Abstract
Background
Jasmonates are important regulators in plant responses to biotic and abiotic stresses as well as in development. Synthesized from lipid-constituents, the initially formed jasmonic acid is converted to different metabolites including the conjugate with isoleucine. Important new components of jasmonate signalling including its receptor were identified, providing deeper insight into the role of jasmonate signalling pathways in stress responses and development.
Scope
The present review is an update of the review on jasmonates published in this journal in 2007. New data of the last five years are described with emphasis on metabolites of jasmonates, on jasmonate perception and signalling, on cross-talk to other plant hormones and on jasmonate signalling in response to herbivores and pathogens, in symbiotic interactions, in flower development, in root growth and in light perception.
Conclusions
The last few years have seen breakthroughs in the identification of JASMONATE ZIM DOMAIN (JAZ) proteins and their interactors such as transcription factors and co-repressors, and the crystallization of the jasmonate receptor as well as of the enzyme conjugating jasmonate to amino acids. Now, the complex nature of networks of jasmonate signalling in stress responses and development including hormone cross-talk can be addressed.
Keywords: Jasmonic acid, oxylipins, enzymes in biosynthesis and metabolism, perception, JA signalling, JAZ, SCF, COI1, responses to herbivores and pathogens, symbiotic interaction, light regulation, JA in development
1. INTRODUCTION
In 2007, an ‘Update on jasmonates’ was published in Annals of Botany covering aspects of biosynthesis, signal transduction and action in plant stress responses, growth and development (Wasternack, 2007). In this previous review, genes and enzymes/proteins involved in biosynthesis, metabolism and signalling were described with respect to the wound response and some developmental processes regulated by jasmonic acid (JA). In 2007, however, there was a breakthrough in analysis of JA signalling with the discovery of the so-called JAZ proteins (JAZMONATE ZIM DOMAIN proteins) as negative regulators in JA-induced gene expression. Three groups identified independently JAZ proteins as targets of the SCFCOI1 complex, where COI1 is the F-box protein as part of the Skp1/Cullin/F-box protein complex which functions as an E3 ubiquitin ligase (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). COI1 (CORONATINE INSENSITIVE1) was identified in Arabidopsis thaliana in 1998, and the corresponding mutant coi1-1 is the most prominent JA signalling mutant (Xie et al., 1998). With the JAZ proteins, however, the first mechanistic explanations were possible on JA perception, including identification of (+)-7-iso-jasmonoyl-l-isoleucine (JA-Ile) as the ligand of a JA receptor (Fonseca et al., 2009). This was complemented by crystallization of the COI1–JAZ co-receptor complex (Sheard et al., 2010), its potentiation by inositol pentakisphosphate (IP5) and identification of the general co-repressor TOPLESS (TPL) and the adaptor protein Novel Interactor of JAZ (NINJA) (Pauwels et al., 2010). Finally, in 2012, JAR1 (JASMONOYL ISOLEUCINE CONJUGATE SYNTHASE1), the essential enzyme in generation of the most bioactive jasmonate compound active as the ligand of the receptor, was crystallized (Westfall et al., 2012).
The identification of these key components in JA perception and signalling allowed identification of downstream targets, the transcription factors (TFs), acting specifically in numerous JA-dependent processes. This led to the first mechanistic explanations of how cross-talk among the different hormones and signalling pathways may occur. That a similar modular principle occurs in jasmonate, auxin, gibberellin (GA) and ethylene (ET) perception and signalling represents one of the most fascinating discoveries in the last few years of plant hormone research.
Beside these fundamental breakthroughs, there has been remarkable improvement in our knowledge on the metabolic fate of JA/JA-Ile, on short- and long-distance signalling, and on cross-talk to other hormones. The role of JA/JA-Ile in plant immunity, herbivory and mycorrhiza has been intensively studied. Several developmentally regulated processes such as seed germination, seedling development, root growth, flower development, seed development, tuber formation and senescence were shown to be regulated by JA/JA-Ile. Finally, the first hints were found for the regulation of JA/JA-Ile signalling by light. Several of these numerous aspects on JA/JA-Ile have been repeatedly discussed in excellent reviews (Katsir et al., 2008a; Kazan and Manners, 2008, 2011, 2012; Browse, 2009a, c; Grant and Jones, 2009; Koo and Howe, 2009; Kuppusamy et al., 2009; Wasternack and Kombrink, 2010; Ballaré, 2011; Pauwels and Goossens, 2011; Robert-Seilaniantz et al., 2011; Dave and Graham, 2012; Kombrink, 2012; Pieterse et al., 2012).
In view of these recent developments, there is an emerging need to complement the earlier update on jasmonates (Wasternack, 2007). Taking new information and fundamental breakthroughs into consideration, we will discuss here in parallel the multifarious roles of jasmonates in plant stress responses and development. However, the amount of published data on various aspects of jasmonates is too exhaustive to cite here due to space limitations.
Furthermore, some subjects such as ‘JA in response to pathogens’, ‘JA in herbivory and plant–insect interactions’ and ‘JA in light signalling’ are not covered in detail because some excellent reviews have been published recently (see above).
2. JA BIOSYNTHESIS
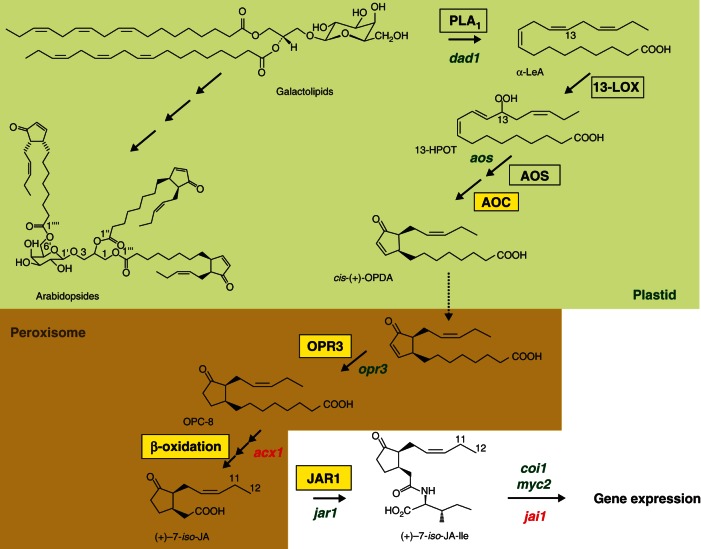
The biosynthesis of JA has been repeatedly and extensively reviewed in recent years (Wasternack, 2007; Browse, 2009a, c; Schaller and Stintzi, 2009; Acosta and Farmer, 2010; Wasternack and Kombrink, 2010; Kombrink, 2012). These reviews present excellent information on reactions, genes, enzymes (including, in several cases, the crystal structures and mechanistic explanations on substrate specificity) and finally regulation of JA biosynthesis. In Fig. 1 we introduce reaction steps, names of enzymes and substrates and refer the reader to the above mentioned reviews for details. Here, we cover only some aspects, where interesting developments have been reported over the last couple of years.
