Preface
Proteasomes are ATP-dependent, multi-subunit proteases found in all eukaryotes, archaea and some bacteria. In eukaryotes, the small protein ubiquitin is post-translationally and covalently attached to proteins targeted for proteasomal degradation. Despite the presence of proteasomes in many prokaryotes, ubiquitin or other post-translational protein modifiers were long presumed absent from these organisms. Recently a prokaryotic ubiquitin-like protein, Pup, was found to target proteins for proteolysis by the Mycobacterium tuberculosis proteasome. The discovery of a ubiquitin-like modifier in prokaryotes opens up the possibility that other bacteria may also have small post-translational protein tagging systems, with the ability to affect cellular processes.
Introduction
Proteasomes regulate a multitude of functions in eukaryotes and are essential for life. The eukaryotic proteasome core (20S) is composed of four rings: two hetero-heptameric rings of beta (β) subunits sandwiched between two hetero-heptameric rings of alpha (α) subunits that restrict access to the catalytic core1 (Fig. 1). Eukaryotic proteasome cores are highly complex, with three of the seven different β-subunits having catalytic activity (Table 1). Entry of substrates into the proteasome core, where they are hydrolyzed, usually requires the activity of a hexameric ring of regulatory particle ATPases (Rpts) that cap the ends of proteasome cores. In addition to the ATPases, numerous accessory factors, including regulatory particle non-ATPases (Rpns) and de-ubiquitinases (DUBs), participate in the recognition, unfolding and degradation of substrates that are post-translationally tagged with chains of ubiquitin (Ub)2.
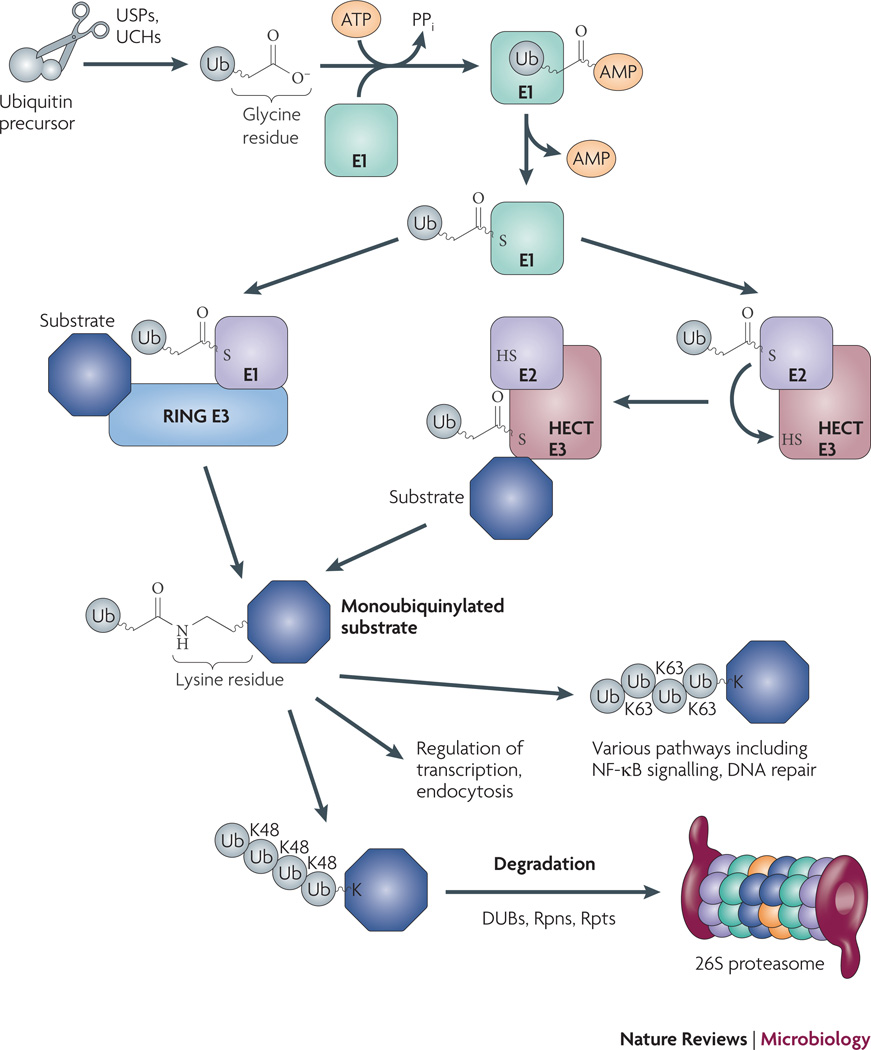
Fig. 1. Overview of the eukaryotic Ub-proteasome system.
