Abstract
Employing mouse skin epidermal JB6 cells and dermal fibroblasts, here we examined the mechanisms of DNA damage by 2-chloroethyl ethyl sulfide (CEES), a monofunctional analog of sulfur mustard (SM). CEES exposure caused H2A.X and p53 phosphorylation as well as p53 accumulation in both cell types, starting at 1 h, that was sustained for 24 h, indicating a DNA-damaging effect of CEES, which was also confirmed and quantified by alkaline comet assay. CEES exposure also induced oxidative stress and oxidative DNA damage in both cell types, measured by an increase in mitochondrial and cellular reactive oxygen species and 8-hydroxydeoxyguanosine levels, respectively. In the studies distinguishing between oxidative and direct DNA damage, 1 h pretreatment with glutathione (GSH) or the antioxidant Trolox showed a decrease in CEES-induced oxidative stress and oxidative DNA damage. However, only GSH pretreatment decreased CEES-induced total DNA damage measured by comet assay, H2A.X and p53 phosphorylation, and total p53 levels. This was possibly due to the formation of GSH–CEES conjugates detected by LC-MS analysis. Together, our results show that CEES causes both direct and oxidative DNA damage, suggesting that to rescue SM-caused skin injuries, pleiotropic agents (or cocktails) are needed that could target multiple pathways of mustard skin toxicities.
Keywords: DNA damage, H2A.X, p53, Oxidative stress, 8-OHdG, CEES, Free radicals
Sulfur mustard (2,2-dichlorodiethyl sulfide; SM) was first introduced as a chemical warfare agent by Germany during World War I against British soldiers in 1917 and has since been used in numerous military conflicts [1,2]. Its wide use as a chemical warfare agent is due to its vesicating or blister-forming property, which causes severe damage to skin, eyes, and respiratory tract [3–5]. Apart from the threat of SM being used in a possible warfare or terrorist attack, there is also a risk of accidental exposure to SM due to the improper disposal of old artillery shells containing SM [6]. On cutaneous exposure, SM is rapidly absorbed by the skin, and 70% of the absorbed SM concentrates in the epidermis, whereas the remaining is found in the basement membrane and dermis [7]. SM exposure is known to attack and cause death of proliferating keratinocytes in the basal epidermal layer of the skin, which might be a contributing factor to the vesication caused by this chemical agent [8–10]. The cytotoxic effects of SM are mainly attributed to its DNA-damaging ability, which could result from alkylation and/or oxidative stress-related mechanisms [11–13].
SM is a bifunctional alkylating agent and, in the presence of water, undergoes nucleophilic substitution to form a cyclic sulfonium ion, which in turn opens to generate a carbenium ion. This intermediate rapidly reacts with most of the cellular macromolecules, including DNA, RNA, and proteins [14]. In addition, SM can also lead to the depletion of cellular glutathione (GSH) and antioxidant enzymes such as superoxide dismutase, catalase, and glutathione peroxidase, resulting in the accumulation of reactive oxygen species (ROS) followed by lipid peroxidation, protein oxidation, and DNA damage [15,16]. 2-Chloroethyl ethyl sulfide (CEES; a monofunctional SM analog) is extensively used to study the mechanisms of SM-induced toxicity, including both direct and oxidative DNA damage [17,18]. Numerous studies have also shown the involvement of oxidative stress and GSH depletion in SM/CEES-induced skin toxicity [16,19,20]. Studies using CEES have shown an increase in oxidative DNA damage, lipid peroxidation, protein oxidation, and ROS production, in both animal and cell culture models [18,21]. CEES-caused DNA damage is also suggested by studies showing activation of ATM/ATR checkpoint kinases as well as the PARP pathway, which activate DNA damage repair by stimulating the DNA damage sensors p53 and H2A.X [22–24].
Taken together, previous studies have shown that CEES exposure could lead to oxidative DNA damage; however, the extent of involvement of oxidative stress in CEES/SM-induced DNA damage is not fully studied. Accordingly, here we first quantified the CEES-induced DNA damage, employing the DNA damage markers H2A.X and p53 and the comet assay, and then deciphered the involvement of oxidative DNA damage. For our studies, we used both skin epidermal cells and dermal fibroblasts, as both the cell types exhibit variable mechanisms of cell death after SM exposure [25]. Our results using the antioxidants Trolox and GSH, the latter also known to form conjugates with SM [26], show that oxidative DNA damage is not the prominent contributor to the overall CEES-induced DNA damage.
Materials and methods
Cell lines and CEES treatment
JB6 cells were obtained from American Type Culture Collection (Manassas, VA, USA) and cultured in minimal essential medium (Gibco BRL, Grand Island, NY, USA) containing 5% heat-inactivated fetal bovine serum (FBS) and 25 µg/ml gentamycin. The SKH-1 fibroblasts were isolated from neonatal SKH-1 hairless mouse skin using the protocol by Hakkinen et al. [27]. The isolated fibroblasts were cultured in Dulbecco's modified Eagle's medium with 10% FBS, 25 mM Hepes and 1% antibiotic and antimycotic (Gibco BRL). Both the cell lines were grown under standard culture conditions at 37 °C in a humidified 5% CO2 incubator. CEES, GSH, Trolox, antimycin A, and valinomycin were obtained from Sigma–Aldrich Chemical Co. (St. Louis, MO, USA). A 1M stock solution of CEES was prepared fresh in dimethyl sulfoxide (DMSO) and the treatment of cells was carried out as described earlier [23]. Cells were exposed to either DMSO (vehicle control) alone or various concentrations of CEES (0.1, 0.25, 0.5, 0.75, and 1 mM), and studies were carried out at the desired time points up to 24 h. For GSH treatments, 100 mM stock was prepared and pH adjusted to 7.4 before treatment. Cells were exposed to either DMSO alone or 0.5mM CEES for 1 h or treated with 10 mM GSH or 800 µM Trolox 1 h before CEES exposure, and desired assays were carried out 1–4 h after these treatments. Unless stated otherwise, the final concentration of DMSO in the culture medium during treatments did not exceed 0.1% (v/v). All CEES preparations were carried out in a continuously operated chemical and biological safety hood, and treatments were done under a safety laminar hood using all required and approved personal protective equipment.
Western blot analysis
After the exposures and treatments, whole-cell lysates were prepared at the desired time points. Protein estimation was done using the Lowry method (Bio-Rad Laboratories, Richmond, CA, USA), and immunoblot analysis using 80 µg of protein per sample was carried out using SDS–PAGE on 12% Tris–glycine gels via Western blotting as reported earlier [21,23]. Membranes were blocked using Odyssey blocking buffer for 1 h at room temperature and probed against primary antibodies for H2A.X Ser139 and p53 Ser15 (Cell Signaling, Beverly, MA, USA) overnight at 4 °C, followed by incubation with IRDye 800CW conjugated, goat anti-rabbit IgG polyclonal secondary antibody for 1 h at room temperature. The membranes were then visualized using an Odyssey infrared imager (Li-Cor Biosciences, Lincoln, NE, USA). To ensure equal protein loading, each membrane was stripped and reprobed with β-actin antibody (Sigma–Aldrich).
Immunofluorescence confocal microscopy
Immunofluorescence studies were done as described earlier in [28]. Briefly, cells were plated on coverslips placed in six-well plates overnight and then exposed to either DMSO or 0.5 mM CEES for 4 h. After the desired exposures, cells were washed gently with phosphate-buffered saline (PBS) and fixed in 3% formaldehyde for 10 min. Cells were again washed twice with PBS, incubated with Triton X-100 for 5 min, and later washed twice with PBS followed by a final wash with 10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.2% v/v Tween 20. Cells were incubated in Cas-block blocking solution for 1 h followed by overnight incubation with H2A.X Ser139 antibody. Cells were then washed three times with PBS and finally incubated with Texas red-tagged goat anti-rabbit secondary antibody for 45 min and counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) for 5 min. Cell images were captured at 400× magnification on a Nikon D Eclipse C1 confocal microscope (Nikon Instruments, Melville, NY, USA), and images were processed by EZC1 Freeviewer software.