Fig. 1.
Synthesis of jasmonic acid (JA)/JA-Ile from α-linolenic acid generated from galactolipids. Enzymes which have been crystallized are given in yellow boxes. Steps impaired in mutants of Arabidopsis (green) or tomato (red) are indicated. acx1, acyl-CoA-oxidase1; AOC, allene oxide cyclase; AOS, allene oxide synthase; coi1, coronatine insensitive1; dad1, delayed anther dehiscence1; 13-HPOT, (13S)-hydroperoxyoctadecatrienoic acid; jai1, jasmonic acid insensitive1; JAR1, JA-amino acid synthetase; α-LeA, α-linolenic acid; 13-LOX, 13-lipoxygenase; myc2, bHLHzip transcription factor MYC2; OPR3, OPDA reductase3; OPC-8, 3-oxo-2-( 2-pentenyl)-cyclopentane-1-octanoic acid; cis-(+)-OPDA; cis-(+)-12-oxophytodienoic acid; PLA1, phospholipase A1.
2.1. Release of linolenic acid from galactolipids involved in JA biosynthesis
The fatty acid substrate of JA biosynthesis is α-linolenic acid (18:3) (α-LeA) released from galactolipids of chloroplast membranes. It is generally accepted that a phospholipase1 (PLA1) releasing α-LeA from the sn1 position of galactolipids is responsible for generation of the JA substrate, whereas the large family of PLA2s are not involved in JA biosynthesis (for nomenclature of phopsholipase A enzymes see Scherer et al., 2010). It was, however, a matter of debate as to which of the PLA1s are involved in JA biosynthesis. Initially, DEFECTIVE IN ANTHER DEHISCENSE 1 (DAD1) was shown to be responsible for JA formation as the mutant dad1 showed reduced JA levels exclusively in flowers and was therefore male-sterile like the coi1 mutant (Ishiguro et al., 2001). This DAD1 function was strongly substantiated by identification of DAD1 as a target of the homeotic protein AGAMOUS (Ito et al., 2007). AGAMOUS binds to the DAD1 genomic region only during late stamen development. In this way, AGAMOUS orchestrates elongation of filaments, maturation of pollen and dehiscence of anthers, the three critical events in late stamen development (Ito et al., 2007). However, this flower-specific action of DAD1 raised doubts regarding the active roles of PLA1s in wound-induced JA formation in leaves. DONGLE (DGL), a PLA1 from A. thaliana, was thought to be involved in wound-induced and basal JA biosynthesis (Yang et al., 2007; Hyun et al., 2008, respectively). But there were still doubts due to highly ambiguous leaf-specific data on DAD1 and DGL lines generated in different laboratories. More recently, DAD1 and DGL RNAi lines were generated, and these lines were similar to the wild-type in the early wound response. The DGL protein was detected in lipid bodies but not in plastids as required for JA biosynthesis (Ellinger et al., 2010), suggesting that both enzymes are not involved in JA biosynthesis. Of an additional 16 lipase mutants screened, only PLA1y1 (At1g06800) had a reduced level of JA in wounded leaves. However, there might still be unidentified lipases involved in wound- and pathogen-induced JA formation (Ellinger et al., 2010). These Arabidopsis data were complemented by data from RNAi lines suppressing the expression of the GALACTOLIPASE A1 (GLA1) of Nicotiana attenuata, which indicated its involvement in JA formation in leaves and roots, but not during Phytophthora parasitica infection (Bonaventure et al., 2011a). It is thus obvious that there are pathway- and stimuli-specific lipases acting in oxylipin formation.
2.2. The LIPOXYGENASE (LOX) gene family members are involved in JA-dependent responses
Oxygenation of α-LeA is the initial step in JA biosynthesis. The oxygen has to be inserted in the C-13 position by a lipoxygenase (LOX) (Fig. 1). Among the six LOXs of Arabidopsis, four of them are 13-LOXs (LOX2, LOX3, LOX4, LOX6) (Bannenberg et al., 2009), although their functions are only partly understood. LOX2 was thought to be involved in the wound response for a long time (Bell et al., 1995) and subsequent studies revealed that LOX2 was responsible for the bulk of JA formation in the first h upon wounding (Glauser et al., 2009; Schommer et al., 2008). Similarly, an involvement of LOX2 in the generation of oxylipins during natural and dark-induced senescence as well as under sorbitol stress was demonstrated by using LOX2-RNAi lines. The LOX2-RNAi lines carry basal levels of cis-12-oxo-phytodienoic acid (OPDA) and JA, but do not show an enhanced accumulation during natural and dark-induced senescence (Seltmann et al., 2010). Therefore, the regulation of LOX2 may be under a COl1-dependent transcriptional control, but the gain-of-function mutant fou2 indicated also a Ca2+-dependent control of LOX2 protein leading to constitutively elevated JA levels (Bonaventure et al., 2007a). The fou2 mutant was initially identified in a screen on elevated fatty acid oxidation and thought to be affected in a vacuolar Ca2+ channel (Bonaventure et al., 2007a, b). However, later FOU2/TPC1 was identified as a Ca2+- and voltage-dependent vacuolar cation channel (Beyhl et al., 2009). Moreover, FOU2/TPC1 itself is a target of the large family of TCP (TEOSINTE BRANCHED/TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1) TFs, which are involved in growth-related processes, such as leaf growth, shoot branching and floral organ morphogenesis (Danisman et al., 2012). Interestingly, several TPCs are targets of miR319. Among them TPC4 is preferentially involved in the control of JA biosynthesis and leaf senescence (Schommer et al., 2008). This control takes place via LOX2, and clearly indicates a developmental regulation of LOX2 expression, which is partially uncoupled from its transcriptional regulation during wounding (Schommer et al., 2008). Meanwhile, LOX2 was identified as a target of additional TCPs such as TCP20, thereby regulating leaf development and senescence (Danisman et al., 2012). Another level of LOX2-mediated control may occur in translation. The availability of eukaryotic initiation factor 4E (elf4E) is modulated by elf4E-binding proteins. AtLOX2 was identified as an elf4E-binding protein, suggesting a translational control via LOX2 activity (Freire et al., 2000). LOX2 is also involved in lipid peroxidation that occurs under abiotic and biotic stresses. Here, a LOX2-mediated double oxygenation of plastid galactolipids leading to arabidopsides was recorded upon pathogen infection, but was not responsible for the pathogen-induced increase in JA (Zoeller et al., 2012). Interestingly, the formation of lipid peroxides was accompanied by the synthesis of azelaic acid, a new signalling compound that has been shown to prime the immune response (Jung et al., 2009; Dempsey and Klessig, 2012; Zoeller et al., 2012) (see section 7.2).
