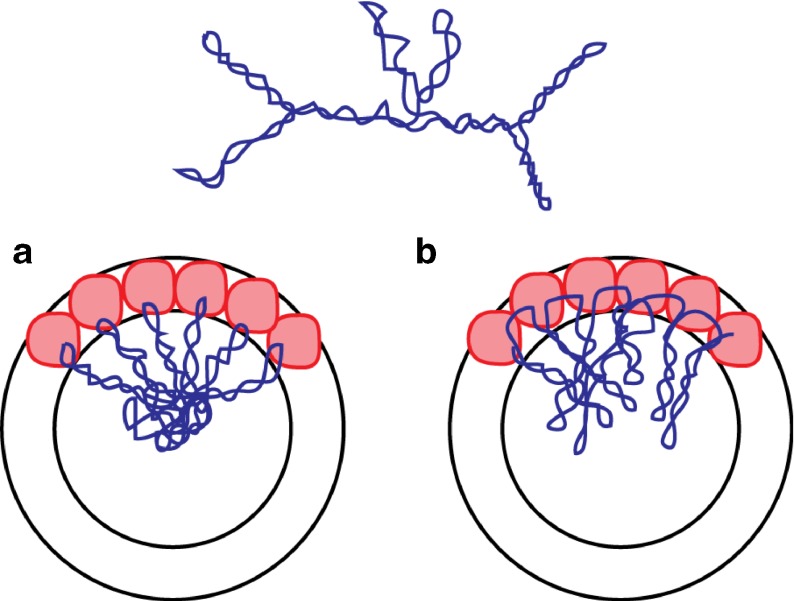
The two models that we developed [11, 12] are based on two fundamentally different assumptions about how the RNA is arranged in the virus. As we will soon explain, the assumptions lead to a “stalactite model” for PaV and a “stalagmite model” for STMV (Fig. 1). In this article, we review previous work on these two viruses and others, asking whether the two types of structures are equally probable in the world of small, icosahedral ssRNA viruses. We also examine the probable implications for viral assembly for the stalactite and stalagmite models.
Fig. 1.
When an RNA molecule (top) is packaged into a virus, it may be organized in either of two ways. a The body of the RNA occupies the centermost part of the virus, and the stem-loops reach outward toward the capsid, much like stalagmites in a grotto. b The body of the RNA is associated with the capsid proteins, and the stem-loops reach inward toward the center of the virus, resembling stalactites in a grotto
The structure of the Pariacoto virus: a stalactite model
Pariacoto virus (PaV) is a T = 3, non-enveloped, icosahedral virus. A member of the Nodaviridae family, it was originally isolated from the Southern Armyworm, Spodoptera eridania, the larva of a Peruvian moth [13]. The PaV genome consists of two positive-sense ssRNAs [14], one of which (RNA1, 3011 nucleotides) codes for protein A, the catalytic subunit for the RNA-dependent RNA replicase, while the other (RNA2, 1311 nucleotides) codes for the capsid precursor protein, designated α. The relatively small size (30 nm diameter) and the ease with which it can be produced in various cell lines [15] make PaV and other members of the Nodaviridae family relatively easy to characterize at the molecular level [16–18].
The crystal structure of PaV [6] revealed an asymmetric unit containing three quasi-equivalent protein subunits (A, B, and C). It also contains 30 segments of double-helical RNA (each 25 base pairs long) just inside the capsid. The sequence does not contain 30 copies of any sequence of that length, so the 30 RNA duplexes certainly have a variety of sequences, and probably some differences in structure, e.g., mismatches and bulges. As a consequence, the double-helical structure seen in the crystal structure represents an average of these duplexes. Taken together, the 1,500 nucleotides in the 30 duplexes represent about 35% of the total genome—but no particular part of the RNA can be assigned to any part of these duplexes.