Comet assay
Single-cell gel electrophoresis (SCGE), also known as the alkaline comet assay (pH ≥ 13), was employed to measure the DNA damage [29,30] after the desired exposures and treatments. Briefly, 300 µl of cell suspension was mixed with 1000 µl of 1% low-melting-point agarose and added to a slide precoated with 1% normal-melting-point agarose. Thereafter, the slides were left at 4 °C in the dark overnight in lysis solution (2.5 M NaCl, 100mM EDTA, 10 mM Tris–base, pH 10.5, to which 1% Triton X-100 and 1% DMSO were freshly added). After lysis, the slides were washed with ddH2O for 1 h, transferred to an electrophoresis unit, covered with fresh electrophoresis buffer (300mM NaOH, 1 mM EDTA, pH ≥ 13), left for unwinding of DNA for 30 min, and electrophoresed for 20 min at 22 V and 200 mA. The slides were then neutralized with neutralization buffer (500mM Tris–HCl, pH 8.0), washed with ddH2O, and stained with 3 µg/ml of propidium iodide. The slides were then dried overnight and scored for comets using fluorescence microscopy under a Nikon inverted microscope (Nikon Eclipse TE300) at 200× magnification. Images were captured using an attached CoolSNAPES CCD camera. One hundred fifty cells, 50 each on triplicate slides, were captured and scored using Komet 5.5 software (ANDOR Technology, South Windsor, CT, USA). The DNA damage is represented as tail extent moment (TEM), which is the product of tail length and percentage tail DNA.
Cellular and mitochondrial superoxide detection
After DMSO or CEES exposure and GSH or Trolox treatment before CEES exposure, cellular and mitochondrial ROS, mainly superoxide , were measured using dihydroethidium (DHE) and MitoSOX red (Invitrogen; Carlsbad, CA, USA), respectively. DHE or MitoSOX red was added to the cells at a final concentration of 5 µM and incubated for 30 min or 1 h, respectively, at 37 °C. At the end of the treatment, the medium was removed, and the cells were washed with warm PBS (500 µl), scraped, and placed into microtiter tubes. JB6 cells, treated with 30 µM antimycin A for 20 min or 200 nM valinomycin for 1 h, were used as positive controls for the selectivity of MitoSOX red and DHE staining, respectively, and 100 µM H2O2 for 10 min was used as a negative control. Fluorescence was measured using a Beckman Coulter Cytomics FC 500 flow cytometer (Beckman Coulter, Brea, CA, USA), and the results were obtained by gating the fluorescence of MitoSOX red or DHE versus linear side scatter (SS Lin) of the homogeneous cell population.
8-Hydroxydeoxyguanosine (8-OHdG) ELISA
After the desired exposure and treatment, cells were collected and DNA was extracted using a DNeasy tissue kit (Qiagen, Valencia, CA, USA). DNA purity was measured using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). Approximately 135 µl of 1mg/ml DNA was incubated at 95 °C for 5 min and rapidly chilled on ice followed by incubation with 5 units of nuclease P1 (Sigma–Aldrich) for 2 h at 37 °C in 20 mM sodium acetate, pH 5.2, and 5 units of alkaline phosphatase (Sigma–Aldrich) for 1 h at 37 °C in 100 mM Tris, pH 7.5. The samples were then analyzed for 8-OHdG levels using an OxiSelect oxidative DNA damage ELISA kit (STA-320 from Cell Biolabs). The absorbance was read at 450 nm using a Spectra Max 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA). The 8-OHdG levels were calculated and expressed in ng/ml.
LC-MS for GSH–CEES conjugate detection
CEES (0.5 mM) in DMSO vehicle was incubated with 10 mM GSH in water, pH 7.4, for 1 h and analyzed for GSH–CEES conjugate formation. The HPLC system used was an Agilent 1100 capillary system (Agilent Technologies, Santa Clara, CA, USA), and GSH–CEES conjugate formation was assayed using a C18 MS column (Vydac). Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. The solvent gradient ramped from 3 to 80% solvent B from 0 to 15 min and was held there for 2 min before ramping back to starting conditions. The flow rate was 50 µl/min. The mass spectrometric analysis was performed using a Waters Quattro LC triple-quadrupole, running MassLynx 4.1. The instrument was run in positive mode with a capillary voltage of 3 kV and a cone voltage of 23 V with a scan range of 50 to 650 m/z over 0.5 s.
Statistical analysis
Data were analyzed using the SigmaStat 2.0 software (Jandel Scientific, San Rafael, CA, USA) employing one-way analysis of variance to determine the statistical significance of the difference between groups, followed by Tukey's test for multiple comparisons (SigmaStat 3.5). Differences were considered significant if the P value was ≤0.05.
Results
CEES exposure causes an increase in the phosphorylation of H2A.X Ser139 and p53 Ser15, accumulation of total p53, and increase in DNA comet tail, indicating DNA damage
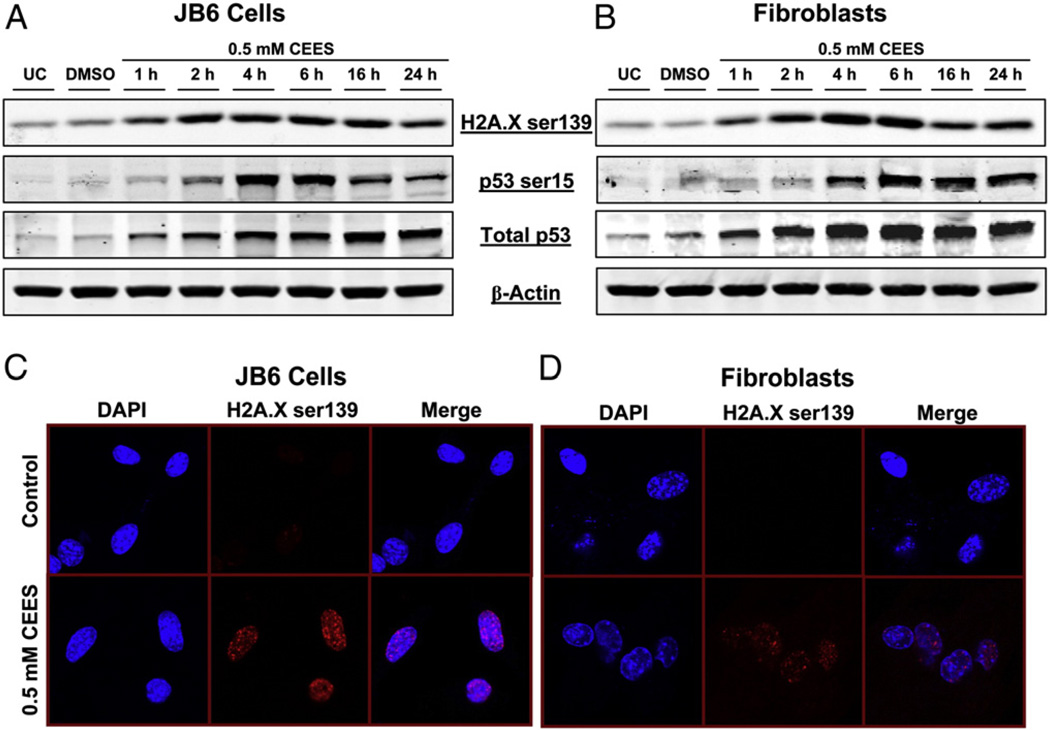
To examine CEES-caused DNA damage, first we assessed the phosphorylation of H2A.X at Ser139 and p53 at Ser15 as well as total p53 levels, which are considered DNA damage markers [31,32]. CEES exposure, in both cell types, caused an increase in the phosphorylation of H2A.X and p53 as well as an increase in total p53 level after 1 h, with a much stronger effect on these parameters between 2 and 4 h that persisted up to 24 h exposure (Figs. 1A and B). The CEES-induced phosphorylation of H2A.X was further confirmed by immunofluorescence analysis after 4 h of CEES exposure. The results in Fig. 1C demonstrate that compared to vehicle controls, CEES exposure caused an increase in the formation of γH2A.X foci (red fluorescence) in the nuclei of both epidermal keratinocytes and fibroblasts, confirming the Western blot results.
Fig. 1.
Effects of CEES exposure on the phosphorylation of H2A.X at Ser139 and p53 at Ser15 in JB6 cells and fibroblasts. (A) JB6 cells and (B) fibroblasts were exposed to DMSO or 0.5 mM CEES and collected at various time points starting within 1 h of CEES exposure. Whole-cell lysates were prepared and subjected to SDS–PAGE followed by Western blot analysis for H2A.X Ser139 and p53 Ser15 using 80 µg protein as detailed under Materials and methods. Total p53 protein levels and β-actin levels were determined by stripping and reprobing the membranes with total p53 and β-actin antibodies. (C and D) 4 h after similar treatments, both cell types were subjected to immunofluorescence to assess the phosphorylation of H2A.X Ser139, which were seen as foci formation in the nucleus (stained using DAPI). Cell images were captured at 400× original magnification on a Nikon D Eclipse C1 confocal microscope as described under Materials and methods. UC, untreated control; DMSO, vehicle control.
