Abstract
Background and Aims Hypericum perforatum
(St. John's wort) is a widespread Eurasian perennial plant species with remarkable variation in its morphology, ploidy and breeding system, which ranges from sex to apomixis. Here, hypotheses on the evolutionary origin of St. John's wort are tested and contrasted with the subsequent history of interspecific gene flow.
Methods
Extensive field collections were analysed for quantitative morphological variation, ploidy, chromosome numbers and genetic diversity using nuclear (amplified fragment length polymorphism) and plastid (trnL-trnF) markers. The mode of reproduction was analysed by FCSS (flow cytometric seed screen).
Key Results
It is demonstrated that H. perforatum is not of hybrid origin, and for the first time wild diploid populations are documented. Pseudogamous facultative apomictic reproduction is prevalent in the polyploids, whereas diploids are predominantly sexual, a phenomenon which also characterizes its sister species H. maculatum. Both molecular markers characterize identical major gene pools, distinguishing H. perforatum from H. maculatum and two genetic groups in H. perforatum. All three gene pools are in close geographical contact. Extensive gene flow and hybridization throughout Europe within and between gene pools and species is exemplified by the molecular data and confirmed by morphometric analyses.
Conclusions Hypericum perforatum
is of a single evolutionary origin and later split into two major gene pools. Subsequently, independent and recurrent polyploidization occurred in all lineages and was accompanied by substantial gene flow within and between H. perforatum and H. maculatum. These processes are highly influenced by the reproductive system in both species, with a switch to predominantly apomictic reproduction in polyploids, irrespective of their origin.
Keywords: Apomixis, evolution, hybridization, Hypericum perforatum, Hypericum maculatum, reproductive mode, St. John's wort
INTRODUCTION
Over the past 2 million years, climatic fluctuations have dramatically influenced Eurasian biota, with significant and repeated changes to environmental conditions occurring during Pleistocene glaciation and deglaciation cycles. Environmental changes may have been most dramatic in and around the glaciated regions, but the climatic fluctuations during the Pleistocene also had strong impacts on adjacent environments, causing drastic sea level variation to open or close migration routes (Randi, 2007), altitudinal shifts of vegetation belts (Parolly et al., 2010) and aridification processes (Rebernik et al., 2010). These have led to range shifts through contractions into refugia and subsequent survival and (re)colonization (Taberlet et al., 1998; Hewitt, 1999) for all species that escaped and migrated, survived and eventually re-colonized (Petit et al., 2001, 2003; Huck et al., 2009; Schmitt, 2009). These processes were often linked to local adaptation (e.g. Manel et al., 2010) and sometimes were followed by allopatric speciation or rapid and extensive radiation (Jordon-Thaden et al., 2010; Valente et al., 2010) and secondary contact of related species and subsequent hybridization (Schmickl and Koch, 2011; Kiefer and Koch, 2012).
Hypericum (Hypericaceae; Stevens, 2007; APG III, 2009) is a species-rich genus centred around the temperate regions of the Northern Hemisphere that underwent rapid radiation during the Pleistocene (Nürk et al., 2013). The genus consists of about 500 species of shrubs, herbs and a few trees (Nürk and Blattner, 2010), which have been grouped into 36 sections (Robson, 1977, 1981, 1987, 1990, 2001, 2010), although the relationships between sections remains speculative. Cladistic analysis of morphological characters (Nürk and Blattner, 2010) largely confirmed Robson's concepts, but found significant differences in the relationships between sections. However, a Palaearctic crown clade (core Hypericum) (sections 9, 9b, 9d, 9e, 10–19, 23, 24, 26 and 27 according to the nomenclature of Robson and summarized in Nürk et al., 2013) has been characterized based on phylogenetic analysis (Nürk et al., 2013), and this largely corresponds to the morphologically defined ‘Euhypericum’ group, including >45 % of the diversity of Hypericum and 207 species, most of which are native to the Old World. They are characterized by the possession of dark glands, the dominance of a herbaceous habit, and stamen arrangement in a 2 + 2 + 1 configuration.
Hypericum perforatum and H. maculatum are representatives of these widespread Eurasian herbaceous perennials in the Hypericum ‘core group’. In the large and phylogenetically unresolved Hypericum ‘core group’, these species form a well-defined clade of four species with European H. undulatum and H. tetrapterum. Hypericum perforatum is distributed across Europe and consists of four subspecies (Robson, 2002, 2003): the widespread and worldwide invasive subsp. perforatum, subsp. songaricum (predominantly Asian), subsp. veronense (from Asia to the Mediterranean and Macaronesia with records from central France and southern Germany) and subsp. chinenese (China) (Robson, 2003). In general the species occur in open habitats, with a strong preference for dry and warm sites in various types of grasslands, ruderal sites and disturbed habitats (e.g. along roads and railways). Wild populations of H. perforatum are composed of polyploid pseudogamous facultative apomicts (e.g. Matzk et al., 2001), and only speculative evidence suggests that putative diploid populations from natural stands are secondarily evolved dihaploids or amphihaploids derived from polyploid apomicts (Matzk et al., 2001; Robson, 2002). Consequently, H. perforatum is assumed to have had a hybrid origin. Morphologically the closest species to H. perforatum are H. maculatum and H. attenuatum. Hypericum maculatum and H. attenuatum have overlapping distributions in western Siberia, and, in contrast to H. perforatum, diploid populations of both species have been recorded from the wild (Robson, 2002). Hypericum maculatum is thought to comprise two diploid subspecies (subsp. maculatum and subsp. immaculatum) and a tetraploid subspecies (subsp. obtusiusculum), which is hypothesized to be autotetraploid as evidenced by chromosome morphology and synthesis by colchicine doubling of subsp. maculatum (Robson, 1958). Hypericum maculatum subsp. immaculatum, which was assumed to be morphologically closest to H. perforatum, is only found in the Balkans. Presently H. maculatum subsp. maculatum and tetraploid facultative apomictic H. perforatum populations are distributed across Europe (Matzk et al., 2003). Hypericum attenuatum has an Asian distribution and occurs only in Russia (eastern Siberia and the Far East), China and Mongolia (Robson, 2002). Based on morphology, H. attenuatum is most closely related to H. maculatum, only differing from that species by the acute sepals. The latter species has been considered as the most likely parental source of H. perforatum (together with H. maculatum), but this is not supported by nuclear-encoded DNA sequence variation (Nürk et al., 2013) demonstrating that there is no close relationship between H. maculatum and H. attenuatum. This species is a diploid perennial herb distributed in fields, pastures, steppes, and grassy and dry stony slopes up to an elevation of 2000 m a.s.l. (China).
Although extensive morphological, anatomical and cytogenetic data have enabled phylogenetic inference at the genus level, little is known about the evolutionary history of individual species or species groups. The evolutionary scenarios of some species, including H. perforatum, are highly complicated due to ongoing hybridization between taxa. For example, morphologically defined hybrids between H. perforatum and H. maculatum have been described and can be broadly summarized under the taxon Hypericum × desetangsii (Robson, 2002). However, their putative hybrid status has never been tested using multivariate, morphometric analyses or molecular markers.
Although there is a broad interest in H. perforatum and its closest relatives, little is known regarding systematic relationships and evolutionary history, the distribution of genetic variation and intra- and interspecific introgression. The aims of this work were thus to shed light upon these aspects of Hypericum evolution. Quantitative and qualitative morphological variation was used to identify taxonomic units independently from nuclear [amplified fragment length polymorphism (AFLPs)] and plastid (DNA sequencing) molecular genetic markers. As hybridization and phylogenetic reticulation have led to the establishment of various polyploids, we contrasted these data with ploidy estimates and detailed analyses of the reproductive mode.
Using these different approaches and considering also its closest relative H. maculatum, we have investigated: (1) the identity and origin of H. perforatum with regard to putative hybrid origination scenarios; (2) the distribution of genetic variation within and between H. perforatum and H. maculatum; and (3) the distribution and ancestral state of sexual and apomictic reproduction within and among genotypes. These insights are used here to present an evolutionary and phylogeographic scenario for the origin of H. perforatum. Finally, we discuss the impact of gene flow within and between species with respect to reproductive variability (e.g. sex vs. apomixis) and identify the importance of this flexible breeding system for the stability of asexual lineages over time.
