Abstract
The second messenger cAMP has been extensively studied for half a century, but the plethora of regulatory mechanisms controlling cAMP synthesis in mammalian cells is just beginning to be revealed. In mammalian cells, cAMP is produced by two evolutionary related families of adenylyl cyclases, soluble adenylyl cyclases (sAC) and transmembrane adenylyl cyclases (tmAC). These two enzyme families serve distinct physiological functions. They share a conserved overall architecture in their catalytic domains and a common catalytic mechanism, but they differ in their sub-cellular localizations and responses to various regulators. The major regulators of tmACs are heterotrimeric G proteins, which transduce extracellular signals via G protein-coupled receptors. sAC enzymes, in contrast, are regulated by the intracellular signaling molecules bicarbonate and calcium. Here, we discuss and compare the biochemical, structural and regulatory characteristics of the two mammalian AC families. This comparison reveals the mechanisms underlying their different properties but also illustrates many unifying themes for these evolutionary related signaling enzymes.
Keywords: cAMP signaling, class III nucleotidyl cyclases, enzyme regulation, soluble adenylyl cyclases, transmembrane adenylyl cyclases
Introduction
The second messenger cyclic adenosine 3′,5′-monophosphate (cAMP) was discovered by Earl Sutherland during his studies of hormonal regulation of metabolism in mammalian heart and liver.1,2 Subsequent studies revealed cAMP to be a prototypical second messenger, modulating physiological processes in all domains and kingdoms of life (its presence in plants remains controversial). The effects of cAMP are mediated by distinct targets in different kingdoms. In mammalian cells there are at least three known types of cAMP effector proteins: protein kinase A (PKA), exchange proteins activated by cAMP (EPACs), and cyclic nucleotide gated ion channels (CNGs and HCNs),3 plus a potential fourth target recently identified, Phosphodiesterase type 10.4 The second messenger is universally generated by adenylyl cyclases (AC; EC 4.6.1.1), enzymes that catalyze the cyclization of ATP to generate cAMP and inorganic pyrophosphate. Nucleotidyl cyclases (i.e. enzymes generating cAMP and the related second messenger cGMP from GTP) are a diverse family of enzymes that can be separated into six classes5,6 (see below), with all known eukaryotic nucleotidyl cyclases belonging to a single class (class III). In many organisms and cell types, several different adenylyl cyclases (ACs) and/or guanylyl cyclases (GCs) are expressed simultaneously.7 Such heterogeneity reflects the multitude of cellular processes that are regulated by cyclic nucleotide second messengers in eukaryotic cells.
In mammals, cAMP is formed by either of two types of widely expressed Class III ACs (Figure 1). A family of transmembrane adenylyl cyclases (tmACs) encoded by nine distinct genes termed type I through type IX was discovered first, and this family represents the more widely studied source of cAMP. tmACs are directly regulated by heterotrimeric G proteins and generate cAMP in response to hormones and neurotransmitters which signal through G protein-coupled receptors (GPCRs).8 As their name implies, tmACs are obligatory membrane proteins. The individual regulatory properties of each isoform have been reviewed extensively, 7,9,10 and specific tmAC isoforms have been linked to a subset of the physiological responses mediated by tmACs; for example, learning and memory,11–14 cardiac myocyte function,15 and olfaction16 are, at least in part, mediated by tmACs types I and VIII, type V, and type III, respectively.
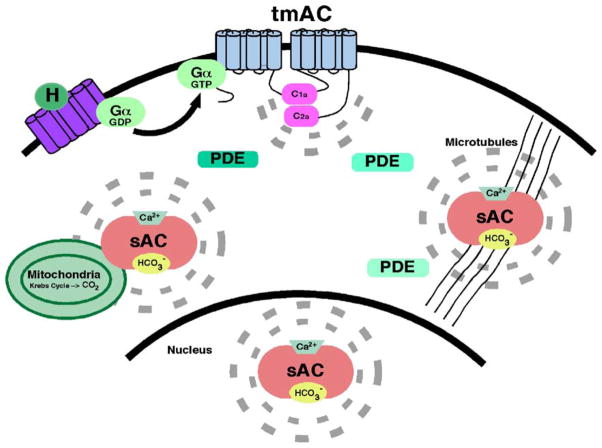
Figure 1.
Revised model for cAMP signaling. The second messenger is formed, acts, and is degraded close to its target, leading to the formation of cAMP micro-domains within the cell. H denotes a receptor-activating hormone and broken lines indicate the limited diffusion sphere of cAMP formed by the respective adenylyl cyclase.
A second, independent source of cAMP in mammalian cells is the more recently discovered, soluble adenylyl cyclase (sAC).17 sAC is uniquely regulated by bicarbonate18 and calcium,19,20 and it is insensitive to heterotrimeric G protein regulation.17 sAC is widely expressed21 and is not strictly a soluble protein; it is present at discrete sub-cellular localizations in a wide variety of cells.22 Isoform diversity from the single confirmed sAC gene in mammalian genomes is generated by complex alternative splicing.23,24 Physiological processes demonstrated to be regulated by sAC include sperm activation,25,26 pH regulation in epididymis,27 and TNF activation of granulocytes.28 Its unique regulation by bicarbonate suggests it also contributes to other processes responsive to carbon dioxide and/or functions as a metabolic sensor in cells.29
The catalytic mechanisms and regulation of both mammalian G protein-regulated tmAC catalytic domains and a bicarbonate-regulated, bacterial sAC-like cyclase has now been studied kinetically and crystallographically. Here, we discuss the biochemical, structural and regulatory characteristics of these two families of mammalian ACs. This comparison reveals the mechanisms underlying their different properties and illustrates many unifying themes for these evolutionary related signaling enzymes.
Classification of Nucleotidyl Cyclases
The known nucleotidyl cyclase sequences have been grouped into six classes based on sequence homology within their catalytic portions.5,6 The AC enzymes from Escherichia coli and a number of related Gram-negative prokaryotes belong to class I. Class II is comprised of the “toxin” ACs from pathogens such as Bordetella pertussis30 and Bacillus anthracis.31 These ACs are secreted enzymes that translocate into host cells where they disrupt intracellular signaling by flooding host cells with supraphysiological amounts of cAMP. Eukaryotic adenylyl and guanylyl cyclases belong to class III, the broadest class that contains members from bacterial and animal kingdoms.32 Classes IV, V and VI are relatively recently defined, and are comprised of one or a few prokaryotic members.6,33,34
Class III adenylyl cyclases domain topologies
Class III catalytic domains are found in a number of different contexts with a variety of regulatory domains, which confer modulation by a wide array of signals. Despite sensitivity to diverse regulators, the several crystal structures of class III ACs revealed a conserved basic architecture of the catalytic centers (see below). These structures, together with a number of biochemical studies, show that activity requires dimerization of two catalytic domains35 (Figure 2). In the case of mammalian receptor GCs36 and many bacterial ACs,37,38 where each protein contains only a single catalytic domain, homodimerization creates two symmetrical active sites at opposite poles of the dimer interface. In contrast, mammalian tmACs, soluble guanylyl cyclases (sGC), and the known isoforms of mammalian sACs form an active cyclase via heterodimerization of structurally similar catalytic domains (C1 and C2).24,36,39 These two catalytic domains can reside on separate polypeptide chains, as in sGCs (true heterodimer), or on a single polypeptide, as in tmACs and sAC (pseudo-heterodimer; from now on also designated as heterodimer). In heterodimeric cyclases, C1 only carries a subset of the active site residues essential for catalysis; the remaining essential residues are contributed by C2. Therefore, as described in detail below, only one of the two sites formed at a heterodimeric interface is active.40 It is assumed that the primordial cyclase was a homodimer, and gene duplication permitted diversification as well as mutation of catalytic residues within the second “active site”. An unusual mycobacterial AC homodimer showed structural asymmetry and half-of-the-sites-reactivity which could be the evolutionary step before degeneration of the second site.41 In tmACs, this pseudo-symmetrical “active” site apparently evolved to serve a regulatory function as part of an effector binding site (see below).
Figure 2.
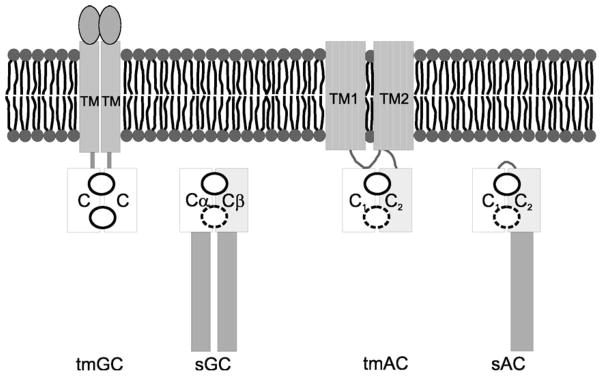
Scheme of domain arrangements in various class III cyclases. The conserved, dimeric catalytic core is formed by association of two domains, which belong either to a single polypeptide or to two protein chains. Intermolecular dimers can be homodimers or heterodimers. The dimers contain either two active sites (homodimers; indicated by circles) or one active site and a degenerate, inactive site (broken circle in heterodimers and pseudoheterodimers). C, catalytic domain; TM, transmembrane domain.
The different topologies and domain interactions of representative class III proteins are shown in Figure 2. Among mammalian adenylyl cyclases, the nine tmAC genes encode a variable N-terminal region, followed by a six transmembrane helices domain (TM1), the first catalytic domain C1, another six transmembrane helices domain (TM2), and the second catalytic domain C2. In contrast, the sAC gene encodes a number of spliced forms that contain no detectable TM domain, and the two N-terminal catalytic domains C1 and C2 are separated by a relatively short stretch of approximately 20 amino acid residues.
Evolution of Mammalian Adenylyl Cyclases
sAC appears to be the most ancient among mammalian cyclases; its sequence is more similar to ACs from cyanobacteria and myxobacteria than to other mammalian cyclases17 (Figure 3). Additionally, mammalian sAC and the CyaC cyclase from the cyanobacteria Spirulina platensis are similarly regulated; both are synergistically activated by bicarbonate and calcium ions.20,42 Bicarbonate-regulated sAC-like cyclases are also found in Anabaena PCC 7120, Stigmatella aurantiaca, Mycobacterium tuberculosis, 43 and the thermophilic bacterium Chloroflexus aurantiacus,44 and sAC-like proteins were cloned and expressed from sea urchin45 and Plasmodium falciparum, the parasite responsible for the most lethal form of malaria.46 Database searches identify sAC-like ACs in the genomes of all mammals studied (human, chimpanzee, dog, cow, rabbit, mouse, and rat), as well as in chicken, ciona intestinalis, and insects (mosquito and honey bee). The human, chimpanzee, dog, and cow genomes predict a second locus encoding a sAC-like gene, but its activity or physiological relevance is unknown. Surprisingly, corresponding genes have yet to be found in the genomes of organisms such as Caenorhabditis elegans and Drosophila melonogaster, and it has been proposed that these organisms lost their sAC-like cyclase during evolution.47
Figure 3.