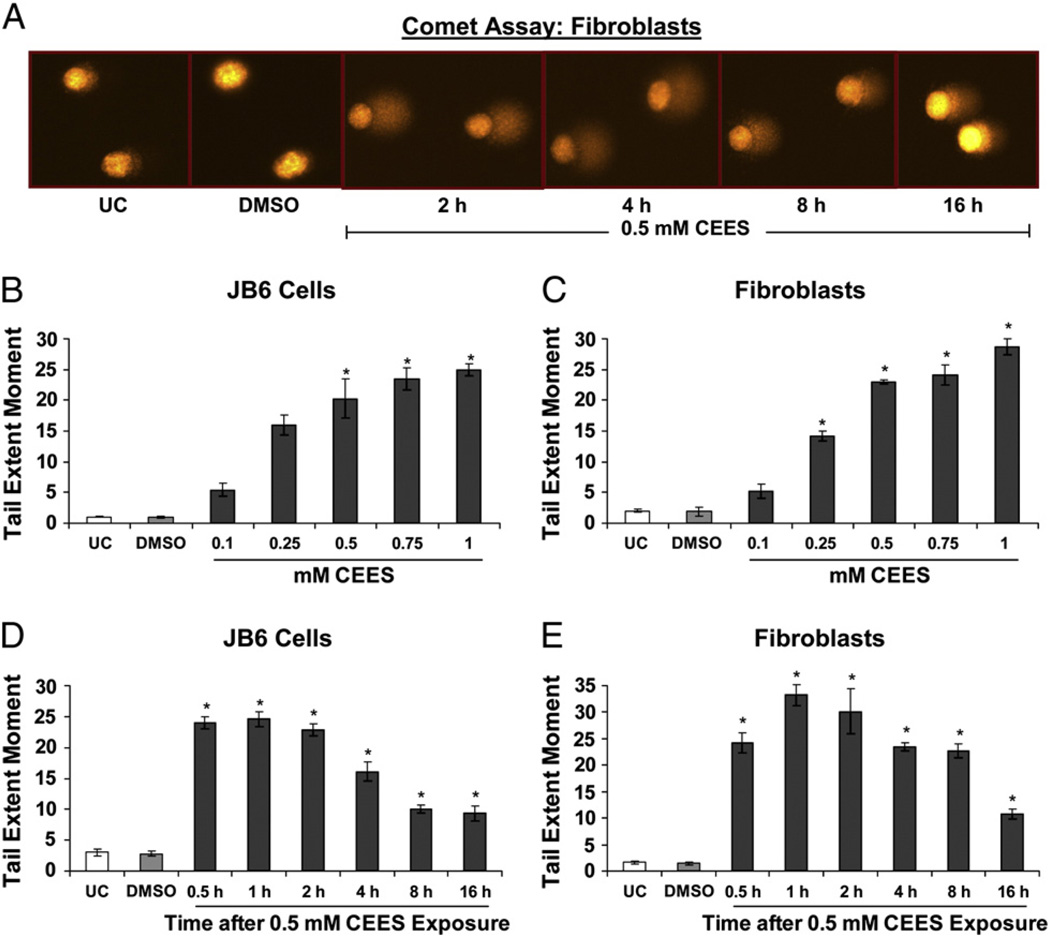
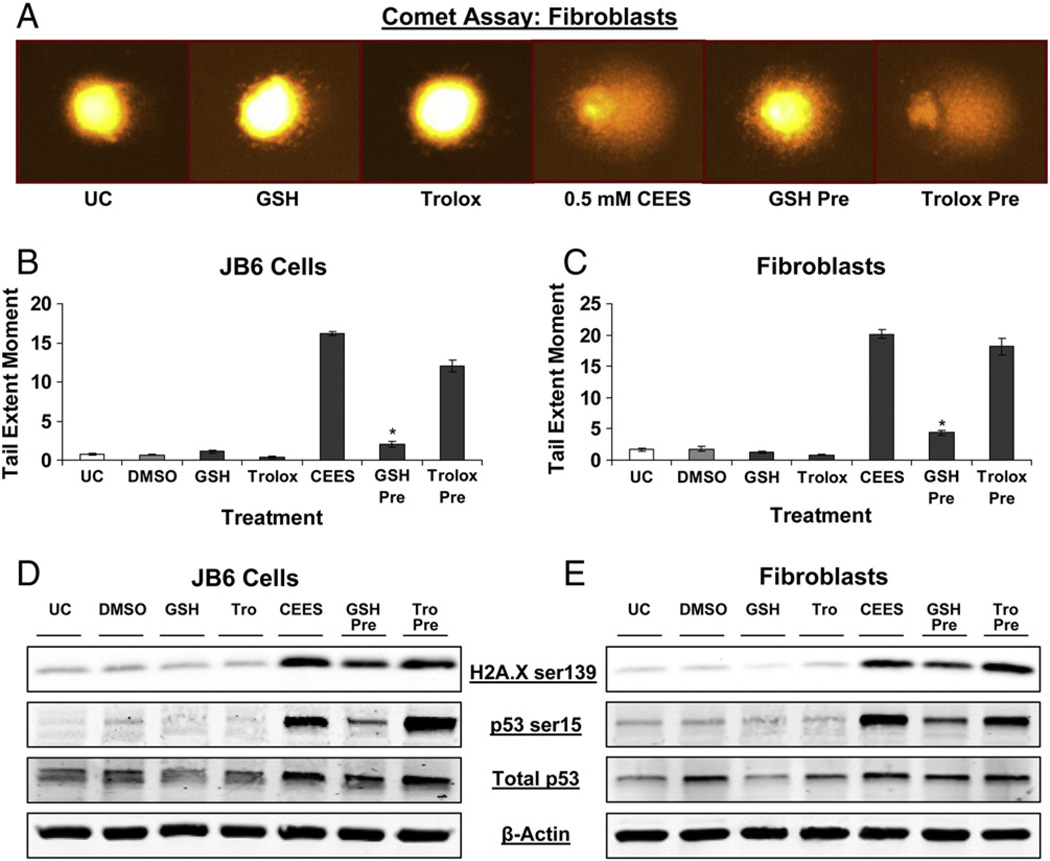
To further quantify CEES-induced DNA damage, we employed the alkaline comet assay. This is a well-known technique for the quantification of damaged DNA, in which SCGE is used to determine the extent of DNA damage via the measurement of the fluorescence of the DNA tail from the cell (Fig. 2). Fig. 2A shows representative pictures of untreated fibroblasts and fibroblasts exposed to DMSO or 0.5 mM CEES for the indicated times and subjected to comet assay, in which damaged DNA is seen as a fluorescent tail attached to the cell. A dose-dependent increase in DNA damage was observed at 2 h of CEES exposure in both JB6 cells and fibroblasts with the maximum DNA damage seen with 1.0 mM CEES (Figs. 2B and C). Next, a time-kinetics study of the DNA damage caused by 0.5 mM CEES showed that the TEM was maximum between 1 and 2 h after exposure in both cell lines (Figs. 2D and E). However, starting at 4 h of CEES exposure, there was a decrease in TEM that was substantial by 16 h of exposure (Figs. 2D and E), suggesting a possible role of repair and rescue mechanisms coming into the effect by this time.
Fig. 2.
Effects of CEES exposure on the extent of DNA damage in JB6 cells and fibroblasts. (A) Comet assay was performed in JB6 cells and fibroblasts, and representative pictures (as shown here for fibroblasts) of the comets formed after CEES exposure at various time points were taken using a fluorescence microscope at 200× original magnification. (B and C) For CEES dose–response study, the cells were exposed to DMSO either alone or with 0.1 to 1.0 mM concentration of CEES for 2 h and subjected to comet assay. (D and E) In a time–response study, cells were exposed to either DMSO or 0.5 mM CEES and collected at various time points from 30 min to 16 h of treatment and subjected to comet assay as described under Materials and methods. Data are presented as means ± SEM, n = 4. *P<0.001 compared to untreated control. UC, untreated control; DMSO, vehicle control.