MATERIALS AND METHODS
Considering the difficulty in distinguishing Hypericum perforatum, H. maculatum and their putative hybrids, and the lack of any comprehensive morphometric study, a morphological analysis was performed on 150 populations from both species. This data set was established to (1) test for the influence of ploidy on morphological variation; (2) analyse the influence of differing growing conditions on diagnostic characters; and (3) identify a set of individuals and populations for H. perforatum and H. maculatum for subsequent molecular and cytogenetic analyses.
Current taxonomic concepts not only describe different subspecies for both taxa, but also provide names for hybrids between H. maculatum and H. perforatum (e.g. broadly summarized in past literature as H. × desetangsii). However, here we do not aim to provide taxonomic names for the different morphologically defined groups of accessions at the subspecies level (or hybrid level) we analysed. Rather, based on the results of the 150 population data set, a sub-set of populations and individuals was subsequently chosen for detailed molecular genetic and cytogenetic analyses (195 individuals in total) to cover the distribution ranges of both species and all ploidies in Europe.
Plant material and morphometric analysis
For H. perforatum and H. maculatum, a total of 150 populations were analysed (476 individuals in total, Supplementary Data Table S1, Supplementary Data Fig. S1). An additional eight accessions (H. attenuatum, H. erectum, H. gracillimum, H. kamtschaticum, H. oliganthum, H. pibairense, H. tetrapterum and H. undulatum) were added for plastid DNA outgroup analysis. The outgroup selection followed an extensive phylogenetic survey which demonstrated that H. perforatum, H. maculatum, H. tetrapterum and H. undulatum are combined in a single clade (Nürk et al., 2013). The remaining four outgroup species are from different and more distinct clades, all within the monophyletic ‘core Hypericum’ (Nürk et al., 2013).
Furthermore, for all individuals collected in the wild and for which seed material was available from the original mother plants, offspring grown in a common garden experiment at the Botanical Gardens in Heidelberg (257 individuals in total) were analysed. This material was used to test the principal discriminatory power of the selected morphological characters, in addition to character variability (or lack of) under different environmental conditions (original habitat vs. common garden experiment) thus to confirm the general reliability of the original morphomatrix. A sub-set of 195 individuals of this plant material was used for the molecular (AFLPs and plastid DNA) and cytogenetic analyses (Supplementary Data Tables S1 and S5).
All morphological analyses were performed with dried herbarium vouchers deposited at the herbarium at Heidelberg Botanic Garden (HEID) and at the Munich Staatssammlung (M). Measurements were made using a high-power binocular microscope to measure dimensions on digital live images at high resolution. The removal of some plant organs (e.g. flowers and flower buds) from the original voucher was necessary for appropriate analysis. Here, 100 µL of a pre-washing solution (washing-up liquid, 0·1 %) was added to the removed plant organ, and further covered with boiling water (total volume 10 mL) for 2 min. The organs were then dissected on a glass slide and, after removal of the water, the samples were fixed and mounted on blotting paper and finally dried for 2 d at 40 °C. In total, 28 characters (continuous, discrete and multistate) were scored (Supplementary Data Table S2) from leaves, stem, flowers and capsules, which have been used in various determination keys to discriminate species and subspecies of H. perforatum and H. maculatum. The coding of character states did not imply or reflect any evolutionary progression (Möller et al., 2007). From these 28 characters, four characters were removed, because of a lack of variation, and from the remaining 24 characters some were combined and translated into a new single character. A final matrix of 18 variables was extracted (Supplementary Data Table S3).
A principal co-ordinate analysis (PCoA) was run to analyse the multivariate data using single individuals as OTUs (operational taxonomic units) with SPSS (version 16·0·1 for Windows). SPSS was also used to compare other variables, such as ploidy, growing conditions (wild origin vs. common garden experiment), genetic differentiation and taxon designation, with the morphometric data. PCoA requires that continuous character states are normally distributed, and, as our tests demonstrated that no character showed significant skewness, data transformation was not necessary.
In order to derive morphology-based indices of intermediacy between H. maculatum and H. perforatum for single individuals with genetic and cytogenetic data available, we conducted multiple discriminate analyses [MDA; also called canonical discriminant analysis (CDA)]. Diploid and genetically ‘pure’ (see AFLP and plastid DNA data) individuals from H. perforatum and H. maculatum served as reference groups of individuals indicative for the most divergent and putatively parental morphotypes. The intermediacy index for each individual analysed genetically was calculated from the two-dimensional MDA bar plot and in comparison with the putatively parental diploid morphotypes, placing the value of the individual to be tested on a re-scaled distance (0 to 1) between the mean values of diploid H. perforatum (left border) and H. maculatum (right border) reference individuals. Consequently, the intermediacy index varies between 0 (pure H. perforatum) and 1 (pure H. maculatum).
AFLP analysis
Amplified fragment length polymorphism determinations were conducted with 195 individuals, a representative sub-set of populations and individuals screened for morphological variation (Supplementary Data Table S5; Fig. 3). Total genomic DNA was extracted from 50–75 mg of dried leaf tissue following the procedure of Doyle and Doyle (1987) with minor modifications (Gong et al., 2008). Leaf tissue was ground in a Precellys 24 (Bertin Technologies) homogenizer. The DNA pellet was washed twice with 70 % ethanol and dissolved in 50 µL of TE buffer. Per reaction, 2 U of RNase A were added and incubated at 37 °C for 1 h. The concentrations of the samples were measured using a NanoDrop ND-1000 spectrophotometer, and each sample was diluted with ddH2O to a final DNA concentration of 100 ng μL−1.
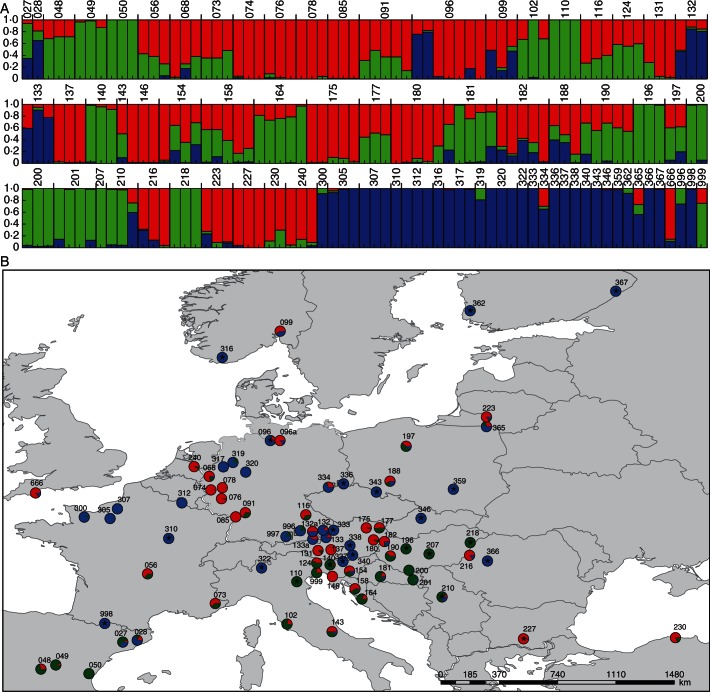
Fig. 3.
(A) Structure analysis of the AFLP data of single individuals (optimal K = 3; n = 195). The population code is given above the bars. The colour code corresponds to the plastid DNA data and distinguishes between Hypericum maculatum and two groups within H. perforatum (blue = H. maculatum; red and green = H. perforatum; Fig. 1). (B) Distribution map of populations. Pie charts combine the data from the individuals on the populational level. Asterisks (*) indicate diploid populations. Colour code follows (A) and Fig. 1 (blue = H. maculatum; red and green = H. perforatum). Additional populations analysed for morphometrics are shown in Supplementary Data Fig. S1.