Phylogenetic relationship between various class III ACs and GCs. Amino acid sequences of the catalytic regions were aligned using ClustalW133 and represented as an unrooted tree using Fitch (Phylip 3.5; Felsenstein, J., Department of Genetics, University of Washington, Seattle) using E. coli CyaA as an outgroup. Numbers represent bootstrap confidence values of 1000 replicates. The individual catalytic domains from cyclases with two catalytic domains, i.e. C1 and C2, on a single polypeptide chain were analyzed separately. Accession numbers for the aligned amino acid sequences are as follows: human ANP-A (hGCA), P16066; human ANP-B (hGCB), P26594; human sGCα2 (hGCa2), P33402; human sGCα3 (hGCa3), Q02108; human sGCβ1 (hGCb1), Q02153; human sGCβ2 (hGCb2), O75343; human sGCβ3 (hGCb3), NP_000848; human GC-2C (hGCCC), P25092; human GC-2D (hGCD), Q02846; human GC-2F (hGCF), P51841; Plasmodium falciparum GCα (PfGCaC1/C2), NP_705488; P. falciparum GCβ (PfGCbC1/C2), AAN35978; Dictyostelium discoideum gca (DdGcaC1/C2), XP_643824; D. discoideum SgcA (DdSgcAC1/C2), XP_643219; Drosophila melanogaster Gyc32E (DmGC32E), Q07553; D. melanogaster Gyc88E (DmGC88E), NP_731974; D. melanogaster CG31183 (CG31183), NP_650505; D. melanogaster CG10738 (CG10738), NP_729905; D. melanogaster CG9783 (CG9783), NP_649477; D. melanogaster CG3216 (CG3216), NP_611532; D. melanogaster 99B (DmGC99B), Q07093; human tmAC-1 (htmAC1C1/ C2), Q08828; human tmAC-2 (htmAC2C1/C2), Q08462; human tmAC-3 (htmAC3C1/C2), O60266; human tmAC-4 (htmAC4C1/C2), Q8NFM4; human tmAC-5 (htmAC5C1/C2), O95622; human tmAC-6 (htmAC6C1/C2), O43306; human tmAC-7 (htmAC7C1/C2), P51828; human tmAC-8 (htmAC8C1/C2), P40145; human tmAC-9 (htmAC9C1/C2), O60503; D. melanogaster AC-A (DmACAC1/C2), NP_620475; D. melanogaster AC-B (DmACBC1/C2), NP_620474; D. melanogaster AC-C (DmACCC1/C2), NP_609593; D. melanogaster AC-D (DmACDC1/C2), NP_620478; D. melanogaster AC-E (DmACEC1/C2), NP_620479; D. melanogaster AC-1 (DmAC1C1/C2); Anopheles gambiae AgaC (AgACC1/ C2), EAA10271; human sAC (hsACC1/C2), NP_060887; P. falciparum ACα (PfAca), NP_701931; P. falciparum ACβ (PfACbC1/C2), NP_704518; D. discoideum Aca (DdAcaC1/C2), AAA33163; D. discoideum AcaA (DdAcaAC1/C2), XP_640636; D. discoideum AcrA (DdAcrA), AAD50121; D. discoideum AcgA (DdAcg), Q03101; Trypanosoma brucei AC-1 (TbbAC1), Q99279; T. brucei AC-2 (TbbAC2), Q99396; T. brucei AC-3 (TbbAC3), Q99280; T. brucei AC-4 (TbbAC4), Q26721; T. congolense AC (TcongAC), Q26896; T. equiperdum AC (TequiAC), P26338; Mycobacterium tuberculosis Rv1264 (Rv1624), CAB00890; M. tuberculosis Rv1319c (Rv1319c), Q10632; M. tuberculosis Rv1625c (Rv1625c), P0A4Y0; Candida albicans Cyr1 (CalbAC), AAG18428; Cryptococcus neoformans AC (CneoACC1/C2), AAG60619; Schizosaccharomyces pombe AC (SpomACC1/C2), P14605; Saccharomyces cerevisiae AC (ScerACC1/C2), P08678; Chloroflexus aurantiacus Chlo1066 (Ch1066C1/C2), ZP_00018085; C. aurantiacus Chlo1187 (Ch1187C1/C2), ZP_00018205; C. aurantiacus Chlo1431 (Ch1431C1/C2), ZP_00018442; Spirulina platensis CyaA (SplatACA), BAA22996; S. platensis CyaC (SplatACC), T17197; Stigmatella aurantiaca CyaA (SaurACA), CAA11549; S. aurantiaca CyaB1 (SaurACB1), T10905; Synecocystis sp. CyaA2 (PCC6803), BAA16969; Anabaena pirulensis CyaA (ApirACA), P43524; A. pirulensis CyaB1 (ApirACB1), NP_486306; A. pirulensis CyaB2 (ApirACB2), BAA13999; A. pirulensis CyaC (ApirACC), BAA14000; Mycobacterium leprae AC (MlepAC), CAA19149; Sinorhizobium melioti AC-1 (SmeliAC1), P19485; S. melioti AC-2 (SmeliAC2), Q52915; S. melioti AC-3 (SmeliAC3), Q9Z3Q0; Mesorhizobium loti AC-3 (MlotiAC3), BAB50205; Myxococcus xanthus CyaA (MxanCyaA), BAC00918; M. xanthus CyaB (MxanCyaB), BAD98264; Rhizobium etli CyaA (RetlCyaA), AAM66143; R. etli CyaB (RetlCyaB), AAM66145; Anenome cylindrica A (AcylACA), P43524; E. coli ACA (EcoliACA), CAA47280.
G protein responsive tmACs first appear in metazoans. Structurally related cyclases have been characterized from M. tuberculosis and protozoans, but these are not thought to be G protein regulated.37,48 Interestingly, the M. tuberculosis tmAC-like cyclase is analogous to half of a mammalian tmAC; it consists of a single six transmembrane helices domain followed by a cytoplasmic loop and has to homodimerize for activity.37 Fungal ACs, although they occupy a separate branch of the nucleotidyl cyclase family tree (Figure 3), appear to combine features found in both tmACs and sAC. The adenylyl cyclases from the yeast Saccharomyces cerevisiae and the pathogenic fungi Cryptococcus neoformans and Candida albicans are G protein regulated49–51 and were recently shown to be bicarbonate responsive.52
Molecular Structure
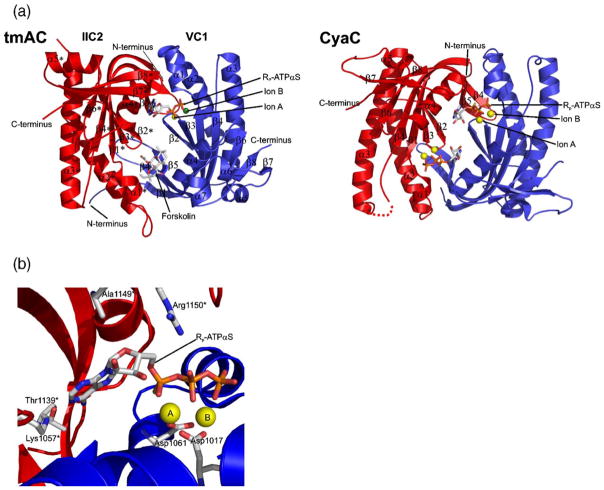
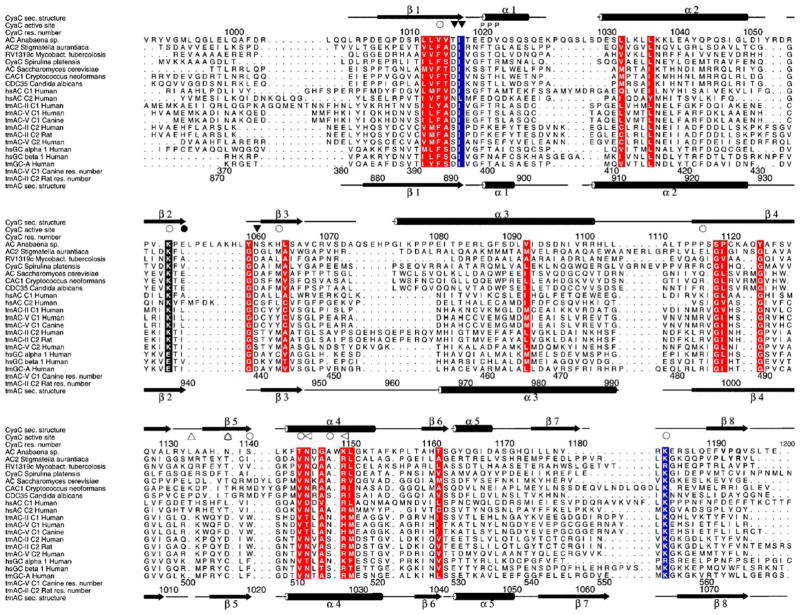
As stated above, the functional catalytic unit of class III enzymes is formed by either homodimerization of a single catalytic domain or heterodimerization of structurally similar C1 and C2 catalytic domain monomers. The first crystal structure of a class III AC catalytic domain to be solved was a non-physiological mammalian C2 homodimer,53 followed by crystal structures of a mammalian C1C2 heterodimer,40,54 two trypanosomal catalytic cores,55 several mycobacterial ACs,41,56,57 and a cyanobacterial sAC homolog.38 These structures confirmed the conserved architecture of class III catalytic domains (Figure 4(a)) previously suggested by their sequence similarities (Figure 5).Within each monomer, a central seven-stranded β-sheet is shielded from the solvent by helices α2, α3, and α5. The two monomers interact head-to-tail and are held together in a wreath-like fashion by an extended clamp formed by β5, the C-terminal part of β4, and the loop in between. The active site of the catalytic entity is located at the dimer interface and formed by residues from both catalytic domains. Two highly conserved Asp residues (Asp396 and Asp440 in Figure 5; residue numbers refer to the tmAC heterodimer described by Tesmer et al.40 if not stated otherwise) and the phosphate tails of the bound substrate, ATP, coordinate two magnesium ions, called ion A and ion B.38,54 The substrate phosphates are further bound by positively charged residues (Arg484, Arg1029*, Lys1065*; the asterisk (*) indicates a C2 residue) and main-chain atoms of the N terminus of helix α1. The ribose moiety of the substrate occupies a mainly hydrophobic pocket with a Ser conserved in tmACs (Ser1028*) but replaced by an Ala conserved in sAC homologs (Ala1149* in CyaC), a difference likely important for the characteristic substrate affinities of the two AC families (see below). The adenine base is bound in a hydrophobic cleft and recognized through interactions with two polar residues. A highly conserved Lys (Lys938*) binds to the ring nitrogen N1 of the adenine, and an Asp (Asp1018*) interacts with the amino group N6; the Asp in tmACs is functionally replaced by a Thr found in sAC-like enzymes at this position.38,43 The Lys and the Asp are replaced in GC enzymes by Glu and Cys, respectively. Making the corresponding amino acid substitutions in GCs converts the substrate specificity to ATP,58,59 indicating that these two residues dominate substrate selection. However, additional factors contribute to substrate selection and turn-over.60 AC enzyme specificity is not switched to GTP when analogous mutations are introduced,57,58,60 perhaps due to an impact on formation of the dimer interface caused by alteration of these residues.57
Figure 4.