Ubiquitin (Ub) is encoded by four different loci in yeast as part of a larger polypeptide49. Processing proteases expose C-terminal Gly-Gly that are activated by adenylation with an E1 enzyme. The E1 enzyme subsequently transfers Ub to an E2 enzyme, where a thioester bond is formed. The E2 then transfers Ub to any number of E3 ligases. The E3 ligase family can be sub-divided into HECT (Homologous to the E6-AP Carboxyl Terminus) and RING (Really Interesting Gene) domain ligases: RING ligases hold both the E2 and substrate, and facilitate the direct transfer of Ub from the E2 to the substrate; in contrast, HECT ligases form a thioester bond with Ub prior to transfer to a substrate lysine. E3 ligases dictate the type of Ub linkages that are formed. Proteins with Lys (K) 48 linked chains are usually targeted for degradation by the 26S proteasome. Other types of Ub linkages (mono- and poly-K63 and others) can result in degradation but generally serve other functions. See text for additional details.
Table 1.
Features of Eukaryotic and Prokaryotic Proteasome Systems=.
| Eukarya | Bacteria/Archaea | |
|---|---|---|
| Core Structure | Two rings of seven different α-subunits; two rings of seven different β-subunits, | Two rings of seven identical α-subunits; two rings of seven identical β-subunits. |
| Active Site(s) | Three β-subunit N-terminal threonines; three activities: tryptic, chymotryptic, post-acidic. | All β-subunit N-terminal threonines: chymotryptic; Mtb proteasome has additional activities. |
| Accessory Factors | Regulatory complex composed of numerous subunits, including Ub-binding proteins, deubiquitylases; hetero-hexamer of AAA ATPases. | Homo-hexamers of AAA ATPases; PafA: required for pupylation in Mtb. |
| Substrates | Numerous, most require Ub conjugation prior to proteolysis. Some substrates do not require Ub (e.g. ornithine decarboxylase). |
Mycobacteria: FabD, Ino1, Mpa, PanB, SodA (all require Pup) Archaea: unknown. |
| Cellular Pathways | Including protein turnover, signal transduction, NFkB regulation, endocytosis, DNA repair, autophagy, cell cycle, transcription. |
Mycobacteria: protein turnover; Archaea: unknown. |
References and additional details are in the text.
Almost all proteins targeted for proteasomal degradation in eukaryotes are tagged with Ub. Immature Ub polypeptides are processed by proteases including ubiquitin-specific proteases (USPs) and ubiquitin C-terminal hydrolases (UCHs), resulting in mature Ub molecules ending in a Gly-Gly motif, a feature common to all conjugatable ubiquitin-like modifiers (Ubls)3–5. The newly exposed C-terminal Gly is subjected to a series of reactions that result in the conjugation of Ub to lysine (Lys) residues in target proteins2, 6, 7 (Fig. 1). Activating (E1) enzymes utilize ATP to adenylate the C-terminal Gly of Ub, which is then passed to an active-site cysteine in the E1 enzyme. From here, Ub is transferred to a Ub-conjugating enzyme (E2) that delivers Ub to a protein ligase (E3), which catalyses the formation of an isopeptide bond with a Lys on the target substrate. E3 ligases are numerous in eukaryotes and contain a variety of substrate binding activities that provide specificity to the Ub-proteasome system by determining which substrates are conjugated with Ub. In general, proteins that are targeted to the proteasome have additional Ub molecules successively attached to Lys48 on other Ub molecules, forming polyubiquitin chains2, 8, 9. Polyubiquitin chains can be recognized by the regulatory complex of the proteasome, then removed and recycled by DUBs for more ubiquitylation reactions (Fig. 1).
Regulated proteolysis has profound implications on eukaryotic physiology, controlling numerous processes including gene expression and cell division. In addition to targeting proteins for degradation, Ub has other activities such as forming scaffolds on which other proteins may interact. Ub has the potential to form poly-Ub chains using any one of its seven Lys residues, or even its amino terminus. For example, the NF-kB pathway utilizes at least two different types of Ub linkage in order to activate gene expression10. NF-κB is a transcription factor that is bound by IκB in the eukaryotic cytosol, where it is unable to activate gene expression. In the IL-1 and TLR pathways of NF-κB activation, the E3 ligase TRAF6 is activated to form K63-linked Ub chains on the regulatory protein, NEMO, as well as on TRAF6 itself. The K63 chains are believed to form scaffolds that recruit additional factors that ultimately activate the kinase IKK. Activation of IKK leads to the phosphorylation of IκB, which results in K48-linked polyubiquitylation of IκB and its degradation. Degradation of IκB releases NF-κB, allowing it to translocate into the nucleus where it can activate gene expression. Thus, two different Ub linkages play critical roles in a single important pathway.
In addition to polyubiquitylation, monoubiquitylation is important for numerous functions, including the regulation of enzyme activity, DNA repair and sub-cellular protein targeting11. Other Ub-like modifiers (Ubls) such as SUMO and NEDD8 also affect various important cellular processes6 (Table 2).
Table 2.