CEES exposure causes an increase in mitochondrial and cellular ROS as well as 8-OHdG levels, indicating oxidative DNA damage
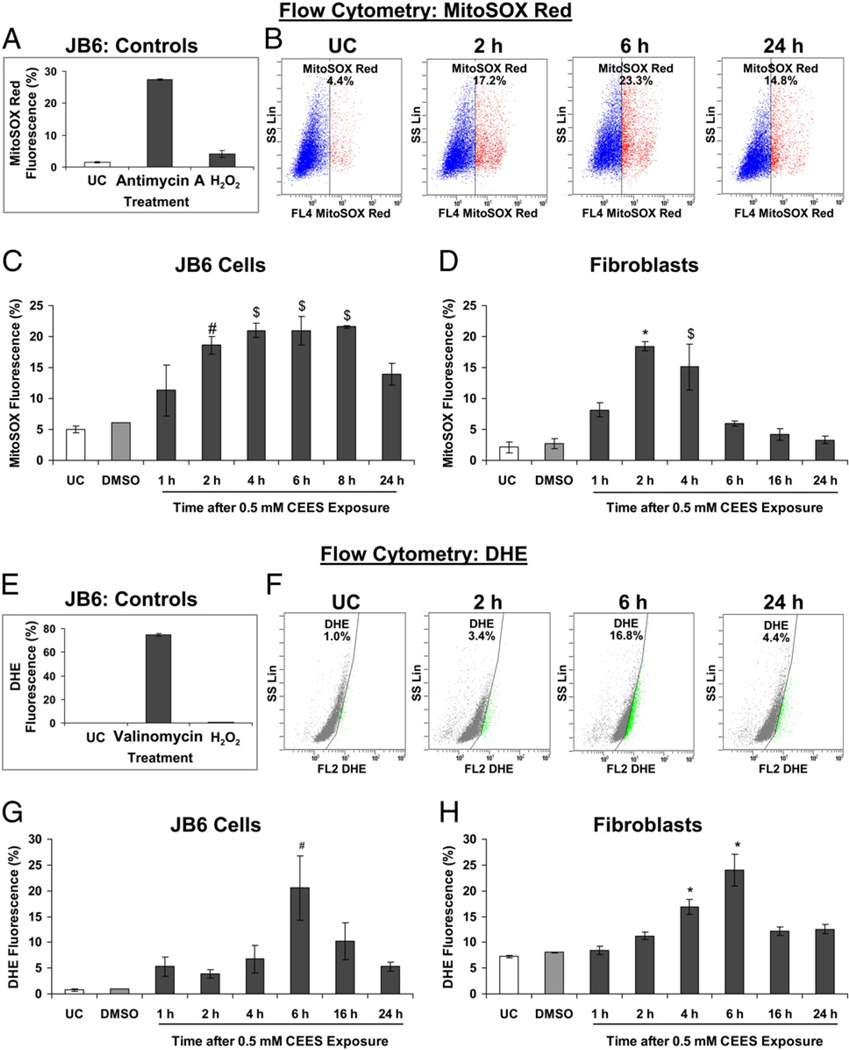
To assess whether and to what extent CEES causes oxidative stress in mouse skin epidermal cells and fibroblasts, we measured both mitochondrial and cellular ROS levels. As mitochondria are the major source of ROS within the cell, we measured mitochondrial production using MitoSOX red, which selectively detects mitochondrial via its oxidation, thus forming a red-fluorescent compound that is measurable in live cells [33]. Treatment of JB6 cells with antimycin A, which induces the production of superoxide radicals, caused an increase in MitoSOX red fluorescence-positive cells. However, after H2O2 treatment there was no significant increase in such fluorescence-positive cells (Fig. 3A), showing the specificity of MitoSOX red in detecting superoxide radicals. Next, cells were exposed to 0.5 mM CEES from 1 to 24 h, incubated with MitoSOX red for 30 min, and measured for live cell fluorescence by flow cytometry. As seen in the representative flow cytograms (Fig. 3B), CEES caused an increase in MitoSOX red fluorescence-positive cells within 1 h of its exposure but the most significant increase was seen at 4 h in JB6 cells and between 2 and 4 h in fibroblasts (Figs. 3C and D). We also sought to determine how cellular ROS production was affected by CEES and, therefore, next used DHE, an indicator of cellular generation. Treatment of JB6 cells with valinomycin, which induces the production of superoxide radicals, caused an increase in DHE fluorescence-positive cells. However, after H2O2 treatment there was no significant increase in such fluorescence-positive cells (Fig. 3E), showing the specificity of DHE in detecting cellular superoxide radicals. As we did during mitochondrial detection, we next exposed both JB6 cells and fibroblasts to 0.5 mM CEES from 1 to 24 h, incubated them with DHE for 20 min, and measured live cell fluorescence by flow cytometry. As seen in the representative flow cytograms (Fig. 3F), both JB6 cells and fibroblasts, relative to control cells, demonstrated a significant increase in DHE fluorescence-positive cells at 6 h, and thereafter, there was a decrease at 16 and 24 h, respectively (Figs. 3G and H). These results show that CEES induces maximum production at 2–6 h after exposure, indicating oxidative stress in both mitochondria and whole cells.
Fig. 3.
Effects of CEES exposure on oxidative stress in JB6 cells and fibroblasts. (A and E) The specificity of the MitoSOX red and DHE toward superoxide was determined by treating JB6 cells with 30 µM antimycin A and 200 nM valinomycin, respectively, and 100 µM H2O2. (B and F) The representative flow cytograms of JB6 cells subjected to MitoSOX red and DHE staining, respectively, after 0.5 mM CEES exposure. (C and G) JB6 cells and (D and H) fibroblasts were exposed to DMSO or 0.5 mM CEES for 1 to 16 h and then incubated for 30 min with the mitochondrial indicator MitoSOX red (C and D) or with the cellular indicator DHE (G and H), and the live cell fluorescence was determined using flow cytometry as described under Materials and methods. Data are presented as means ± SEM, n = 3. *P<0.001, $P<0.005, #P<0.05 compared to untreated control. UC, untreated control; DMSO, vehicle control.