The AFLP analysis was performed according to Vos et al. (1995) and modified by Gong et al. (2008). The analysis was carried out using EcoRI-A and MseI-C as pre-selective primers, followed by three selective primer combinations (Percifield et al., 2007): EcoRI-AAC (FAM)/MseI-CAC, EcoRI-ACG (TET)/MseI-CAC and EcoRI-AGC (HEX)/MseI-CAA. The three differentially fluorescence-labelled primer pairs were multiplexed (2 µL of TET, 2 µL of FAM and 6 µL of HEX) and diluted 30 times with ddH2O, of which 1 µL was taken and mixed with 0·2 µL of ET-ROX 550 size standard and 5·8 µL of ddH2O. After 2 min denaturation at 95 °C, samples were run on a 48-capillary MegaBase 500 automated sequencer (Amersham Biosciences). Raw data were scored and exported as a presence/absence matrix using Genemarker v1·6 (SoftGenetics LLC). In order to maximize the reliability of the data set, scored fragment size was restricted from 60 to 350 nucleotides. In each sequencer run, one standard sample was placed in two plate positions (to check for consistency within and among different runs) and a negative control was run in a single position.
Plastid DNA sequence variation
All individuals used for the AFLP analysis were assessed for plastid haplotype variation. A potential complicating factor in Hypericum is occasional biparental plastid inheritance (reviewed in Harris and Ingram, 1991), and hence plastid DNA variation is not indicative of maternal gene flow only. After an initial screening of various plastid regions (trnL intron, trnL-trnF intergenic spacer, trnF, trnH, trnH-psbA intergenic spacer, psbA, trnC, trnC-ycf6 intergenic spacer, ycf6, psbA, psbA-matK intergenic spacer, matK, rps16 intron, trnT, trnT-trnL intergenic spacer, trnL, trnS, trnS-psbZ intergenic spacer, psbZ, psbZ-trnG intergenic spacer, trnG, rpoC1), three DNA regions were chosen which provided measureable levels of DNA sequence variation: psbB-psbH (psbB, psbT, psbT-psbN intergenic spacer, psbN, psbN-psbH intergenic spacer), the trnL intron and trnS-ycf9 (trnS, trnS-psbZ intergenic spacer, psbZ, ycf9).
For DNA amplification, the following primer pairs were used, each of which containined a 5′-M13 sequencing primer sequence (indicated in lower case letters): psbB-psbH, 5′-gcatgttttcccagtcacgacTCCAAAAANKKGGAGATCCAAC-3′ (forward psbB)-5′-acttcaggaaacagctatgacTCAAYRGTYTGTGTAGCCAT-3′ (reverse psbH); trnL intron, 5′-gcatgttttcccagtcacgacCGAAATCGGTAGACGCTACG-3′ (forward trnL-L)-5′-acttcaggaaacagctatgacGGGGATAGAGGGACTTGAAC-3′ (reverse trnL-L), 5′-gcatgttttcccagtcacgacGAGAGAGAGGGATTCGAACC-3′; trnS; (forward trnS uga)-5′-acttcaggaaacagctatgacCAAAMACAGCCAATTGGAAAGC-3′ (reverse ycf9P) (Taberlet et al., 1991; Shaw et al., 2005; Heinze, 2007, with only minor modifications). The amplifications were performed on a PTC 200 Peltier Thermal Cycler (MJ Research) using the following conditions: 2 min initial denaturation at 95 °C, 30 cycles of amplification with 20 s at 95 °C, 45 s at 61 °C and 1 min at 72 °C, and 10 s of final elongation at 72 °C. PCR success was checked with electrophoresis in a 1 % agarose gel in TAE buffer and stained with GelRed (Biotium Inc., Hayward, CA, USA). PCR product clean-up was carried out with a NucleoFast Kit (Macherey-Nagel, Germany). Sequencing was performed by GATC GmbH (Konstanz, Germany) using an M13 sequencing primer (forward, 5′-GCATGTTTTCCCAGTCACGAC-3′; reverse, 5′-ACTTCAGGAAACAGCTATGAC-3′). Sequences were checked and trimmed using DNASTAR Lasergene (GATC Biotech, Konstanz, Germany).
Cytological and cytogenetic analysis
Ploidy estimates were based on either flow cytometric genome size measurements or direct counts of chromosome numbers from root tips. For karyotyping, root tips were collected from plants cultivated in the Botanical Garden and immediately put in 2 mm 8-hydroxychinolin for 2–3 h at room temperature, followed by fixation in ethanol–glacial acetic acid (3:1) (Carnoy fixative). For long-term storage at –20 °C, fixed root tips were finally transferred to 70 % ethanol. Before cell spread preparation, root tips were washed in water followed by enzymatic digestion (0·1 % cytohelicase and 0·1 % pectohelicase in 0·1 % cellulase-citrate buffer) for 90 min at 37 °C. The cell suspension was mixed on a microscope slide with one drop of 45 % acetic acid for 1–2 min at 45 °C on a hot plate. For fixation, a ring of Carnoy fixative was dropped on the slide and left to dry on the hot plate. Objects were finally stained with Vectashield (DAPI solution, Axxora, Germany) and sealed with clear nail varnish for permanent use. Chromosome counts were determined and documented as digital images.
For all cultivated accessions, genome size estimates were performed using fresh leaf material to determine ploidy independently. Small amounts of leaf tissue of a sample and internal standards were chopped together in 0·5 mL of extraction buffer (Partec CyStain PI kit, Partec, Münster) and filtered. The samples were then stained by adding 2 mL of Partec CyStain PI solution containing DNase-free RNase following the manufacturer's instructions (Partec) and incubated for 4 h at 4 °C in the dark. Analyses were performed with a CyFlow Space (Partec) flow cytometer (green laser with 532 nm), and data evaluation was carried out using the FloMax analysis program (Partec). Solanum lycopersicum ‘Stupicke’ (1·48 pg/1C; Doležel et al., 1992) and Zea mays ‘CE-777’ (2·71 pg/1C; Lysak and Doležel, 1998) were used as primary standards, and the following Hypericum accessions were chosen as external standards: diploid Hyp1410 and tetraploid Hyp1517 (chromosome numbers were counted). Based upon published data, the expected value for H. perforatum was 0·325 pg/1Cx (Bennett and Leitch, 2010).
The reproductive mode for 68 accessions with sufficient seed material available was analysed using the flow cytometric seed screen (FCSS; Matzk et al., 2000, 2001, 2003), in which embryo and endosperm ploidy are compared with the known ploidy of the mother plant. In a normally reproducing sexual plant the ploidy ratio of embryo to endosperm is 2:3 and any deviation from 2:3 can be interpreted as non-sexual (e.g. apomictic) reproduction. These 68 analysed samples represent diploid and polyploid accessions of all identified genetic clusters and hybrid combinations identified by molecular analyses. The analyses followed the protocol as described by Matzk et al. (2003) and used the diploid accession Hyp1410 as external standard and reference to calculate relative C-values. For any of the analysed accessions, 24 randomly selected seeds per individual were chosen, cut and incubated individually with 0·5 mL of extraction buffer, and prepared as described above.
Analysis of AFLP and plastid DNA sequence data
The error rate of AFLP genotyping was calculated following the procedure described by Bonin et al. (2004), whereby fragments occurring with a frequency <1·5 % were excluded from the data set. Ten individuals were selected randomly for replication and calculation of the error rate. The AFLP data were analysed using a PCoA running SYN-TAX 2000 (Exeter software, Setauket, NY, USA) and visualized with SPSS 16·0·1.
The genetic structure of the data set was additionally examined by genetic admixture analysis using the program STRUCTURE vers. 2·3·3 (Pritchard et al., 2000; Falush et al., 2007). The data were analysed with STRUCTURE, with K ranging from 2 to 10, with ten replicate runs for each K, and a burn-in period of 1 × 105 and 1 × 106 iterations. The ‘admixture model’ was chosen combined with uncorrelated and with correlated allele frequencies. The likelihood of Ks ranging from 1 to 10 was calculated using the R-script Structure-sum (Ehrich, 2006), which compares the likelihood of the runs (Rosenberg et al., 2002), the similarity coefficient between the runs, and delta K as defined by Evanno et al. (2005). The optimal K was chosen from the model with the highest probability. In the visualization of Evanno's delta K, a peak has to appear in the optimal fitting model and the results over multiple runs had to be consistent (Evanno et al., 2005; Ehrich et al., 2007).
