Abstract
Ewing family tumors (EFTs) and prostate carcinomas (PCa) are characterized by rearrangement of ETS genes, most commonly FLI1 (EFTs) and ERG (PCa). Previously, we characterized an antibody against ERG (EPR3864) for detecting ERG-rearranged PCa. EPR3864 also cross reacts with FLI1, thus, here we evaluated the utility of EPR3864 for discriminating EFTs from other small round blue cell tumors (SRBCTs) by immunohistochemistry. Of 57 evaluable EFTs, 47 (82%) demonstrated at least moderate, diffuse, nuclear ERG/FLI1 staining (including 89% and 100% of cases with confirmed EWSR1:FLI1 and EWSR1:ERG fusions, respectively), of which 1, 3 and 43 showed negative, cytoplasmic or membranous CD99 staining, respectively. Amongst other SRBCTs (n=61 cases, 6 types), at least moderate, diffuse, nuclear EPR3864 staining was seen in all precursor-B-lymphoblastic lymphomas/leukemias and subsets of Burkitt’s lymphomas (10%) and synovial sarcomas (45%). In summary, EPR3864 may have utility for detecting EWSR1:FLI1 and EWSR1:ERG rearranged EFTs, in addition to PCa.
Keywords: EPR3864, EWSR1:FLI1, EWSR1:ERG, Ewing’s tumor
Introduction
Ewing family tumors (EFTs), which encompass Ewing sarcomas/peripheral nueroectodermal tumors, are characterized by chromosomal rearrangements fusing EWSR1 to members of the ETS transcription factor family. Although most commonly fused to the ETS gene FLI1 (~90%) through t(11;22)(q24;q12), EWSR1 can also fuse to ERG (~5–10%) and rarely ETV1, FEV, ETV4 and ETV51–4. EFTs and other small round blue cell tumors, including neuroblastomas, rhabdomyosarcomas, synovial sarcomas (poorly differentiated and monophasic variants), lymphoblastic lymphomas/leukemias, desmoplastic small round cell tumors and nephroblastomas (Wilm’s tumors), can be morphologically indistinguishable and definitive diagnosis commonly involves immunohistochemistry, typically against CD99 and FLI1, and molecular tests2,5–15.
CD99, also known as MIC2, encodes an integral membrane glycoprotein and shows diffuse membranous staining in >90% EFTs by immunohistochemistry using a variety of monoclonal antibodies (including 12E7, HBA71 and O13)13,16–19. Additionally, less specific cytoplasmic staining can also be observed. However, CD99 is not specific for EFTs, as it also stains lymphoblastic lymphomas/leukemias19,20, anaplastic large cell lymphomas21, synovial sarcomas22,23, some rhabdomyosarcomas24,25, as well as a variety of other tumors26–30.
The EWSR1:FLI1 gene fusions results in the fusion of the N-terminus of EWSR1 to the C-terminus of FLI1, which preserves the ETS DNA binding domain, and transforms NIH 3T3 cells31,32. FLI1 is normally expressed in endothelial and hematopoietic cells5, and consistent with its role as a transcription factor, both FLI1 and the EWSR1:FLI1 product show nuclear localization5,33. Both polyclonal and monoclonal antibodies against FLI1 have been shown to have diagnostic utility in EFTs, with staining of 63–89% (median 81%)5,6,9,10,34–36 and 75–100% (median 91%)7–9,37,38 of EFTs, respectively. In addition to EFTs, both monoclonal and polyclonal antibodies against FLI1 have been reported to also stain vascular tumors, lymphoblastic lymphomas and Merkel cell carcinomas, as well as a fraction of other small round blue cell tumors including poorly differentiated synovial sarcomas, and other non-Hodgkin lymphomas5–7,9,20,35,37,39. Polyclonal antibodies against FLI1 have also been reported to stain at least some olfactory neuroblastomas, desmoplastic small round cell tumors, and a variety of carcinomas (but not prostate carcinomas)6,35. Similarly, monoclonal antibodies against FLI1 have been reported to stain haemangiopericytomas, neuroendocrine carcinomas, melanomas, lung adenocarcinoma, and a variety of normal tissues, including prostate, breast, and colon epithelium7,9. In the only head to head comparison we are aware of, Mhawech-Fauceglia et al. reported that monoclonal antibodies against FLI1 were more sensitive for EFTs, while polyclonal antibodies were more specific, consistent with other published studies (summarized above)9.
Like EFTs, prostate carcinoma is characterized by chromosomal rearrangements involving ETS transcription factor family members, which are fused to the 5′ untranslated regions of androgen regulated genes and occur in approximately half of prostate carcinomas40–42. Fusions involving ERG (most commonly TMPRSS2:ERG) represent approximately 90% of all ETS fusions in prostate carcinoma, with less frequent fusions involving ETV1, ETV4, ETV5 and one reported case involving FLI140–44. Recently we and others have demonstrated the utility of a novel rabbit monoclonal antibody raised against the c-terminus of ERG (clone EPR3864), which demonstrates high sensitivity and specificity (>95%) for the detection of ERG rearranged prostate carcinoma45–53. As Mohamed et al. recently demonstrated that EPR3864 also reacts with exogenous FLI1 by Western blotting54, we hypothesized that EPR3864 may also have utility in the discrimination of EFTs from other small round blue cell tumors. Thus, here we characterized EPR3864 staining of ERG/FLI1 by immunohistochemistry in the discrimination of EFTs.
Methods
EFTs and SRBCTs tissues
A tissue microarray was constructed using formalin-fixed paraffin-embedded blocks from 105 EFT cases (each case represented by triplicate cores) from 85 patients, which includes a mixture of primary diagnostic specimens, primary samples post chemotherapy, recurrences and metastases (Table 1), and includes multiple cases from 16 patients (range 2–4 cases). Single sections from one EFT case each from two additional patients, who did not have samples on the tissue microarray, were also evaluated and are included in the results. The tissue microarray also contained cores representing normal ovary, spleen, lung, spinal cord, colon, kidney, tonsil, liver and testes.