Structure of class III catalytic units. (a) Overall structures of the catalytic domains of a mammalian tmAC (left) and the cyanobacterial sAC-like enzyme CyaC (right), both in complex with ATP analogs. The two subunits of the catalytic cores are colored red and blue, respectively, and secondary structure elements are labeled according to Steegborn et al.,38 and Tesmer et al.40 (b) Active site of the sAC-like cyclase CyaC in complex with the ATP analog Rp-ATPαS and two magnesium ions A and B (yellow spheres). The ion coordinating Asp residues 1017 and 1061, the residues recognizing the adenine ring (1139* and 1057*), the phosphate binding Arg (1150*), and the Ala conserved in sACs and replaced by Ser in tmACs (1149*) are labeled. This Figure was generated with PyMol.134
Figure 5.
Structure-based sequence alignment of representative members of nucleotidyl cyclase class III. Included are bicarbonate responsive cyclases (top 9 sequences) from bacteria, fungi, and human; the human G protein-regulated tmACs II and V and the structurally characterized tmACs from rat and dog; and two guanylyl cyclases, the homodimeric transmembrane receptor GC-A and the heterodimeric soluble guanylyl cyclases sGC1 (bottom three sequences). Secondary structure elements of CyaC are shown on top and those of tmAC II C2 at the bottom. Ion binding residues (▽) and residues binding the substrate (○) or the transition state (◁) are labeled with filled and empty symbols indicating C1 and C2 residues, respectively. CyaC Thr1139*, which is characteristic for sAC enzymes and replaced by Asp in tmACs and Cys in GCs, is indicated (△). Homologous regions are shaded red, and highly conserved positions are shown in blue. Residue numbering on the top refers to CyaC and at the bottom to human tmAC VC1 and IIC2, respectively. The Figure was generated with Alscript.135
For both mammalian tmACs and sAC, active cyclase is formed via heterodimerization of two similar catalytic domains. Differences between the two catalytic domains within either cyclase result in a non-symmetrical dimer interface; i.e. the two centers formed are not identical. In fact, only one site possesses all the residues essential for catalysis.40 The site at the other pole of the dimer center is degenerate, and in tmACs, this degenerate center is part of the binding site for the tmAC specific regulator forskolin40,53 (see below). Thus, because C1 and C2 contribute distinct essential residues to the lone catalytic center, even though their overall structure is the same, the two catalytic domains serve different functions.
The Catalytic Mechanism of Class III Cyclases
The overall structure as well as most residues involved in substrate binding and catalysis are conserved among class III cyclases. The positions with variations mostly show functionally conserved substitutions (see above) which define a limited number of subfamilies within class III.32 Therefore, it is likely that class III cyclases share a common catalytic mechanism. The small differences between individual cyclases or subfamilies, such as the Δ-loop in trypanosomal ACs,55 mostly confer unique mechanisms of regulation upon these enzymes.
The simplest reaction mechanism compatible with stereochemical studies on class III cyclases61 is the now widely accepted in-line attack of the 3′-hydroxyl group of the ribose at Pα occurring simultaneously to the release of the Pβγ pyrophosphate (Figure 6); i.e. a pseudo-bimolecular nucleophilic substitution (SN2) mechanism. The function of ion B seems to be a stabilization of the bound substrate as well as the subsequently produced pyrophosphate through interactions with Pβ and Pγ.54 Ion A, in contrast, serves to stabilize the pentacovalent transition state at Pα and to activate the 3′-hydroxyl of the ribose as attacking group, either directly or through a magnesium-activated base such as a magnesium-bound water or hydroxyl ion.
Figure 6.
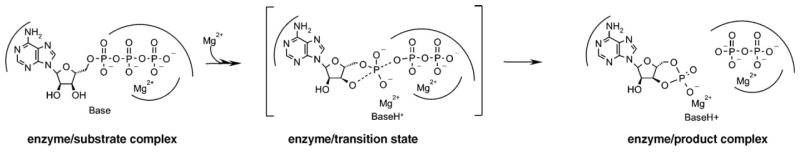
Catalytic mechanism of class III AC enzymes. Schematic view of the two-ion catalyzed reaction, which starts with the substrate ATP and yields the products cAMP and pyrophosphate. The reaction proceeds through a magnesium-stabilized penta-covalent transition state with an in-line arrangement of the incoming and the leaving group.
Tesmer et al. proposed a substrate-induced active site closure as part of the catalytic cycle, based on a more closed form observed for tmAC after, e.g. the ATP analog ATPαS had been soaked into the active site.40,54,62 The closure is due to movements of α1 and an “arm” sub-domain formed by β7, β8, and the loop in between them toward the dimer center (Figure 7(a)). High-resolution data for a sAC-like enzyme revealed, however, that at least in this enzyme ATPαS binds in a conformation not suitable for the reaction to occur. In contrast, α,β-Me-ATP binds in a productive conformation with the 3′-hydroxyl and the bond to be broken arranged in a straight line42 (Figure 6). Interestingly, sAC complexed to α,β-Me-ATP has an even more open active site which is again due to movements of α1 and β7/ 8. Based on these results, we proposed a modified model for the catalytic role of the α1 and β7/8 movements: substrate binding fully opens the active site, and the subsequent closure drives catalysis by separating the products cAMP and pyrophosphate. 42 Consistent with this model, the tmAC–Gsα complexes with P-site inhibitors, which presumably resemble an enzyme/product complex, are similar to the closed enzyme conformation.63 During active site closure, Pγ and Pβ would be shifted toward the active site exit. Pyrophosphate dissociation would finally enable the active site to open again by releasing interactions to α1 and β7/8. These interactions explain why P-site ligands (see below) require either a polyphosphate tail or pyrophosphate for efficient inhibition; the interactions of pyrophosphate are essential for trapping the enzyme in its closed conformation.
Figure 7.
Modulation of class III AC catalytic activity. (a) Complex of the tmAC catalytic core with Gsα. Displayed is an overlay of the tmAC/Gsα complex in the absence (grey) and in the presence (red) of the ATP analog Rp–ATPαS, showing the large movement of α1 upon ligand binding, accompanied by a smaller shift of the β7/β8 loop. (b) Active site of a tmAC occupied by two Mg2+ (yellow spheres) and the inhibitor MANT–GTP. The MANT fluorophor extends from the active site into the dimer interface patch formed by α1 and the β7/β8 loop, the structural elements, which are responsible for the open–closed transition of the enzyme. (c) Bicarbonate inducible closure of the sAC active site. Overlay of the sAC–α,β-Me-ATP structure (open state; darkest gray, α1 and β7–β8 in blue), the sAC–Rp-ATPαS complex (partially closed; middle gray and red), and the bicarbonate-soaked Rp-ATPαS structure (closed; lightest gray and yellow). This Figure was reproduced from Steegborn et al.38 (d) Active site region of CyaC in complex with the substrate analog α,β-Me-ATP and the non-competitive inhibitor catechol estrogen (CE). The inhibitor is bound next to the active site and acts as chelator on the catalytic magnesium ion (yellow sphere), thereby distorting the active site and the substrate analog (green sphere: calcium). Two inhibitor molecules are bound due to the symmetry of the homodimeric sAC homolog CyaC used in this study. This Figure was generated with PyMol134 and Setor (c).136
Although class II cyclases are unrelated to class III enzymes in topology and quarternary structure, it appears that they utilize a similar catalytic mechanism. The monomeric catalytic cores of two class II enzymes, edema factor from Bacillus anthracis and CyaA from Bordetella pertussis, comprise a single active site at the interface between two structurally distinct domains.31,64 Recent studies revealed that this active site, although non-homologous to the class III catalytic center, provides two ion binding sites which seem to correspond functionally to ion A and ion B sites.65,66 This similarity, together with the lack of structural homology might indicate that these two cyclase classes are a result of convergent evolution of a two-ion active site for this substitution reaction at Pα. This conclusion might also apply to the DNA polymerase ‘palm domains’ which catalyze an intermolecular substitution at Pα. Part of the palm domain shows a topology also found in class III cyclases67 and a very similar arrangement of two magnesium ions catalyzing its related reaction.68
Regulation of Adenylyl Cyclases
In mammals, cAMP is involved in a multitude of physiological processes.9 In fact, this single second messenger can modulate seemingly disparate functions within a single cell.69,70 The co-existence of sAC and tmACs, along with a broad family of cAMP degrading phosphodiesterases (PDEs),71 permitted a revision of the original models for cAMP signal transduction in order to explain this paradox. These original models relied upon diffusion through the cytoplasm of the second messenger generated at the plasma membrane to elicit cAMP-dependent responses at intracellular organelles and structures. However, modern methods for measuring cAMP in situ (for example, using FRET-based and conductance-based sensors) revealed that diffusion is restricted.72–74 This observation suggests the existence of additional sources of cAMP localized closer to the second messenger site of action. In the revised model (Figure 1), cAMP is formed, acts, and is degraded in independently regulated cAMP signaling micro-domains at the plasma membrane or at intracellular sites.75,76
Spatial restriction and independent modulation of the cyclases in different micro-domains would be achieved by the plethora of regulatory mechanisms of sACs and tmACs. Both families are represented by multiple isoforms and their activities are modulated through protein regulators and small-molecule ligands. We discuss here physiologically relevant regulatory mechanisms as well as the modulation of AC activity with synthetic compounds.