For plastid data and network analysis, several outgroup taxa composed of a representative set of the closest relatives of H. perforatum and H. maculatum were included for the DNA sequence analysis (H. tetrapterum, H. attenuatum, H. pibairense, H. erectum, H. kamtschaticum, H. gracillimum, H. undulatum and H. oliganthum). The alignment of the plastid DNA sequences was made manually, with subsequent adjustment in PhyDE version 0·9971 (Müller et al., 2005). Maximum parsimony analysis was performed running PAUP 4·0b10 (Swofford, 2002). The parsimony heuristic search was performed with the following settings: gaps were treated as missing data and, using the gap-based coded 0/1-matrix, multistate taxa were interpreted as uncertainty, tree construction was via stepwise addition, and with tree-bisection-reconnection (TBR) as the branch-swapping algorithm, MaxTrees limit was set to 10 000, and the MulTrees option was selected (saving all minimal trees found during branch swapping). For bootstrapping, 1000 replicates with a tree maximum of 500 retained trees were run.
A network analysis was performed using TCS v.1·21, with an acceptance limit set to 95 % and a connection limit of 17 steps (Clement et al., 2000). Gaps were coded separately and added to the final data matrix, and length variation in poly(A) or poly(T) regions was excluded from the analysis.
RESULTS
Morphology can distinguish H. maculatum and H. perforatum but not cytotypes
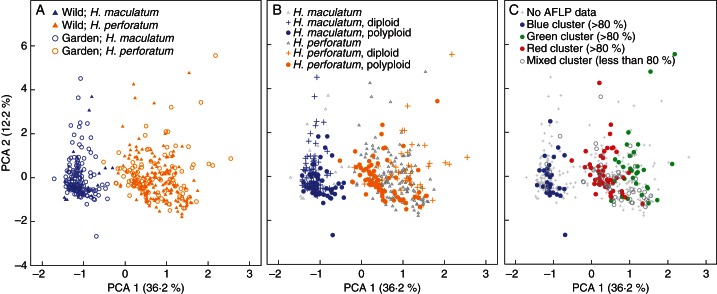
In the PCoA of the morphological data set of 150 populations/accessions (476 individuals), two major groups representing H. maculatum and H. perforatum could be distinguished with 48·4 % of the total variance (Fig. 1A–C). We found no significant difference when specimens cultivated in the Botanical Garden were compared with the corresponding data set of individuals collected directly in the wild (Fig. 1A). Based on the PCoA, we also observed no obvious and significant morphological differentiation associated with ploidy and cytotype (diploid vs. polyploid) for H. perforatum and for H. maculatum (Fig. 1B). Consequently morphology is a weak predictor for ploidy, and vice versa. This indicates that, on average, the characters measured are stable under environmentally different conditions.
Fig. 1.
Principal co-oordinate analysis (PCA) of the whole data set including 476 individuals (Supplementary Data Table S3) and comparing H. maculatum with H. perforatum. (A) Phenotypic constancy of morphological characters: evaluation of discriminating power of morphological characters comparing morphology-based characters obtained from individuals directly collected in the wild with the respective material cultivated in a common garden experiment at Heidelberg Botanical Garden. (B) Combination of the morphological differentiation with ploidy. Unknown cytotypes are also indicated. (C) Combination of the morphological differentiation and affiliation with one out of the three genetic clusters as revealed by AFLP analysis. A colour signature (blue = H. maculatum; green/red = H. perforatum) follows the results as summarized in Supplementary Data Table S5. A colour is assigned only if the respective proportion of the genetic variation (blue, red or green) contributes >80 %. The remaining individuals scored for AFLPs but with no contribution of any genetic cluster >80 % are shown by open circles.
Plastid DNA sequence variation is taxon specific
The plastid DNA data for the data set of 195 individuals with three accessions (for additional haplotypes) and outgroup individuals representing eight additional taxa were joined into an alignment of 1602 bp representing 21 haplotypes (alignment, Supplementary Data Fig. S2; GenBank accession codes, Supplementary Data Table S4). Parsimony analysis running PAUP revealed the 12 shortest trees each 85 steps in length, with consistency (CI) and retention (RI) indices of 0·66 and 0·87, respectively. In summary, 63 variable characters were identified, with 39 potentially parsimony informative characters (ten gaps, 29 nucleotide polymorphisms), and 24 parsimony uninformative characters (eight gaps, 16 nucleotide polymorphisms). In H. perforatum, 16 polymorphic characters were found which separated the species into ten haplotypes (HP01–HP10), whereas in H. maculatum 20 polymorphic characters were screened which separated the species into four haplotypes (HM01–HM04).
The parsimony network analysis running TCS was characterized by 90 mutational steps and clearly separated H. maculatum from H. perforatum plastid haplotypes (Fig. 2).
Fig. 2.
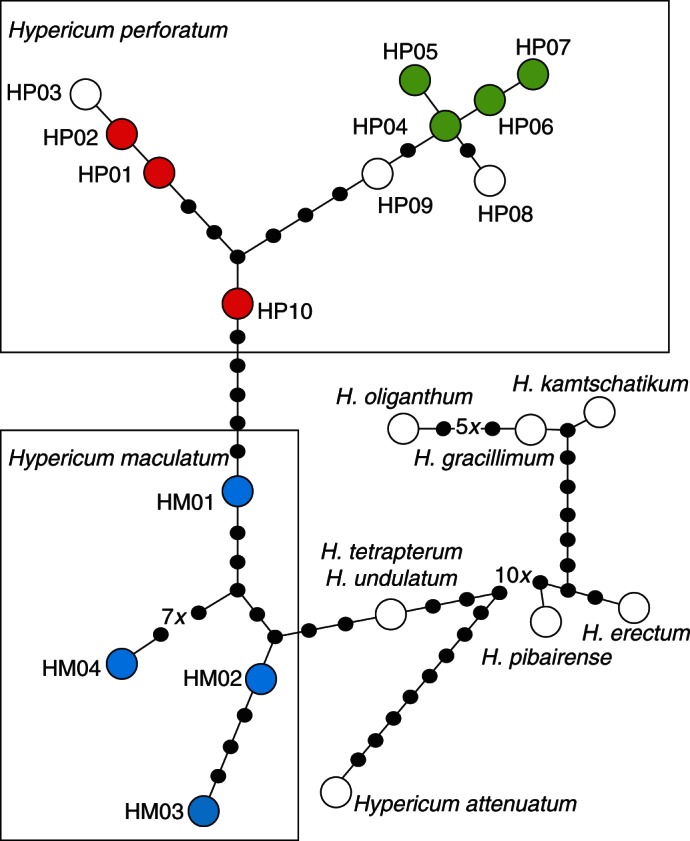
Plastid DNA parsimony network of Hypericum perforatum and H. maculatum, and including various outgroup taxa. Hypericum tetrapterum and H. undulatum accessions shared an identical plastid DNA haplotype. The different colour codes correspond to those used for AFLP gene pool recognition (blue = H. maculatum; red and green = H. perforatum). Haplotype coding (HP01–HP10, HM01–HM04; and outgroup taxa) and respective sequence accession data are provided in Supplementary Data Table S4.
Hypericum undulatum and H. tetrapterum, which are know to be phylogenetically closely related (Nürk et al., 2013), are indeed closely related to H. maculatum, and H. undulatum shares its haplotype with H. tetrapterum. All other outgroup taxa and their plastid DNA haplotypes are placed much more distantly to these four species (Fig. 2) in accordance with phylogenetic inference (Nürk et al., 2013).
Among H. perforatum haplotypes, two separate groups can be distinguished (Fig. 2): one ‘green cluster’ consists of six haplotypes (HP04–HP09) (with corresponding haplotypes listed in Supplementary Data Table S5). The second ‘red cluster’ consists of four haplotypes (HP01, HP02, HP03 and HP10). Haplotype HP10 is placed in an intermediate position, but is genetically closer to HP01 than to any other haplotype. Hypericum maculatum was characterized by four haplotypes (HM01–HM04) with a similar level of genetic variation to those of the single H. perforatum clusters. All remaining species, including the closely related H. tetrapterum, H. undulatum and H. attenuatum, are well separated.