Table 1.
Demographics of patients (and cases) with EFTs evaluable for ERG and CD99 staining
| Parameter1 | Total number (n) | Median (IQR) |
|---|---|---|
| Age at diagnosis: | 49 | 18 (13–30) |
|
| ||
| Parameter1 | Total number (n) | Number of patients (%) |
|
| ||
| Sex: | 49 | |
| Male | 29 (59%) | |
| Female | 20 (41%) | |
| Stage: | 57 | |
| Primary | 37 (65%) | |
| Primary; s/p chemo | 4 (7%) | |
| Recurrence | 6 (11%) | |
| Metastasis | 10 (18%) | |
| Location: | 57 | |
| Osseous; axial | 18 (32%) | |
| Osseous; extra-axial | 19 (33%) | |
| Extra-osseous; axial | 18 (32%) | |
| Extra-osseous; extra-axial | 2 (4%) | |
| Molecular Confirmation2: | 49 | |
| EWSR1:FLI1 | 29 (59%) | |
| EWSR1:ERG | 3 (6%) | |
| No EWSR1 rearrangement | 6 (12%) | |
| NA | 11 (22%) | |
Total number of patients with at least one evaluable core used for age at diagnosis and sex. Total number of cases with at least one evaluable core used for stage and location.
Patients who had at least one case confirmed by two of three molecular tests (FISH for EWSR1 breakapart, cytogenetics [t(11;22) or t(21;22)] and RT-PCR for EWSR1:FLI1 or EWSR1:ERG). Those without available molecular data (NA) are indicated.
Single formalin-fixed paraffin-embedded sections from 61 other SRBCTs, which include 11 nephroblastomas (Wilm’s tumors), 11 neuroblastomas, 7 rhabdomyosarcomas (4 alveolar, 2 embryonal, and 1 indeterminate, favor alveolar), 10 Burkitt’s lymphomas, 4 desmoplastic small round cell tumors, 11 monophasic synovial sarcomas and 7 precursor-B-lymphoblastic lymphomas/leukemias, were also evaluated for ERG/FLI staining.
EFTs and small round blue cell tumors included a mixture of primary diagnostic specimens, primary samples post chemotherapy, recurrences and metastases. All cases were diagnosed at the University of Michigan Health Systems, with EFTs diagnosed based on characteristic morphology and immunohistochemistry staining, with some cases undergoing molecular confirmation (cytogenetics, fluorescence in situ hybridization and/or reverse transcription-PCR) as part of the diagnostic workup. Cases were also assessed for EWSR1 breakapart by fluorescence in situ hybridization and reverse transcription PCR for EWSR1:FLI1 and EWSR1:ERG if not performed as part of the diagnostic workup. Cases were considered molecularly confirmed (for EWSR1:FLI1 or EWSR1:ERG) if two of the three tests (cytogenetics, fluorescence in situ hybridization and reverse transcription-PCR) were concordant. All tissues were obtained with prior Institutional Review Board approval.
Immunohistochemistry for ERG/FLI1 on the tissue microarray and single sections of EFTs and other SRBCTs was performed as described, using a ready-to-use, pre-diluted monoclonal antibody raised against ERG, clone EPR3864 (Ventana Medical Systems, Tucson, AZ)45,53,55. Staining of vessels was used as a positive control and cores or sections without staining of vessels were excluded from further analysis. Nuclear ERG/FLI1 staining intensity was scored as 0 (absent), 1+ (weak), 2+ (moderate) or 3+ (strong). Unless otherwise indicated, staining was diffuse (>80% of tumor). Immunohistochemistry for CD99 was performed on the tissue microarray using the rabbit monoclonal antibody EPR3097 (BioCare Medical, catalog #CME392), at 1:200 dilution for 30 min with Envision+ horseradish peroxidase detection. Epitope retrieval was performed using 10mM citrate buffer (pH 6) in a microwave for 10 min. Immunohistochemistry for CD99 was performed previously on the single sections of EFTs during the diagnostic workup and were re-reviewed. Staining for CD99 was scored as negative, cytoplasmic, or membranous. Unless otherwise indicated, staining was diffuse. EFT presence and viability, and ERG/FLI1 and CD99 staining, were evaluated by S.A.T, J.N.S. and L.P.K., with discrepancies resolved by D.R.L.
Analysis
EFT cases where no viable tumor was present in any of the three cores were excluded from further analysis. In cases where variable expression in two or more cores was observed, the greatest staining in any core was reported as the overall score and the variable expression was noted. Association between ERG/FLI1 and CD99 staining was evaluated using a two-tailed Fisher’s exact test using GraphPad Prism v. 5 (GraphPad Software).
Results
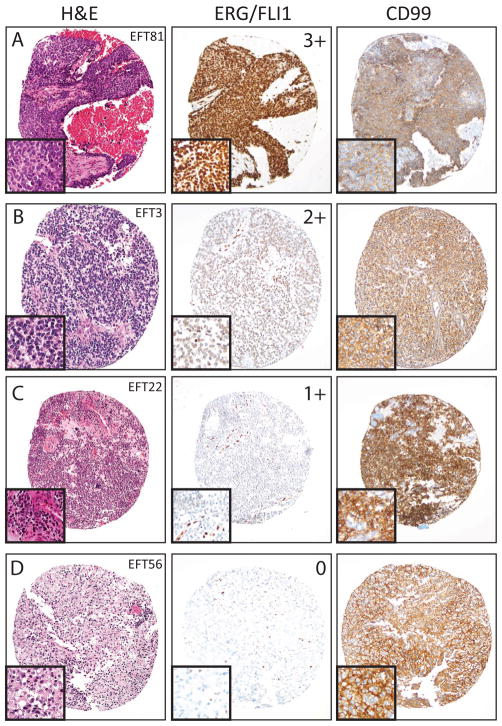
55 EFT cases from 47 patients had at least one core with viable tumor and were evaluable for FLI1/ERG and CD99 staining (from a tissue microarray with 105 cases from 85 patients). Single formalin-fixed paraffin-embedded sections from 2 additional EFTs (from patients not represented on the tissue microarray) were evaluable for ERG/FLI1 staining as well as CD99 staining performed at diagnosis. Thus, in total, our final evaluable cohort consisted of 57 EFT cases from 49 patients, as summarized in Table 1. ERG/FLI1 staining was scored as strong (3+), moderate (2+), weak (1+) or negative (0), while CD99 staining was scored as membranous or cytoplasmic (both positive) or negative. Examples of ERG/FLI1 and CD99 staining are shown in Figure 1. Amongst control cores of normal tissue on the tissue microarray, normal spleen and tonsil showed 3+ ERG/FLI1 staining, while normal ovary, lung, spinal cord, colon, kidney, liver and testes were negative (0+).