Isoform Diversity
In mammals, nine genes encode tmACs. These are numbered AC I through IX, according to their time of discovery, and each displays unique regulatory properties.7,9,10,77 Besides the different tissue distribution of these isoforms, there are also reports of alternative splicing of some of the tmAC genes, increasing their isoform complexity. Three tmAC VIII variants were identified in rat brain.78 In addition to a “full-length” isoform, one isoform is missing potential glycosylation sites from an extracellular loop and is postulated to be targeted to distinct membrane compartments. A third isoform is missing a region, which seems to be important for calcium/calmodulin regulation. Alternatively spliced forms of tmAC III in mammalian germ cells,79 tmAC IV in uterine myometrium,80 and of tmAC V and VI have been identified,81–83 but their biochemical properties or physiological significance have not yet been elucidated.
sAC is present in all tissues thus far examined.18,24 Although encoded by a single confirmed gene (a second potential sAC gene in humans has not yet been confirmed to be functional), multiple sAC isoforms with distinct regulatory properties are generated by alternative splicing. The longest known cDNAs encode the full-length sAC protein of approximately 187 kDa (sACfl). The activity, originally purified from rat testis resided in a protein of approximately 50 kDa,17,18 which was later found to be generated by alternatively splicing23 (sACt). This truncated protein comprises little more than the two N-terminal catalytic domains,23 and much of what we know about the biochemical activity of sAC has been determined from this more easily purified sACt isoform. Both protein products, sACfl and sACt, are expressed in mouse testis,26 and while the two isoforms are similarly regulated by small molecules (see below), they display distinct intrinsic specific activities.17,19 The truncated isoform has a tenfold higher specific activity due to the presence of an auto-inhibitory domain within sACfl,84 suggesting sACfl will be subject to additional modes of regulation.
Additional sAC isoforms generated by alternative splicing have been identified in somatic tissues.24 At least two distinct human sAC cDNAs predicted proteins with an altered or removed C1 catalytic domain. One splice variant contained a 37 base-pair insertion between exons 3 and 4 leading the authors to postulate a new start codon in exon 4. Such a sAC isoform would possess only one catalytic domain (C2) and would be predicted to require homodimerization or heterodimerization with an as yet unknown isoform (possessing C1) for activity. Another variant skipped exon 4, splicing from exon 3 in-frame to exon 5; this protein would be missing one of the essential magnesium binding aspartates provided by C1, suggesting it too would have to dimerize to produce an active adenylyl cyclase. Preliminary studies in our laboratory confirm the widespread distribution of these spliced forms in human tissues and predict the existence of a complex pattern of alternative splicing of sAC in a number of human and rodent somatic tissues (J. Farrell & Y. Chen, unpublished results).
Protein Regulators
The major regulators of tmACs are heterotrimeric G-proteins. The α-subunit of Gs (Gsα) stimulates most if not all the tmAC isoforms. In contrast, Giα selectively inhibits tmACs I, V, and VI, and individual tmAC isoforms also display unique regulation by G protein βγ subunits.7,10 Despite their structural similarities (described above), the three-dimensional structures reveal differences that might explain sAC's insensitivity to heterotrimeric G-protein regulation. A region implicated in Gβγ binding, specifically the extended loop between β3 and α3 in tmAC II C2,85 is missing in sAC (Figure 5). A second and possibly more important region for Gβγ responsiveness maps to the C1b domain next to but outside of the structurally conserved catalytic core domain.86 The binding surface for Gsα is again located on tmAC C2 and is formed by the α1/α2 loop, the N terminus of α2, and the C terminus of α3 (Figures 4(a) and 7(a)).40 sAC C2 contains a 21 residue insertion between α1 and α2 which likely blocks Gsα access sterically, and a shortened C terminus in α3 (Figure 5). It remains to be seen whether the large insertion between α1 and α2 of sAC is responsible for an unknown sAC specific regulatory mechanism. A similar situation is encountered with the potential Giα binding site which was mapped to the α1/α2 and the α3/β4 region of C1 by mutagenesis:87 in addition to sequence differences, sAC contains a larger loop between α1 and α2 than tmACs (Figure 5) which should prevent Giα binding.
A second major protein regulator of tmACs is the calcium receptor protein calmodulin. Calcium-loaded calmodulin directly stimulates tmAC isoforms type I and type VIII,78,88–90 and possibly also tmAC III.91 Activation in type I proceeds via interaction with the C1b regulatory domain located between the first catalytic domain and the second set of transmembrane domains92,93 while calcium/ calmodulin regulation of tmAC VIII occurs via interaction with the C-terminal C2b domain.94 Calcium/calmodulin can also modulate tmAC I and III activity indirectly via calcium/calmodulin regulation of CaM kinases.95,96
tmACs can also be regulated by other protein kinases. PKA regulates types V, and VI97,98 and PKC regulates types II, V, and VI.99–101 tmACs are also subject to regulation via other post-translational modifications, including the NO-dependent inhibition of types V and VI102 likely mediated by S-nitrosylation; and the N-linked glycosylation of tmAC VIII.78,103 Surprisingly, an RGS protein, RGS2, which would have been predicted to modulate tmAC activity indirectly by acting as GTPase activator on heterotrimeric G proteins, directly regulates tmAC III, V and VI.104 More regulation mechanisms for tmACs await to be identified; the transmembrane regions, for example, also seem to have a regulatory role.105
In contrast to tmACs, no regulatory proteins or post-translational modifications have yet been identified for sAC, but there is evidence for intramolecular protein modulation in particular sAC isoforms. As mentioned above, sACfl displays diminished specific activity relative to the sACt isoform; the difference has been mapped to a putative auto-inhibitory domain located just downstream from the second catalytic domain.84 Neither the structural consequences of this domain nor any mechanism of disinhibition of cyclase activity are yet known. Generally, the molecular mechanisms of mammalian AC regulation through their regulatory domains as well as through other regulatory proteins, including Gsα, Giα, Gβγ and calmodulin, are poorly understood. Gsα increases the affinity of C1 for C2,39,106 but in addition to stabilization of the heterodimer, more subtle rearrangements are necessary to explain the observed levels of stimulation and the negligible influence of the activator on substrate affinity.40,107 Gsα stimulation of tmACs was therefore proposed to proceed via an induced rotation of the two catalytic domains into a proper arrangement,40 but due to the lack of a mammalian tmAC structure in absence of Gsα, this idea was based on a comparison with the non-physiological IIC2 homodimer.53 A similar mechanism was proposed for regulation of a mycobacterial class III AC through its pH sensing domain. In the inactive state, the pH-sensing domain prevents proper orientation of the two catalytic domains. Activation is mediated by a helix-to-loop transition in the linker between regulatory and catalytic domains which enables a large repositioning of the two catalytic domains relative to each other as well as major conformational changes of active site regions.56 The only other structurally characterized regulatory domain of a class III AC is the isolated GAF-region of a the cyanobacterial CyaB2 AC,108 which could not reveal how it translates ligand binding into a change of activity of the catalytic domain. Taken together, repositioning of the two domains of the class III catalytic core appears to constitute a mechanism for regulating its activity, but a firm and detailed understanding of the regulation of mammalian AC enzymes through regulatory domains and proteins will require further structural and mechanistic studies.
Regulation by Small Molecules
Forskolin
Forskolin is a diterpene compound isolated from plants that activates all mammalian tmACs10,109 with the exception of tmAC IX.110,111 It occupies part of the second, degenerated “active site” in tmACs (Figure 4(a)),40 and it has been speculated to exploit the binding site of an as yet unidentified endogenous regulatory ligand. Forskolin has been suggested to activate tmACs by inducing dimerization and/or active site rearrangements,40 but this idea remains speculative because the structure of a tmAC in the absence of forskolin is not yet known. Mammalian sAC is insensitive to forskolin,17,112 and two major changes in sAC, an insertion of four residues in the loop between β2 and β3 and an Ala to Arg exchange at the ribose binding site, seem to render its second, degenerated active site too small to accommodate forskolin (Figure 5).
Substrate ATP, ATP-analogs and P-site inhibitors
Comparison of the tmAC and sAC/CyaC active sites reveals striking similarity;38,40 all 18 residues important for substrate binding and catalysis are either conserved or show small and mostly functionally conservative variations (Figure 5). Two of these small differences appear to distinguish tmACs and sAC-like enzymes. An Asp conserved in tmACs (position 1018*) and involved in recognition of the adenine base of the substrate ATP is functionally replaced by a threonine (CyaC Thr1139*) in sAC-like enzymes (Figure 4(b)). Secondly, a Ser in tmACs (Ser1028* in tmAC IIC2) is replaced by a conserved Ala(Ala1149*inCyaC)insAC-likeenzymes.Ser1028* appears to be a polar interaction partner for the ribose oxygen atoms in an otherwise hydrophobic environment; this difference likely explains the higher substrate affinity of tmACs (10−4 – 10−5 M)113 compared to sAC-like enzymes (10−3 M).20,38
The relatively low affinity for ATP displayed by mammalian sAC may represent a physiological adaptation. Its Km value in the millimolar range implies sAC would be sensitive to physiologically relevant fluctuations in intracellular ATP levels (found to be 1–3 mM in most cell types) and function as a cellular ATP sensor. Interestingly, longer isoforms of mammalian sAC contain a Walker A (“P-loop”) motif often found in nucleotide binding pockets. This motif is dispensable for catalytic activity,17,84 but it might be involved in an as yet unknown nucleotide-dependent mechanism of sAC regulation. The mechanism of substrate inhibition observed for the catalytic cores of sAC enzymes at high ATP levels20 is also unknown.
P-site inhibitors are adenosine analogs that potently inhibit tmACs.114 Crystal structures of a tmAC in complex with P-site inhibitors revealed they bind to the active site, along with the reaction product pyrophosphate.40,63 Binding to the active site was initially surprising; kinetic studies had suggested that P-site ligands behave as uncompetitive inhibitors.115 However, Dessauer et al. elegantly explained this behavior by demonstrating that in the presence of pyrophosphate, the enzyme can be trapped in the inactive, closed conformation, the form with greatest binding affinity for the inhibitor.63,114,116 In contrast, the substrate binds preferably to the active, open form of the enzyme. The same differences between the adenine and sugar binding pockets of sAC and tmACs, which decreases AC's affinity for ATP, should also explain its diminished sensitivity to inhibition by P-site ligands.117 Interestingly, a new series of P-site inhibitors, such as the compoundPMC-6 (1R,4R-3-(6-aminopurin-9-yl)-cyclopentanecarboxylic acid hydroxyamide), contain a metal chelating moiety118,119 and can discriminate between some tmAC isoforms;119 PMC-6 inhibits type II and III modestly, but tmAC V with high potency.