AFLP and plastid DNA analysis detects three major gene pools
After deleting invariable fragments and those occurring in fewer than three individuals, 408 fragments were scored (Table 1), with an error rate of 3·1 % (Bonin et al., 2004; 16 differences were observed in 604 total phenotype comparisons). The mean number of bands differed significantly (P < 0·01) between compared sets of individuals; in summary, polyploids carried slightly more fragments than diploids, and in total H. perforatum displayed more fragments than H. maculatum (Table 1).
Table 1.
Summary table of the number of AFLP fragments scored when dividing the total sample (195 individuals, Supplementary Data Table S5) into different subgroups (according to taxonomy, considering ploidy or AFLP/plastid DNA gene pool designation)
| No. of bands (s.d.) | |
|---|---|
| Species comparisons (no. of individuals) | |
| H. perforatum (156) vs. H. maculatum (39) | 124 (23) vs. 94 (18) |
| Within-species comparisons | |
| H. maculatum diploids (13) vs. polyploids (21) | 82 (12) vs. 105 (11) |
| H. perforatum diploids (20) vs. polyploids (63) | 112 (15) vs. 132 (23) |
| Genepool comparisons | |
| Polyploids: ‘red cluster’ (45) vs. ‘green cluster’ (11) | 140 (18) vs. 120 (24) |
| Diploids: ‘red cluster’ (3) vs. ‘green cluster’ (15) | 122 (4) vs. 110 (16) |
For the structure analysis, K = 3 was more optimal than K = 4 considering the highest mean delta K in the run with correlated allele frequencies, similarity coefficient and lnP(D), whereas the highest mean delta K in the independent (uncorrelated) allele model was K = 4 (Supplementary Data Fig. S3). K = 3 was chosen because it showed the most consistent data over all runs with both models, and results from the structure analysis are shown in Fig. 3. A comparable result was obtained after a PCoA of the same data (data not shown).
The structure analysis distinguished three major genetic clusters, with the blue cluster typically characterizing H. maculatum, and the red and green clusters characterizing different gene pools of H. perforatum (Fig. 3; Supplementary Data Table S5). The geographical distribution of the different gene pools of H. perforatum demonstrates that Central Europe is divided, with a red gene pool primarily found in the north and a green gene pool found primarily in the south (Fig. 3).
Further correlations between morphological identity and genetic variation provided more insights into the genetic architecture of these gene pools, as follows.
(1) In general there is significant morphological differentiation at the species level correlated with these three gene pools, and in Fig. 2C all individuals with >80 % genetic variation attributable to one of the three gene pools are indicated.
(2) If we assume that definitely all individuals characterized by a ‘H. maculatum’ AFLP gene pool, but carrying a H. perforatum plastid haplotype (and vice versa), are hybrids and introgressed individuals, we could also expect some intermediate phenotype for these individuals (CDA; Supplementary Data Table S5). Of the 20 putative hybrids or introgressed individuals between H. maculatum and H. perforatum (based on AFLP gene pools compared with species-specific plastid DNA haplotypes), only one individual from population 099 showed an intermediacy index of 0·5, which one might expect from a recent hybrid origin between both species. Conversely, a significant number of H. perforatum individuals (nine individuals) with no genetic signature of hybridization or introgression exhibited an intermediacy index of around 0·5. Consequently, we further tested if there was a correlation between the green and red AFLP gene pools of genetically pure H. perforatum and the CDA intermediacy value using all remaining H. perforatum (with H. maculatum and its hybrids also excluded). Since the ratio of green and red gene pools within a single individual varied, we tested two different data selections of >90 and >75 % (representation of the respective gene pool in a single individual), respectively. Comparing all individuals with >90 % of the genetic diversity assigned to one of the two gene pools (52 individuals) revealed significant differentiation according to CDA and morphology, a result which was also found at the 75 % level for 69 individuals (P < 0·005 in both cases; comparing the CDA values with the respective gene pool association using a t-test after confirmation of normality of the sample means using a Kolmogorov–Smirnov test using SPSS). This clearly demonstrates that the two gene pools of H. perforatum are also correlated with morphotypes if obvious hybrids between H. maculatum and H. perforatum are excluded. This result is in accordance with the finding shown in Fig. 2C.
(3) The above-described data also indicate high levels of gene flow between the two H. perforatum gene pools, since 59 and 42 H. perforatum individuals (54 and 38 %) could not be assigned to one of the two gene pools at the 90 and 75 % levels, respectively.
Considering the north–south distribution of the H. perforatum AFLP gene pool (Fig. 3), we propose that the genetic and morphological distinctness is reflective of two taxa within a broadly defined H. perforatum.
Cytogenetic analysis identifies ancestral diploids with sexual reproduction
Ploidy estimations were primarily based on flow cytometry, although in some cases these were independently confirmed by chromosome counts (Supplementary Data Tables S1 and S5). Importantly, we identified eight diploid H. perforatum accessions and, with one exception, all diploids were found in southern populations in the ‘green AFLP gene pool’ and had plastid DNA haplotypes from the ‘green haplotype cluster’. These are the first reports of diploid H. perforatum from wild populations. The only exception is population 227 from Bulgaria, which is from the ‘red AFLP gene pool’ and has plastid DNA haplotype HP02 from the ‘red haplotype cluster’. These observations lead to the conclusion that for both identified H. perforatum gene pools and morphotypes, there is also an ancestral diploid cytotype. Similarly, in H. maculatum, 32 diploid accessions were detected, in addition to 15 polyploid accessions with a contribution from all three main genetic clusters. Hybrids of any combination were always identified as polyploids, with the exception of two diploid H. maculatum × H. perforatum individuals (found in populations 334 and 220).
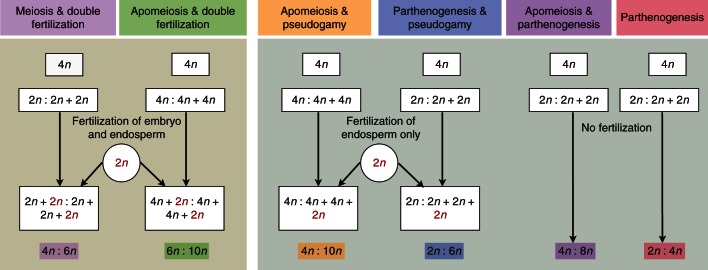
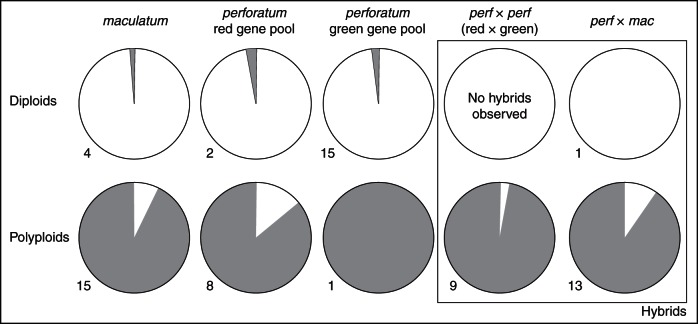
A representative subset of 68 individuals (diploid H. perforatum and H. maculatum from all three major AFLP gene pools, polyploid H. perforatum and H. maculatum and combinations of mixed AFLP gene pools containing H. maculatum × H. perforatum, H. perforatum × H. perforatum) were analysed for their reproductive behaviour using the flow cytometric seed screen, and show a complex and highly polymorphic pattern. The original measurements and the interpretation of the endosperm:embryo C-value ratios are provided in Supplementary Data Table S6. A detailed interpretation, indicating a complex mix of apomeiosis, apomixis, parthenogenesis and pseudogamy, is shown in Fig. 4. From the initial 1632 measurements (68 accessions × 24 seeds), 532 measurements provided no C-value ratio, an expected result considering that Hypericum often produces seeds without endosperm (Robson, 2002). In summary, it is shown that the reproductive mode in H. maculatum is similar to that in H. perforatum. For the purpose of this study, the results (Supplementary Data Table S6) were scored to differentiate between fully sexual and asexual reproduction only; they indicate a clear dichotomy between almost exclusively sexual diploids and asexual polyploids, irrespective of hybrid origin (Fig. 5).