Figure 1. ERG/FLI1 staining in Ewing family tumors (EFTs).
EFTs were evaluated for ERG/FLI1 and CD99 staining by immunohistochemistry. ERG/FLI1 staining (diffuse nuclear) was scored as negative (0), weak (1+), moderate (2+), or strong (3+), and CD99 staining was scored as negative, cytoplasmic or membranous. Representative hematoxylin and eosin (H&E left panels), ERG/FLI1 (middle panel) and CD99 (right panel) staining from cases showing (A) 3+, (B) 2+, (C) 1+ and (D) 0 ERG/FLI1 staining and membranous CD99 staining are shown. All images are 10x original magnification with 20x insets.
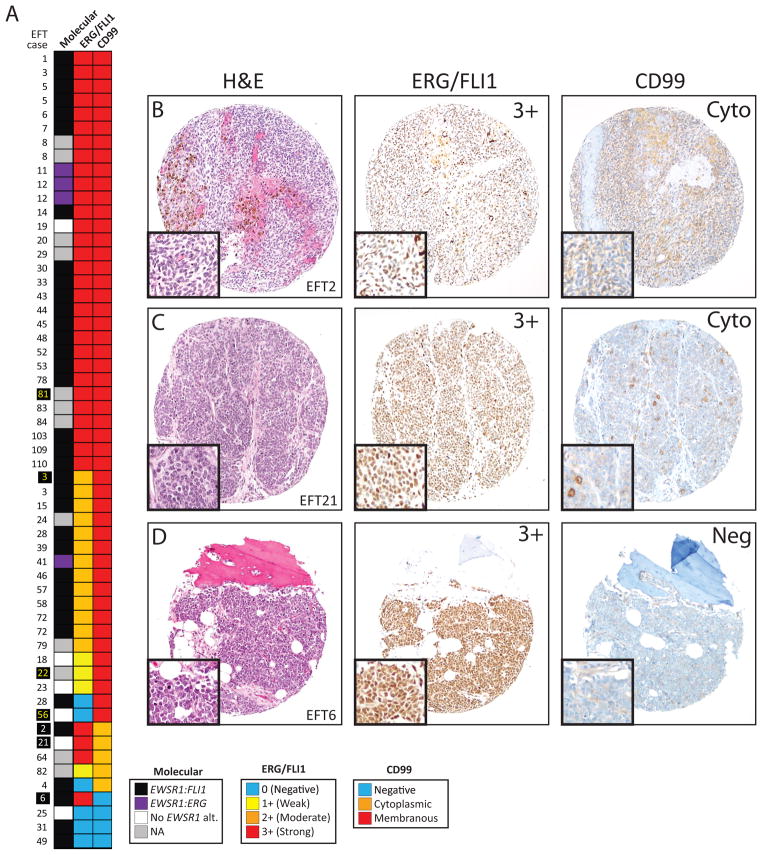
Of the 55 evaluable cases on the tissue microarray, 54 (98%) showed homogenous ERG/FLI1 staining between evaluable cores, and thus all cases were scored based on the highest staining intensity. The primary case from patient #8 showed 2 cores with 3+ ERG/FLI1 staining, and one core with 1+ staining, while a metastatic lesion from this patient showed 3 cores with 3+ ERG/FLI1 staining (Figure 2). Of 6 additional patients with more than one evaluable case on the tissue microarray, 2 showed different ERG/FLI1 staining intensity between cases. Patient #3 had three evaluable metastatic cases, with two showing 3+ERG/FLI staining in all three cores each, while one showed 2+ ERG/FLI1 staining in all three cores. The primary case from patient #28 showed negative ERG/FLI1 staining in all three cores, while a recurrence showed 2+ staining in all three cores (Figure 2).
Figure 2. ERG/FLI1 and CD99 staining in Ewing family tumors (EFTs).
A. Heat map of molecular status and ERG/FLI1 and CD99 staining for 57 evaluable EFT cases. Cases with confirmed EWSR1:FLI1 (black) or EWSR1:ERG (purple) rearrangements are indicated, along with cases without evidence of an EWSR1 rearrangement (white) or those not assessed (gray). ERG/FLI1 staining (diffuse nuclear) and CD99 staining were scored as in Figure 1 (indicated in the legend). Cases shown in Figure 1 are indicated by yellow names. B–D. Representative hematoxylin and eosin (H&E left panels), ERG/FLI1 (middle panel) and CD99 (right panel) cores from cases showing 3+ ERG/FLI1 expression and (B&C) cytoplasmic or negative (D) CD99 staining are shown. Cases shown are indicated by white names in A. All images are 10x original magnification with 20x insets.
All evaluable cases on the tissue microarray showed homogenous CD99 staining within evaluable cores, and one patient had two cases with discordant CD99 staining. Patient #6 had one case (a lung metastasis) showing membranous CD99 expression in one evaluable core, while a separate case (a femur metastasis) showed negative CD99 staining (Figure 2).