The RNA duplexes are perpendicular to the twofold crystallographic axes, and they lie on the 30 edges of a dodecahedron, forming a “dodecahedral cage” (note that the dodecahedron and the icosahedron form a pair of complementary Platonic solids: the former has 12 faces and 20 vertices, while the latter has 20 faces and 12 vertices; each has 30 edges, but the edges of the icosahedron are perpendicular to those of the dodecahedron, and vice versa).
We were challenged to develop an all-atom model for PaV by Jack Johnson, Liang Tang, and Annette Schneemann, who had done the original crystallographic and cryo-EM studies on PaV [6]. Their examination of the density maps led them to conclude that the remaining RNA (the 65% not seen in the crystal structure) must occupy the centermost region of the virus, and that it is connected to the dodecahedral cage at the vertices. This led us to jointly postulate a stalactite conformation (Fig. 1b) for the RNA.
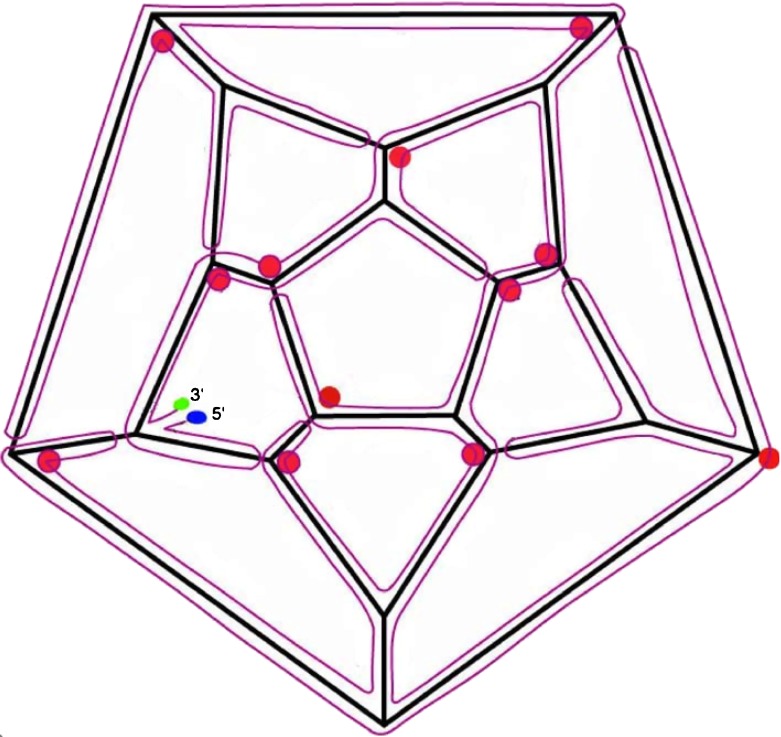
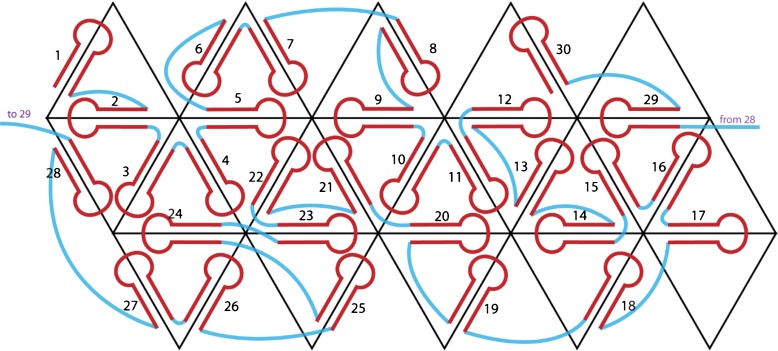
The RNA secondary structure is not known, nor is it known whether all base pairing in RNA1 and RNA2 is intramolecular, or whether there are base pairs between RNA1 and RNA2. In the absence of such information, we chose to represent the bipartite genome as one single strand. With this assumption, and in the absence of data on the RNA secondary structure, we had to postulate a secondary structure that is consistent with the crystal structure (30 anti-parallel duplexes whose axes define the dodecahedral cage) and with the stalactite model. It is easily shown that there are many possible ways to satisfy these constraints. We chose a representative solution [9], as shown in Fig. 2.