Fig. 4.
Diagram summarizing the various modes of sexual and non-sexual reproduction in Hypericum maculatum and H. perforatum, and expected ratios in DNA content (embryo:endosperm) as measured here with FCSS (flow cytometric seed screen). The diagram uses a tetraploid (4n) mother plant as an example. The male gamete is indicated in red if contributing to endosperm fertilization and/or egg cell fertilization. The colour code corresponds to the categorization of our FCSS data summarized in Supplementary Data Table S6.
Fig. 5.
The reproductive system of diploid and polyploid Hypericum perforatum, H. maculatum and hybrids. White, proportion of sexual reproduction; grey, proportion of any kind of asexual and apomictic seed production (see also Fig. 4). Diploids and polyploids are separated and further differentiated according to their genotypic designation (gene pool as indicated with AFLP and plastid DNA data, Figs 1 and 3; Supplementary Data Table S5). The numbers indicate the number of individuals investigated with seed screen analysis and providing endosperm:embryo DNA ratios.
DISCUSSION
Evolutionary origin of Hypericum perforatum
In recent decades several hypotheses concerning the evolutionary origin and history of H. perforatum have been put forward, including either an allopolyploid origin involving H. attenuatum and H. maculatum (Robson, 2002) or autopolyploidization from an ancestor closely related to diploid H. maculatum (Brutovská et al., 2000). Evidence for these contrasting hypotheses originates from (1) morphological data indicating similarities between H. perforatum, H. attenuatum and H. maculatum; and (2) cytogenetic, crossing and segregation analyses (Brutovská et al., 2000). Before our study, there had been no comprehensive survey of the distribution of diploids and polyploids in any of these almost exclusively morphologically defined species and subspecies (e.g. three H. maculatum and four H. perforatum subspecies; Robson, 2001). Furthermore, genetic and phylogenetic–systematic data are largely lacking or fragmental (e.g. Barcaccia et al., 2006), and karyological data are also largely missing for any hybrid combination (summarized in Robson, 2002). Finally, no systematic and broad-ranging quantitative morphological studies exist which have the power to discriminate between hybridization, introgression and reticulate evolution. In contrast, taxonomic work has established a series of taxa and hybrids within and between almost all subspecies of H. perforatum and H. maculatum; these have been broadly treated as H. × desetangsii or as distinct taxa (e.g. Robson, 2002). The molecular data presented here and in a recent study (Nürk et al., 2013) shed light on the hypothetical origins of H. perforatum. Nuclear-encoded internal transcribed spacer (ITS) sequence data showed that it is unlikely that H. attenuatum is involved in the evolution of H. perforatum, and this taxon is grouped into a distinct group of taxa outside the H. perforatum/maculatum/undulatum/tetrapterum clade (Nürk et al., 2013). Our corresponding plastid DNA haplotype network is in full agreement with this finding and also does not indicate any close relationship of H. attenuatum with any of these four taxa (Fig. 1), but it favours a sister relationship between H. perforatum and H. maculatum. Consequently, we could exclude an allopolyploid origin involving only distantly related taxa from Asia as supposed by Robson (2002). The autopolyploidization hypotheses (e.g. Brutovská et al., 2000) can also be rejected for various reasons: diploid wild populations have been characterized not only for H. maculatum but also for H. perforatum, and all can be attributed to separate gene pools revealed by AFLP analysis (Fig. 3). This correlates with plastid DNA variation, which is mostly maternally inherited, and provides clear evidence for ancestral and cladogenetic evolution of diploid H. perforatum, with H. maculatum as a sister group. Furthermore, polyploid H. perforatum accessions showed different signatures of origin, either autopolyploidization or secondary hybridization and introgression between all three detected gene pools.
Following its cladogenetic origin and sister relationship to H. maculatum, H. perforatum subsequently differentiated into at least two ‘cryptic’ ancestral diploid taxa, as is congruently shown by all data (AFLPs, plastid DNA, morphology) collected here (‘red’ vs. ‘green’ gene pools). Regardless of potential gene flow between these two H. perforatum taxa (or with H. maculatum), the two are similar to the description of the previously described subsp. perforatum (more northerly distributed) and subsp. veronense (more southerly distributed and extending to the Eastern Mediterranean and Asia). In general, subsp. veronense can be distinguished from subsp. perforatum by its sessile leaves (at least on the main stem) and the petal laminar glands usually being all pale. The taxa are in close contact and apparently hybridize frequently, as our AFLP and plastid DNA data indicate. Consequently, we suggest that an assessment encompassing genetics, cytology and morphology is required to assign H. perforatum accessions accurately to either of the H. perforatum subspecies. However, future studies are needed to study the relationships of these two European subspecies to the other two subspecies (subsp. songaricum concentrated in Asia, and subsp. chinenese from China; Robson, 2003).
The scenario is further complicated by the fact that additional hybridization and introgression with H. maculatum is easily possible, and the data here imply that this may not always be detectable at the morphological level. Taken together, these data suggest that past observations implying an allopolyploid origin of H. perforatum could alternatively be best explained by multiple secondary independent hybridization and introgression events within and between already existing taxa and gene pools.
Hybridization and polyploidization
The phylogenetic relationships among Central European Hypericum spp. have long been the subject of speculation. Recently, significantly more insight into these relationships has been provided by a comprehensive classification scheme (Robson, 2002) which has been largely confirmed by independent morphometric and molecular phylogenetic analyses (Nürk and Blattner, 2010; Nürk et al., 2013). According to morphological studies, various European species can be grouped into section Hypericum sub-section Hypericum, including H. maculatum and H. perforatum, European species (H. elegans, H. tetrapterum, H. triquetrifolium, H. undulatum) and non-European species (H. scouleri, H. yezoense, H. momoseanum, H. iwatelittorale, H. tosaense) (Robson, 2002). As H. perforatum and H. maculatum share a diploid chromosome number of 2n = 16 in addition to an equal genome size (H. perforatum 1C = 0·36 pg, SD 0·46 %; H. maculatum 1C = 0·36 pg, SD 1·8 %), we hypothesize that hybridization between the species is easily possible.
Other taxa, for example H. attenuatum, are placed in different sub-sections and series in section Hypericum (Robson, 2002). However, molecular phylogenetic analyses do not support these sub-sectional and series classifications and show relatively closer genetic relationships between H. perforatum, H. maculatum, H. tetrapterum and H. undulatum (Nürk et al., 2013). This is also well reflected by the various hybrids described which involve H. perforatum, as interspecific hybridization between Central European Hypericum spp. is frequently observed (Robson, 2002). Many putative hybrids have been described mostly based on morphology. For example, among the relatives of H. perforatum, putative parental hybridization partner species include H. maculatum, H. tetrapterum and H. undulatum, and hybrids between them have been described in various combinations (Robson, 2002). Among these described hybrids in the Central European flora, H. × desetangsii (H. perforatum × H. maculatum) is of special interest (Robson, 2002). Additional hybrids between H. perforatum and H. maculatum have been described in parallel as separate species, e.g. H. carpaticum (Mártonfi, 2001), following an apomictic species concept (Hörandl, 1998). It may be possible that indeed such apomictic ‘species’ can be identified as morphologically stable units, since our data obtained from the cultivation experiment showed that diagnostic morphological characters are not significantly influenced by environmental factors (Fig. 2A). Further natural hybridization with related H. tetrapterum (diploids only) has been described rarely [H. maculatum × H. tetrapterum = H. × laschii; Fröhlich (1911) and H. perforatum × H. tetrapterum = H. × medium; Robson (2002)]. For H. undulatum (diploids and tetraploids) hybrids are also known involving H. tetrapterum (Robson). The low frequency of these hybrids is also reflected in our study, since we were not able, for example, to detect any H. undulatum/H. tetrapterum plastid haplotype in our sampling of H. perforatum and H. maculatum and their various hybrids.