Of the 57 total evaluable EFT cases, 6 (11%) demonstrated negative (0) ERG/FLI1 staining, 4 (7%) demonstrated weak (1+) staining, 13 (23%) demonstrated moderate (2+) staining, and 34 (60%) demonstrated strong (3+) staining (Figure 2). All EFTs with positive ERG/FLI1 staining showed diffuse nuclear ERG/FLI1 expression. Of the 47 (82%) EFTs with at least moderate (2+) ERG/FLI1 staining, 1 (2%) showed negative CD99 staining, 3 (6%) showed cytoplasmic staining, and 43 (91%) showed membranous staining. Of the remaining 10 (18%) EFTs with negative to weak (0–1+) ERG/FLI1 staining, 3 (30%) showed negative CD99 staining, 2 (20%) showed cytoplasmic staining, and 5 (50%) showed membranous staining (Figure 2). Overall, at least moderate (2+) ERG/FLI1 staining and membranous CD99 staining were significantly associated, (43 of 57 evaluable cases, p=0.005, Fisher’s exact test), and 52 of 57 (91%) of cases showed either at least moderate (2+) ERG/FLI1 staining or membranous CD99 staining.
Of the 57 cases, 45 (79%) had evaluable molecular data (See Methods). Of evaluable cases, 35 (78%) harbored EWSR1:FLI1 fusions, 4 (9%) harbored EWSR1:ERG fusions, and 6 (13%) lacked evidence of EWSR1 rearrangements. Amongst the 35 cases with EWSR1:FLI1 fusions, 31 (89%) showed at least moderate ERG/FLI1 staining, and 30 (86%) showed membranous CD99 staining. All 4 cases with EWSR1:ERG fusions showed at least moderate ERG/FLI staining and membranous CD99 staining. Lastly, amongst the 6 cases without evidence of EWSR1 rearrangement, 2 (33%) showed at least moderate ERG/FLI1 staining and 4 (67%) showed membranous CD99 staining. Importantly, these results confirm the ability of EPR3864 to detect the products of both EWSR1:FLI1 and EWSR1:ERG gene fusions.
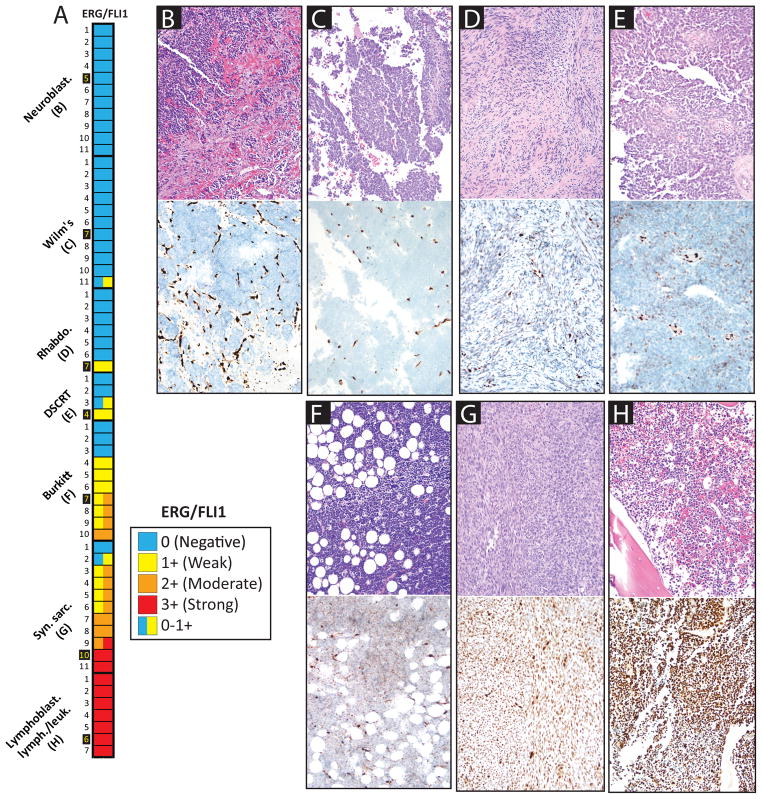
In addition to EFTs, we also evaluated ERG/FLI1 staining using single sections from 61 other SRBCTs (Figure 3). Amongst other SRBCTs, at least 2+ focal nuclear staining was observed in 0 of 11 (0%) nephroblastomas (Wilm’s tumors), 0 of 11 (0%) neuroblastomas, 0 of 7 (0%) alveolar/embryonal rhabdomyosarcomas, 0 of 4 (0%) desmoplastic small round cell tumors, 4 of 10 (40%) Burkitt’s lymphomas, 9 of 11 (82%) synovial sarcomas (10 monophasic, 1 poorly differentiated), and 7 of 7 (100%) precursor-B-lymphoblastic lymphomas/leukemias. Of all non EFTs stained for ERG/FLI1, at least 2+ diffuse nuclear staining was seen in 1 of 10 (10%) Burkitt’s lymphomas, 5 of 11 synovial sarcomas (45%) and 7 of 7 (100%) of precursor-B-lymphoblastic lymphomas/leukemias. A heat map of ERG/FLI1 staining in all small round blue cell tumors is shown in Figure 3.
Figure 3. ERG/FLI1 staining in small round blue cell tumor (SRBCT) mimickers of Ewing family tumors (EFTs).
Heat map of ERG/FLI1 staining (nuclear) in 61 non-EFT SRBCTs., ERG/FLI1 staining was scored as in Figures 1 & 2. In cases with heterogeneous staining, the variable intensity is indicated by multiple colors in the heat map cell. Cases shown are indicated by yellow names. B–H. Hematoxylin and eosin (H&E, top panels) and ERG/FLI1 (bottom panels) staining for representative cases are shown (20x original magnification).
Discussion
The diagnosis of EFTs from other small round blue cell tumors often requires immunohistochemistry, in addition to morphology, cytogenetics and/or molecular techniques. CD99 shows high sensitivity for EFTs, although it is not entirely specific. A combination of CD99, FLI1, HNK1 and CAV1, show high specificity and sensitivity for EFTs and has been proposed as an immunohistochemistry panel for the differential diagnosis of SRBCTs10. Both polyclonal and monoclonal antibodies against FLI1 have been employed, each with described limitations.