Comparison of the biochemistry of ubiquitin and several ubiquitin-like modifiers6
| Processing enzymes: |
Conjugating enzymes (number)=: |
Linkage: | |
|---|---|---|---|
| Eukaryotes: | |||
| Ubiquitin | DUBs, USPs, UCHs | E1(2)50; E2(>30); E3(~600) | GG~K |
| SUMO-1–4 | SENP/Ulps | E1(1, heterodimer), E2(1), E3(3) | GG~K |
| NEDD8/Rub1 | Cop9, Yuh1 | E1(1, heterodimer), E2(1), E3(1) | GG~K |
| ISG15 | Ubp43 | E1(1), E2(1), E3(2) | GG~K |
| Urm1 | -- | E1(1) | GG~K |
| FAT10 | -- | E1(1)51 | GG~K |
| Prokaryotes: | |||
| Pup | De-amidase? De-pupylase? | PafA? PafD? | GGE~K |
Based on estimates for humans.
Unlike their eukaryotic counterparts, the roles of proteasomes on prokaryotic physiology are largely unknown12. Furthermore, despite the presence of bacterial proteasomes, Ub and Ubls had never been successfully identified in prokaryotes, leading to the conclusion that they were absent from this domain of life. This presumption was recently over-turned with the report of a small protein modifier in bacteria that targets proteins for degradation by a bacterial proteasome13. The discovery of a post-translational tagging system in bacteria has opened up the possibility that small protein modifiers, like those in eukaryotes, could have far-reaching implications on prokaryotic physiology and even pathogenesis. Here, I discuss what is currently known about bacterial proteasomes and Ubl biology, with focus on the pathogen Mycobacterium tuberculosis (Mtb).
Prokaryotic proteasomes
Any one bacterial species usually has a collection of ATP-dependent proteases including ClpP, HslV, Lon, or FtsH14. In addition to these proteases, some bacteria and all archaea have proteasomes. Like their eukaryotic counterparts, prokaryotic proteasome 20S cores are self-compartmentalized proteases composed of 14 α subunits and 14 β subunits, with the amino-terminal threonines of the β subunits providing the protease activity15. In contrast to eukaryotic proteasomes, core particles from archaea and bacteria are far simpler structures with homo-heptameric rings of catalytic β subunits flanked by homo-heptameric rings of α subunits16–21 (Table 1, Fig. 2). To date, only bacteria found in the class Actinomycetes are known to have proteasomes19, 22–24. Multi-subunit regulatory complexes similar to those in eukaryotes have not been identified in prokaryotes, suggesting the mechanisms by which proteins are targeted for degradation are different, or that regulatory complex interactions with cores are transient or weak. The best evidence so far that a proteasome-type ATPase can interact with a prokaryotic proteasome was reported by Smith and co-workers, where they showed the archaeal proteasome-activating nucleotidase (PAN) from Methanococcus janaschii could interact with Thermoplasma acidophilum proteasome cores to stimulate degradation of an unfolded, non-native protein25. In contrast, no one has ever demonstrated degradation of polypeptides by bacterial proteasomes in vitro.
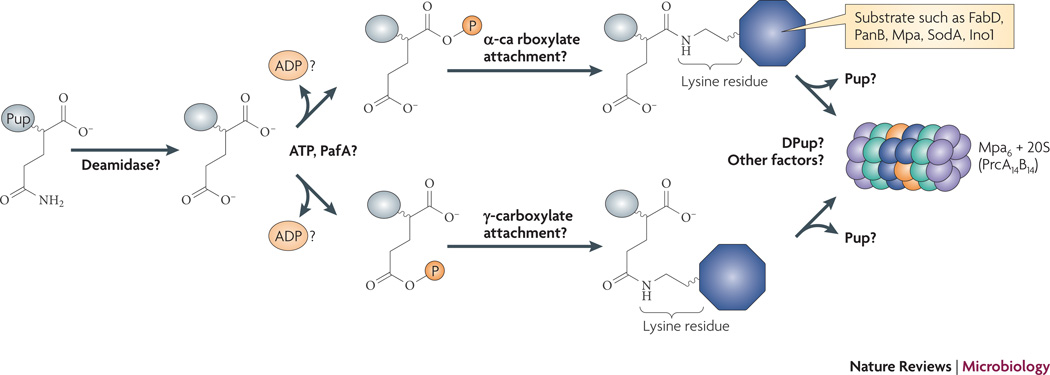
Fig. 2. Proposed model of the Pup-proteasome pathway in Mtb.
Unlike Ub, Pup is not processed proteolytically from a larger precursor protein. Pup appears to be de-amidated at the C-terminal Gln. From this point, it has been proposed that PafA phosphorylates the γ-carboxylate of the C-terminus of Pup, but this has not been established. The attachment of Pup to the substrate Lys can potentially be via either the α- or γ-carboxylate. It is not known if poly-pupylation occurs, nor is it known if Pup is removed by a de-pupylase (“DPUP”) prior to degradation, and recycled like Ub.