In the lox2-1 mutant, however, JA and JA-Ile are still synthesized in the first 5 min upon wounding (Glauser et al., 2009) indicating the activity of other 13-LOXs. Moreover, a detailed proteome analysis of JA-induced proteins in A. thaliana showed a marked increase in LOX3 protein (Gfeller et al., 2011). These data and recent work revealed that all four 13-LOX forms contribute to JA formation at least in the wound response (Caldelari et al., 2011; Chauvin et al., 2013). Among them LOX-6 showed a preferential role in the wound response in the early stage of leaf cell differentiation. Using single and different combinations of double, triple and quadruple mutants of lox2-1, lox3B, lox4A and lox6A as well as LOX6-promoter GUS lines, the dominant role of LOX6 in early wound-induced JA formation was confirmed. The LOX6 promoter was specifically active in and near the xylem cells of young tissues which complement promoter activity of LOX3 and LOX4 in vascular tissues (Vellosillo et al., 2007), where other JA biosynthesis genes such as AOS and AOC are also expressed (Kubigsteltig et al., 1999; Stenzel et al., 2012).
In contrast to the wound response, the role of 13-LOX forms in fertility and flower development is different. Both processes are clearly JA-dependent, but fertility does not require LOX2. In contrast, the double mutant lox3lox4 is male sterile, accompanied by abnormal anther maturation, defective dehiscence and non-viable pollen. Additionally, the mutant has a global proliferative arrest as evident by an abnormal carpelloid flower (Caldelari et al., 2011). The remaining LOXs of A. thaliana, LOX1 and LOX5, are 9-LOXs and are not involved in JA biosynthesis. Their products are active in local and systemic defence mechanisms against bacterial pathogens (Vicente et al., 2012). LOXs of fungi are different from plant and mammalian LOXs, and generate 9- and 13-hydroperoxides (reviewed by Brodhun and Feussner, 2011).
2.3. ALLENE OXIDE CYCLASE (AOC)
Recently, differential expressions of the four AOCs of A. thaliana were demonstrated by corresponding promoter:: GUS lines (Stenzel et al., 2012). In leaves, AOC1, AOC2 and AOC3 were expressed in all leaf tissues, whereas the AOC4 promoter was preferentially active in the main veins of fully developed leaves. In roots, promoters of all AOCs were highly active in the meristematic tissues and the elongation zone, including the lateral root primordia. Results obtained in distinct flower organs indicated redundant and non-redundant expression of AOCs. An additional level of regulation of AOCs was indicated by interaction studies using BiFC, where homo- and hetero-dimerization of all the four AOCs were detected (Stenzel et al., 2012). In soybean, where six genes encode AOCs, initial data showed functional diversity in terms of expression for stress responses (Wu et al., 2011). Recently, the crystal structure of AOC1 and AOC2 of Physcomitrella patens revealed new mechanistic insights into AOC catalysis, including tight binding of the substrate, accompanied by conformational changes within the binding pocket (Neumann et al., 2012). Both PpAOCs are similar in structure and oligomeric to the AtAOC2 crystalized previously as a trimer (Hofmann et al., 2006).
AOC and other enzymes in JA biosynthesis such as LOX and ALLENE OXIDE SYNHASE (AOS) are partially associated with chloroplast membranes (Farmaki et al., 2007). For AOS the level of the protein within the envelope is affected by rhomboids, a family of intra-membrane serine proteases of inner envelope membrane (Knopf et al., 2012). The association of LOX, AOS and AOC with chloroplast membranes implies that the esterified OPDAs, called arabidopsides, may be formed from fatty acids esterified in galactolipids by membrane-bound enzymes, as indicated by recent labelling experiments (Nilsson et al., 2012). Arabidopsides that occur exclusively in Arabidopsis are a diverse group of compounds of the galactolipids MGDG and DGDG, where OPDA is esterified in the sn-1 and/or sn-2 position. The occurrence of the different types of arabidopsides, their formation and putative function has been reviewed (Göbel and Feussner, 2009; Mosblech et al., 2009).
2.4. OPDA REDUCTASE3 (OPR3)
Among the six OPRs of A. thaliana only OPR3 is involved in JA biosynthesis, which was substantiated by mechanistic studies with the crystal structure of OPR3 and OPR1 (Breithaupt et al., 2001, 2006) (reviewed by Schaller and Stintzi, 2009; Wasternack and Kombrink, 2010; Kombrink, 2012). OPR1 might be involved in reduction of phytoprostanes, a group of non-enzymatically formed compounds with structural similarity to OPDA (Mueller et al., 2008).
Initially, opr3, a JA-deficient and OPDA-accumulating mutant carrying a 17-kb T-DNA insertion in an OPR3 intron, showed resistance to Alternaria brassicicola, which was discussed as a direct role of OPDA in pathogen defence (Stintzi et al., 2001). In many studies, opr3 was permanently used to distinguish between JA- and OPDA-dependent signalling. Recently, JA accumulation in opr3 upon infection with Botrytis cinerea has also been reported (Chehab et al., 2011). opr3 is not a null mutant, and is able to generate mature full-length OPR3 transcript upon splicing of the T-DNA containing intron under specific conditions, such as B. cinerea infection leading to JA formation. Therefore, at least under some conditions, opr3 is not an ideal platform for dissecting OPDA-specific signalling.
The important and versatile role of OPRs was recently illustrated for maize. The opr7opr8 double mutant has dramatically reduced levels of JA in all organs tested, accompanied by strong defects in development, including sex determination leading to feminized tassels and the elongation of ear shanks (Yan et al., 2012) (see section 9.12). This double mutant was highly susceptible to root-rotting oomycetes (Pythium spp.) and herbivory. In rice, an OPR involved in JA biosynthesis is encoded by OsOPR7, whereas the other 12 members of this gene family belong to another subgroup, which is not involved in JA formation (Tani et al., 2008). OsOPR7 is expressed upon wounding or drought stress, and can complement the opr3 phenotype in A. thaliana (Tani et al., 2008). The OsOPR7 protein can convert both enantiomeric forms of cis-OPDA, (+)-cis-OPDA and (–)-cis-OPDA.