A class of synthetic nucleotide analogs identified as competitive AC inhibitors are 2′(3′)-O-(N-methylanthraniloyl)-substituted nucleotides (MANT-nucleotides). 120 The crystal structure of a tmAC catalytic domain in complex with MANT–GTP showed the nucleotide bound to the ATP binding site but with guanine in a reversed orientation relative to the adenine ring (Figure 7(b)).121 The MANT fluorophore binds to a hydrophobic pocket at the interface between C1 and C2 and prevents the “open” to “closed” domain rearrangement, thereby identifying a novel target site for inhibitor development. However, similar to most P-site ligands, MANT compounds are more potent against tmACs than against sAC but display limited tmAC isoform selectivity.117
Catalysis by class III ACs proceeds via an in-line attack of the 3′-hydroxyl group in concert with the release of pyrophosphate. Therefore, it should have been surprising that ACs are inhibited only by Rp–ATPαS, whereas the Sp form serves as substrate.38,61 Comparing the conformation of CyaC-bound Rp–ATPαS and α,β-Me–ATP reveals that replacement of the pro-R oxygen will cause ATPαS to bind in a nonproductive conformation and, therefore, be an inhibitor.38 Pα is turned around so that the pro-S oxygen, which is more favorable for magnesium coordination than sulfur, coordinates ion A (Figure 4(b)). This rotation moves Pα away from the position suitable for in-line attack by the 3′-hydroxyl. In contrast, Sp-ATPαS can bind in a productive conformation by coordinating ion A with its unmodified pro-R oxygen.
Bicarbonate
Bicarbonate directly and specifically activates sAC enzymes; tmACs are insensitive.18 Indeed, sAC-like ACs are the only known signaling proteins directly regulated by bicarbonate. Crystallographic studies on CyaC did not expose the bicarbonate-binding site, but flash soaking experiments revealed an induced active site closure through a ~4–5 Å tilt of β-strands 7 and 8 and the intervening loop, accompanied by a ~3 Å shift of α1 (Figure 7(c)).38 These shifts push the β/γ-phosphates of the bound ATP analog and could facilitate separation of the ultimate products, cAMP and pyrophosphate, during catalysis (see above). The bicarbonate-induced closed conformation resembles the conformation of tmAC in complex with product analogs.63 Thus, bicarbonate appears to increase sAC activity by facilitating the open/ closed transition assumed to occur during catalysis, which is consistent with kinetic studies revealing an effect on kcat.20
The precise bicarbonate binding site in sAC-like ACs remains unclear. A substrate binding residue, Thr1139* in CyaC, which is conserved in bicarbonate-regulated sAC-like enzymes and is replaced by an Asp in tmACs (Asp1018* in tmAC IIC2), was postulated to be important for bicarbonate recognition.43 However, mutagenesis at this position severely reduced the enzyme's activity, preventing a clear interpretation of the experiments. Crystallographic identification of the bicarbonate binding site is severely hampered by the high EC50 value of 10–25 mM.18,20 Such a low affinity, which is necessary for sensing physiologically relevant bicarbonate levels (bicarbonate concentrations in cells and body fluids vary between 5 mM and 25 mM), reflects a weak and possibly transient interaction that is difficult to study.
Calcium
The activities of the different mammalian tmACs display both positive and negative regulation by calcium.10,122 The isoform specific activation is mediated by calcium-loaded calmodulin and was discussed above. All tmAC isoforms are directly inhibited by high micromolar concentrations of calcium.123 Presumably, at these concentrations, calcium non-productively competes with magnesium for the tmAC ion A site.123,124 Specific tmAC isoforms (types V and VI) are also inhibited by sub-micromolar concentrations of calcium.123- Although the mechanism is not understood in detail, calcium and magnesium appear to bind to two distinct enzyme conformations and compete either by binding to the same ion site or by binding to allosterically coupled sites.123,124
In contrast to tmACs, calcium supports sAC activity even when it is the only divalent cation available.20 Therefore, although the ion binding residues are conserved between sAC and tmACs, sAC is uniquely catalytically active with calcium in the ion A site. In the presence of magnesium, high micromolar concentrations of calcium stimulate the activity of sAC-like ACs by increasing the enzyme's affinity for the substrate ATP from a KM of about 10 mM to KM=1 mM.20 X-ray crystallography revealed that calcium replaces magnesium at the ion B pocket of the active site, coordinating the β and γ-phosphates of the bound substrate analog.38 Therefore, while sAC is active with either magnesium or calcium occupying both ion sites, its activity is highest with magnesium in the ion A site and calcium in the ion B site. All groups in contact with ion B are conserved between sAC and tmACs, yet this type of calcium effect has not been described in tmACs.
Calcium may play a second modulatory role on sAC. Sub-micromolar concentrations of calcium were observed to stimulate native sAC protein immunoprecipitated from testis.19 Stimulation was independent of calmodulin and appeared to be due to an increase of vmax, not substrate affinity. These results suggest that post-translational modifications or interaction with an unknown regulatory protein may alter the calcium affinity of the enzyme or confer some additional modulatory mechanism.
Atypical AC inhibitors
In addition to the nucleotide analog inhibitors described above, a number of other, unrelated compounds have been described which inhibit mammalian AC activity. Catechol estrogens (CE) and tyrphostins with two vicinal hydroxyl groups were independently identified as non-competitive cyclase inhibitors,125–127 yet they seem to share a common binding site and inhibitory mechanism.42 For CE, the mechanism was identified from a crystal structure of the trimeric complex of the sAC-homolog CyaC with a substrate analog and the inhibitor.42 CE is bound to a hydrophobic patch near the active site, and its vicinal hydroxyl groups chelate the catalytic magnesium ion, distorting the active site and trapping the enzyme–substrate complex in a non-productive conformation (Figure 7(d)). CEs inhibit both sAC and tmACs with comparable affinities, but sequence variations in the binding pocket indicate that more specific ligands for this site might exist.42 Although catechol estrogens are known to be enriched in certain human tissues,128,129 it remains unclear whether they represent physiological AC regulators or whether more specific physiological regulators exist for this binding site. In either event, CEs and tyrphostins represent promising starting compounds for the development of more specific cyclase inhibitors.
Another non-physiological compound acting as non-competitive inhibitor of tmACs is calmidazolium, 130 which is a known chelator for divalent ions. It is tempting to speculate that calmidazolium exploits the CE interaction site and inhibitory mechanism.
Because of similarities in catalytic cleft topology and the nature of the reactions catalyzed,67 inhibitors of DNA polymerase and HIV reverse transcriptase were tested for effects on adenylyl cyclases. Foscarnet131 and antiviral acyclic nucleoside derivatives of 9-(2-phosphonylmethoxyethyl)adenine132 proved to be potent inhibitors of nucleotidyl cyclases, and these compounds may provide novel ideas for the development of specific inhibitors.
Finally, novel sAC inhibitors were recently identified in a high-throughput chemical library screen.26,75 One particular compound, KH7,26,28 is non-competitive with ATP, but its structure bears little similarity to CEs or P-site inhibitors, suggesting it may define yet another inhibitory principle. It shows a pronounced selectivity for sAC relative to tmACs or guanylyl cyclases, yet it appears to potently inhibit all thus far identified sAC-like enzymes in a wide range of organisms throughout evolution (K. Hess, E.M.B., L.R.L. & J.B., unpublished results).
Concluding Remarks
Even though cAMP has been extensively studied for over 50 years, the plethora of regulatory mechanisms controlling cAMP synthesis is just beginning to be uncovered. In mammalian cells this important second messenger can be produced by two related families of adenylyl cyclases, sAC and tmACs. These enzymes share a conserved catalytic mechanism but differ in their specific physiological functions due to altered regulation and distinct intracellular localization. Additional modes of regulation await discovery. Many questions remain about tmAC isoform divergence and individual cellular functions, and thus far, studies exploring sAC regulation focused on the catalytic domains, leaving the remainder of the protein largely unexplored.
Acknowledgments
We gratefully acknowledge Melanie Gertz and Drs Martin Cann, Joachim Schultz, and Wei-Jen Tang for critical reading of the manuscript and valuable suggestions. We thank Lucy Skrabanek for assistance with generating the phylogenetic tree. Recent work from the authors laboratories described in this review has been funded by the Ellison Medical Foundation (to J.B.), American Diabetes Association (to L.R.L.), Hirschl-Weil Cauler Trust (to L.R.L.), National Institutes of Health (AI64842, GM62328, HD42060, and HD38722 to L.R.L. and J.B.; DA007274 to M.K.), and Deutsche Forschungsgemeinschaft (STE1701/1-1 to C.S.).