Fig. 2.
Organization of a hypothetical secondary structure for PaV RNA, in the stalactite conformation [9]. This is just one solution for mapping the RNA onto the dodecahedral cage in a fashion that is consistent with the data from X-ray crystallography and cryo-electron microscopy. The 5′ and 3′ ends are indicated. Each edge of the dodecahedral cage contains an antiparallel RNA double helix. Red circles indicate three- and four-way junctions at those vertices where RNA stalactites drop down into the interior of the virus to connect with that part of the RNA not on the dodecahedral cage
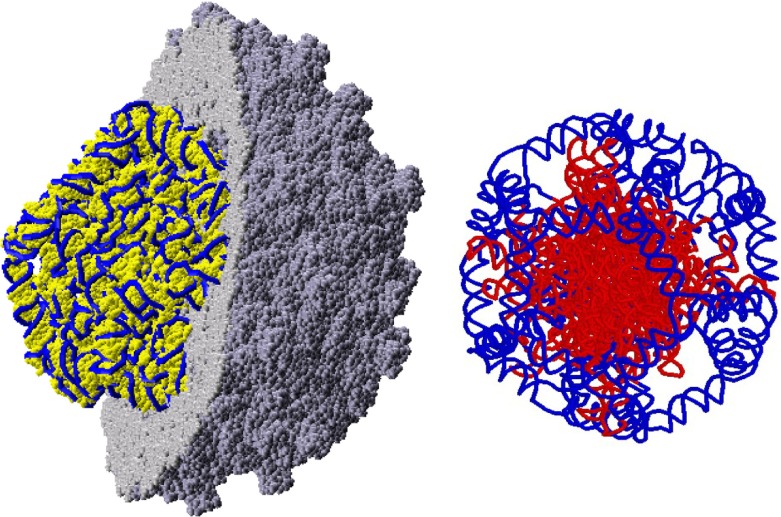
To build a three-dimensional model, we modeled the stalactites with pieces taken from the 23 S ribosomal RNA of E. coli, and we invented a sequence compatible with the secondary structure for those parts of the RNA on the dodecahedral cage and the vertices. The final all-atom model [11] contains every residue of the genome and every residue of the capsid proteins, so it was the first complete model of any virus-like particle (we say “virus-like particle” because it contains an RNA that is the same size as the genomic RNA, but that does not have the genomic sequence). The three-dimensional model is shown in Fig. 3.
Fig. 3.
All-atom stalactite model for Pariacoto virus (PaV) [11]. On the left is a view with half of the capsid (grey) cut away to reveal the RNA genome (yellow, with the backbone highlighted in blue). On the right is a representation of the RNA backbone: the blue regions are those parts in contact with the capsid, while the stalactites are colored in red
The structure of satellite tobacco mosaic virus: a stalagmite model
Satellite tobacco mosaic virus (STMV) is a T = 1 icosahedral virus with a diameter of 17 nm and a single-stranded RNA genome containing 1,058 nucleotides [19]. The 1.8 Å X-ray structure of this virus showed 30 RNA double helices, each one centered on a twofold symmetry axis [4]. Each helix has nine base pairs, plus an unpaired nucleotide at the 3′ end of each strand and 57% of the genome is visible in the crystal structure [4]. Since the viral particle can enter the crystal lattice in any of 60 orientations, the RNA density is averaged among these orientations, so the crystal structure provides no information on the sequence of each double helix.
Larson and McPherson proposed that the RNA double helices represent 30 hairpin loops, and that they are connected by single-stranded linkers [5]. Subsequent atomic force microscopic studies on RNA released from the capsid are consistent with this linear arrangement of structural domains [20]. Their model for the organization of the genome in the mature virus suggests an efficient pathway for viral assembly. It is a stalagmite model (Fig. 1a).