2.5. Regulation of JA biosynthesis
As described previously (Wasternack, 2007; Browse, 2009a, c), the regulation of JA biosynthesis is determined by a positive feedback loop, substrate availability and tissue specificity. Additional regulation is provided by the concurrent action of the branches in the LOX pathway. Among the seven different branches known for the LOX pathway (Feussner and Wasternack, 2002) the AOS and HYDROPEROXIDE LYASE (HPL) reactions are concurrent on the same substrate, the product of a 13-LOX. The HPL branch leads to volatile and non-volatile oxylipins, e.g. the leaf aldehydes and leaf alcohols (Andreou et al., 2009). Many of them are defence compounds and are formed upon herbivore attack (Matsui et al., 2006; Schuman et al., 2012). The HPL branch involved in the formation of green leafy volatiles (GLVs) is selectively suppressed by chewing herbivores, which might be an evolutionary advantage (Savchenko et al., 2013). One of the three HPLs of rice positively regulates the formation of GLVs but negatively regulates JA biosynthesis by substrate competition (Tong et al., 2012). Consequently, the direct and indirect defence is modulated. Non-volatile oxylipins, such as various traumatic acids and azelaic acid, are formed upon stress in pea seedlings (Mukhtarova et al., 2011), suggesting, for the first time, a central role of azelaic acid as a defence signal (Jung et al., 2009; Dempsey and Klessig, 2012; Zoeller et al., 2012) (see section 7.2.).
Additional components of regulation were obtained from characterization of JAZ proteins, Ca2+-related signalling, JA-related transcription factors and mitogen-activated protein kinases (MAPKs). The positive feedback loop in JA biosynthesis can be explained now by the SCFCOI1–JAZ regulatory module that is known to be active in the expression of LOX, AOS, AOC, OPR3 and ACX. The formation of JA/JA-Ile will subject the negative regulator JAZ to proteasomal degradation, which allows MYC2 to activate the JA-responsive promoters of JA biosynthesis genes. JAZ and MYC genes are, however, JA/JA-Ile responsive, allowing a permanent replenishment of the negative (JAZs) and positive (MYC2) regulators that result in an adjustment of the expression of JA biosynthesis genes (Chung et al., 2008). The Arabidopsis microarray datasets from various developmental stages and stress conditions reveal transcriptional regulation of all JA biosynthesis genes (Pauwels et al., 2009; van Verk et al., 2011). There are, however, indications for post-translational regulation of enzyme activities. The OPR3 activity seems to result from a monomer/dimer equilibrium including a self-inhibition by dimerization (Breithaupt et al., 2006; Schaller and Stintzi, 2009). The above interaction studies with all four AOCs of A. thaliana revealed interaction among them all. The observed homo- and hetero-dimerization led at least partially to altered enzyme activity (Stenzel et al., 2012).
Ca2+ and MAPK cascades are also involved in the regulation of JA biosynthesis. In A. thaliana, MKK3 and MPK6 are activated by JA leading to negative regulation of MYC2 expression and repression of JA biosynthesis genes (Takahashi et al., 2007). In a parallel pathway, however, there is an MKK3/MPK6-independent activation of MYC2 by JA, and the MKK3/MPK6 cascade is epistatic to MYC2 (Takahashi et al., 2007). There exists a link between JA biosynthesis and MAPK pathways, as revealed by co-expression analyses of microarray datasets in A. thaliana (van Verk et al., 2011). Here, it became obvious that OPR3 and to a minor extent AOS are co-expressed with MYC2, MEK1, MEKK1, MKK4 and MPK3.
For wound- and herbivore-induced JA accumulation in Nicotiana attenuata, the Ca2+-dependent protein kinases CDPK4 and CDPK5 are negative regulators (Yang D-H et al., 2012), whereas a wound-induced protein kinase (WIPK) is rapidly activated near the wound region thereby activating JA biosynthesis (Wu et al., 2007). In tomato, a MPK1, MPK2 and MPK3 are involved in expression of JA biosynthesis genes (Kandoth et al., 2007). Here, the activation of MPKs is systemin-dependent (see section 6). Further control of JA biosynthesis is mediated by the COP9 (CONSTITUTIVE PHOTOMORPHOGENESIS 9) signalosome (CSN), a multi-protein complex involved in the regulation of CULLIN-RING E3 ubiquitin ligases. CSN not only is required for optimum plant development, but is also involved in plant defence against herbivores and pathogens by its modulation of JA levels (Hind et al., 2011).
Ca2+ is an early acting second messenger in response to many biotic and abiotic stimuli (Kudla et al., 2010). Although several of these stimuli are associated with increased JA biosynthesis, the involvement of Ca2+ upstream of JA biosynthesis is poorly understood. Besides Ca2+-mediated control of LOX2 (Bonaventure et al., 2007a) as discussed earlier (see section 2.2), three additional examples will be given here to elucidate, how Ca2+ is involved in the regulation of JA biosynthesis and signalling:
In the family of Ca2+/CaM-binding TFs, AtSR1 is required for down-regulation of salicylic acid (SA) levels in plant immune responses (Du et al., 2009). Upon wounding, however, the negative impact of SA in both basal and induced JA biosynthesis is abolished by AtSR1 (Qiu et al., 2012).
The calmodulin-like protein CLM42 negatively regulates the defence response during herbivory by decreasing the COI1-mediated JA sensitivity (Vadassery et al., 2012). The cytosolic and nuclear located protein CLM42 is active downstream of herbivore-induced Ca2+ elevation but is upstream of COI1-mediated JA-Ile perception.
In A. thaliana, the overexpression of a plasma membrane-located glutamate receptor results in increased glutamate-mediated Ca2+ influx and resistance to necrotrophic pathogens (Kang et al., 2006). A putative link to JA biosynthesis is lacking, but is suggested by the up-regulation of VSP1, LOX2 and other JA-responsive genes.
Ca2+ is clearly a key player in plant responses to environmental stimuli, leading to context-dependent Ca2+ fluctuations upstream and downstream of JA biosynthesis or in parallel to JA generation, and is a part of the regulatory network of evolutionary divergent metabolic pathways (Pauwels et al., 2009).