Abbreviations used
- AC
adenylyl cyclase
- α, β-Me-ATP
α,β-methylene-adenosine-5′-triphosphate
- ATPαS
adenosine-5′-α-thio-triphosphate
- FRET
fluorescence resonance energy transfer
- GC
guanylyl cyclase
- Gβγ
G protein, β and γ subunits
- Gsα
stimulatory G protein, α subunit
- Giα
inhibitory G protein, α subunit
- Km
Michaelis constant
- MANT
2′(3′)-O-(N-methylanthraniloyl)
- PDE
phosphodiesterase
- PKA
protein kinase A
- PMC-6
1R,4R-3-(6-aminopurin-9-yl)-cyclopentanecarboxylic acid hydroxyamide
- sAC
soluble adenylyl cyclase
- sACfl
full-length soluble adenylyl cyclase
- sACt
truncated form of soluble adenylyl cyclase
- tmAC
transmembrane adenylyl cyclase
- TM
transmembrane
- vmax
maximum reaction velocity
References
- 1.Rall TW, Sutherland EW, Berthet J. The relationship of epinephrine and glucagon to liver phosphorylase. IV Effect of epinephrine and glucagon on the reactivation of phosphorylase in liver homogenates. J Biol Chem. 1957;224:463–475. [PubMed] [Google Scholar]
- 2.Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–1091. [PubMed] [Google Scholar]
- 3.Kopperud R, Krakstad C, Selheim F, Doskeland SO. cAMP effector mechanisms. Novel twists for an ‘old’ signaling system. FEBS Letters. 2003;546:121–126. doi: 10.1016/s0014-5793(03)00563-5. [DOI] [PubMed] [Google Scholar]
- 4.Gross-Langenhoff M, Hofbauer K, Weber J, Schultz A, Schultz JE. cAMP is a ligand for the tandem GAF domain of human phosphodiesterase 10 and cGMP for the tandem GAF domain of phosphodiesterase 11. J Biol Chem. 2006;281:2841–2846. doi: 10.1074/jbc.M511468200. [DOI] [PubMed] [Google Scholar]
- 5.Danchin A. Phylogeny of adenylyl cyclases. Adv Second Messenger Phosphoprotein Res. 1993;27:109–162. [PubMed] [Google Scholar]
- 6.Tellez-Sosa J, Soberon N, Vega-Segura A, Torres-Marquez ME, Cevallos MA. The Rhizobium etli cyaC product: characterization of a novel adenylate cyclase class. J Bacteriol. 2002;184:3560–3568. doi: 10.1128/JB.184.13.3560-3568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Defer N, Best-Belpomme M, Hanoune J. Tissue specificity and physiological relevance of various isoforms of adenylyl cyclase. Am J Physiol Renal Physiol. 2000;279:F400–F416. doi: 10.1152/ajprenal.2000.279.3.F400. [DOI] [PubMed] [Google Scholar]
- 8.Taussig R, Gilman AG. Mammalian membrane-bound adenylyl cyclases. J Biol Chem. 1995;270:1–4. doi: 10.1074/jbc.270.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Hanoune J, Defer N. Regulation and role of adenylyl cyclase isoforms. Annu Rev Pharmacol Toxicol. 2001;41:145–174. doi: 10.1146/annurev.pharmtox.41.1.145. [DOI] [PubMed] [Google Scholar]
- 10.Sunahara RK, Taussig R. Isoforms of mammalian adenylyl cyclase: multiplicities of signaling. Mol Interv. 2002;2:168–184. doi: 10.1124/mi.2.3.168. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Pineda VV, Chan GC, Wong ST, Muglia LJ, Storm DR. Type 8 adenylyl cyclase is targeted to excitatory synapses and required for mossy fiber long-term potentiation. J Neurosci. 2003;23:9710–9718. doi: 10.1523/JNEUROSCI.23-30-09710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storm DR, Hansel C, Hacker B, Parent A, Linden DJ. Impaired cerebellar long-term potentiation in type I adenylyl cyclase mutant mice. Neuron. 1998;20:1199–1210. doi: 10.1016/s0896-6273(00)80500-0. [DOI] [PubMed] [Google Scholar]
- 13.Wu ZL, Thomas SA, Villacres EC, Xia Z, Simmons ML, Chavkin C, et al. Altered behavior and long-term potentiation in type I adenylyl cyclase mutant mice. Proc Natl Acad Sci USA. 1995;92:220–224. doi: 10.1073/pnas.92.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villacres EC, Wu Z, Hua W, Nielsen MD, Watters JJ, Yan C, et al. Developmentally expressed Ca(2+)-sensitive adenylyl cyclase activity is disrupted in the brains of type I adenylyl cyclase mutant mice. J Biol Chem. 1995;270:14352–14357. doi: 10.1074/jbc.270.24.14352. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa Y, Iwatsubo K, Tsunematsu T, Okumura S. Genetic manipulation and functional analysis of cAMP signalling in cardiac muscle: implications for a new target of pharmacotherapy. Biochem Soc Trans. 2005;33:1337–1340. doi: 10.1042/BST0331337. [DOI] [PubMed] [Google Scholar]
- 16.Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, et al. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 17.Buck J, Sinclair ML, Schapal L, Cann MJ, Levin LR. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc Natl Acad Sci USA. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science. 2000;289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- 19.Jaiswal BS, Conti M. Calcium regulation of the soluble adenylyl cyclase expressed in mammalian spermatozoa. Proc Natl Acad Sci USA. 2003;100:10676–10681. doi: 10.1073/pnas.1831008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litvin TN, Kamenetsky M, Zarifyan A, Buck J, Levin LR. Kinetic properties of “soluble” adenylyl cyclase. Synergism between calcium and bicarbonate. J Biol Chem. 2003;278:15922–15926. doi: 10.1074/jbc.M212475200. [DOI] [PubMed] [Google Scholar]
- 21.Sinclair ML, Wang XY, Mattia M, Conti M, Buck J, Wolgemuth DJ, Levin LR. Specific expression of soluble adenylyl cyclase in male germ cells. Mol Reprod Dev. 2000;56:6–11. doi: 10.1002/(SICI)1098-2795(200005)56:1<6::AID-MRD2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Zippin JH, Chen Y, Nahirney P, Kamenetsky M, Wuttke MS, Fischman DA, et al. Compartmentalization of bicarbonate-sensitive adenylyl cyclase in distinct signaling microdomains. FASEB J. 2003;17:82–84. doi: 10.1096/fj.02-0598fje. [DOI] [PubMed] [Google Scholar]
- 23.Jaiswal BS, Conti M. Identification and functional analysis of splice variants of the germ cell soluble adenylyl cyclase. J Biol Chem. 2001;276:31698–31708. doi: 10.1074/jbc.M011698200. [DOI] [PubMed] [Google Scholar]
- 24.Geng W, Wang Z, Zhang J, Reed BY, Pak CY, Moe OW. Cloning and characterization of the human soluble adenylyl cyclase. Am J Physiol Cell Physiol. 2005;288:C1305–C1316. doi: 10.1152/ajpcell.00584.2004. [DOI] [PubMed] [Google Scholar]
- 25.Esposito G, Jaiswal BS, Xie F, Krajnc-Franken MA, Robben TJ, Strik AM, et al. Mice deficient for soluble adenylyl cyclase are infertile because of a severe sperm-motility defect. Proc Natl Acad Sci USA. 2004;101:2993–2998. doi: 10.1073/pnas.0400050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess KC, Jones BH, Marquez B, Chen Y, Ord TS, Kamenetsky M, et al. The “soluble” adenylyl cyclase in sperm mediates multiple signaling events required for fertilization. Dev Cell. 2005;9:249–259. doi: 10.1016/j.devcel.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, et al. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem. 2003;278:49523–49529. doi: 10.1074/jbc.M309543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han H, Stessin A, Roberts J, Hess K, Gautam N, Kamenetsky M, et al. Calcium-sensing soluble adenylyl cyclase mediates TNF signal transduction in human neutrophils. J Exp Med. 2005;202:353–361. doi: 10.1084/jem.20050778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zippin JH, Levin LR, Buck J. CO(2)/ HCO(3)(-)-responsive soluble adenylyl cyclase as a putative metabolic sensor. Trends Endocrinol Metab. 2001;12:366–370. doi: 10.1016/s1043-2760(01)00454-4. [DOI] [PubMed] [Google Scholar]
- 30.Ladant D, Ullmann A. Bordatella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol. 1999;7:172–176. doi: 10.1016/s0966-842x(99)01468-7. [DOI] [PubMed] [Google Scholar]
- 31.Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, et al. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 32.Linder JU, Schultz JE. The class III adenylyl cyclases: multi-purpose signalling modules. Cell Signal. 2003;15:1081–1089. doi: 10.1016/s0898-6568(03)00130-x. [DOI] [PubMed] [Google Scholar]
- 33.Cotta MA, Whitehead TR, Wheeler MB. Identification of a novel adenylate cyclase in the ruminal anaerobe, Prevotella ruminicola D31d. FEMS Microbiol Letters. 1998;164:257–260. doi: 10.1111/j.1574-6968.1998.tb13095.x. [DOI] [PubMed] [Google Scholar]
- 34.Sismeiro O, Trotot P, Biville F, Vivares C, Danchin A. Aeromonas hydrophila adenylyl cyclase 2: a new class of adenylyl cyclases with thermophilic properties and sequence similarities to proteins from hyperthermophilic archaebacteria. J Bacteriol. 1998;180:3339–3344. doi: 10.1128/jb.180.13.3339-3344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang WJ, Hurley JH. Catalytic mechanism and regulation of mammalian adenylyl cyclases. Mol Pharmacol. 1998;54:231–240. doi: 10.1124/mol.54.2.231. [DOI] [PubMed] [Google Scholar]
- 36.Wedel B, Garbers D. The guanylyl cyclase family at Y2K. Annu Rev Physiol. 2001;63:215–233. doi: 10.1146/annurev.physiol.63.1.215. [DOI] [PubMed] [Google Scholar]
- 37.Guo YL, Seebacher T, Kurz U, Linder JU, Schultz JE. Adenylyl cyclase Rv1625c of Mycobacterium tuberculosis: a progenitor of mammalian adenylyl cyclases. EMBO J. 2001;20:3667–3675. doi: 10.1093/emboj/20.14.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steegborn C, Litvin TN, Levin LR, Buck J, Wu H. Bicarbonate activation of adenylyl cyclase via promotion of catalytic active site closure and metal recruitment. Nature Struct Mol Biol. 2005;12:32–37. doi: 10.1038/nsmb880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whisnant RE, Gilman AG, Dessauer CW. Interaction of the two cytosolic domains of mammalian adenylyl cyclase. Proc Natl Acad Sci USA. 1996;93:6621–6625. doi: 10.1073/pnas.93.13.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha. GTPgammaS. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 41.Sinha SC, Wetterer M, Sprang SR, Schultz JE, Linder JU. Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c. EMBO J. 2005;24:663–673. doi: 10.1038/sj.emboj.7600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steegborn C, Litvin TN, Hess KC, Capper AB, Taussig R, Buck J, et al. A novel mechanism for adenylyl cyclase inhibition from the crystal structure of its complex with catechol estrogen. J Biol Chem. 2005;280:31754–31759. doi: 10.1074/jbc.M507144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cann MJ, Hammer A, Zhou J, Kanacher T. A defined subset of adenylyl cyclases is regulated by bicarbonate ion. J Biol Chem. 2003;278:35033–35038. doi: 10.1074/jbc.M303025200. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi M, Buck J, Levin LR. Conservation of functional domain structure in bicarbonate-regulated “soluble” adenylyl cyclases in bacteria and eukaryotes. Dev Genes Evol. 2004;214:503–509. doi: 10.1007/s00427-004-0432-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nomura M, Beltran C, Darszon A, Vacquier VD. A soluble adenylyl cyclase from sea urchin spermatozoa. Gene. 2005;353:231–238. doi: 10.1016/j.gene.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 46.Muhia DK, Swales CA, Eckstein-Ludwig U, Saran S, Polley SD, Kelly JM, et al. Multiple splice variants encode a novel adenylyl cyclase of possible plastid origin expressed in the sexual stage of the malaria parasite Plasmodium falciparum. J Biol Chem. 2003;278:22014–22022. doi: 10.1074/jbc.M301639200. [DOI] [PubMed] [Google Scholar]