The presented data do not provide any detailed insights into past Pleistocene refuge areas, or where the three gene pools (H. maculatum and the two subspecies of H. perforatum) might have originated. However, diploid populations are found consistently with phylogeographic literature in three major European refuge areas, namely the Iberian Peninsula, Italy and the Balkans (Comes and Kadereit, 1998; Taberlet et. al., 1998).
Post-glacial range expansion, in combination with the invasive potential of H. perforatum (in particular the ‘red gene pool’), greatly increased the possibility of hybridization and introgression not only in evolutionarily but also in historical time scales spanning the last few hundred years.
Breeding system evolution and apomixis
The switch from sexual to asexual (i.e. apomictic) reproduction has been associated with hybridization and/or polyploidy in both plants and animals (Suomolainen, 1950; Richards, 2003). Here, we have identified that H. maculatum is composed mostly of diploids and a single gene pool, whereas H. perforatum is mostly composed of polyploids from two distinct gene pools. Gene flow is possible between all three gene pools, and hence both interspecific (i.e. between H. maculatum and H. perforatum) and intraspecific hybridization (i.e. between red and green H. perforatum) is evident (Fig. 3; Supplementary Data Fig. S5). Taken together, the data here show a stronger association between polyploidy and expression of apomixis than between hybridization (i.e. introgression) and apomixis. More recently identification and characterization of the APOSPORY locus in H. perforatum unravelled the genetic basis of apospory as part of the complex trait apomixes and highlighted a dosage dependency (and high correlation with ploidy) of the respective gene products (Schallau et al., 2010). This finding has also been confirmed independently by studying expression of apomixis in tetraploid compared with hexaploid H. perforatum and testing for differences in its colonization ability and invasiveness (A. P. Molins, J. M. Corral, O. M. Aliya, M. A. Koch, A. Landau, J. L. Maron and T. F. Sharbel, unpubl. res.).
On a broader phylogenetic scale, it would appear that apomixis is an ancestral trait which is older than the H. perforatum–maculatum dichotomy, as the trait is also found in many other species. The other two species of the clade, H. undulatum and H. tetrapterum, have been shown to comprise sexual diploids (Matzk et al., 2003), but various other polyploid members of ‘core Hypericum’ such as H. kamschaticum or H. yezoense, are also facultative apomicts (Matzk et al., 2003), whereas other diploids (e.g. H. elegans and H. erectum) turned out to be obligate sexual.
Aposporous apomixis is variable in Hypericum, on both the individual and embryological levels (Galla et al., 2011), and the level of developmental and molecular genetic (or epigenetic) homology for this trait across Hypericum is unclear. Assuming a single ancestral origin of an apomixis factor in Hypericum, it is furthermore unknown why the trait is manifested in some species and not in others. Hence, the data presented here support gene dosage as the factor associated with apomixis expression, whereas the distribution of apomixis on a broader scale is consistent with a genetic background effect (A. P. Molins, J. M. Corral, O. M. Aliya, M. A. Koch, A. Landau, J. L. Maron and T. F. Sharbel, unpubl. res.). A similar evolutionary scenario on a continental scale has been demonstrated for North American Boechera (Brassicaceae) (Kiefer and Koch, 2012), although the type and genetic basis of agamospermy are different (Schranz et al., 2005; Dobes et al., 2007; Kiefer et al., 2009).
The biogeographical data presented here are inconclusive with respect to the advantages of polyploidy, hybridization and reproductive mode for the post-glacial recolonization patterns exhibited by H. perforatum and H. maculatum across Europe. Although asexuality and/or polyploidy could have resulted in more efficient recolonization properties (Hörandl, 2009) in ancestral Hypericum from southern glacial refugia into northern Europe, the tight correlation between apomixis and polyploidy confounds our ability to differentiate between their relative advantages based upon the data presented here.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Xiaorui Sun for the collaboration on morphological investigations; the gardening team of the Botanical Garden Heidelberg, and in particular Torsten Jakob, for continuous support with plant cultivation; Susanne Ball for her substantial help in the laboratory and handling all the material and germplasm collections; and Peter Sack, who organized and helped with the voucher exchange. We also appreciate a critical discussion and many comments on the final draft version provided by Graham Muir. Finally we thank the curators of Munich (M, MSU), Tokyo (TOK), Frank Blattner and Nikolai Nürk (GAT), and Marta Puente Molins (IPK Gatersleben) and John Maron (University of Montana) for providing additional vouchers. This work was supported by DFG research framework HYPEVOL and respective grants to M.A.K. (KO2302/7) and T.F.S. (SH 337/1-1).
LITERATURE CITED
- APG III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Barcaccia G, Arzenton F, Sharbel TF, Varotto S, Parrini P, Lucchin M. Genetic diversity and reproductive biology in local populations of the facultative apomict Hypericum perforatum L. Heredity. 2006;96:322–334. doi: 10.1038/sj.hdy.6800808. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Angiosperm DNA C-values database. 2010 (release 7·0, December 2010). http://www.kew.org/cvalues/ [Google Scholar]
- Bonin A, Bellemain E, Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Brutovská R, Cellárová E, Schubert I. Cytogenetic characterization of three Hypericum species by in situ hybridization. Theoretical and Applied Genetics. 2000;101:46–50. [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Comes HP, Kadereit JW. The effect of Quaternary climatic changes on plant distribution and evolution. Trends in Plant Sciences. 1998;3:432–438. [Google Scholar]
- Dobeš C., Sharbel TF, Koch MA. Towards understanding the dynamics of hybridization and apomixis in the evolution of genus Boechera (Brassicaceae) Systematics and Biodiversity. 2007;5:321–332. [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. Comparison of three fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiologia Plantarum. 1992;85:625–631. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Ehrich D. AFLPdat: a collection of R functions for convenient handling of AFLP data. Molecular Ecology Notes. 2006;6:603–604. [Google Scholar]
- Ehrich D, Gaudeul M, Assefa A, et al. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Molecular Ecology. 2007;16:2542–2559. doi: 10.1111/j.1365-294X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich A. Der Formenkreis der Arten Hypericum perforatum L., H. maculatum Cr. und H. acutum Mönch, nebst deren Zwischenformen innerhalb des Gebietes von Europa. Sitzungsberichte der Kaiserlichen Akademie der Wissenschaft Wien, Mathematisch-Naturwissenschaftliche Klasse. 1911;120:505–599. [Google Scholar]
- Galla G, Barcaccia G, Schallau A, Puente Molins M, Bäumlein H, Sharbel TF. The cytohistological basis of apospory in Hypericum perforatum L. Sexual Plant Reproduction. 2011;24:47–61. doi: 10.1007/s00497-010-0147-7. [DOI] [PubMed] [Google Scholar]
- Gong W, Chuan C, Dobeš C, Fu C, Koch MA. Phylogeography of a living fossil: Pleistocene glaciations forced Ginkgo biloba L. (Ginkgoaceae) into two refuge areas in China with limited subsequent postglacial expansion. Molecular Phylogenetics and Evolution. 2008;48:1094–1105. doi: 10.1016/j.ympev.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Harris SA, Ingram R. Chloroplast DNA and biosystematics: the effects of interspecific diversity and plastid transmission. Taxon. 1991;40:393–412. [Google Scholar]
- Heinze B. A database of PCR primers for the chloroplast genomes of higher plants. Plant Methods. 2007;3(4) doi: 10.1186/1746-4811-3-4. http://dx.doi.org/10.1186/1746-4811-3-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM. Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society. 1999;68:87–112. [Google Scholar]