2.6. JAR1 catalysing the final step in the generation of ligand
The cloning of JAR1 as a member of the GH3 gene family, which belongs to the large group of enzymes forming acyl-adenylate/thioester intermediates, was a breakthrough in the JA field. This enzyme catalyses the final step in the formation of the bioactive JA compound (Staswick and Tiryaki, 2004). The identification of (+)-7-iso-JA-Ile as the ligand of the COI1–JAZ co-receptor complex (Fonseca et al., 2009) and the crystallization of the receptor complex (Sheard et al., 2010) provided mechanistic explanations for JA/JA-Ile perception (see section 4). Meanwhile, the JA-specific JAR1/AtGH3.11 and the benzoate-specific PBS3/AtGH3.12 have been crystallized (Westfall et al., 2012). For crystallization of JAR1, a racemic mixture of JA was used, but only (–)-JA-Ile was found in the structure. The authors assumed that (–)-JA is accepted as substrate by JAR1 and is converted to (+)-JA-Ile (Westfall et al., 2012). However, the initial in vivo product in JA biosynthesis is (+)-7-iso-JA, and its conjugate with l-Ile is the ligand of the receptor (Fonseca et al., 2009; Sheard et al., 2010). Although the JA epimer used by JAR1 still continues to be ambiguous, the water-mediated hydrogen bond to the cyclopentanone ring of JA and the hydrophobic binding pocket for the pentenyl side observed in the crystal structure of JAR1 (Westfall et al., 2012) explain now mechanistically the repeatedly recorded structure/activity relationships for numerous JA compounds (for a review see Wasternack, 2007). The crystal structures of PBS3 and JAR1 define the role of conformational changes in the carboxy-terminal domain for conjugation of amino acids to various acyl acid substrates and illustrates how a promiscuous enzyme might evolve by a highly adaptable structure (Westfall et al., 2012). For a long time, equilibration between the enantiomers of JA and of JA-Ile was assumed (Wasternack, 2007), and epimerization was suggested as a mechanism to sustain the most bioactive JA compound, (+)-7-iso-JA-Ile (Fonseca et al., 2009). Meanwhile, an assay has been developed for quantification of (+)-7-iso-JA-Ile from tomato extracts, indicating that the compound is less unstable than assumed earlier (Suza et al., 2010). These data indicate that (+)-7-iso-JA-lle is exclusively formed upon wounding and by a recombinant JAR1, with a strong preference for Ile compared with other amino acids. In SlJAR1-RNAi lines, wound-induced formation of (+)-7-iso-JA-Ile was down-regulated by 50–75 % suggesting the existence of other JA-conjugating enzymes than JAR1 (Suza et al., 2010). Note that this must be taken into consideration while evaluating the jar1 mutant data. The homeostasis of JA-Ile is highly dependent on its hydrolysis in vitro. In JA-Ile-hydrolase 1-silenced plants of N. attenuata, the herbivore-induced burst in JA-Ile and its following reactions in direct and indirect defence responses are strongly attenuated (Woldemariam et al., 2012).
3. THE METABOLIC FATE OF JA
In an earlier update, only four enzymes involved in JA metabolism were described in terms of enzymatic properties and cloning of their cDNAs (Wasternack, 2007). Meanwhile, new JA metabolites have been identified, with additional enzymes having been cloned and characterized.
Due to the central role of JA-Ile in JA signalling and the parallel occurrence of JA and JA-lle as sustained by JAR1, we will combine them in the subsequent sections as ‘JA/JA-Ile’, this being active as a signalling module. However, there are three important caveats: (1) Are active JA metabolites involved in specific responses that are not directly caused by JA/JA-lle? (2) Is JA/JA-Ile signalling switched off by metabolic conversion? (3) Do JA metabolites function as a storage form of JA?
3.1. Profiles of JA/JA-Ile metabolites
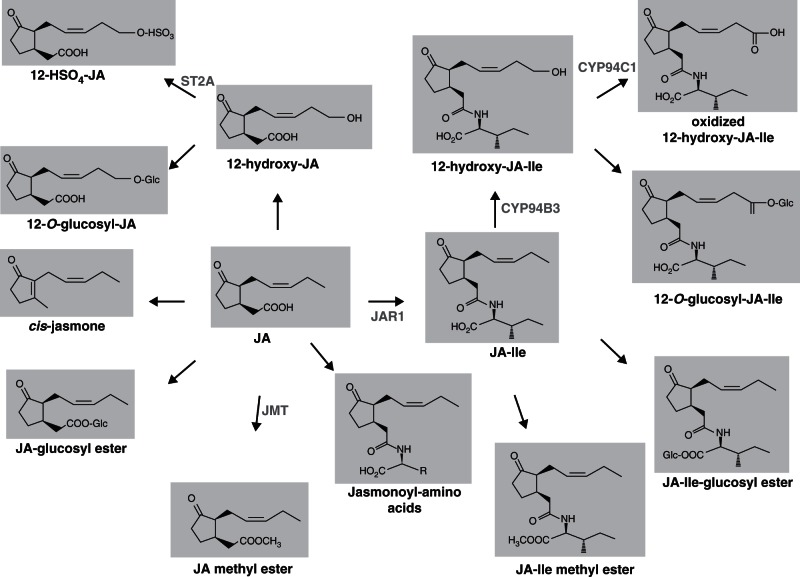
In the early days of JA research, numerous JA compounds were identified as constituents of distinct plant tissues or as volatiles emitted from flowers (reviewed by Wasternack et al., 2013). The profiles of JA-related compounds are presented in Fig. 2. Many of them were already known in 2007. Meanwhile, glucosylated forms of JA, JA-Ile, 12-OH-JA and 12-OH-JA-Ile have been described (Chung et al., 2008; Glauser et al., 2008, 2009). Most of them accumulate very rapidly (within minutes) in wounded Arabidopsis or tomato leaves. Corresponding wound-induced formation of 11-OH-JA, 12-OH-JA, 12-OH-JA-Ile, 12-COOH-JA-Ile and 12-HSO4-JA were also recorded (Gidda et al., 2003; Guranowski et al., 2007; Glauser et al., 2008, 2009; Miersch et al., 2008). A large-scale screening for different JA/JA-Ile metabolites in different organs of various plant species showed their relative abundance up to three orders of magnitude higher than that of JA or OPDA (Miersch et al., 2008). Immature seeds and leaves of Glycine max contain high levels of 12-OH-JA, 12-HSO4-JA and 12-O-Glc-JA. In most cases, however, it is not known whether these abundantly occurring JA/JA-Ile metabolites are biologically active or function as storage forms of JA/JA-Ile (Miersch et al., 2008). It has been suggested that higher levels of 12-OH-JA, 12-HSO4-JA and 12-O-Glc-JA in the tassels of Zea mays may be associated with sex determination during development of this male reproductive structure in monoecious species (Acosta et al., 2009; Browse, 2009b). Support for the involvement of a JA compound in sex determination also came from the maize double mutant opr7opr8 (Yan et al., 2012) (see section 9.12).
Fig. 2.
Metabolic fate of jasmonic acid (JA) and JA-Ile. Enzymes which have been cloned are given in grey. JAR1, JA-amino acid synthetase; JMT, JA methyl transferase; ST2A, sulfotransferase 2A.