- 47.Roelofs J, Van Haastert PJ. Deducing the origin of soluble adenylyl cyclase, a gene lost in multiple lineages. Mol Biol Evol. 2002;19:2239–2246. doi: 10.1093/oxfordjournals.molbev.a004047. [DOI] [PubMed] [Google Scholar]
- 48.Weber JH, Vishnyakov A, Hambach K, Schultz A, Schultz JE, Linder JU. Adenylyl cyclases from Plasmodium, Paramecium and Tetrahymena are novel ion channel/enzyme fusion proteins. Cell Signal. 2004;16:115–125. doi: 10.1016/s0898-6568(03)00129-3. [DOI] [PubMed] [Google Scholar]
- 49.Uno I, Mitsuzawa H, Matsumoto K, Tanaka K, Oshima T, Ishikawa T. Reconstitution of the GTP-dependent adenylate cyclase from products of the yeast CYR1 and RAS2 genes in Escherichia coli. Proc Natl Acad Sci USA. 1985;82:7855–7859. doi: 10.1073/pnas.82.23.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, et al. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maidan MM, De Rop L, Serneels J, Exler S, Rupp S, Tournu H, et al. The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol Biol Cell. 2005;16:1971–1986. doi: 10.1091/mbc.E04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schroppel K, Naglik JR, et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang G, Liu Y, Ruoho AE, Hurley JH. Structure of the adenylyl cyclase catalytic core. Nature. 1997;386:247–253. doi: 10.1038/386247a0. [DOI] [PubMed] [Google Scholar]
- 54.Tesmer JJ, Sunahara RK, Johnson RA, Gosselin G, Gilman AG, Sprang SR. Two-metal-Ion catalysis in adenylyl cyclase. Science. 1999;285:756–760. doi: 10.1126/science.285.5428.756. [DOI] [PubMed] [Google Scholar]
- 55.Bieger B, Essen LO. Structural analysis of adenylate cyclases from Trypanosoma brucei in their monomeric state. EMBO J. 2001;20:433–445. doi: 10.1093/emboj/20.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tews I, Findeisen F, Sinning I, Schultz A, Schultz JE, Linder JU. The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science. 2005;308:1020–1023. doi: 10.1126/science.1107642. [DOI] [PubMed] [Google Scholar]
- 57.Ketkar AD, Shenoy AR, Ramagopal UA, Visweswariah SS, Suguna K. A structural basis for the role of nucleotide specifying residues in regulating the oligomerization of the Rv1625c adenylyl cyclase from M. tuberculosis. J Mol Biol. 2006;356:904–916. doi: 10.1016/j.jmb.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 58.Sunahara RK, Beuve A, Tesmer JJ, Sprang SR, Garbers DL, Gilman AG. Exchange of substrate and inhibitor specificities between adenylyl and guanylyl cyclases. J Biol Chem. 1998;273:16332–16338. doi: 10.1074/jbc.273.26.16332. [DOI] [PubMed] [Google Scholar]
- 59.Tucker CL, Hurley JH, Miller TR, Hurley JB. Two amino acid substitutions convert a guanylyl cyclase, RetGC-1, into an adenylyl cyclase. Proc Natl Acad Sci USA. 1998;95:5993–5997. doi: 10.1073/pnas.95.11.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linder JU. Substrate selection by class III adenylyl cyclases and guanylyl cyclases. IUBMB Life. 2005;57:797–803. doi: 10.1080/15216540500415636. [DOI] [PubMed] [Google Scholar]
- 61.Eckstein F, Romaniuk PJ, Heideman W, Storm DR. Stereochemistry of the mammalian adenylate cyclase reaction. J Biol Chem. 1981;256:9118–9120. [PubMed] [Google Scholar]
- 62.Tesmer JJ, Sprang SR. The structure, catalytic mechanism and regulation of adenylyl cyclase. Curr Opin Struct Biol. 1998;8:713–719. doi: 10.1016/s0959-440x(98)80090-0. [DOI] [PubMed] [Google Scholar]
- 63.Tesmer JJ, Dessauer CW, Sunahara RK, Murray LD, Johnson RA, Gilman AG, Sprang SR. Molecular basis for P-site inhibition of adenylyl cyclase. Biochemistry. 2000;39:14464–14471. doi: 10.1021/bi0015562. [DOI] [PubMed] [Google Scholar]
- 64.Guo Q, Shen Y, Lee YS, Gibbs CS, Mrksich M, Tang WJ. Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin. EMBO J. 2005;24:3190–3201. doi: 10.1038/sj.emboj.7600800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo Q, Shen Y, Zhukovskaya NL, Florian J, Tang WJ. Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate. J Biol Chem. 2004;279:29427–29435. doi: 10.1074/jbc.M402689200. [DOI] [PubMed] [Google Scholar]
- 66.Shen Y, Zhukovskaya NL, Guo Q, Florian J, Tang WJ. Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. EMBO J. 2005;24:929–941. doi: 10.1038/sj.emboj.7600574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Artymiuk PJ, Poirrette AR, Rice DW, Willett P. A polymerase I palm in adenylyl cyclase? Nature. 1997;388:33–34. doi: 10.1038/40310. [DOI] [PubMed] [Google Scholar]
- 68.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 69.Buxton IL, Brunton LL. Compartments of cyclic AMP and protein kinase in mammalian cardiomyocytes. J Biol Chem. 1983;258:10233–10239. [PubMed] [Google Scholar]
- 70.Hayes JS, Bowling N, King KL, Boder GB. Evidence for selective regulation of the phosphorylation of myocyte proteins by isoproterenol and prostaglandin E1. Biochim Biophys Acta. 1982;714:136–142. doi: 10.1016/0304-4165(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 71.Soderling SH, Beavo JA. Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr Opin Cell Biol. 2000;12:174–179. doi: 10.1016/s0955-0674(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 72.Jurevicius J, Fischmeister R. cAMP compartmentation is responsible for a local activation of cardiac Ca2+ channels by beta-adrenergic agonists. Proc Natl Acad Sci USA. 1996;93:295–299. doi: 10.1073/pnas.93.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rich TC, Tse TE, Rohan JG, Schaack J, Karpen JW. In vivo assessment of local phosphodiesterase activity using tailored cyclic nucleotide-gated channels as cAMP sensors. J Gen Physiol. 2001;118:63–78. doi: 10.1085/jgp.118.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 75.Zippin JH, Farrell J, Huron D, Kamenetsky M, Hess KC, Fischman DA, et al. Bicarbonateresponsive “soluble” adenylyl cyclase defines a nuclear cAMP microdomain. J Cell Biol. 2004;164:527–534. doi: 10.1083/jcb.200311119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bundey RA, Insel PA. Discrete intracellular signaling domains of soluble adenylyl cyclase: camps of cAMP? Sci STKE. 2004:pe19. doi: 10.1126/stke.2312004pe19. [DOI] [PubMed] [Google Scholar]
- 77.Cooper DM. Regulation and organization of adenylyl cyclases and cAMP. Biochem J. 2003;375:517–529. doi: 10.1042/BJ20031061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cali JJ, Parekh RS, Krupinski J. Splice variants of type VIII adenylyl cyclase. Differences in glycosylation and regulation by Ca2+/calmodulin. J Biol Chem. 1996;271:1089–1095. doi: 10.1074/jbc.271.2.1089. [DOI] [PubMed] [Google Scholar]
- 79.Gautier-Courteille C, Salanova M, Conti M. The olfactory adenylyl cyclase III is expressed in rat germ cells during spermiogenesis. Endocrinology. 1998;139:2588–2599. doi: 10.1210/endo.139.5.5967. [DOI] [PubMed] [Google Scholar]
- 80.Emala CW, Kumasaka D, Hirshman CA, Lindeman KS. Adenylyl cyclase messenger ribonucleic acid in myometrium: splice variant of type IV. Biol Reprod. 1998;59:169–175. doi: 10.1095/biolreprod59.1.169. [DOI] [PubMed] [Google Scholar]
- 81.Iwami G, Akanuma M, Kawabe J, Cannon PJ, Homcy CJ, Ishikawa Y. Multiplicity in type V adenylylcyclase: type V-a and type V-b. Mol Cell Endocrinol. 1995;110:43–47. doi: 10.1016/0303-7207(95)03514-8. [DOI] [PubMed] [Google Scholar]
- 82.Katsushika S, Kawabe J, Homcy CJ, Ishikawa Y. In vivo generation of an adenylylcyclase isoform with a half-molecule motif. J Biol Chem. 1993;268:2273–2276. [PubMed] [Google Scholar]
- 83.Premont RT, Chen J, Ma HW, Ponnapalli M, Iyengar R. Two members of a widely expressed subfamily of hormone-stimulated adenylyl cyclases. Proc Natl Acad Sci USA. 1992;89:9809–9813. doi: 10.1073/pnas.89.20.9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chaloupka JA, Bullock SA, Iourgenko V, Levin LR, Buck J. Autoinhibitory regulation of soluble adenylyl cyclase. Mol Reprod Dev. 2006;73:361–368. doi: 10.1002/mrd.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen J, DeVivo M, Dingus J, Harry A, Li J, Sui J, et al. A region of adenylyl cyclase 2 critical for regulation by G protein beta gamma subunits. Science. 1995;268:1166–1169. doi: 10.1126/science.7761832. [DOI] [PubMed] [Google Scholar]
- 86.Diel S, Klass K, Wittig B, Kleuss C. Gbetagamma activation site in adenylyl cyclase type II. Adenylyl cyclase type III is inhibited by Gbetagamma. J Biol Chem. 2006;281:288–294. doi: 10.1074/jbc.M511045200. [DOI] [PubMed] [Google Scholar]
- 87.Dessauer CW, Tesmer JJ, Sprang SR, Gilman AG. Identification of a Gialpha binding site on type V adenylyl cyclase. J Biol Chem. 1998;273:25831–25839. doi: 10.1074/jbc.273.40.25831. [DOI] [PubMed] [Google Scholar]
- 88.Smigel MD. Purification of the catalyst of adenylate cyclase. J Biol Chem. 1986;261:1976–1982. [PubMed] [Google Scholar]
- 89.Tang WJ, Krupinski J, Gilman AG. Expression and characterization of calmodulin-activated (type I) adenylylcyclase. J Biol Chem. 1991;266:8595–8603. [PubMed] [Google Scholar]