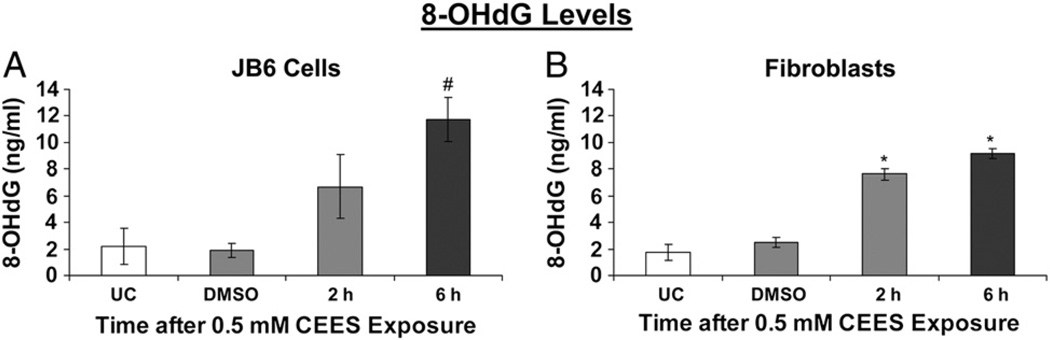
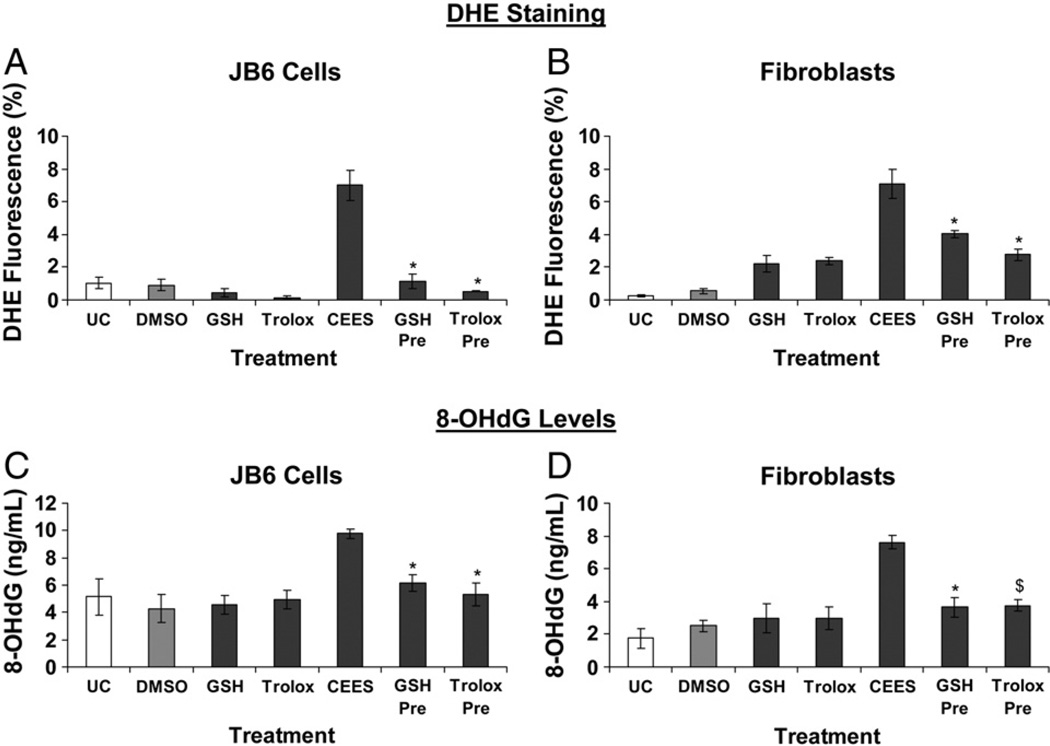
Next, to identify the role of oxidative stress in the CEES-induced DNA damage in both these cell types, we measured the levels of 8-OHdG, a ubiquitous marker of oxidative DNA damage, which measures the oxidation of deoxyguanosine (one of the constituents of DNA), leading to the formation of 8-OHdG (Fig. 4). There was an increase in 8-OHdG levels within 2 h after 0.5 mM CEES exposure in both cell types, with the levels further increasing by 6 h, thus indicating the role of oxidative stress in CEES-induced DNA damage (Fig. 4). In JB6 cells, compared to 1.9 ng/ml 8-OHdG levels in the vehicle-treated cells, 6 h of CEES exposure resulted in a significant increase (11.7 ng/ml) in 8-OHdG levels (Fig. 4A). Similarly, in fibroblasts, 2 and 6 h CEES exposure resulted in 7.6 and 9.2 ng/ml 8-OHdG levels, respectively; compared to 2.5 ng/ml 8-OHdG in vehicle controls (Fig. 4B).
Fig. 4.
Effects of CEES exposure on oxidative DNA damage in JB6 cells and fibroblasts. (A) JB6 cells and (B) fibroblasts were treated with DMSO or 0.5 mM CEES for 2 and 6 h. DNA was isolated from the cells and 8-OHdG levels were measured using indirect ELISA as described under Materials and methods. Data are presented as means ± SEM, n = 3. *P<0.001, #P<0.05 compared to untreated control. UC, untreated control; DMSO, vehicle control.
GSH and Trolox treatments protect the cells from CEES-induced ROS generation and oxidative DNA damage
Our previous studies have shown that CEES exposure causes depletion of intracellular GSH levels and that replenishing the GSH levels exogenously helped in reversing CEES-induced cytotoxicity as reflected by cell viability and DNA synthesis and also by measurement of S- and G2/M-phase arrest in the cell cycle progression [20]. Herein, we further assessed the protective effect of GSH on CEES-induced oxidative stress and oxidative DNA damage. GSH is known for its antioxidant and conjugating properties that could affect CEES-induced DNA damage through both of the pathways [34,35]. To distinguish the effect of GSH's conjugating properties from its antioxidant properties we also used the antioxidant Trolox, a cell-permeative analog of vitamin E [36], which could help identify the effect of its antioxidant properties alone on CEES-induced DNA damage.
Cells were treated with 10 mM GSH or 800 µM Trolox for 1 h before 4 h of CEES exposure and the live cell fluorescence was measured with DHE. In JB6 cells, GSH and Trolox treatments caused 96% and complete decrease in CEES-induced cellular production, respectively (Fig. 5A). Similar GSH and Trolox treatments in fibroblasts resulted in 47 and 66% decrease in CEES-induced cellular production, respectively (Fig. 5B). These results showed that both GSH and Trolox prevented CEES-induced cellular production of ROS. Next, to determine the effects of the antioxidant properties of GSH and Trolox on CEES-induced oxidative DNA damage, we measured the 8-OHdG levels in cells treated for 1 h with 10 mM GSH or 800 µM Trolox before 0.5 mM CEES exposure (Figs. 5C and D). Results showed that both GSH and Trolox treatments caused 68 and 82% (JB6 cells) and 78 and 76% (fibroblasts) attenuation of CEES-caused increase in 8-OHdG levels, respectively (Figs. 5C and D). These data show that both GSH and Trolox cause a significant decrease in oxidative DNA damage caused by CEES.
Fig. 5.
Effects of GSH or Trolox treatment on CEES-induced oxidative stress and oxidative DNA damage in JB6 cells and fibroblasts. (A and B) JB6 cells and fibroblasts were treated with 10 mM GSH or 800 µM Trolox for 1 h before 0.5 mM CEES exposure for 4 h and the cellular ROS were determined by incubation with DHE for 30 min followed by flow cytometry as described under Materials and methods. (C and D) To assess the effect of 1 h pretreatment with 10 mM GSH or 800 µM Trolox on the oxidative DNA damage caused by 0.5 mM CEES exposure, DNA was isolated after the desired treatments and 8-OHdG ELISA was conducted as described under Materials and methods. Data are presented as means ± SEM, n = 3. *P<0.001, $P<0.005 compared to 0.5 mM CEES exposure. UC, untreated control; DMSO, vehicle control.
GSH treatment has a greater protective effect than Trolox in attenuating CEES-induced DNA damage
Because we observed that both GSH and Trolox can decrease CEES-induced ROS levels and oxidative DNA damage, we sought to determine if this effect was reflected in total DNA damage seen after CEES exposure. Both JB6 cells and fibroblasts were subjected to comet assay after treatment for 1 h with 10 mM GSH or 800 µM Trolox before 0.5 mM CEES exposure (Fig. 6A). GSH 1-h pretreatment caused a 92 and 86% reversal of the CEES-induced increase in DNA damage (measured as TEM in comet assay) in JB6 cells and fibroblasts, respectively (Figs. 6B and C). However, Trolox 1-h pretreatment resulted in only 27 and 10% reversal of the CEES-induced increase in TEM in JB6 cells and fibroblasts, respectively (Figs. 6B and C). We also used Western blotting to determine this variable effects of GSH and Trolox on DNA damage by measuring the phosphorylation of H2A.X and p53 as well as total p53 levels after similar treatments of GSH or Trolox 1 h before 4 h of 0.5 mM CEES exposure (Figs. 6D and E). Comparable to the results obtained from measuring the DNA damage by comet assay, Western blotting also showed that GSH but not Trolox pretreatment significantly reduced the CEES-induced H2A.X and p53 phosphorylation as well as total p53 levels (Figs. 6D and E).
Fig. 6.
Effects of GSH or Trolox treatment on CEES-induced DNA damage in JB6 cells and fibroblasts. (A–C) JB6 cells and fibroblasts were treated with 10 mM GSH or 800 µM Trolox for 1 h before 0.5 mM CEES exposure for 1 h; cells were thereafter collected and subjected to comet assay as described under Materials and methods. (A) Representative pictures of the comet showing the effects of the GSH or Trolox treatment in fibroblasts. (D and E) JB6 cells and fibroblasts were treated with 10 mM GSH or 800 µM Trolox for 1 h before 0.5 mM CEES exposure for 4 h; whole-cell lysates were prepared at the end of the treatments and subjected to SDS–PAGE followed by Western blot analysis as described under Materials and methods to detect H2A.X Ser139 and p53 Ser15 phosphorylation. Membranes were stripped and reprobed with total p53 and β-actin antibodies. Data are presented as means ± SEM, n = 3. *P<0.001 compared to 0.5 mM CEES treatment. UC, untreated control; DMSO, vehicle control.