3.2. cis-Jasmone (CJ)
CJ is a volatile compound and represents the main constituent of the floral bouquet of different plants thereby attracting insect pollinators. It is emitted in response to herbivory, application of insect oral secretions or JA treatment. However, the biosynthetic route leading to the formation of CJ is still unclear. Initially, CJ was regarded as a decarboxylated product of JA, being responsible for the disposal of JA due to its high volatility (Koch et al., 1997). Isomerization of cis-(+)-OPDA into iso-OPDA, however, allows a direct route to CJ via β-oxidation to 3,7-didehydro-JA and decarboxylation (Dabrowska and Boland, 2007; Schulze et al., 2007; Dabrowska et al., 2009). CJ is clearly biologically active, preferentially in plant–insect interactions as summarized by Matthes et al. (2010). Most evidence derives from the microarray-based transcriptome analysis of CJ-treated Arabidopsis plants (Matthes et al., 2010). The set of CJ-induced genes was different from those induced by JA, and CJ-induced gene expression was independent of that induced by COI1 and JAR1. Furthermore, key components that are not involved in JA signalling are assumed to have distinct roles in CJ signalling; for example, TFs TGA 2, 5 and 6, and SCARECROW-like 14 have been shown to play a key role for CJ in indirect defence (Matthes et al., 2010).
3.3. CYP94 enzymes generate hydroxylated and carboxylated JA-Ile
Most recently, three groups independently identified the cytochrome P450 enzyme CYP94B3 that hydroxylates JA-Ile at the terminal carbon atom of the pentenyl side chain (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012). Additionally, Heitz et al. (2012) characterized the enzyme CYP94C1, which is active in the subsequent oxidation step to the oxidized 12-OH-JA-Ile (Fig. 2). Heterologous expression in yeast showed substrate preference of CYP94B3 for JA-Ile (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012). Typical JA-lle-deficient phenotypes as observed in the CYP94B3 over-expressors that show higher susceptibility to insect attack provided further evidence for the involvement of both enzymes (Koo et al., 2011). Accordingly, the wounded cyp94b3 mutant exhibited increased accumulation of JA-Ile (Koo et al., 2011; Koo and Howe, 2012). These data together with the fact that hydroxylated JA-Ile was less effective in the COI1–JAZ interaction assay (Koo et al., 2011) support the assumption that hydroxylation and carboxylation of JA-Ile may switch off JA/JA-Ile signalling. Such a role of hydroxylation is also known for other hormones, and was initially shown for hydroxylation of JA to 12-OH-JA (Gidda et al., 2003; Miersch et al., 2008). Here, typical JA responses, such as expression of JA-inducible genes, root growth inhibition or seed germination inhibition were compromised by treatment with 12-OH-JA. It will be interesting to examine the role of other members of the CYP94 gene family, e.g. a putative JA hydroxylase. Six members are known for the CYP94 gene family, which seems to have evolved rapidly and active in conversion of fatty acid-derived compounds (Nelson and Werck-Reichhart, 2011; Koo and Howe, 2012). The broad specificity of CYP94s for fatty acyl substrates in vitro (Kandel et al., 2007; Pinot and Beisson, 2011) and the activity of other hydroxylases, such as CYP709C1 active for long-chain fatty acids, illustrate the diversity in hydroxylated fatty acid-derived compounds.
3.4. Methyl jasmonate (MeJA)
Prior to 2007 when there was not much information about JAZ proteins, any discussion on bioactivity of JA and MeJA was controversial. JA levels were always recorded and correlated to JA responses. Furthermore, transgenic lines of Arabidopsis over-expressing the JA carboxy methyl transferase (JMT) led to the assumption that MeJA is the preferentially active signal in JA responses (Seo et al., 2001). An ectopic expression of JMT in N. attenuata, however, negatively affected the formation of JA-Ile, and the biological activity of MeJA was only apparent when MeJA was converted to JA followed by its conjugation to JA-Ile (Stitz et al., 2011). The specificity of JA-Ile in the COI1–JAZ interaction (see section 4.3) was the final proof that there is no direct bioactivity of JA and MeJA.
3.5. Sulfated jasmonates
Among the 18 sulfotransferases of A. thaliana, the gene AtST2a has been cloned, and its recombinant protein has been shown to be specific for the conversion of 11-OH-JA and 12-OH-JA to the corresponding sulfated derivatives (Gidda et al., 2003). Besides OPDA, JA and JA-lle, 12-OH-JA also mediates the expression of AtST2a. Subsequent cloning of the homologous gene from tomato showed similar properties, and transgenic lines over-expressing or repressing SlST2a showed a dramatic shift among the three involved compounds 12-OH-JA, 12-HSO4-JA and 12-O-Glc-JA (J. Heise and C. Wasternack, unpubl. res.). Interestingly, in the adenosine 5′-phosphosulfate kinase gene family consisting of four members and being involved in generation of active sulfate for the sulfotransferase reaction, the apk1apk2 double mutant exhibits a five-fold decrease in 12-OH-JA and 12-HSO4-JA accompanied with a concomitant increase in 12-O-Glc-JA (Mugford et al., 2009). This indicates that conversions of 12-OH-JA into either 12-O-Glc-JA or 12-HSO4-JA are concurrent reactions (see section 3.6).
Further cross-talk between 12-OH-JA and sulfate metabolism was demonstrated by using the mutant fou8, which was identified in a screen for mutants with altered fatty acid oxidation. In fou8 plants, an increment in the LOX2 level is attributed to increased fatty acid oxidation. Consequently, the JA pathway is permanently activated, as indicated by the appearance of JA-related phenotypes in fou8 plants (Rodriguez et al., 2010b). In fou8 plants, the conversion of 3′phospho-adenosine-5′-phosphate (PAP) to AMP, the byproduct of the sulfotransferase reaction, is also affected (Lee et al., 2012), and as a result sulfur metabolism including sulfation of glucosinolates and 12-OH-JA is dramatically altered (Lee et al., 2012). However, the most convincing evidence for the cross-talk between sulfur metabolism and JA biosynthesis is the fact that in the triple mutant fou8apk1apk2, the fou8 phenotypes are genetically suppressed, indicating that a component of the sulfur futile cycle affects the LOX activity necessary for JA biosynthesis (Rodriguez et al., 2010b).
3.6. Glucosylated jasmonates
The plethora of jasmonate compounds is enormous. Besides the compounds mentioned above, JA also occurs conjugated to 1-aminocyclopropane-1-carboxylic acid (ACC), the ET precursor (Staswick and Tiryaki, 2004). However, there is no information on its biological activity. Another group of jasmonate compounds are the glucosylated derivatives. They may occur as glucosyl esters, which are presumably inactive compounds, as the conjugation with amino acids by JAR1 required for most JA-like activities cannot take place. Initially, 12-OH-JA as tuberonic acid (TA) and its O-glucoside (TAG) were identified in potato leaflets and shown to have tuber-inducing properties (Yoshihara and Greulich, 1998) (see section 9.5).