Proteasomes and pathogenesis
The first known function associated with any prokaryotic proteasome was discovered in the bacterial pathogen Mtb. Mtb is one of the top three leading causes of death in the world, and new and improved therapies are desperately needed. One approach to develop new antimicrobial drugs is to find compounds that attack pathogen pathways that normally protect bacteria against host immunity. Work from Carl Nathan’s laboratory demonstrated that the production of nitric oxide (NO) by the inducible nitric oxide synthase (iNOS) in macrophages is essential to control Mtb growth in mice26. NO has numerous activities that restrict microbial growth, including the ability to damage nucleic acids, proteins, and lipids27. Although studies have shown that mice wild type for iNOS survived much longer after Mtb infection than iNOS-deficient mice, bacteria were never completely sterilized from the animals. This suggested that Mtb had mechanisms to resist eradication by NO. In an effort to find new targets for anti-mycobacterial drug development, a screen for Mtb mutants sensitized to NO was performed. Mutations in genes encoding the putative proteasome accessory factors Mpa (Mycobacterium proteasome ATPase) and PafA (proteasome accessory factor A) sensitized Mtb to NO in vitro and, importantly, severely attenuated Mtb growth in mice22.
Mpa and PafA were hypothesized to participate in proteasome function because they are encoded near proteasome core genes and usually only found in proteasome-bearing bacteria28. Mpa has sequence similarity to eukaryotic proteasomal ATPases and is proposed to bind, unfold and deliver degradation substrates into the proteasome core. At the time, PafA did not resemble any protein of known function but nonetheless appeared to be in the same pathway as Mpa and the proteasome; pafA mutants were similarly sensitive to NO in vitro, and had the same attenuated phenotype in mice22, 29. Unlike most eukaryotic proteasomal accessory factors, Mpa and PafA are not essential for growth under normal conditions. Chemical inhibition or genetic repression of proteasome protease activity also sensitized Mtb to NO, linking proteasome activity to Mpa and PafA22, 30. However, Mtb proteasome protease activity was required for normal growth under non-stressed conditions, suggesting the proteasome core has functions independent of both Mpa and PafA.
Bacterial “ubiquitin”
One of the mysteries of the bacterial proteasome system was how proteins were targeted for degradation in the apparent absence of Ub or Ubls. To begin to answer this question, natural substrates of the Mtb proteasome were first identified. Two biosynthetic enzymes, FabD (malonyl co-A acyl carrier protein) and PanB (ketopantenoate hydroxymethyltransferse), were found as degradation substrates31. Both proteins accumulated in proteasome-defective Mtb, however, unlike proteins conjugated with Ub, FabD and PanB did not appear to be modified because they appeared to migrate through protein gels only at their predicted molecular weight. This observation, in addition to the lack of apparent protein modifiers in bacteria, led to the hypothesis that proteasomal degradation signals were inherent to the substrates.
Despite the identification of endogenous Mtb substrates, attempts to degrade FabD with recombinant proteasomes and Mpa in vitro were unsuccessful, suggesting that other co-factors were required for full proteasome function. To address the hypothesis that Mpa needed to interact with other degradation co-factors, a bacterial two-hybrid screen in E. coli was performed with Mpa as bait. The screen resulted in the identification of Rv2111c, a protein of unknown function encoded with the proteasome core genes of Mtb13. Purified Rv2111c non-covalently interacted with Mpa but did not promote FabD degradation by proteasomes and Mpa in vitro.
At this point, numerous presumed players of the bacterial proteasome system had been identified, but how they interacted with each other remained ambiguous. Only circumstantial evidence suggested there were contacts between several of the proteins, and no stable interactions between any bacterial proteasome with cognate ATPases had been reported. It was possible that yet additional proteins specific to proteasome-bearing bacteria were necessary to facilitate degradation. The development of a new two-hybrid system allowed this hypothesis to be tested by looking for interactions between proteasome components and degradation substrates in mycobacteria32. A positive interaction was detected between the substrate FabD and Rv2111c13. Surprisingly, upon validation of this result, epitope-tagged FabD and Rv2111c co-immunopreciptated from mycobacterial lysates as a covalently-linked complex, and not as separate proteins. Mass spectrometry (MS) revealed that Rv2111c formed an isopeptide bond between its caroboxyl (C) terminus and the ε-amino group of a specific lysine (Lys173) in FabD.
Although Rv2111c is not predicted to have a Ub-fold, a feature common among almost all Ubls, it nonetheless has a penultimate C-terminal Gly-Gly motif. The C-terminus of Rv2111c is Gly-Gly-Gln, thus it was predicted that the Gln might be removed in a manner similar to the proteolytic processing of Ubls. However, high-resolution tandem MS/MS revealed that not only was the terminal Gln retained, but it was converted to a glutamate (Glu). When unconjugated Rv2111c purified from mycobacteria was analyzed by MS, nearly all molecules were de-amidated13. This result suggested that de-amidation preceded covalent attachment to substrate proteins.
Because this covalent modification was reminiscent of ubiquitylation in eukaryotes, Rv2111c was named prokaryotic ubiquitin-like protein (Pup). Modification with Pup, or “pupylation”, was required for the proteasome-dependent degradation of FabD; mutagenesis of FabD’s modified lysine, Lys173, nearly abolished pupylation, and dramatically stabilized this proteasome substrate in wild type mycobacteria13.