- Hörandl E. Species concepts in agamic complexes: applications in the Ranunculus auricomus complex and general perspectives. Folia Geobotanica. 1998;33:335–348. [Google Scholar]
- Hörandl E. Geographical parthenogenesis: opportunities for asexuality. In: Schön I, Martens K, Van Dijk P, editors. Lost sex. Heidelberg: Springer; 2009. pp. 161–186. [Google Scholar]
- Huck S, Büdel B, Kadereit JW, Printzen C. Range-wide phylogeography of the European temperate–montane herbaceous plant Meum athamanticum Jacq.: evidence for periglacial persistence. Journal of Biogeography. 2009;36:1588–1599. [Google Scholar]
- Jordon-Thaden I, Hase I, Al-Shehbaz IA, Koch MA. Molecular phylogeny and systematics of the genus Draba (Brassicaceae) and identification of its closest related genera. Molecular Phylogenetics and Evolution. 2010;55:524–540. doi: 10.1016/j.ympev.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Kiefer C., Koch MA. A continental-wide perspective: the genepool of nuclear encoded ribosomal DNA and single-copy gene sequences in North American Boechera (Brassicaceae) PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0036491. e36491. http://dx.doi.org/10.1371/journal.pone.0036491 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, Dobes C, Sharbel TF, Koch MA. Phylogeographic structure of the chloroplast DNA gene pool in North American Boechera – a genus and continental wide perspective. Molecular Phylogenetics and Evolution. 2009;52:303–311. doi: 10.1016/j.ympev.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Dolezel J. Estimation of nuclear DNA content in Sesleria (Poaceae) Caryologia. 1998;51:123–132. [Google Scholar]
- Manel S, Poncet BN, Legendre P, Gugerli F, Holderegger R. Common factors drive adaptive genetic variation at different scales in Arabis alpina. Molecular Ecology. 2010;19:3824–3835. doi: 10.1111/j.1365-294X.2010.04716.x. [DOI] [PubMed] [Google Scholar]
- Mártonfi P. New species of the genus Hypericum sect Hypericum (Guttiferae) from Slovakia. Folia Geobotanica. 2001;36:371–384. [Google Scholar]
- Matzk F, Meister A, Schubert I. An efficient screen for reproductive pathways using mature seeds of monocots and dicots. The Plant Journal. 2000;21:97–108. doi: 10.1046/j.1365-313x.2000.00647.x. [DOI] [PubMed] [Google Scholar]
- Matzk F, Meister A, Brutovská R, Schubert I. Reconstruction of reproductive diversity in Hypericum perforatum L. opens novel strategies to manage apomixis. The Plant Journal. 2001;26:275–282. doi: 10.1046/j.1365-313x.2001.01026.x. [DOI] [PubMed] [Google Scholar]
- Matzk F, Hammer K, Schubert I. Coevolution of apomixis and genome size within the genus Hypericum. Sexual Plant Reproduction. 2003;16:51–58. [Google Scholar]
- Möller M, Gao LM, Mill RR, Li DZ, Hollingworth ML, Gibby M. Morphometric analysis of the Taxus wallichiana-complex based on herbarium material. Botanical Journal of the Linnean Society. 2007;155:307–335. [Google Scholar]
- Müller K, Quandt D, Müller J, Neinhuis C. PhyDE 0·9971: phylogenetic data editor. 2005 www.phyde.de . [Google Scholar]
- Nürk NM, Blattner FR. Cladistic analysis of morphological characters in Hypericum (Hypericaceae) Taxon. 2010;59:1495–1507. [Google Scholar]
- Nürk NM, Madriňán S, Carine MA, Chase MW, Blattner FR. Molecular phylogenetics and morphological evolution of St. John's wort (Hypericum) Molecular Phylogenetics and Evolution. 2013;66:1–16. doi: 10.1016/j.ympev.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Parolly G, Nordt B, Bleeker W, Mummenhoff K. Heldreichia Boiss. (Brassicaceae) revisited: a morphological and molecular study. Taxon. 2010;59:187–202. [Google Scholar]
- Percifield R, Hawkins J, McCoy J, Widrlechner M, Wendel J. Genetic diversity in Hypericum and AFLP markers for species-specific identification of H. perforatum L. Planta Medica. 2007;73:1614–1621. doi: 10.1055/s-2007-993749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Bialozyt R, Brewer S, Cheddadi R, Comps B. From spatial patterns of genetic diversity to postglacial migration processes in forest trees. In: Silvertown J, Antonovics J, editors. Integrating ecology and evolution in a spatial context. Oxford: Blackwell Science Ltd; 2001. pp. 295–318. [Google Scholar]
- Petit RJ, Aguinagalde I, de Beaulieu JL, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi E. Phylogeography of South European mammals. In: Weiss S, Ferrand N, editors. Phylogeography of southern European refugia. Heidelberg: Springer; 2007. pp. 110–126. [Google Scholar]
- Rebernik CA, Weiss-Schneeweiss H, Schneeweiss GM, et al. Quaternary range dynamics and polyploid evolution in an arid brushland plant species (Melampodium cinereum, Asteraceae) Molecular Phylogenetics and Evolution. 2010;54:594–606. doi: 10.1016/j.ympev.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Richards AJ. Apomixis in flowering plants: an overview. Philosophical Transactions of the Royal Society B: Biological Sciences. 2003;358:1085–1093. doi: 10.1098/rstb.2003.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson NKB. Hypericum maculatum in Britain and Europe. Proceedings of the Botanical Society of the British Isles. 1958;3:99–100. [Google Scholar]
- Robson NKB. Studies in the genus Hypericum L. (Guttiferae): 1. Infrageneric classification. Bulletin of the British Museum (Natural History), Botany. 1977;5:291–355. [Google Scholar]
- Robson NKB. Studies in the genus Hypericum L. (Guttiferae): 2. Characters of the genus. Bulletin of the British Museum (Natural History), Botany. 1981;8:55–226. [Google Scholar]
- Robson NKB. Studies in the genus Hypericum L. (Guttiferae): 7. Section 29. Brathys (part 1) Bulletin of the British Museum (Natural History), Botany. 1987;16:1–106. [Google Scholar]
- Robson NKB. Studies in the genus Hypericum L. (Guttiferae): 8. Sections 29. Brathys (part 2) and 30. Trigynobrathys. Bulletin of the British Museum (Natural History), Botany. 1990;20:1–151. [Google Scholar]
- Robson NKB. Studies in the genus Hypericum L. (Guttiferae) 4(1). Sections 7. Roscyna to 9. Hypericum sensu lato (part 1) Bulletin of the British Museum (Natural History), Botany. 2001;31:37–88. [Google Scholar]
- Robson NKB. Studies in the genus Hypericum L. (Guttiferae) 4 (2). Section 9. Hypericum senso lato (part 2): subsection 1. Hypericum series 1. Hypericum. Bulletin of the Natural History Museum London, Botany. 2002;32:61–123. [Google Scholar]
- Robson NKB. Hypericum botany. In: Ernst E, editor. Hypericum: the genus Hypericum. London: Taylor and Francis; 2003. pp. 1–22. [Google Scholar]
- Robson NKB. Studies in the genus Hypericum L. (Hypericaceae) 5(1). 1302 Sections 10. Olympia to 15/16. Crossophyllum. Phytotaxa. 2010;4:5–126. [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Schallau A, Arzenton F, Johnston AJ, et al. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. The Plant Journal. 2010;62:773–784. doi: 10.1111/j.1365-313X.2010.04188.x. [DOI] [PubMed] [Google Scholar]
- Schmickl R, Koch MA. Arabidopsis hybrid speciation processes. Proceedings of the National Academy of Sciences, USA. 2011;108:14192–14197. doi: 10.1073/pnas.1104212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt T. Biogeographical and evolutionary importance of the European high mountain systems. Frontiers in Zoology. 2009;6 doi: 10.1186/1742-9994-6-9. e9. http://dx.doi.org/10.1186/1742-9994-6-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz E, Dobes C, Koch MA, Mitchell-Olds T. Sexual reproduction, hybridization, apomixis and polyploidization in the genus Boechera (Brassicaceae) American Journal of Botany. 2005;92:1797–1810. doi: 10.3732/ajb.92.11.1797. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, et al. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Stevens PF. Hypericaceae. In: Kubitzki K, editor. The families and genera of vascular plants. XI. Heidelberg: Springer Verlag; 2007. pp. 194–201. [Google Scholar]
- Suomalainen E. Parthenogenesis in animals. Advances in Genetics. 1950;3:193–253. [PubMed] [Google Scholar]
- Swofford D.L. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Valente LM, Savolainen V, Vargas P. Unparalleled rates of species diversification in Europe. Proceedings of the Royal Society B: Biological Sciences. 2010;277:1489–1496. doi: 10.1098/rspb.2009.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.