The O-glucosylated jasmonates modified at C-11 and C-12 of hydroxylated JA accumulate rapidly upon leaf wounding (see above) (Glauser et al., 2008; Miersch et al., 2008). Jasmonates with other sugar moieties such as gentiobiose were also detected during the cell cycle of tobacco BY2 cells (Swiatek et al., 2004). In unwounded leaves of Glycine max, the accumulation of 12-O-Glc-JA has been shown to be up to three orders of magnitude higher than that of JA (Miersch et al., 2008). In wounded tomato leaves, 12-O-Glc-JA accumulates subsequently to JA and 12-OH-JA (Miersch et al., 2008; O. Miersch, unpubl. res.). In transgenic tomato lines constitutively over-expressing the gene ST2a, accumulation of 12-O-Glc-JA upon wounding has been shown to be much less due to its shift to the sulfated derivative (J. Heise et al., unpubl. res.). However, the biological role of 12-O-Glc-JA in the wound response is not clear. Possibly, 12-O-Glc-JA is a transport form of 12-OH-JA, or it represents a sequestration of JA as known for the glucosides of SA and benzoic acid.
12-O-Glc-JA was identified as a leaf closing factor (LCF) in motor cells of nyctinastic plants, such as Albizzia and Samanea saman (Nakamura et al., 2011) (see section 9.8). As with the JA-Ile receptor, only a specific enantiomer, here the (–) form, of LCF is active. In addition to the enantiomer specificity of the jasmonoyl moiety, the d/l-stereochemistry of the glucon moiety is important (Ueda et al., 2012). This accords with the weak activity of 12-OH-JA and inactivity of JA and JA-Ile in leaf closing. The LCF was inactive in all classical JA responses such as LOX2 expression or leaf volatile emission, and is perceived in a COI1/JAZ-independent manner (Nakamura et al., 2011). The involvement of JA-related compounds in nyctinastic leaf movement was confirmed by the gene expression data from a Medicago truncatula mutant with a defective pulvinus that is required for nyctinasty (Zhou et al., 2012). This mutant, called petiolule-like pulvinus, showed down-regulation of genes involved in JA biosynthesis and metabolism.
From rice cell cultures, a putative SA glucosyl transferase (OsSGT) has been purified that shows glucosylation not only of SA but also of 12-OH-JA (Seto et al., 2009). The OsSGT mRNA accumulating in cell cultures upon treatment with JA, 12-OH-JA and SA as well as in leaves after wounding is indicative of its putative role in the wound response.
4. JA PERCEPTION AND SIGNALLING
4.1. SCF complexes
The ubiquitin-proteasome system is the central regulator in plant hormone sensing and signalling. It consists of an Skp1/Cullin/F-box (SCF) complex that functions as an E3 ubiquitin ligase, where the F-box protein recognizes a target protein which is ubiquitinated and subsequently subjected to proteasomal degradation. For JA perception and signalling, COI1 acts as an F-box protein (Xie et al., 1998). One of the most interesting aspects in plant hormone research is that several of them are perceived by an SCF complex with similar modules, where the F-box protein confers the hormone specificity. Since these facets have been extensively reviewed over the past couple of years (Katsir et al., 2008a; Chini et al., 2009a; Santner and Estelle, 2010; Kelley and Estelle, 2012; Shan et al., 2012) only JA-related aspects will be discussed here.
4.2. JAZ proteins
In 2007, members of a new protein family of Arabidopsis were discovered by chance and called JASMONATE ZIM DOMAIN (JAZ) proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Initially observed to be early up-regulated by wounding or JA treatment, JAZ proteins were recognized as targets of the SCFCOI1 complex. The degradation of JAZ allows the release of positively acting TFs, such as MYC2 that binds to JA-responsive elements occurring in promoters of JA-responsive genes, thereby initiating transcription. This basic scheme (Fig. 3) has been independently developed in three different laboratories (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007) and was subsequently extended by identification of new up- and downstream components. Among the upstream components, the RING-type ubiquitin ligases, RING DOMAIN LIGASE3 (RGLG3) and RGLG4, were identified as modulators of JA/JA-Ile signalling in response to various stimuli (Zhang X et al., 2012b). As downstream components the general co-repressors TOPLESS (TPL) and TPL-related proteins and their interaction with the adaptor protein ‘Novel Interactor of JAZ’ (NINJA) were identified (Pauwels et al., 2010). Furthermore, while searching the JAZ targets numerous new TFs and JAZ interactors were discovered (Pauwels and Goossens, 2011; Wager and Browse, 2012).
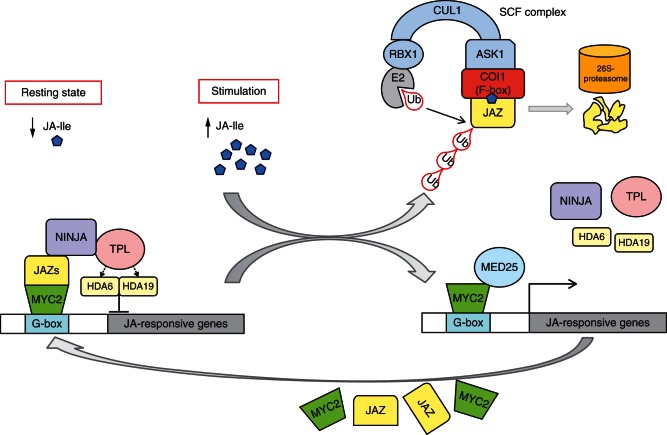
Fig. 3.
Jasmonic acid (JA) perception via the COI1–JAZ co-receptor complex – mechanisms in JA-induced gene expression. In the resting state (left, low JA-Ile level), the binding of MYC2 to a G-box within the promoter of a JA-responsive gene does not activate transcription due to binding of the repressors Jasmonate ZIM domain proteins (JAZs) to MYC2. The co-repressors Novel Interactor of JAZ (NINJA) bound to JAZs, and TOPLESS (TPL) repress transcription via HISTONE DEACETYLASE 6 (HDA6) and HDA19. Upon stimulation (right, high JA-Ile level), JAZs are recruited by COI1 and subjected to ubiquitinylation and subsequent degradation by the 26S proteasome. Subsequently, MYC2 can activate transcription of early JA-responsive genes such as those encoding JAZ and MYC2. Transcription is mediated by the subunit 25 of Mediator complex (MED25; see section 4). ASK1, Arabidopsis SKP1 (S-phase kinase-associated protein 1) homologue; CUL, CULLIN; E2, ubiquitin-conjugating enzyme; MYC2, bHLHzip transcription factor; RBX, RING-H2 protein; SCF-complex, complex consisting of Skp1, Cullin-1 and F-box protein; Ub, ubiquitin.