GSH forms conjugates with CEES in aqueous medium
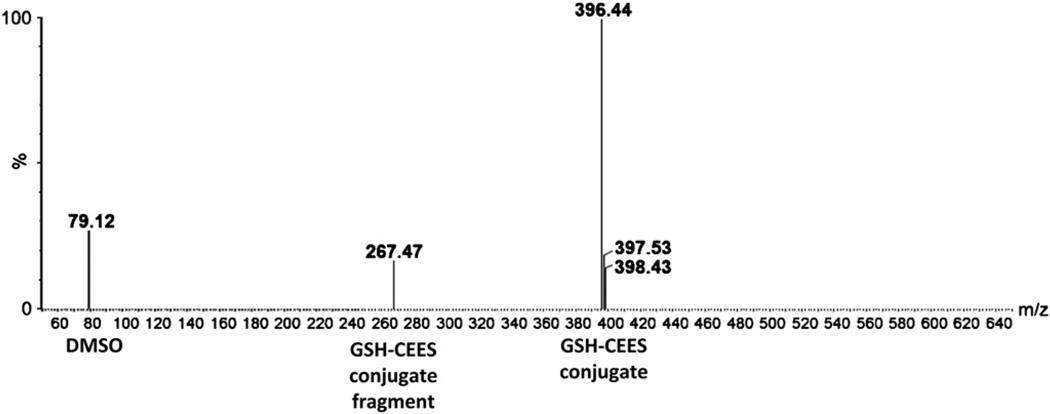
We next employed LC-MS analysis, to determine if the protective effect of GSH pretreatment on CEES-induced DNA damage could be due to its conjugation with CEES. Incubation of 0.5 mM CEES with 10 mM GSH for 1 h resulted in the LC-MS detection of peaks with m/z 79, m/z 267, and m/z 396 (Fig. 7). The m/z 79 peak corresponds to DMSO, which is the vehicle for CEES; m/z 267 peak corresponds to CEES conjugated to a fragment of glutathione, formed from the loss of a neutral glutamic acid [37]; and m/z 396 peak corresponds to GSH–CEES conjugate. These results show that a decrease in CEES-induced DNA damage by GSH pretreatment could be due to its conjugation ability with CEES compared to its antioxidant properties.
Fig. 7.
Detection of GSH–CEES conjugates through LC-MS. 0.5 mM CEES prepared in DMSO was incubated for 1 h with 10 mM GSH prepared in water, with pH adjusted to 7.4, and subjected to LC-MS analysis as described under Materials and methods. The formation of GSH–CEES conjugate was detected as a peak with m/z 396.
Discussion
Earlier studies have reported that SM-induced DNA damage and subsequent skin toxicity involve alkylating properties of SM as well as oxidative stress [38–42]. However, this study is the first comprehensive report on the SM analog CEES-induced DNA damage, oxidative stress, and oxidative DNA damage and the effects of antioxidants GSH and Trolox on these consequences in both skin epidermal cells and dermal fibroblasts. Results from this study show that oxidative stress is involved in CEES-induced DNA damage in both the skin cell types; however, a decrease in CEES-caused DNA damage by GSH but not the antioxidant Trolox suggests that oxidative stress does not play a major role in CEES-induced DNA damage under these conditions. Additionally, our results demonstrate the conjugation of GSH with CEES in an aqueous environment, which possibly contributes to the ability of GSH to reduce the total DNA damage induced by CEES in the epidermal cells and fibroblasts.
Unlike SM, which forms cross-links and adducts with biological molecules, its monofunctional analog CEES is capable of forming only adducts because of the presence of one chlorine atom, compared to the two chlorine atoms in SM. Nonetheless, because of the high resemblance to the biological effects of SM and formation of DNA adducts similar to SM, CEES has been widely used to study the mechanism of action of SM [43,44]. In addition to the effect of SM exposure mainly on epidermal keratinocytes, higher doses of SM are reported to cause damage to dermal fibroblasts and related inflammatory responses in the dermis [45]. Accordingly, we have investigated the mechanisms of CEES-induced DNA damage in both mouse skin epidermal (JB6) cells and mouse skin primary fibroblasts and found that CEES had similar effects on both these cell types. Our results suggested an important role for DNA damage as well as the activation of DNA damage repair pathways in CEES-induced toxic effects (Table 1). This is in agreement with other studies showing that DNA is an important target of SM- and CEES-induced skin injury [22,24,45]. Other data in this study showing that both H2A.X and p53 are phosphorylated within 1 h of CEES exposure suggest that DNA damage is an early event after CEES exposure (Table 1). In addition, a substantial decrease in DNA damage after 16 h of CEES exposure suggests the possibility of the start of DNA repair at this time point. This is consistent with our recent findings showing that CEES causes the activation of DNA-damage-sensing pathways such as ATM and ATR, which are involved in cell cycle arrest for DNA repair [23]. Earlier studies have also shown the activation of p53 and its role in SM-induced skin toxicity [46–48]. H2A.X phosphorylation, an indicator of direct and indirect formation of the DNA double-strand breaks (via single-strand breaks, adducts, and cross-linking) [49], is also shown to be activated after SM exposure [24,50]. Taken together, our results and those published earlier suggest that the activation of DNA repair molecules is an early event in CEES-induced DNA damage and is comparable among SM and CEES exposures. Hence they could be important biomarkers in the efficacy studies of the rescue agents.
Table 1.
Effect of CEES on the biomarkers of DNA damage and oxidative stress in JB6 epidermal cells and dermal fibroblasts.
| Biomarkers | Time after CEES exposure | |||||
|---|---|---|---|---|---|---|
| JB6 Cells | Fibroblasts | |||||
| 1–2 h | 4–6/8 h | 16–24 h | 1–2 h | 4–6/8 h | 16–24 h | |
| 1. DNA Damage | ||||||
| A. Comet Assay | +++ | ++ | + | +++ | ++ | + |
| B. Western Immunoblotting | ||||||
| i. Phospho H2AX | ++ | +++ | ++ | ++ | +++ | ++ |
| ii. Phospho P53 | + | +++ | ++ | + | +++ | +++ |
| iii. Total P53 | + | ++ | +++ | + | +++ | ++ |
| 2. Oxidative stress | ||||||
| A. MitoSOX Red | ++ | +++ | ++ | +++ | ++ | + |
| B. DHE | ++ | +++ | ++ | + | +++ | + |
| 3. Oxidative DNA damage | ||||||
| (8-OHdG) | ++ | +++ | ND | +++ | +++ | ND |
(+++), high, (++), medium or (+) low effect of CEES on DNA damage, oxidative stress and oxidative DNA damage following 1–24 h of its exposure in epidermal and dermal cells.
ND, not determined.
SM- and CEES-induced ROS generation has been shown to induce inflammatory responses in the skin [17,21,45]. The role of oxidative stress in SM- and CEES-induced skin injury is also shown by the depletion of GSH and thioredoxin, ROS generation, and supplementation of antioxidants and antioxidant enzymes to reduce the toxic effects of these agents [16,18,20,51]. The oxidative stress thus induced by exposure to vesicating agents leads to lipid peroxidation, protein adduct formation, and oxidative DNA damage in skin [18,21]. However, the extent of the involvement of oxidative stress in CEES-induced DNA damage is not fully defined. Our present study using MitoSOX red and DHE dyes for the first time demonstrates that CEES induces mitochondrial and cellular oxidative stress, mainly due to the production of , not only in epidermal cells but also in primary dermal fibroblasts. The occurrence of peak mitochondrial oxidative stress before peak cellular oxidative stress could be due to the lipophilic nature of CEES, which allows it to freely enter mitochondria, causing mitochondrial DNA damage and dysfunction [18,52]. Mitochondria, being the major source of ROS in the cell, may lead to an earlier increase in mitochondrial oxidative stress compared to the total cellular oxidative stress. Our results also showed for the first time that CEES-induced oxidative stress plays a critical role in the observed oxidative DNA damage by CEES in both skin epidermal cells and dermal fibroblasts. Notably, CEES-induced oxidative stress and oxidative DNA damage responses were similar in both these cell types (Table 1). Taken together, these findings implicate the possible involvement of fibroblast (dermal layer)-mediated oxidative stress, together with that of epidermal cells, in CEES-induced inflammatory responses in skin.