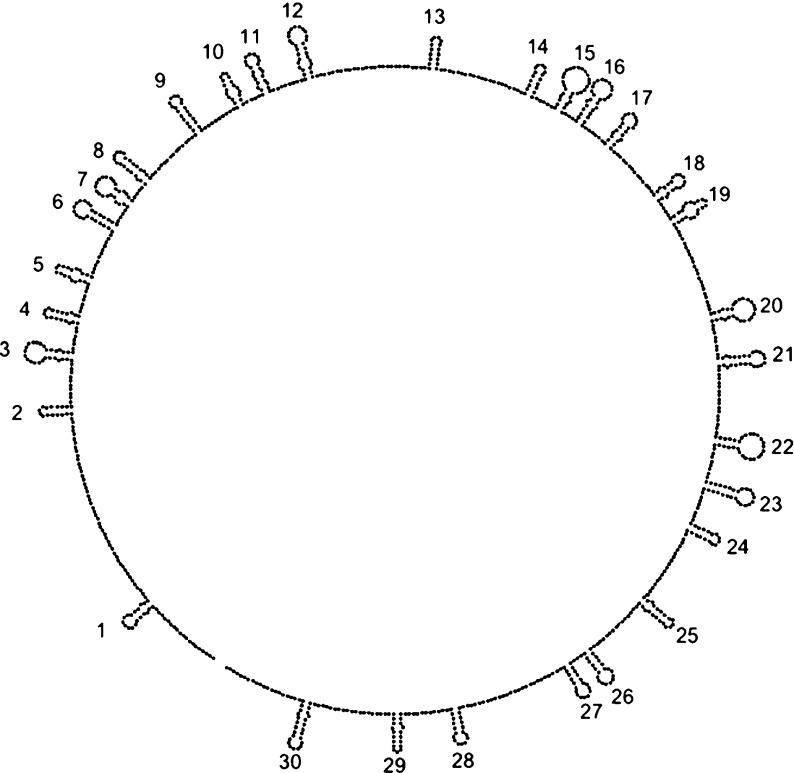
At the time the Larson-McPherson model was built, there were no data to permit assignment of specific regions of the genomic RNA to each of the stem-loops. Schroeder et al. later did chemical probing of the RNA inside the virus; they combined the resulting data with helix search algorithms to identify possible combinations of stem-loops, allowing them to propose a genomic secondary structure consistent with the organization originally proposed by Larson and McPherson; they emphasized that this is a representative secondary structure from a large ensemble of possible structures [21]. The model of Schroeder et al. is shown in Fig. 4.
Fig. 4.
The secondary structure of STMV RNA proposed by Schroeder et al. [21], based on chemical probing and the assumption of Larson and McPherson that the RNA is organized into a series of stem-loops
Figure 5 shows how the 30 stem-loops in the Schroeder secondary structure model can be organized so that each edge of the icosahedral structure is covered by one hairpin loop [12]. We arranged these according to the lengths of the connections between successive stem-loops. With linkers of less than eight nucleotides, the neighboring hairpins adopt a tail-to-tail conformation, while head-to-tail or head-to-head conformations can be used when the linker is longer than 13 nucleotides. Of course, the arrangement shown in Fig. 5 is just one out of a very large ensemble of possible arrangements.
Fig. 5.