In another study, Burns and co-workers also noticed the Gly-Gly motif in Pup33. Using epitope-tagged Pup, they purified and identified from M. smegmatis two covalently linked proteins, super oxide dismutase (SodA) and myo-inositol-1-phosphate synthase (Ino1) with the same Pup~substrate GGE~K linkage. In addition, the group showed deletion of the C-terminal Gln abrogated pupylation. Consistent with the Pearce and co-workers study, they showed that pupylated proteins were more stable in a proteasome-defective mutant when compared to wild type M. smegmatis.
Taken together these results revealed for the first time that the post-translational modification of a polypeptide by a small protein modifier can occur in bacteria in a manner akin to Ubl modification in eukaryotes. So far, it does not appear that Pup forms chains like Ub, but it is still possible that certain substrates are poly-pupylated. It is likely that Pup serves to target proteins for degradation in all proteasome-bearing bacteria, but it is important to remember that eukaryotic Ub has functions in addition to regulating protein stability (Table 1). Thus, we cannot rule out that Pup, too, may have degradation independent functions.
A Pup ligase?
Currently the only protein known to be essential for pupylation is PafA. A disruption mutation in pafA resulted in failed substrate pupylation in Mtb13. PafA has no homology with eukaryotic E1, E2, or E3 enzymes, suggesting the activity of PafA is different from canonical Ubl systems. Structure analysis using HHpred prediction software34 revealed that PafA is similar to an E. coli protein of unknown function, YbdK, that is homologous to glutamine synthetases (GS) with γ-glutamate cysteine ligase activity (γ-GCS)35 (S. Hubbard, personal communication). Lehmann and co-workers structural analysis suggested that YbdK does not bind ammonia like other GS because it lacks several amino acids involved in GS catalysis. They demonstrated that YbdK had ligase activity between glutamate and L-cysteine (Cys), but they did not know if Cys was the normal biological substrate. No ligase activity was detected with the other 19 amino acids or ammonia.
Using bioinformatics, Iyer and co-workers reported similar predictions and proposed a model where PafA uses ATP to phosphorylate Pup’s C-terminal Glu γ-carboxylate, which then reacts with the target Lys amino group to form an isopeptide bond36. However, it is not yet established whether Pup’s Glu γ-carboxylate is indeed the site of substrate attachment: the C-terminal Glu has two carboxylate groups that have the potential to be attached to substrate Lys (Fig. 2). This model also presumes that Pup is already de-amidated, and does not suggest that PafA is involved in this process.
YbdK was shown to form dimers35, and Iyer and co-workers predicted that PafA may also form dimers with itself or with its homologue PafD (Rv2112c)36. This along with other hypotheses about PafA structure and function remain to be tested.
Pupylation and Mtb pathogenesis
The failure to degrade pupylated proteins results in NO-sensitivity and attenuated virulence, however, which degradation substrates are linked to these phenotypes are not yet known. There are several possible reasons why proteasome function is protective, none of which is mutually exclusive from the others. One hypothesis is that the bacterial proteasome is required to degrade NO-damaged proteins that would otherwise be toxic to the cell. If this were true, it would be expected that damaged proteins would accumulate in proteasome-defective bacteria treated with NO. Preliminary data show that protein oxidation increases in Mtb treated with acidified nitrite, a source of NO, but the amount and number of oxidized protein species do not appear different between wild type and Mpa-deficient Mtb (K. H. Darwin, unpublished). This result may not be surprising given that the Mtb proteasome is not required for protection against several other stresses that are expected to result in protein oxidation or misfolding. Interestingly, mpa, pafA and proteasome core mutants are more resistant to hydrogen peroxide, further suggesting the proteasome is not needed to combat all oxidant stresses22, 30.
Another hypothesis is that a limited number of proteins, or even a single protein, becomes particularly toxic upon exposure to NO. These may include metal-binding proteins, or proteins that tend to aggregate when misfolded. The accumulation of these substrates in a proteasome-defective strain under normal growth conditions may not have a deleterious effect. However, in the presence of NO, metal ions could be displaced or protein misfolding could occur, resulting in cell death.
Yet another alternative explanation that links proteasome function with pathogenesis is that the Mtb proteasome regulates anti-oxidant or virulence gene expression. All ATP-dependent proteases, including ClpP, Lon, FtsH, and HslV, degrade transcription factors in numerous bacterial species14. In a simple scenario, it is possible that the proteasome degrades transcription factors that de-repress anti-oxidant genes, or other genes required for tuberculosis pathogenesis. Indirect changes in gene expression due to lack of proteasome function could also impact the ability of Mtb to resist NO or cause disease.
It is possible that more than one of the above scenarios explains why Mtb defective for protein degradation is attenuated in vivo. The lack of protein turnover in any organism would not surprisingly render it less competitive for growth under stressful conditions. Thus targeting the proteasome pathway, including putative post-translational modification enzymes like PafA, may be an effective approach for battling various infectious diseases.