SM and its analogs cause depletion of GSH via their alkylating properties or oxidative stress, and addition of extracellular GSH or N-acetylcysteine and sulforaphane, which increases the intracellular GSH levels, causes a reversal in CEES-induced skin injury [16,20,39,53]. Results of this study showed that treating cells with GSH or the antioxidant Trolox before CEES exposure significantly reduces the oxidative stress and oxidative DNA damage induced by CEES. Importantly however, unlike GSH, Trolox was unable to reverse the CEES-induced DNA damage. Our additional studies using LC-MS analysis suggested that, whereas antioxidant properties of GSH could decrease CEES-induced oxidative stress, the major contribution to the observed protective effect of GSH was possibly due to its ability to form conjugates with CEES. More studies are needed in the future to further support this notion; however, GSH has been shown to form conjugates with SM [26], supporting our present observation with CEES. Taken together, our data indicate that oxidative stress is an early event after CEES exposure and is involved in CEES-induced oxidative DNA damage in both skin epidermal cells and dermal fibroblasts. However, oxidative DNA damage is not the prominent contributor to the overall CEES-caused DNA damage. Thus, pleiotropic agents (or cocktails) that target the direct attack of CEES as well as counteract the consequences of CEES-induced oxidative stress are needed for the rescue of vesicating agent-induced skin injuries.
Acknowledgments
This work was supported by the Countermeasures against Chemical Threats (CounterACT) Program, National Institutes of Health Office of the Director, and the National Institute of Environmental Health Sciences (Grant U54ES-015678). The authors thank the University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences Mass Spectrometry Facility for analyzing the samples.
Abbreviations
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia-Rad3-related
- CEES
2-chloroethyl ethyl sulfide
- DAPI
4′,6′-diamidino-2-phenylindole
- DHE
dihydroethidium
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- GSH
glutathione
- 8-OHdG
8-hydroxydeoxyguanosine
- PARP
poly(ADP-ribose) polymerase
- ROS
reactive oxygen species
- SCGE
single-cell gel electrophoresis
- SM
sulfur mustard
- TEM
tail extent moment
References
- 1.Shohrati M, Peyman M, Peyman A, Davoudi M, Ghanei M. Cutaneous and ocular late complications of sulfur mustard in Iranian veterans. Cutan. Ocul. Toxicol. 2007;26:73–81. doi: 10.1080/15569520701212399. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson WS. Delayed keratitis due to mustard gas (dichlorodiethyl sulfide burns): report of two cases. Arch. Ophthalmol. 1948;40:291–301. doi: 10.1001/archopht.1948.00900030297007. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson WS. Delayed mustard gas keratitis (dichlorodiethyl sulfide): a report of two cases. Trans. Am. Ophthalmol. Soc. 1947;45:81–92. [PMC free article] [PubMed] [Google Scholar]
- 4.Ghabili K, Agutter PS, Ghanei M, Ansarin K, Shoja MM. Mustard gas toxicity: the acute and chronic pathological effects. J. Appl. Toxicol. 2010;30:627–643. doi: 10.1002/jat.1581. [DOI] [PubMed] [Google Scholar]
- 5.Ghanei M, Poursaleh Z, Harandi AA, Emadi SE, Emadi SN. Acute and chronic effects of sulfur mustard on the skin: a comprehensive review. Cutan. Ocul. Toxicol. 2010;29:269–277. doi: 10.3109/15569527.2010.511367. [DOI] [PubMed] [Google Scholar]
- 6.Ruhl CM, Park SJ, Danisa O, Morgan RF, Papirmeister B, Sidell FR, Edlich RF, Anthony LS, Himel HN. A serious skin sulfur mustard burn from an artillery shell. J. Emerg. Med. 1994;12:159–166. doi: 10.1016/0736-4679(94)90693-9. [DOI] [PubMed] [Google Scholar]
- 7.Smith KJ, Hurst CG, Moeller RB, Skelton HG, Sidell FR. Sulfur mustard: its continuing threat as a chemical warfare agent, the cutaneous lesions induced, progress in understanding its mechanism of action, its long-term health effects, and new developments for protection and therapy. J. Am. Acad. Dermatol. 1995;32:765–776. doi: 10.1016/0190-9622(95)91457-9. [DOI] [PubMed] [Google Scholar]
- 8.Papirmeister B, Gross CL, Meier HL, Petrali JP, Johnson JB. Molecular basis for mustard-induced vesication. Fundam. Appl. Toxicol. 1985;5:S134–S149. [PubMed] [Google Scholar]
- 9.Monteiro-Riviere NA, Inman AO. Ultrastructural characterization of sulfur mustard-induced vesication in isolated perfused porcine skin. Microsc. Res. Tech. 1997;37:229–241. doi: 10.1002/(SICI)1097-0029(19970501)37:3<229::AID-JEMT8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Mellor SG, Rice P, Cooper GJ. Vesicant burns. Br. J. Plast. Surg. 1991;44:434–437. doi: 10.1016/0007-1226(91)90202-u. [DOI] [PubMed] [Google Scholar]
- 11.Brookes P, Lawley PD. The reaction of mono- and di-functional alkylating agents with nucleic acids. Biochem. J. 1961;80:496–503. doi: 10.1042/bj0800496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korkmaz A, Yaren H, Topal T, Oter S. Molecular targets against mustard toxicity: implication of cell surface receptors, peroxynitrite production, and PARP activation. Arch. Toxicol. 2006;80:662–670. doi: 10.1007/s00204-006-0089-x. [DOI] [PubMed] [Google Scholar]
- 13.Kehe K, Szinicz L. Medical aspects of sulphur mustard poisoning. Toxicology. 2005;214:198–209. doi: 10.1016/j.tox.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Fidder A, Moes GW, Scheffer AG, van der Schans GP, Baan RA, de Jong LP, Benschop HP. Synthesis, characterization, and quantitation of the major adducts formed between sulfur mustard and DNA of calf thymus and human blood. Chem. Res. Toxicol. 1994;7:199–204. doi: 10.1021/tx00038a013. [DOI] [PubMed] [Google Scholar]
- 15.Husain K, Dube SN, Sugendran K, Singh R, Das Gupta S, Somani SM. Effect of topically applied sulphur mustard on antioxidant enzymes in blood cells and body tissues of rats. J. Appl. Toxicol. 1996;16:245–248. doi: 10.1002/(SICI)1099-1263(199605)16:3<245::AID-JAT339>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Paromov V, Suntres Z, Smith M, Stone WL. Sulfur mustard toxicity following dermal exposure: role of oxidative stress, and antioxidant therapy. J. Burns Wounds. 2007;7:e7. [PMC free article] [PubMed] [Google Scholar]
- 17.Tewari-Singh N, Rana S, Gu M, Pal A, Orlicky DJ, White CW, Agarwal R. Inflammatory biomarkers of sulfur mustard analog 2-chloroethyl ethyl sulfide-induced skin injury in SKH-1 hairless mice. Toxicol. Sci. 2009;108:194–206. doi: 10.1093/toxsci/kfn261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould NS, White CW, Day BJ. A role for mitochondrial oxidative stress in sulfur mustard analog 2-chloroethyl ethyl sulfide-induced lung cell injury and antioxidant protection. J. Pharmacol. Exp. Ther. 2009;328:732–739. doi: 10.1124/jpet.108.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskin JD, Black AT, Jan YH, Sinko PJ, Heindel ND, Sunil V, Heck DE, Laskin DL. Oxidants and antioxidants in sulfur mustard-induced injury. Ann. N. Y. Acad. Sci. 2010;1203:92–100. doi: 10.1111/j.1749-6632.2010.05605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tewari-Singh N, Agarwal C, Huang J, Day BJ, White CW, Agarwal R. Efficacy of glutathione in ameliorating sulfur mustard analog-induced toxicity in cultured skin epidermal cells and in SKH-1 mouse skin in vivo. J. Pharmacol. Exp. Ther. 2011;336:450–459. doi: 10.1124/jpet.110.173708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal A, Tewari-Singh N, Gu M, Agarwal C, Huang J, Day BJ, White CW, Agarwal R. Sulfur mustard analog induces oxidative stress and activates signaling cascades in the skin of SKH-1 hairless mice. Free Radic. Biol. Med. 2009;47:1640–1651. doi: 10.1016/j.freeradbiomed.