A model for the organization of the 30 stem-loops of the STMV genomic RNA in STMV, following the proposal of Larson and McPherson [5], and incorporating the secondary structure proposed by Schroeder et al. [21]
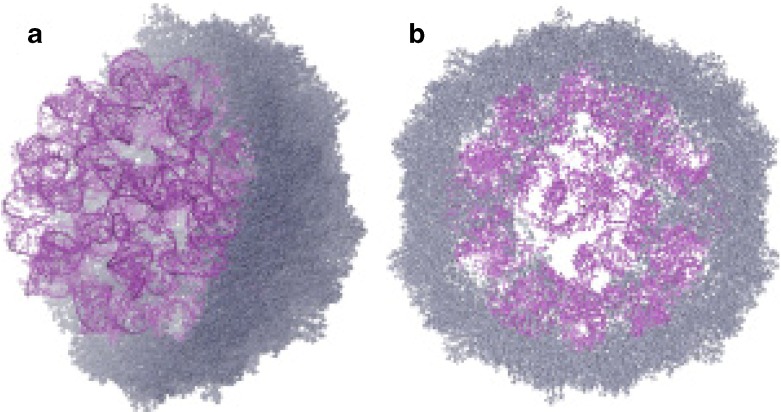
We built and refined a model for the STMV RNA, which we combined with the the X-ray structure of the capsid [4]. The capsid proteins have 147 residues, but the 12 amino-terminal amino acids are not seen in the crystal structure, so we also modeled those. The final model [12] is shown in Fig. 6. We believe that this is the first complete all-atom model for any virus that incorporates the actual sequence of the genomic RNA.
Fig. 6.
An all-atom model for STMV [12], with the capsid in grey and the RNA genome in violet. On the left is a cutaway view showing half the capsid and the complete RNA genome, while on the right is a 50 Å slice through the center of the virus
(For completeness, we should note that we have recently determined the secondary structure of the STMV genome by chemical probing of an in vitro transcript [22], finding a very extended structure that is quite different from the Schroeder model; we do not yet know if this structure can be packaged into the virus without refolding. We have argued that if the RNA in vitro does have the structure proposed by Schroeder et al., then it is either packaged co-transcriptionally, or there is extensive refolding upon packaging.)
MS2: a recent addition
MS2 is a T = 3 icosahedral single-stranded RNA bacteriophage with a genome of 3,569 nucleotides. The crystal structure of the wild-type virion has been determined at 2.8 Å [7, 23]. Capsid assembly can be triggered in vitro by an RNA stem-loop containing only 19 nucleotides (the TR stem-loop) [24, 25], and the structure of a virus-like particle assembled from capsid proteins and 60 copies of the TR stem-loop has been determined by cryo-EM [10]. Instead of TR, one can also trigger assembly by using RNA aptamers selected for their binding affinity to the capsid protein [26]. One of these (F5) can be modified to give tighter binding by replacing a single adenosine with 2′-deoxy-2-aminopurine; the 2.8 Å crystal structure of this aptamer soaked into pre-crystallized recombinant capsids revealed the details of the RNA–protein interaction [8].
The combination of these experimental results clearly shows that MS2 genomic RNA is organized into a stalagmite structure inside the mature virus (we note here that we have developed an all-atom model for MS2, in collaboration with the research groups of Peter Stockley (University of Leeds) and Reidun Twarock (University of York), and we expect to submit that model for publication in the near future).
Discussion and conclusions
Structural studies have produced two classes of models for the organization of RNA genomes inside small icosahedral viruses. The combination of data from X-ray crystallography and cryo-EM on PaV [6] led us to suggest that the genome has a stalactite structure [9]. In contrast, the crystal structure of STMV [4] was interpreted as favoring the organization of the genome into a stalagmite structure [5]. To date, there are no concrete data to prove that either of these is correct, but we have shown that it is possible to build all-atom models where the genome is organized into either the stalactite structure [11] or the stalagmite structure [12].