- 90.Yeager RE, Heideman W, Rosenberg GB, Storm DR. Purification of the calmodulin-sensitive adenylate cyclase from bovine cerebral cortex. Biochemistry. 1985;24:3776–3783. doi: 10.1021/bi00335a054. [DOI] [PubMed] [Google Scholar]
- 91.Choi EJ, Xia Z, Storm DR. Stimulation of the type III olfactory adenylyl cyclase by calcium and calmodulin. Biochemistry. 1992;31:6492–6498. doi: 10.1021/bi00143a019. [DOI] [PubMed] [Google Scholar]
- 92.Levin LR, Reed RR. Identification of functional domains of adenylyl cyclase using in vivo chimeras. J Biol Chem. 1995;270:7573–7579. doi: 10.1074/jbc.270.13.7573. [DOI] [PubMed] [Google Scholar]
- 93.Vorherr T, Knopfel L, Hofmann F, Mollner S, Pfeuffer T, Carafoli E. The calmodulin binding domain of nitric oxide synthase and adenylyl cyclase. Biochemistry. 1993;32:6081–6088. doi: 10.1021/bi00074a020. [DOI] [PubMed] [Google Scholar]
- 94.Gu C, Cooper DM. Calmodulin-binding sites on adenylyl cyclase type VIII. J Biol Chem. 1999;274:8012–8021. doi: 10.1074/jbc.274.12.8012. [DOI] [PubMed] [Google Scholar]
- 95.Wayman GA, Impey S, Storm DR. Ca2+ inhibition of type III adenylyl cyclase in vivo. J Biol Chem. 1995;270:21480–21486. doi: 10.1074/jbc.270.37.21480. [DOI] [PubMed] [Google Scholar]
- 96.Wayman GA, Wei J, Wong S, Storm DR. Regulation of type I adenylyl cyclase by calmodulin kinase IV in vivo. Mol Cell Biol. 1996;16:6075–6082. doi: 10.1128/mcb.16.11.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Y, Harry A, Li J, Smit MJ, Bai X, Magnusson R, et al. Adenylyl cyclase 6 is selectively regulated by protein kinase A phosphorylation in a region involved in Galphas stimulation. Proc Natl Acad Sci USA. 1997;94:14100–14104. doi: 10.1073/pnas.94.25.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iwami G, Kawabe J, Ebina T, Cannon PJ, Homcy CJ, Ishikawa Y. Regulation of adenylyl cyclase by protein kinase A. J Biol Chem. 1995;270:12481–12484. doi: 10.1074/jbc.270.21.12481. [DOI] [PubMed] [Google Scholar]
- 99.Jacobowitz O, Iyengar R. Phorbol ester-induced stimulation and phosphorylation of adenylyl cyclase 2. Proc Natl Acad Sci USA. 1994;91:10630–10634. doi: 10.1073/pnas.91.22.10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoshimasa T, Sibley DR, Bouvier M, Lefkowitz RJ, Caron MG. Cross-talk between cellular signalling pathways suggested by phorbolester-induced adenylate cyclase phosphorylation. Nature. 1987;327:67–70. doi: 10.1038/327067a0. [DOI] [PubMed] [Google Scholar]
- 101.Yoshimura M, Cooper DM. Type-specific stimulation of adenylylcyclase by protein kinase C. J Biol Chem. 1993;268:4604–4607. [PubMed] [Google Scholar]
- 102.Hill J, Howlett A, Klein C. Nitric oxide selectively inhibits adenylyl cyclase isoforms 5 and 6. Cell Signal. 2000;12:233–237. doi: 10.1016/s0898-6568(99)00082-0. [DOI] [PubMed] [Google Scholar]
- 103.Gu C, Sorkin A, Cooper DM. Persistent interactions between the two transmembrane clusters dictate the targeting and functional assembly of adenylyl cyclase. Curr Biol. 2001;11:185–190. doi: 10.1016/s0960-9822(01)00044-6. [DOI] [PubMed] [Google Scholar]
- 104.Sinnarajah S, Dessauer CW, Srikumar D, Chen J, Yuen J, Yilma S, et al. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–1055. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- 105.Cooper DM, Crossthwaite AJ. Higher-order organization and regulation of adenylyl cyclases. Trends Pharmacol Sci. 2006;27:426–431. doi: 10.1016/j.tips.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 106.Sunahara RK, Dessauer CW, Whisnant RE, Kleuss C, Gilman AG. Interaction of Gsalpha with the cytosolic domains of mammalian adenylyl cyclase. J Biol Chem. 1997;272:22265–22271. doi: 10.1074/jbc.272.35.22265. [DOI] [PubMed] [Google Scholar]
- 107.Dessauer CW, Gilman AG. Purification and characterization of a soluble form of mammalian adenylyl cyclase. J Biol Chem. 1996;271:16967–16974. doi: 10.1074/jbc.271.28.16967. [DOI] [PubMed] [Google Scholar]
- 108.Martinez SE, Bruder S, Schultz A, Zheng N, Schultz JE, Beavo JA, Linder JU. Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: modes of ligand binding and dimerization. Proc Natl Acad Sci USA. 2005;102:3082–3087. doi: 10.1073/pnas.0409913102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seamon KB, Daly JW. Forskolin: its biological and chemical properties. Adv Cyclic Nucl Protein Phosphoryl Res. 1986;20:1–150. [PubMed] [Google Scholar]
- 110.Yan SZ, Huang ZH, Andrews RK, Tang WJ. Conversion of forskolin-insensitive to forskolin-sensitive (mouse-type IX) adenylyl cyclase. Mol Pharmacol. 1998;53:182–187. doi: 10.1124/mol.53.2.182. [DOI] [PubMed] [Google Scholar]
- 111.Hacker BM, Tomlinson JE, Wayman GA, Sultana R, Chan G, Villacres E, et al. Cloning, chromosomal mapping, and regulatory properties of the human type 9 adenylyl cyclase (ADCY9) Genomics. 1998;50:97–104. doi: 10.1006/geno.1998.5293. [DOI] [PubMed] [Google Scholar]
- 112.Forte LR, Bylund DB, Zahler WL. Forskolin does not activate sperm adenylate cyclase. Mol Pharmacol. 1983;24:42–47. [PubMed] [Google Scholar]
- 113.Johnson RA, Salomon Y. Assay of adenylyl cyclase catalytic activity. Methods Enzymol. 1991;195:3–21. doi: 10.1016/0076-6879(91)95150-i. [DOI] [PubMed] [Google Scholar]
- 114.Dessauer CW, Tesmer JJ, Sprang SR, Gilman AG. The interactions of adenylate cyclases withP-siteinhibitors. Trends Pharmacol Sci. 1999;20:205–210. doi: 10.1016/s0165-6147(99)01310-3. [DOI] [PubMed] [Google Scholar]
- 115.Johnson RA, Shoshani I. Kinetics of “P”-site-mediated inhibition of adenylyl cyclase and the requirements for substrate. J Biol Chem. 1990;265:11595–11600. [PubMed] [Google Scholar]
- 116.Dessauer CW, Gilman AG. The catalytic mechanism of mammalian adenylyl cyclase. Equilibrium binding and kinetic analysis of P-site inhibition. J Biol Chem. 1997;272:27787–27795. doi: 10.1074/jbc.272.44.27787. [DOI] [PubMed] [Google Scholar]
- 117.Gille A, Lushington GH, Mou TC, Doughty MB, Johnson RA, Seifert R. Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides. J Biol Chem. 2004;279:19955–19969. doi: 10.1074/jbc.M312560200. [DOI] [PubMed] [Google Scholar]
- 118.Levy DE, Bao M, Cherbavaz DB, Tomlinson JE, Sedlock DM, Homcy CJ, Scarborough RM. Metal coordination-based inhibitors of adenylyl cyclase: novel potent P-site antagonists. J Med Chem. 2003;46:2177–2186. doi: 10.1021/jm0205604. [DOI] [PubMed] [Google Scholar]
- 119.Iwatsubo K, Minamisawa S, Tsunematsu T, Nakagome M, Toya Y, Tomlinson JE, et al. Direct inhibition of type 5 adenylyl cyclase prevents myocardial apoptosis without functional deterioration. J Biol Chem. 2004;279:40938–40945. doi: 10.1074/jbc.M314238200. [DOI] [PubMed] [Google Scholar]
- 120.Gille A, Seifert R. 2′(3′)-O-(N-methylanthraniloyl)-substituted GTP analogs: a novel class of potent competitive adenylyl cyclase inhibitors. J Biol Chem. 2003;278:12672–12679. doi: 10.1074/jbc.M211292200. [DOI] [PubMed] [Google Scholar]
- 121.Mou TC, Gille A, Fancy DA, Seifert R, Sprang SR. Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2′(3′)-O-(N-methylanthraniloyl)-guanosine 5′-triphosphate. J Biol Chem. 2005;280:7253–7261. doi: 10.1074/jbc.M409076200. [DOI] [PubMed] [Google Scholar]
- 122.Cooper DM. Molecular and cellular requirements for the regulation of adenylate cyclases by calcium. Biochem Soc Trans. 2003;31:912–915. doi: 10.1042/bst0310912. [DOI] [PubMed] [Google Scholar]
- 123.Guillou JL, Nakata H, Cooper DM. Inhibition by calcium of mammalian adenylyl cyclases. J Biol Chem. 1999;274:35539–35545. doi: 10.1074/jbc.274.50.35539. [DOI] [PubMed] [Google Scholar]
- 124.Hu B, Nakata H, Gu C, De Beer T, Cooper DM. A critical interplay between Ca2+ inhibition and activation by Mg2+ of AC5 revealed by mutants and chimeric constructs. J Biol Chem. 2002;277:33139–33147. doi: 10.1074/jbc.M112373200. [DOI] [PubMed] [Google Scholar]
- 125.Braun T. Inhibition of the soluble form of testis adenylate cyclase by catechol estrogens and other catechols. Proc Soc Exp Biol Med. 1990;194:58–63. doi: 10.3181/00379727-194-43055. [DOI] [PubMed] [Google Scholar]
- 126.Paul SM, Skolnick P. Catechol oestrogens inhibit oestrogen elicited accumulation of hypothalamic cyclic AMP suggesting role as endogenous anti-oestrogens. Nature. 1977;266:559–561. doi: 10.1038/266559a0. [DOI] [PubMed] [Google Scholar]
- 127.Jaleel M, Shenoy AR, Visweswariah SS. Tyrphostins are inhibitors of guanylyl and adenylyl cyclases. Biochemistry. 2004;43:8247–8255. doi: 10.1021/bi036234n. [DOI] [PubMed] [Google Scholar]
- 128.Liehr JG, Ricci MJ. 4-Hydroxylation of estrogens as marker of human mammary tumors. Proc Natl Acad Sci USA. 1996;93:3294–3296. doi: 10.1073/pnas.93.8.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Paul SM, Axelrod J. Catechol estrogens: presence in brain and endocrine tissues. Science. 1977;197:657–659. doi: 10.1126/science.877577. [DOI] [PubMed] [Google Scholar]
- 130.Haunso A, Simpson J, Antoni FA. Small ligands modulating the activity of mammalian adenylyl cyclases: a novel mode of inhibition by calmidazolium. Mol Pharmacol. 2003;63:624–631. doi: 10.1124/mol.63.3.624. [DOI] [PubMed] [Google Scholar]
- 131.Kudlacek O, Mitterauer T, Nanoff C, Hohenegger M, Tang WJ, Freissmuth M, Kleuss C. Inhibition of adenylyl and guanylyl cyclase isoforms by the antiviral drug foscarnet. J Biol Chem. 2001;276:3010–3016. doi: 10.1074/jbc.M007910200. [DOI] [PubMed] [Google Scholar]
- 132.Shoshani I, Laux WH, Perigaud C, Gosselin G, Johnson RA. Inhibition of adenylyl cyclase by acyclic nucleoside phosphonate antiviral agents. J Biol Chem. 1999;274:34742–34744. doi: 10.1074/jbc.274.49.34742. [DOI] [PubMed] [Google Scholar]
- 133.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucl Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.DeLano W. The PyMol User’s Manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 135.Barton GJ. ALSCRIPT a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 136.Evans SV. SETOR: hardware lighted three-dimensional solid model representations of macro-molecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]