Other bacteria with Pup
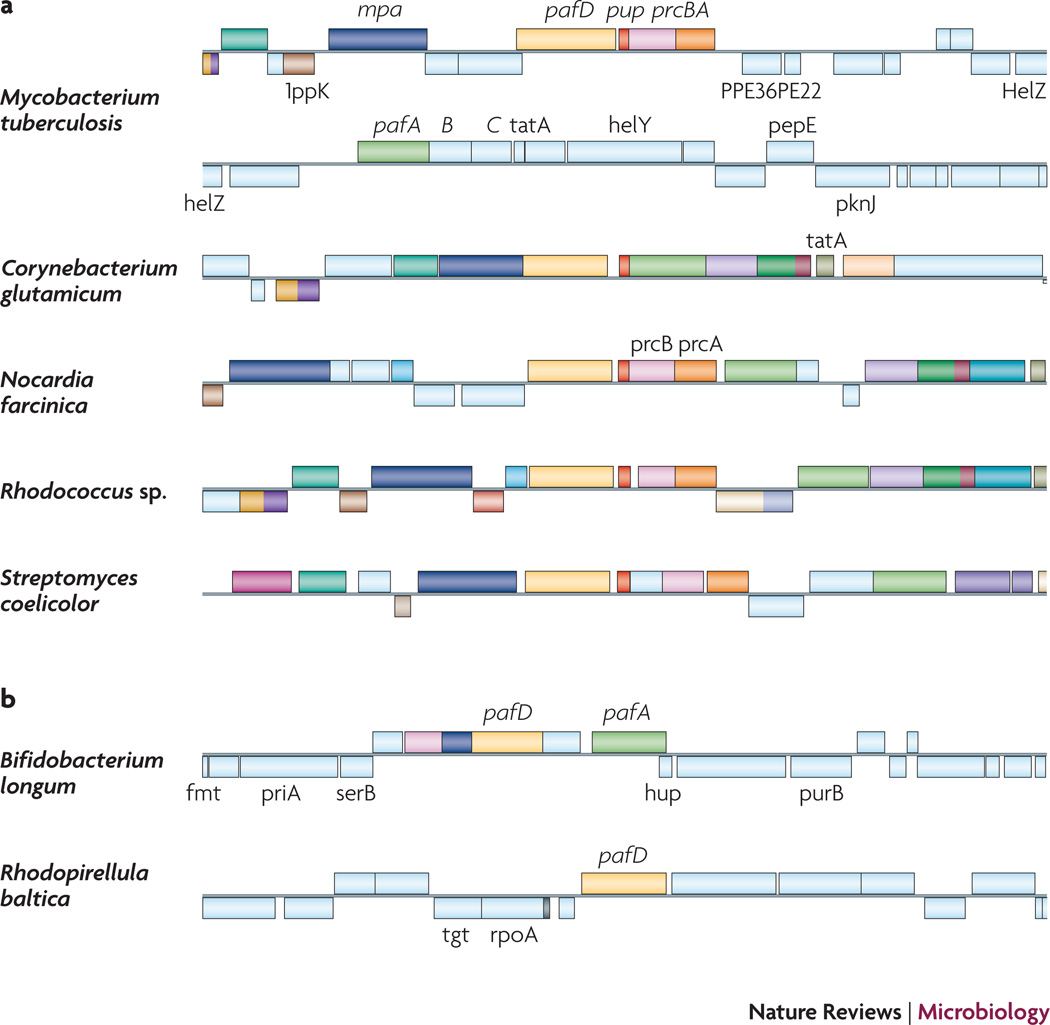
Pup, PafA and Mpa are found in numerous genera of pathogenic and non-pathogenic Actinobacteria, and almost always in strains that encode proteasomes (exceptions include Bifidobacterium and Corynebacterium) (Fig. 3). All sequenced Corynebacterium strains show a Pup homologue based on BLAST analysis37, but it was previously noted that they do not encode proteasomes38. Inspection of the C. glutamicum pup region reveals that mpa, pup and pafA are conserved, and pup appears to be transcriptionally and translationally coupled with pafA, an organization that is not observed in proteasome-bearing bacteria (Fig. 3). The organization of this region when compared to the same locus in Mtb suggests a deletion of the proteasome core genes, leaving the remaining proteasome accessory factor genes intact. Interestingly the corynebacterial Mpa homologues lack a C-terminal extension with a penultimate tyrosine that is essential for function31, 39. Site-directed mutagenesis of the penultimate tyrosine of Mtb Mpa resulted in failed substrate degradation and reduced protection against NO39. Furthermore, a transposon mutation that deleted of the last two amino acids of Mpa (YL) attenuated Mtb virulence in mice as much as a null mutation in mpa31. The archaeal ATPase PAN also has a conserved penultimate Tyr that is required to open proteasome cores for protein degradation25. Taken together, the Mpa C-terminus likely interacts with the proteasome core to activate degradation. Because Corynebacteria do not appear to have proteasomes or have proteases that have dramatically diverged from proteasomes, it is possible that Corynebacterium Mpa homologues have evolved different C-termini. In any case, it remains to be determined if mpa, pafA, and pup are expressed in this or other bacterial genera.