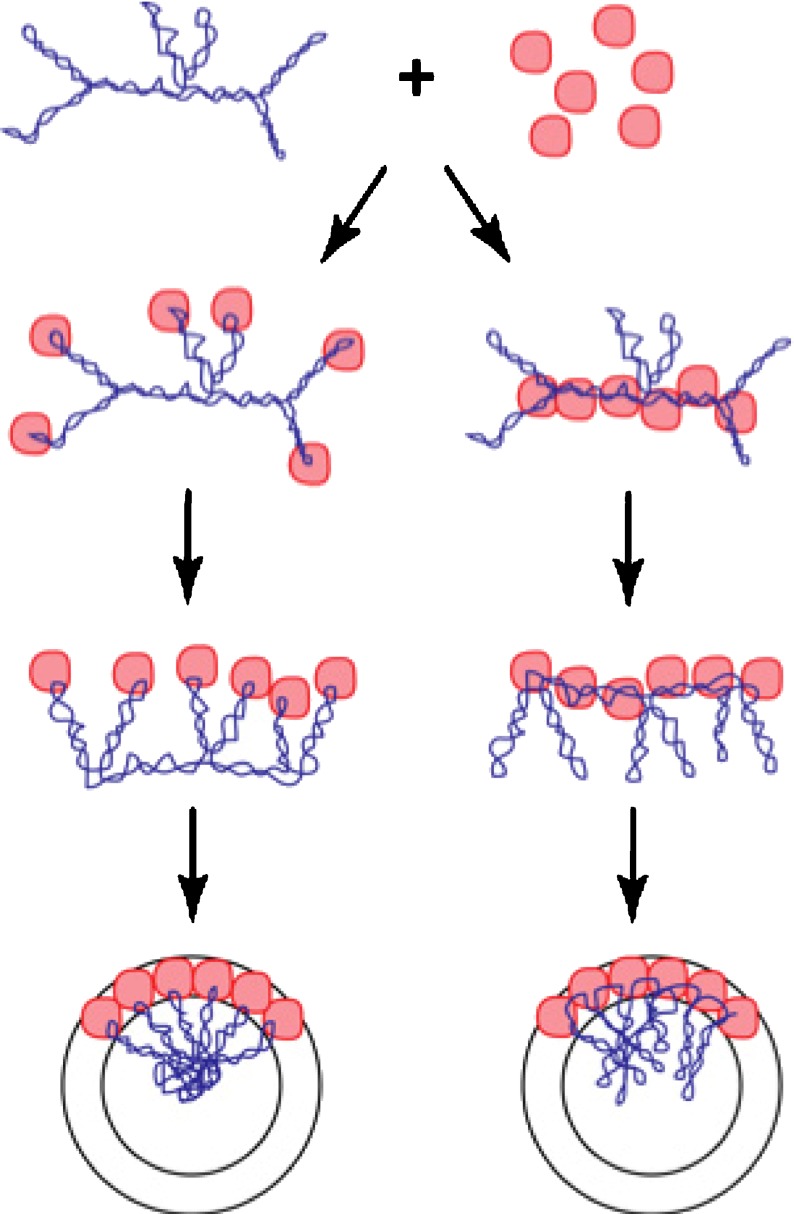
The one case where the genome organization is known for certain is MS2, where the combination of X-ray crystallography [8] and cryo-EM [10] clearly demonstrates the stalagmite structure. Assembly requires the existence of specific stem-loop structures, which is consistent with the proposed assembly pathway for stalagmites and inconsistent with the pathway for stalactite structures (Fig. 7 and Table 1).
Fig. 7.
Alternative pathways for the formation of small icosahedral RNA viruses. RNA (blue) and capsid proteins (red) can interact in either of two ways. In the left-hand pathway, the proteins bind to the tips of RNA stem-loops. Protein–protein interactions lead toward the formation of the mature virus with the RNA in the stalagmite conformation. Alternatively, capsid proteins might bind predominantly to the body of the RNA (right-hand pathway), leading to the stalactite conformation (only a small fragment of the RNA and a fraction of the capsid proteins are shown here, for graphical clarity. In the mature virus, the capsid proteins would form an icosahedral or spherical structure)
Table 1.
Experimental search for alternative packaging pathways

Observations are highlighted in red, and predictions are highlighted in blue
Other viruses are also known to require specific packaging signals, including brome mosaic virus (BMV) [27], which is another T = 3 single-stranded RNA virus (with a tetrapartite genome). In contrast, cowpea chlorotic mottle virus does not require a specific packaging signal [28], even though CCMV and BMV are structurally and genetically very similar [29]. It will be interesting to see whether the stalagmite pattern turns out to be universal in cases that do have packaging signals and to determine the organization of RNA genomes in those viruses that do not require such signals.