Previously we identified EPR3864, a monoclonal antibody raised against ERG, as showing utility for the detection of gene fusions involving ERG in prostate cancer53 (most commonly TMPRSS2:ERG), which occur in approximately half of all prostate cancers identified by prostate specific antigen screening40–42. More recently, Mohamed et al. showed that EPR3864 also detects FLI154, while another recently developed monoclonal antibody against ERG does not react with FLI154 and stains only 7% of EFTs56. FLI1 cross-reactivity of EPR3864 does not appear to be relevant in prostate cancer, given the >95% sensitivity and specificity of EPR3864 for detecting ERG-rearranged prostate cancer and the >99.99% reported specificity for cancer45–48,50–53,55,57. However, we hypothesized that this antibody may show utility for the discrimination of EFTs, which harbor both FLI1 and ERG rearrangements.
By immunohistochemistry with EPR3864, we show that 82% (47 of 57) of EFTs (including 89% of cases with confirmed EWSR1:FLI1 fusions and 100% of cases with confirmed EWSR1:ERG fusions) show at least moderate nuclear staining of ERG/FLI1, which was always diffuse. This rate is comparable to those reported using other polyclonal and monoclonal antibodies against FLI15–10,34–38. Additionally, at least moderate ERG/FLI1 staining and membranous CD99 staining were significantly associated in our study, with 91% of cases showed either at least moderate ERG/FLI1 staining or membranous CD99 staining.
Amongst 61 other SRBCTs, no Wilm’s tumors, neuroblastomas, rhabdomyosarcomas or desmoplastic small round cell tumors showed at least focal moderate (2+) ERG/FLI1 staining. However, we observed at least focal, moderate ERG/FLI1 staining in 40% of Burkitt’s lymphomas, 82% of monophasic synovial sarcomas and 100% of precursor-B-lymphoblastic lymphomas. Unlike EFTs, which always showed diffuse ERG/FLI1, heterogeneous absent-weak (0–1+), or weak-moderate (1–2+) staining was observed in 25% of desmoplastic small round cell tumors, 9% of Wilm’s tumors and 30% of Burkitt’s lymphomas, suggesting that only diffuse moderate-strong staining supports the diagnosis of EFT. In our study, the majority of monophasic synovial sarcomas and precursor-B-lymphoblastic lymphomas showed at least moderate nuclear ERG/FLI1 staining. Previous studies have reported occasional reactivity of FLI1 monoclonal and polyclonal antibodies with poorly differentiated synovial sarcomas (more relevant to the differential diagnosis of EFTs). In our cohort, only one synovial sarcoma was poorly differentiated (which showed focal CD99 staining), while of the remaining 10 monophasic sarcomas, all 6 cases evaluated for CD99 staining were strongly positive. As we did not have available additional poorly differentiated synovial sarcomas for evaluation, additional studies will be required to characterize EPR3864 staining in that entity. Similarly, CD99 and FLI1 staining in precursor-B-lymphoblastic lymphomas/leukemias is well characterized and represents an important differential diagnostic consideration in EFT evaluation58,59. Differentiating EFTs from these entities, which may show cross-reactivity with all FLI1 antibodies, will likely continue to require a combination of morphology, immunohistochemistry and molecular studies.
While this study was in preparation, Minner et al. evaluated EPR3864 staining in 11,483 tumors and 72 normal tissue types and reported strong staining nearly exclusively in prostate carcinoma and vascular tumors, however they reported no staining in 17 evaluable PNETs60. Wang et al. also recently evaluated EPR3864 staining in 32 EFTs, including 22 with EWSR1:FLI1, 8 with EWSR1:ERG and 2 with EWSR1-NFATC2. They observed predominantly diffuse, moderate to strong staining in 7 of 8 ERG rearranged cases, but only 3 of 24 non ERG rearranged cases showed staining with EPR3864 (all very weak)61. Importantly, Minner et al. and Wang et al. used substantially different antibody dilutions (1:400 and 1:2000) compared to our current study which used ready-to-use, pre-diluted EPR3864 antibody (1:50–1:100). Our study also used the same pretreatment conditions and automated staining and detection instrumentation as we recently validated in prostate cancer biopsies45 and use clinically at our institution.
Although the availability of molecular techniques has reduced the need for additional immunohistochemistry markers to identify EFTs, EPR3864 shows similar sensitivity to other polyclonal and monoclonal antibodies against FLI1, and has the advantage of being well characterized (in the context of prostate cancer) in its ability to detect ERG, with minimal background staining. Similarly, like other FLI1/ERG antibodies5,9,56, EPR3864 is a highly sensitive vascular marker53,60,62, supporting an additional area of diagnostic utility (S.A.T., D.R.L. and L.P.K., unpublished observations).
Additional studies will be needed to directly compare EPR3864 with other FLI1 antibodies, and we are not aware of whether studies have investigated whether other currently used antibodies against FLI1 also cross-react with ERG, however we hypothesize that this is unlikely given the lack of reported utility for detection of prostate cancer.
In summary, we demonstrate that EPR3864 shows utility for detecting EFTs harboring both EWSR1:FLI1 and EWSR1:ERG gene fusions. By immunohistochemistry, EPR3864 detection of ERG/FLI1 shows high sensitivity for EFTs (>80% with diffuse moderate-strong nuclear staining) and complements CD99 staining. EPR3864 also stains a substantial proportion of Burkitt’s lymphomas, monophasic synovial sarcomas and precursor-B-lymphoblastic lymphomas/leukemias, and in cases where these entities remain in the differential diagnosis based on morphology, molecular confirmation of EFTs will likely be required. Our results suggest that EPR3864, which has demonstrated utility in the diagnosis and molecular subtyping of prostate cancer (which also harbor ETS gene fusions), may also have utility in the diagnosis of EFTs from other SRBCTs.
Acknowledgments
Funding: Supported in part by the NIH S.P.O.R.E. (P50 CA69568).