Fig. 3. Comparison of the pup regions of bacteria with and without proteasomes.
(A) pup-containing bacteria. Mycobacterium tuberculosis: pup (red); proteasome core genes (β-subunit gene prcB; pink; α-subunit gene prcA; orange); proteasome accessory factor A (pafA; green); and Mycobacterium proteasomal ATPase (mpa; cyan). Homologues in other bacteria are shaded in the same color schemes. pafB and pafC do not appear to be involved in pupylation or degradation in Mtb13, 29. PafD (hatched green) is 40% identical and 60% similar to PafA (e-value 10−73) but its role in proteasome function or pupylation has not been established. (B) Bacteria that have paf homologues but no apparent pup or proteasome genes. Data were collected from http://mbgd.genome.ad.jp/
Are there other Pup-like proteins in Mtb? Based on homology searches there do not appear to be additional Pup-like proteins in mycobacteria. However, as discussed earlier, genetic or chemical inhibition of proteasome protease activity results in a severe growth defect under normal growth conditions, a phenotype that is not observed with a pupylation-defective (pafA) mutant13, 22, 30. This suggests that the proteasome is able to degrade proteins targeted in a Pup-independent manner. It is possible that other tags, like Ub, may be processed from larger proteins. Degradation signals may also be inherent to the substrate, or include other types of modifications, like phosphorylation.
Other bacterial Ubls?
ThiS and MoaD are proteins with a Ub fold, conserved in most bacteria, and are involved in thiamine and molybdopterin co-factor biosynthesis, respectively40. ThiS and MoaD have C-terminal Gly-Gly motifs that undergo a series of chemistries that closely resemble Ub activation, however, these proteins transfer sulfur rather than conjugate to other proteins. Recently the eukaryotic ubiquitin-related modifier 1 (Urm1), which is similar to ThiS and MoaD, was shown to have a sulfur carrier function, in addition to its known ability to conjugate to proteins41–43. Although Urm1 is similar to ThiS and MoaD, its function is more related to the E. coli tRNA sulfuration pathway44, 45, and not thiamine or molybdopterin metabolism. One of the proteins involved in the E. coli tRNA sulfuration is TusA, which is proposed to activate the desulfurase activity of IscS. TusA is thought to accept a persulfur from IscS on one of its two cysteines to form a disulfide bond44. Intriguingly, although TusA is not predicted to have Ubl properties, it has a C-terminal Gly-Gly motif. Perhaps TusA, like ThiS or MoaD, forms an adenylated intermediate to accept sulfur for the bacterial tRNA sulfuration pathway. More intriguingly, perhaps these bacterial sulfur transfer proteins, like Urm1, can also covalently conjugate to proteins.
Examination of bacterial genome sequences has revealed that several species encode proteins highly similar to Ub. For example, the gut commensal organism Bacteroides fragilis encodes a protein nearly identical to human Ub46. Curiously, it does not have a Gly-Gly motif, thus it is not clear if it could have a true Ub-like function. Alternatively, Bacteroides may have a conjugation mechanism that does not require Gly-Gly. In addition to Bacteroides, the gastrointestinal pathogen Helicobacter pylori encodes fragments of Ub47. It is not known if either Bacteroides or Helicobacter make these proteins. An intriguing possibility is that these commensal bacteria may have acquired Ub genes from their hosts. It is tempting to propose that they could be used for intra-bacterial purposes, or even subvert normal mammalian cell functions. The idea that bacterial Ubls can be translocated into eukaryotes may not be so radical as several groups have shown that bacteria inject enzymes with DUB and E3 ligase activity into host cells to disrupt normal cell signaling48.
Prospects
A question that often comes to mind: Why did it take so long to identify a bacterial Ubl? It is perhaps not surprising that homology searches using Ub or Ubls did not find Pup. Pup is only “ubiquitin-like” in that it has a Gly-Gly motif, attaches to lysines, and targets proteins to a proteasome for degradation. Furthermore, it does not have a predicted Ub fold like canonical Ubls. Another reason why it might be difficult to identify bacterial Ubls is that the Ubl-modified form of a protein may be rapidly turned over or is transient. In Mtb the pupylated form of a proteasome substrate is by far the least abundant species and not observable unless affinity purified13.
The discovery of Pup has opened up the possibility that small protein modifiers may be present in other bacteria, including those without proteasomes. The function of these modifiers does not have to be limited to proteolysis. Much like eukaryotes, prokaryotes presumably need signals that target proteins for secretion or sub-cellular sorting. Now that we know bacteria can have Ubls, investigators can look more closely for new post-translational modifications.
Acknowledgements
I am grateful to A. Darwin, I. Mohr and H. Ovaa for critical review of this manuscript. I thank the anonymous referees for excellent suggestions and important corrections. K.H.D. is supported by NIH grants AI065437 and HL092774 and a Center For AIDS Research Pilot Project grant (NIH S P30 A1027742-17).