The question of how the RNA genome is organized is of critical importance for elucidating the assembly pathway. As shown in Fig. 7, the assembly pathways are necessarily quite different for viruses whose genomes have the stalactite conformation in the mature virus vs. those with the stalagmite conformation. If the capsid proteins bind to the stem-loops of the RNA (left-hand pathway), then the body of the RNA will lie in toward the center of the mature virus, with the stem-loops reaching out toward the capsid-like stalagmites. On the other hand, if the capsid proteins bind to the body of the RNA, holding it just under the capsid (right-hand pathway), then the stem-loops will hang downward from the capsid toward the center of the virus, like stalactites.
It should be possible to experimentally determine which of the two conformations is obtained by the RNA genome in a given icosahedral virus. Table 1 outlines the kinds of experiments that should be able to distinguish between the stalactite and stalagmite conformations.
Acknowledgements
This work was first presented at the Workshop on Physical Virology at the Abdus Salam International Centre for Theoretical Physics in Trieste, Italy (September, 2012). SCH is grateful to ICTP and the organizers for providing travel support to that meeting. Use of the word “grotto” in the title was inspired by the Grotta Gigante, near Trieste. Supported by grants from the US National Institutes of Health (GM 070785 to SCH and GM 083621 to CEH).
References
- 1.Knobler CM, Gelbart WM. Physical chemistry of DNA viruses. Annu. Rev. Phys. Chem. 2009;60:367–383. doi: 10.1146/annurev.physchem.59.032607.093728. [DOI] [PubMed] [Google Scholar]
- 2.Jardine PJ, Anderson DL. DNA packaging in double-stranded DNA phages. In: Calendar R, editor. The Bacteriophages. 2. Oxford: Oxford University Press; 2006. pp. 49–65. [Google Scholar]
- 3.Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophage. Curr. Opin. Struck. Biol. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Larson SB, Day J, Greenwood A. McPherson, A.: Refined structure of satellite tobacco mosaic virus at 1.8 Å resolution. J. Mol. Biol. 1998;277:37–59. doi: 10.1006/jmbi.1997.1570. [DOI] [PubMed] [Google Scholar]
- 5.Larson SB, McPherson A. Satellite tobacco mosaic virus RNA: structure and implications for assembly. Curr. Opin. Struck. Biol. 2001;11:59–65. doi: 10.1016/S0959-440X(00)00166-4. [DOI] [PubMed] [Google Scholar]
- 6.Tang L, Johnson KN, Ball LA, Lin T, Yeager M, Johnson JE. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 7.Valegard K, Liljas L, Fridborg K, Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 8.Horn WT, Convery MA, Stonehouse NJ, Adams CJ, Liljas L, Phillips SE, Stockley PG. The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand–RNA interactions. RNA. 2004;10:1776–1782. doi: 10.1261/rna.7710304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tihova M, Dryden KA, Le TV, Harvey SC, Johnson JE, Yeager M, Schneemann A. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J. Virol. 2004;78:2897–2905. doi: 10.1128/JVI.78.6.2897-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 11.Devkota B, Petrov AS, Lemieux S, Boz MB, Tang L, Schneemann A, Johnson JE, Harvey SC. Structural and electrostatic characterization of Pariacoto virus: Implications for viral assembly. Biopolymers. 2009;91:530–538. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Y, Larson SB, Heitsch CE, McPherson A, Harvey SC. A model for the structure of satellite tobacco mosaic virus. J. Struct. Biol. 2012;180:110–116. doi: 10.1016/j.jsb.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeddam JL, Rodriguez JL, Ravallec M, Lagnaoui A. A Noda-like virus isolated from the sweetpotato pest Spodoptera eridania (Cramer) (Lep.; Noctuidae) J. Invertebr. Pathol. 1999;74:267–274. doi: 10.1006/jipa.1999.4881. [DOI] [PubMed] [Google Scholar]