The authors thank Amy Gursky and Angela Fullen for technical assistance, and Gary Pestano (Ventana Medical Systems) for providing the ERG antibody and immunohistochemistry reagents. Supported in part by the NIH S.P.O.R.E. (P50 CA69568). J.C.B. was supported by a Young Investigator Award from the Prostate Cancer Foundation.
The authors thank Gary Pestano (Ventana Medical Systems) for providing the ERG antibody and immunohistochemistry reagents.
Footnotes
Conflict of interest:
The University of Michigan has been issued a patent on the detection of ETS gene fusions in prostate cancer, on which A.M.C. and S.A.T. are listed as co-inventors. The University of Michigan licensed the diagnostic field of use to Gen-Probe, Inc, which sublicensed rights to Ventana Medical Systems, Inc. Neither company played a role in data collection, interpretation or analysis, and did not participate in the study design, review of the manuscript or the decision to submit for publication. N.P. has served as a consultant for Ventana Medical Systems. A.M.C. has served as consultant to Gen-Probe, Inc. and Ventana Medical Systems. S.A.T. has received honoraria and served as a consultant to Ventana Medical Systems.
References
- 1.de Alava E, Gerald WL. Molecular biology of the Ewing’s sarcoma/primitive neuroectodermal tumor family. J Clin Oncol. 2000;18:204–213. doi: 10.1200/JCO.2000.18.1.204. [DOI] [PubMed] [Google Scholar]
- 2.Khoury JD. Ewing sarcoma family of tumors. Adv Anat Pathol. 2005;12:212–220. doi: 10.1097/01.pap.0000175114.55541.52. [DOI] [PubMed] [Google Scholar]
- 3.Sankar S, Lessnick SL. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011;204:351–365. doi: 10.1016/j.cancergen.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene. 2010;29:4504–4516. doi: 10.1038/onc.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folpe AL, Hill CE, Parham DM, et al. Immunohistochemical detection of FLI-1 protein expression: a study of 132 round cell tumors with emphasis on CD99-positive mimics of Ewing’s sarcoma/primitive neuroectodermal tumor. Am J Surg Pathol. 2000;24:1657–1662. doi: 10.1097/00000478-200012000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Llombart-Bosch A, Navarro S. Immunohistochemical detection of EWS and FLI-1 proteinss in Ewing sarcoma and primitive neuroectodermal tumors: comparative analysis with CD99 (MIC-2) expression. Appl Immunohistochem Mol Morphol. 2001;9:255–260. doi: 10.1097/00129039-200109000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Rossi S, Orvieto E, Furlanetto A, et al. Utility of the immunohistochemical detection of FLI-1 expression in round cell and vascular neoplasm using a monoclonal antibody. Mod Pathol. 2004;17:547–552. doi: 10.1038/modpathol.3800065. [DOI] [PubMed] [Google Scholar]
- 8.Folpe AL, Goldblum JR, Rubin BP, et al. Morphologic and immunophenotypic diversity in Ewing family tumors: a study of 66 genetically confirmed cases. Am J Surg Pathol. 2005;29:1025–1033. [PubMed] [Google Scholar]
- 9.Mhawech-Fauceglia P, Herrmann F, Penetrante R, et al. Diagnostic utility of FLI-1 monoclonal antibody and dual-colour, break-apart probe fluorescence in situ (FISH) analysis in Ewing’s sarcoma/primitive neuroectodermal tumour (EWS/PNET). A comparative study with CD99 and FLI-1 polyclonal antibodies. Histopathology. 2006;49:569–575. doi: 10.1111/j.1365-2559.2006.02535.x. [DOI] [PubMed] [Google Scholar]
- 10.Llombart-Bosch A, Machado I, Navarro S, et al. Histological heterogeneity of Ewing’s sarcoma/PNET: an immunohistochemical analysis of 415 genetically confirmed cases with clinical support. Virchows Arch. 2009;455:397–411. doi: 10.1007/s00428-009-0842-7. [DOI] [PubMed] [Google Scholar]
- 11.Gamberi G, Cocchi S, Benini S, et al. Molecular diagnosis in Ewing family tumors: the Rizzoli experience--222 consecutive cases in four years. J Mol Diagn. 2011;13:313–324. doi: 10.1016/j.jmoldx.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downing JR, Head DR, Parham DM, et al. Detection of the (11;22)(q24;q12) translocation of Ewing’s sarcoma and peripheral neuroectodermal tumor by reverse transcription polymerase chain reaction. Am J Pathol. 1993;143:1294–1300. [PMC free article] [PubMed] [Google Scholar]
- 13.Ladanyi M, Lewis R, Garin-Chesa P, et al. EWS rearrangement in Ewing’s sarcoma and peripheral neuroectodermal tumor. Molecular detection and correlation with cytogenetic analysis and MIC2 expression. Diagn Mol Pathol. 1993;2:141–146. [PubMed] [Google Scholar]
- 14.Yamaguchi U, Hasegawa T, Morimoto Y, et al. A practical approach to the clinical diagnosis of Ewing’s sarcoma/primitive neuroectodermal tumour and other small round cell tumours sharing EWS rearrangement using new fluorescence in situ hybridisation probes for EWSR1 on formalin fixed, paraffin wax embedded tissue. J Clin Pathol. 2005;58:1051–1056. doi: 10.1136/jcp.2004.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian X, Jin L, Shearer BM, et al. Molecular diagnosis of Ewing’s sarcoma/primitive neuroectodermal tumor in formalin-fixed paraffin-embedded tissues by RT-PCR and fluorescence in situ hybridization. Diagn Mol Pathol. 2005;14:23–28. doi: 10.1097/01.pdm.0000140192.27878.97. [DOI] [PubMed] [Google Scholar]
- 16.Kovar H, Dworzak M, Strehl S, et al. Overexpression of the pseudoautosomal gene MIC2 in Ewing’s sarcoma and peripheral primitive neuroectodermal tumor. Oncogene. 1990;5:1067–1070. [PubMed] [Google Scholar]