REFERENCES
- 1.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Ventii KH, Wilkinson KD. Protein partners of deubiquitinating enzymes. Biochem J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. Faseb J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 5.Marfany G, Denuc A. To ubiquitinate or to deubiquitinate: it all depends on the partners. Biochem Soc Trans. 2008;36:833–838. doi: 10.1042/BST0360833. [DOI] [PubMed] [Google Scholar]
- 6.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 7.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 8.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 9.Hough R, Rechsteiner M. Ubiquitin-lysozyme conjugates. Purification and susceptibility to proteolysis. J Biol Chem. 1986;261:2391–2399. [PubMed] [Google Scholar]
- 10.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haglund K, Dikic I. Ubiquitylation and cell signaling. Embo J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Mot R, Nagy I, Walz J, Baumeister W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 1999;7:88–92. doi: 10.1016/s0966-842x(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 13.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler SM, Festa RF, Pearce MJ, Darwin KH. Self-compartmentalized Bacteria Proteases and Pathogenesis. Mol. Microbiol. 2006;60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- 15.Zwickl P, Baumeister W, Goldberg AL. In: Proteasomes: The World of Regulatory Proteolysis. Hilt W, Wolf DH, editors. Georgetown, TX: 2000. pp. 8–18. Eurekah.com. [Google Scholar]
- 16.Lin G, et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome. Mol. Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 17.Hu G, et al. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol. Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 18.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268 doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 19.Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J. Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J Mol Biol. 2004;335:233–245. doi: 10.1016/j.jmb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 21.Tamura T, et al. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol. 1995;5:766–774. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- 22.Darwin KH, Ehrt S, Weich N, Gutierrez-Ramos J-C, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 23.Pouch M-N, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete Frankia. Mol. Microbiol. 2000;35:368–377. doi: 10.1046/j.1365-2958.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 24.Knipfer N, Shrader TE. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–383. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- 25.Smith DM, et al. ATP Binding to PAN or the 26S ATPases Causes Association with the 20S Proteasome, Gate Opening, and Translocation of Unfolded Proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 26.MacMicking JD, et al. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiloh MU, Nathan CF. Reactive nitrogen intermediates and the pathogenesis of Salmonella and mycobacteria. Curr Opin Microbiol. 2000;3:35–42. doi: 10.1016/s1369-5274(99)00048-x. [DOI] [PubMed] [Google Scholar]
- 28.Nagy I, Geert S, Vanderleyden J, De Mot R. Further sequence analysis of the DNA regions with the Rhodococcus 20S proteasome structural genes reveals extensive homolgy with Mycobacterium leprae. DNA Seq. 1997;7:225–228. doi: 10.3109/10425179709034040. [DOI] [PubMed] [Google Scholar]
- 29.Festa RA, Pearce MJ, Darwin KH. Characterization of the proteasome accessory factor (paf) operon in Mycobacterium tuberculosis. J Bacteriol. 2007;189:3044–3050. doi: 10.1128/JB.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearce MJ, et al. Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh A, Mai D, Kumar A, Steyn AJ. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc Natl Acad Sci U S A. 2006;103:11346–11351. doi: 10.1073/pnas.0602817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd Proteasomal protein degradation in mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2008 doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann C, et al. YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays. Proteins. 2004;56:376–383. doi: 10.1002/prot.20103. [DOI] [PubMed] [Google Scholar]
- 36.Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, et al. The N-terminal coiled coil of the Rhodococcus erythropolis ARC AAA ATPase is neither necessary for oligomerization nor nucleotide hydrolysis. J Struct Biol. 2004;146:155–165. doi: 10.1016/j.jsb.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 39.Darwin KH, Lin G, Chen Z, Li H, Nathan C. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 40.Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol Rev. 2006;30:825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 41.Pedrioli PG, Leidel S, Hofmann K. Urm1 at the crossroad of modifications. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Rep. 2008;9:1196–1202. doi: 10.1038/embor.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leidel S, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009 doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 43.Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A. 2008;105:18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Numata T, Ikeuchi Y, Fukai S, Suzuki T, Nureki O. Snapshots of tRNA sulphuration via an adenylated intermediate. Nature. 2006;442:419–424. doi: 10.1038/nature04896. [DOI] [PubMed] [Google Scholar]
- 46.Cerdeno-Tarraga AM, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 47.Giannakis M, Chen SL, Karam SM, Engstrand L, Gordon JI. Helicobacter pylori evolution during progression from chronic atrophic gastritis to gastric cancer and its impact on gastric stem cells. Proc Natl Acad Sci U S A. 2008;105:4358–4363. doi: 10.1073/pnas.0800668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rytkonen A, Holden DW. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe. 2007;1:13–22. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. Embo J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- 51.Chiu YH, Sun Q, Chen ZJ. E1-L2 activates both ubiquitin and FAT10. Mol Cell. 2007;27:1014–1023. doi: 10.1016/j.molcel.2007.08.020. [DOI] [PubMed] [Google Scholar]