- 14.Krishna NK, Schneemann A. Formation of an RNA heterodimer upon heating of nodavirus particles. J. Virol. 1999;73:1699–1703. doi: 10.1128/jvi.73.2.1699-1703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneemann A, Zhong W, Gallagher TM, Rueckert RR. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher AJ, McKinney BR, Wery JP, Johnson JE. Crystallization and preliminary data analysis of Flock House virus. Acta Crystallogr. B. 1992;48(Pt 4):515–520. doi: 10.1107/S0108768192000053. [DOI] [PubMed] [Google Scholar]
- 17.Mori K, Nakai T, Muroga K, Arimoto M, Mushiake K, Furusawa I. Properties of a new virus belonging to Nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology. 1992;187:368–371. doi: 10.1016/0042-6822(92)90329-N. [DOI] [PubMed] [Google Scholar]
- 18.Reinganum C, Bashiruddin JB, Cross GF. Boolarra virus: a member of the Nodaviridae isolated from Oncopera intricoides (Lepidoptera: Hepialidae) Intervirology. 1985;24:10–17. doi: 10.1159/000149613. [DOI] [PubMed] [Google Scholar]
- 19.Mirkov TE, Mathews DM, Du Plessis DH, Dodds JA. Nucleotide sequence and translation of satellite tobacco mosaic virus RNA. Virology. 1989;170:139–146. doi: 10.1016/0042-6822(89)90361-9. [DOI] [PubMed] [Google Scholar]
- 20.Kuznetsov YG, Dowell JJ, Gavira JA, Ng JD, McPherson A. Biophysical and atomic force microscopy characterization of the RNA from satellite tobacco mosaic virus. Nucleic Acids Res. 2010;38:8284–8294. doi: 10.1093/nar/gkq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroeder SJ, Stone JW, Bleckley S, Gibbons T, Mathews DM. Ensemble of secondary structures for encapsidated satellite tobacco mosaic virus RNA consistent with chemical probing and crystallography constraints. Biophys. J. 2011;101:167–175. doi: 10.1016/j.bpj.2011.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Athavale SS, Gossett JJ, Bowman JC, Hud NV, Williams LD, Harvey SC. Secondary structure of the free RNA of satellite tobacco mosaic virus. PLoS One. 2013;8:e54384. doi: 10.1371/journal.pone.0054384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golmohammadi R, Valegard K, Fridborg K, Liljas L. The refined structure of bacteriophage MS2 at 2.8 A resolution. J. Mol. Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- 24.Beckett D, Uhlenbeck OC. Ribonucleoprotein complexes of R17 coat protein and a translational operator analog. J. Mol. Biol. 1988;204:927–938. doi: 10.1016/0022-2836(88)90052-6. [DOI] [PubMed] [Google Scholar]
- 25.Beckett D, Wu HN, Uhlenbeck OC. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J. Mol. Biol. 1988;204:939–947. doi: 10.1016/0022-2836(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 26.Hirao I, Spingola M, Peabody D, Ellington AD. The limits of specificity: an experimental analysis with RNA aptamers to MS2 coat protein variants. Mol. Divers. 1998;4:75–89. doi: 10.1023/A:1026401917416. [DOI] [PubMed] [Google Scholar]
- 27.Choi YG, Rao AL. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 2003;77:9750–9757. doi: 10.1128/JVI.77.18.9750-9757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Annamalai P, Rao AL. Dispensability of 3′ tRNA-like sequence for packaging cowpea chlorotic mottle virus genomic RNAs. Virology. 2005;332:650–658. doi: 10.1016/j.virol.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Osman F, Choi YG, Grantham GL, Rao AL. Molecular studies on bromovirus capsid protein. Virology. 1998;251:438–448. doi: 10.1006/viro.1998.9421. [DOI] [PubMed] [Google Scholar]