- 17.Ambros IM, Ambros PF, Strehl S, et al. MIC2 is a specific marker for Ewing’s sarcoma and peripheral primitive neuroectodermal tumors. Evidence for a common histogenesis of Ewing’s sarcoma and peripheral primitive neuroectodermal tumors from MIC2 expression and specific chromosome aberration. Cancer. 1991;67:1886–1893. doi: 10.1002/1097-0142(19910401)67:7<1886::aid-cncr2820670712>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton G, Fellinger EJ, Schratter I, et al. Characterization of a human endocrine tissue and tumor-associated Ewing’s sarcoma antigen. Cancer Res. 1988;48:6127–6131. [PubMed] [Google Scholar]
- 19.Weidner N, Tjoe J. Immunohistochemical profile of monoclonal antibody O13: antibody that recognizes glycoprotein p30/32MIC2 and is useful in diagnosing Ewing’s sarcoma and peripheral neuroepithelioma. Am J Surg Pathol. 1994;18:486–494. [PubMed] [Google Scholar]
- 20.Lin O, Filippa DA, Teruya-Feldstein J. Immunohistochemical evaluation of FLI-1 in acute lymphoblastic lymphoma (ALL): a potential diagnostic pitfall. Appl Immunohistochem Mol Morphol. 2009;17:409–412. doi: 10.1097/PAI.0b013e3181972b6d. [DOI] [PubMed] [Google Scholar]
- 21.Buxton D, Bacchi CE, Gualco G, et al. Frequent expression of CD99 in anaplastic large cell lymphoma: a clinicopathologic and immunohistochemical study of 160 cases. Am J Clin Pathol. 2009;131:574–579. doi: 10.1309/AJCPE68HZXCGWTKK. [DOI] [PubMed] [Google Scholar]
- 22.Hartel PH, Fanburg-Smith JC, Frazier AA, et al. Primary pulmonary and mediastinal synovial sarcoma: a clinicopathologic study of 60 cases and comparison with five prior series. Mod Pathol. 2007;20:760–769. doi: 10.1038/modpathol.3800795. [DOI] [PubMed] [Google Scholar]
- 23.Machen SK, Fisher C, Gautam RS, et al. Utility of cytokeratin subsets for distinguishing poorly differentiated synovial sarcoma from peripheral primitive neuroectodermal tumour. Histopathology. 1998;33:501–507. doi: 10.1046/j.1365-2559.1998.00562.x. [DOI] [PubMed] [Google Scholar]
- 24.Fellinger EJ, Garin-Chesa P, Triche TJ, et al. Immunohistochemical analysis of Ewing’s sarcoma cell surface antigen p30/32MIC2. Am J Pathol. 1991;139:317–325. [PMC free article] [PubMed] [Google Scholar]
- 25.Ramani P, Rampling D, Link M. Immunocytochemical study of 12E7 in small round-cell tumours of childhood: an assessment of its sensitivity and specificity. Histopathology. 1993;23:557–561. doi: 10.1111/j.1365-2559.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Li J, Hao C, et al. Immunohistochemical evaluation of solid pseudopapillary tumors of the pancreas: the expression pattern of CD99 is highly unique. Cancer Lett. 2011;310:9–14. doi: 10.1016/j.canlet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 27.McCluggage WG, Kennedy K, Busam KJ. An immunohistochemical study of cervical neuroendocrine carcinomas: Neoplasms that are commonly TTF1 positive and which may express CK20 and P63. Am J Surg Pathol. 2010;34:525–532. doi: 10.1097/PAS.0b013e3181d1d457. [DOI] [PubMed] [Google Scholar]
- 28.Chen G, Folpe AL, Colby TV, et al. Angiomatoid fibrous histiocytoma: unusual sites and unusual morphology. Mod Pathol. 2011;24:1560–1570. doi: 10.1038/modpathol.2011.126. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Zeng XW, Wu JS, et al. Solitary fibrous tumor of the central nervous system: a clinicopathologic study of 24 cases. Acta Neurochir (Wien) 2011 doi: 10.1007/s00701-011-1160-9. [DOI] [PubMed] [Google Scholar]
- 30.Magro G, Caltabiano R, Kacerovska D, et al. Vulvovaginal myofibroblastoma: expanding the morphological and immunohistochemical spectrum. A clinicopathologic study of 10 cases. Hum Pathol. 2011 doi: 10.1016/j.humpath.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 31.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 32.May WA, Gishizky ML, Lessnick SL, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potikyan G, France KA, Carlson MR, et al. Genetically defined EWS/FLI1 model system suggests mesenchymal origin of Ewing’s family tumors. Lab Invest. 2008;88:1291–1302. doi: 10.1038/labinvest.2008.99. [DOI] [PubMed] [Google Scholar]
- 34.Jimenez RE, Folpe AL, Lapham RL, et al. Primary Ewing’s sarcoma/primitive neuroectodermal tumor of the kidney: a clinicopathologic and immunohistochemical analysis of 11 cases. Am J Surg Pathol. 2002;26:320–327. doi: 10.1097/00000478-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Mhawech-Fauceglia P, Herrmann FR, Bshara W, et al. Friend leukaemia integration-1 expression in malignant and benign tumours: a multiple tumour tissue microarray analysis using polyclonal antibody. J Clin Pathol. 2007;60:694–700. doi: 10.1136/jcp.2006.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson G, Wang M, Wejde J, et al. Detection of EWS/FLI-1 by Immunostaining. An Adjunctive Tool in Diagnosis of Ewing’s Sarcoma and Primitive Neuroectodermal Tumour on Cytological Samples and Paraffin-Embedded Archival Material. Sarcoma. 1999;3:25–32. doi: 10.1080/13577149977839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee AF, Hayes MM, Lebrun D, et al. FLI-1 distinguishes Ewing sarcoma from small cell osteosarcoma and mesenchymal chondrosarcoma. Appl Immunohistochem Mol Morphol. 2011;19:233–238. doi: 10.1097/PAI.0b013e3181fd6697. [DOI] [PubMed] [Google Scholar]