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jowsey PA, Williams FM, Blain PG. DNA damage, signalling and repair after exposure of cells to the sulphur mustard analogue 2-chloroethyl ethyl sulphide. Toxicology. 2009;257:105–112. doi: 10.1016/j.tox.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Tewari-Singh N, Gu M, Agarwal C, White CW, Agarwal R. Biological and molecular mechanisms of sulfur mustard analogue-induced toxicity in JB6 and HaCaT cells: possible role of ataxia telangiectasia-mutated/ataxia telangiectasia-Rad3-related cell cycle checkpoint pathway. Chem. Res. Toxicol. 2010;23:1034–1044. doi: 10.1021/tx100038b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kehe K, Balszuweit F, Steinritz D, Thiermann H. Molecular toxicology of sulfur mustard-induced cutaneous inflammation and blistering. Toxicology. 2009;263:12–19. doi: 10.1016/j.tox.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 25.Rosenthal DS, Simbulan-Rosenthal CM, Liu WF, Velena A, Anderson D, Benton B, Wang ZQ, Smith W, Ray R, Smulson ME. PARP determines the mode of cell death in skin fibroblasts, but not keratinocytes, exposed to sulfur mustard. J. Invest. Dermatol. 2001;117:1566–1573. doi: 10.1046/j.0022-202x.2001.01578.x. [DOI] [PubMed] [Google Scholar]
- 26.Noort D, Benschop HP, Black RM. Biomonitoring of exposure to chemical warfare agents: a review. Toxicol. Appl. Pharmacol. 2002;184:116–126. [PubMed] [Google Scholar]
- 27.Hakkinen L, Koivisto L, Larjava H. An improved method for culture of epidermal keratinocytes from newborn mouse skin. Methods Cell Sci. 2001;23:189–196. doi: 10.1023/a:1016385109922. [DOI] [PubMed] [Google Scholar]
- 28.Kaur M, Velmurugan B, Tyagi A, Agarwal C, Singh RP, Agarwal R. Silibinin suppresses growth of human colorectal carcinoma SW480 cells in culture and xenograft through down-regulation of beta-catenin-dependent signaling. Neoplasia. 2010;12:415–424. doi: 10.1593/neo.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 30.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 31.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 32.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay P, Rajesh M, Hasko G, Hawkins BJ, Madesh M, Pacher P. Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat. Protoc. 2007;2:2295–2301. doi: 10.1038/nprot.2007.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haddad JJ. The involvement of L-gamma-glutamyl-L-cysteinyl-glycine (glutathione/GSH) in the mechanism of redox signaling mediating MAPK (p38)-dependent regulation of pro-inflammatory cytokine production. Biochem. Pharmacol. 2002;63:305–320. doi: 10.1016/s0006-2952(01)00870-x. [DOI] [PubMed] [Google Scholar]
- 35.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 36.Cort WM, Scott JW, Araujo M, Mergens WJ, Cannalonga MA, Osadca M, Harley H, Parrish DR, Pool WR. Antioxidant activity and stability of 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid. J. Am. Oil Chem. Soc. 1975;52:174–178. doi: 10.1007/BF02672164. [DOI] [PubMed] [Google Scholar]
- 37.Burford N, Eelman MD, Groom K. Identification of complexes containing glutathione with As(III), Sb(III), Cd(II),Hg(II), Tl(I), Pb(II) or Bi(III) by electrospray ionization mass spectrometry. J. Inorg. Biochem. 2005;99:1992–1997. doi: 10.1016/j.jinorgbio.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 38.Gross CL, Innace JK, Hovatter RC, Meier HL, Smith WJ. Biochemical manipulation of intracellular glutathione levels influences cytotoxicity to isolated human lymphocytes by sulfur mustard. Cell Biol. Toxicol. 1993;9:259–267. doi: 10.1007/BF00755604. [DOI] [PubMed] [Google Scholar]
- 39.Langford AM, Hobbs MJ, Upshall DG, Blain PG, Williams FM. The effect of sulphur mustard on glutathione levels in rat lung slices and the influence of treatment with arylthiols and cysteine esters. Hum. Exp. Toxicol. 1996;15:619–624. doi: 10.1177/096032719601500804. [DOI] [PubMed] [Google Scholar]
- 40.Naghii MR. Sulfur mustard intoxication, oxidative stress, and antioxidants. Mil. Med. 2002;167:573–575. [PubMed] [Google Scholar]
- 41.Walker IG. Intrastrand bifunctional alkylation of DNA in mammalian cells treated with mustard gas. Can. J. Biochem. 1971;49:332–336. doi: 10.1139/o71-049. [DOI] [PubMed] [Google Scholar]
- 42.Vasilescu D, Adrian-Scotto M, Fadiel A, Hamza A. Ab initio study of alkylation of guanine–cytosine base pair by sulfur and nitrogen mustards. J. Biomol. Struct. Dyn. 2009;27:465–476. doi: 10.1080/07391102.2010.10507331. [DOI] [PubMed] [Google Scholar]
- 43.Ng ET, Sim MK, Loke WK. Protective actions of des-aspartate-angiotensin I in mice model of CEES-induced lung intoxication. J. Appl. Toxicol. 2010;31:568–578. doi: 10.1002/jat.1599. [DOI] [PubMed] [Google Scholar]
- 44.Blaha M, Bowers W, Jr, Kohl J, DuBose D, Walker J, Alkhyyat A, Wong G. Effects of CEES on inflammatory mediators, heat shock protein 70A, histology and ultrastructure in two skin models. J. Appl. Toxicol. 2000;20(Suppl. 1):S101–S108. doi: 10.1002/1099-1263(200012)20:1+<::aid-jat672>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 45.Shakarjian MP, Heck DE, Gray JP, Sinko PJ, Gordon MK, Casillas RP, Heindel ND, Gerecke DR, Laskin DL, Laskin JD. Mechanisms mediating the vesicant actions of sulfur mustard after cutaneous exposure. Toxicol. Sci. 2010;114:5–19. doi: 10.1093/toxsci/kfp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruff AL, Dillman JF., III Sulfur mustard induced cytokine production and cell death: investigating the potential roles of the p38, p53, and NF-kappaB signaling pathways with RNA interference. J. Biochem. Mol. Toxicol. 2010;24:155–164. doi: 10.1002/jbt.20321. [DOI] [PubMed] [Google Scholar]
- 47.Rosenthal DS, Simbulan-Rosenthal CM, Iyer S, Smith WJ, Ray R, Smulson ME. Calmodulin, poly(ADP-ribose)polymerase and p53 are targets for modulating the effects of sulfur mustard. J. Appl. Toxicol. 2000;20(Suppl. 1):S43–S49. doi: 10.1002/1099-1263(200012)20:1+<::aid-jat691>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 48.Minsavage GD, Dillman JF., III Bifunctional alkylating agent-induced p53 and nonclassical nuclear factor kappaB responses and cell death are altered by caffeic acid phenethyl ester: a potential role for antioxidant/electrophilic response-element signaling. J. Pharmacol. Exp. Ther. 2007;321:202–212. doi: 10.1124/jpet.106.116145. [DOI] [PubMed] [Google Scholar]
- 49.Toyooka T, Ishihama M, Ibuki Y. Phosphorylation of histone H2AX is a powerful tool for detecting chemical photogenotoxicity. J. Invest. Dermatol. 2011;131:1313–1321. doi: 10.1038/jid.2011.28. [DOI] [PubMed] [Google Scholar]
- 50.Jowsey PA, Williams FM, Blain PG. The role of homologous recombination in the cellular response to sulphur mustard. Toxicol. Lett. 2010;197:12–18. doi: 10.1016/j.toxlet.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 51.Black AT, Joseph LB, Casillas RP, Heck DE, Gerecke DR, Sinko PJ, Laskin DL, Laskin JD. Role of MAP kinases in regulating expression of antioxidants and inflammatory mediators in mouse keratinocytes following exposure to the half mustard, 2-chloroethyl ethyl sulfide. Toxicol. Appl. Pharmacol. 2010;245:352–360. doi: 10.1016/j.taap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shahin S, Cullinane C, Gray PJ. Mitochondrial and nuclear DNA damage induced by sulphur mustard in keratinocytes. Chem. Biol. Interact. 2001;138:231–245. doi: 10.1016/s0009-2797(01)00275-7. [DOI] [PubMed] [Google Scholar]
- 53.Amir A, Chapman S, Gozes Y, Sahar R, Allon N. Protection by extracellular glutathione against sulfur mustard induced toxicity in vitro. Hum. Exp. Toxicol. 1998;17:652–660. doi: 10.1177/096032719801701202. [DOI] [PubMed] [Google Scholar]