- 38.Terrier-Lacombe MJ, Guillou L, Chibon F, et al. Superficial primitive Ewing’s sarcoma: a clinicopathologic and molecular cytogenetic analysis of 14 cases. Mod Pathol. 2009;22:87–94. doi: 10.1038/modpathol.2008.156. [DOI] [PubMed] [Google Scholar]
- 39.Folpe AL, Chand EM, Goldblum JR, et al. Expression of Fli-1, a nuclear transcription factor, distinguishes vascular neoplasms from potential mimics. Am J Surg Pathol. 2001;25:1061–1066. doi: 10.1097/00000478-200108000-00011. [DOI] [PubMed] [Google Scholar]
- 40.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlins SA, Bjartell A, Chinnaiyan AM, et al. ETS Gene Fusions in Prostate Cancer: From Discovery to Daily Clinical Practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 42.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulo P, Barros-Silva JD, Ribeiro FR, et al. FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes Cancer. 2012;51:240–249. doi: 10.1002/gcc.20948. [DOI] [PubMed] [Google Scholar]
- 44.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 45.Tomlins SA, Palanisamy N, Siddiqui J, et al. Antibody Based Detection of ERG Rearrangementsin Prostate Core Biopsies, Including Diagnostically Challenging Cases: ERG Staining in Prostate Core Biopsies. Arch Pathol Lab Med. 2012;136:935–946. doi: 10.5858/arpa.2011-0424-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braun M, Goltz D, Shaikhibrahim Z, et al. ERG protein expression and genomic rearrangement status in primary and metastatic prostate cancer-a comparative study of two monoclonal antibodies. Prostate Cancer Prostatic Dis. 2012 doi: 10.1038/pcan.2011.67. [DOI] [PubMed] [Google Scholar]
- 47.Yaskiv O, Zhang X, Simmerman K, et al. The Utility of ERG/P63 Double Immunohistochemical Staining in the Diagnosis of Limited Cancer in Prostate Needle Biopsies. Am J Surg Pathol. 2011;35:1062–1068. doi: 10.1097/PAS.0b013e318215cc03. [DOI] [PubMed] [Google Scholar]
- 48.van Leenders GJ, Boormans JL, Vissers CJ, et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Mod Pathol. 2011;24:1128–1138. doi: 10.1038/modpathol.2011.65. [DOI] [PubMed] [Google Scholar]
- 49.Minner S, Enodien M, Sirma H, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–5888. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 50.Hoogland AM, Jenster G, van Weerden WM, et al. ERG immunohistochemistry is not predictive for PSA recurrence, local recurrence or overall survival after radical prostatectomy for prostate cancer. Mod Pathol. 2011 doi: 10.1038/modpathol.2011.176. [DOI] [PubMed] [Google Scholar]
- 51.He H, Magi-Galluzzi C, Li J, et al. The diagnostic utility of novel immunohistochemical marker ERG in the workup of prostate biopsies with “atypical glands suspicious for cancer”. Am J Surg Pathol. 2011;35:608–614. doi: 10.1097/PAS.0b013e31820bcd2d. [DOI] [PubMed] [Google Scholar]
- 52.Falzarano SM, Zhou M, Carver P, et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. VirchowsArch. 2011;459:441–447. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 53.Park K, Tomlins SA, Mudaliar KM, et al. Antibody-Based Detection of ERG Rearrangement-Positive Prostate Cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohamed AA, Tan SH, Mikhalkevich N, et al. Ets family protein, erg expression in developing and adult mouse tissues by a highly specific monoclonal antibody. J Cancer. 2010;1:197–208. doi: 10.7150/jca.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young A, Palanisamy N, Siddiqui J, et al. Correlation of Urine TMPRSS2:ERG and PCA3 to ERG+ and Total Prostate Cancer Burden. Am J Clin Pathol. 2012;138:685–696. doi: 10.1309/AJCPU7PPWUPYG8OH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miettinen M, Wang ZF, Paetau A, et al. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am J Surg Pathol. 2011;35:432–441. doi: 10.1097/PAS.0b013e318206b67b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosen P, Sesterhenn IA, Brassell SA, et al. Clinical potential of the ERG oncoprotein in prostate cancer. Nat Rev Urol. 2012 doi: 10.1038/nrurol.2012.10. [DOI] [PubMed] [Google Scholar]
- 58.Lucas DR, Bentley G, Dan ME, et al. Ewing sarcoma vs lymphoblastic lymphoma. A comparative immunohistochemical study. Am J Clin Pathol. 2001;115:11–17. doi: 10.1309/K1XJ-6CXR-BQQU-V255. [DOI] [PubMed] [Google Scholar]
- 59.Ozdemirli M, Fanburg-Smith JC, Hartmann DP, et al. Differentiating lymphoblastic lymphoma and Ewing’s sarcoma: lymphocyte markers and gene rearrangement. Mod Pathol. 2001;14:1175–1182. doi: 10.1038/modpathol.3880455. [DOI] [PubMed] [Google Scholar]
- 60.Minner S, Luebke AM, Kluth M, et al. High level of Ets-related gene expression has high specificity for prostate cancer: a tissue microarray study of 11 483 cancers. Histopathology. 2012 doi: 10.1111/j.1365-2559.2012.04240.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang WL, Patel NR, Caragea M, et al. Expression of ERG, an Ets family transcription factor, identifies ERG-rearranged Ewing sarcoma. Mod Pathol. 2012;25:1378–1383. doi: 10.1038/modpathol.2012.97. [DOI] [PubMed] [Google Scholar]
- 62.Yaskiv O, Rubin BP, He H, et al. ERG Protein Expression in Human Tumors Detected With a Rabbit Monoclonal Antibody. Am J Clin Pathol. 2012;138:803–810. doi: 10.1309/AJCP3K5VUFALZTKC. [DOI] [PubMed] [Google Scholar]