In addition to the F-box protein COI1, JAZ interactors are: (1) bHLH TFs (MYC2, MYC3, MYC4, GL3, EGL3 and TT8), (2) R2R3 MYB TFs (PAP, GL1, MYB 21 and MYB 24), (3) TFs of other hormone signalling pathways (EIN3, EIL, GAI, RGA and RGL1), (4) co-repressor proteins (NINJA, TPL, HDA6 and HDA19) and (5) JAZ proteins due to their homo- and hetero-dimerizations (Chini et al., 2009b; Pauwels and Goossens, 2011).
There are 12 JAZ proteins in A. thaliana (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2009; Pauwels and Goossens, 2011; Wager and Browse, 2012). They contain a weakly conserved N-terminal domain, a highly conserved C-terminal Jas domain that mediates the interaction with the transcription factors, and the conserved ZIM (TIFY) domain responsible for JAZ dimerization and interaction with NINJA (Vanholme et al., 2007; Chung et al., 2009; Pauwels and Goossens, 2011). The Jas domain is exclusively required for the repressive activity of JAZ proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). The expression of truncated JAZs lacking the Jas domain was associated with dominant insensitivity to exogenous JA. The initial assumption that individual JAZ proteins act specifically with different targets was subsequently revised by numerous interaction studies and the fact that there is a common occurrence of the ZIM and the Jas domain; for example, all 12 JAZs interact with MYC2, and JAZ1 interacts with nearly all target proteins mentioned above. The JA signalling is, however, mediated by a JAZ-regulatory network that entails interaction with multiple transcription factors, formation of homo- and hetero-dimers, alternative splicing of JAZ-encoding genes and differential stability of JAZs (Pauwels and Goossens, 2011; Kazan and Manners, 2012; Shyu et al., 2012). All these processes may result in a large repertoire and combinatorial diversity in JAZ–JAZ interactions, the in vivo function of which is not known (Chung et al., 2009, 2010).
The alternative splicing of JAZ genes can form dominant JAZ variants leading to JA-insensitive plants, if the Jas domain is abolished during splicing. The Jas domain is absolutely required for binding the downstream components, the TFs, and for intact JA signalling. For JAZ10, there are naturally occurring splice variants lacking parts of the Jas domain (JAZ10.3) or the complete Jas domain (JAZ10.4) (Yan et al., 2007; Chung et al., 2009, 2010). The JAZ proteins are localized in the nucleus (Chini et al., 2007; Thines et al., 2007; Grunewald et al., 2009), but an assumed involvement of the Jas domain is not completely clear because the splice variants JAZ10.3 and JAZ10.4 affected in the Jas domain are still localized in the nucleus (Chung et al., 2009). Recently, the nuclear targeting of JAZ1 and JAZ9 has been shown to be dependent on physical interaction with MYC2 via a highly conserved region of the Jas domain (Withers et al., 2012).
There were several hints at the transcriptional repression by JAZs, but generation of mutants with the expected JA-hypersensitive phenotype was upset by the obvious redundancy among the JAZ proteins. Only the T-DNA insertion mutant jaz10-1 and RNAi lines of JAZ1 and JAZ10 exhibited enhanced JA sensitivity (Grunewald et al., 2009), whereas other JAZ mutants and the T-DNA insertion mutant jaz1-1 did not show such a phenotype (Demianski et al., 2012). Most recently, however, transcriptional repression by full-length JAZ8 has been described (Shyu et al., 2012), which is based on increased stability of JAZ8 due to lack of the conserved LPIARR motif. This hexapeptide within the Jas domain represents the conserved degron motif, and is required for closing off the binding pocket of JA-Ile within the receptor complex (Sheard et al., 2010). Due to its absence in JAZ8, a strong interaction with COI1 in the presence of JA-Ile is excluded, leading to the increased stability of JAZ8. However, the consequences of JAZ8 removal from cells are unknown. The residual interaction between JAZ8 and COI1 occurs only at the higher JA-Ile concentration, whereas JAZ1–COI1 interaction takes place when the JA-Ile concentration is low. Such a scheme, in which the COI1–JAZ interaction is determined by the concentration of the ligand, is quite similar to the auxin TIR1-Aux/indole-3-acetic acid (IAA) receptor system (Katsir et al., 2008a; Kelley and Estelle, 2012). Besides this new feature on JAZ function via protein stability, the JAZ8-mediated repressor function was shown to depend on an LxLxL-type EAR (ERF associated amphiphilic repression) motif at the N terminus (Shyu et al., 2012). This motif of JAZ8 can directly bind the co-repressor TPL. In that event, however, the ZIM domain is not required, in contrast to other JAZs, which recruit TPL through the EAR-motif containing adaptor NINJA (see below).
The JAZ gene expression is JA responsive (Chung et al., 2008). Consequently, there is a futile cycle which may contribute to a fine-tuning of JA signalling. MYC2 is involved in the expression of many JA-responsive genes (Dombrecht et al., 2007). For JAZ gene expression, however, other components might be involved, as in myc2 mutants most JAZ genes are expressed upon infection with Pseudomonas syringae, which is known to be a JA- mediated process (Demianski et al., 2012). One candidate could be the MEDIATOR25 (MED25) subunit of the eukaryotic Mediator complex (Fig. 3). MED25 has been recently identified as an integrative hub in JA-mediated gene expression (Çevik et al., 2012). In the pft1/med25 mutant, pathogen-responsive JAZ9 expression is diminished (Kidd et al., 2009), while the JA-induced expression of JAZ6 and JAZ8 is significantly reduced in the med25 mutant lines (Chen et al., 2012).
4.3. COI1–JAZ co-receptor complex
Ten years after cloning of the F-box protein COI1 (Xie et al., 1998), its function as a JA receptor was finally established. Initially, COI1 was assumed to function as a receptor due to its analogy to the auxin receptor TIR1 (Woodward and Bartel, 2005). Photoaffinity-based cross-linking of JA-Ile to COI1 substantiated this idea (Yan et al., 2009). However, the requirement of the SCFCOI1–JAZ complex for JA perception is now generally accepted. Since the identification of JAZs in 2007 followed by the crystallization of the COI1–JAZ co-receptor complex (Sheard et al., 2010), it is now possible to establish a mechanistic view on JA-Ile perception. In this complex, the Jas domain of JAZ proteins interacts with COI1, if the ligand JA-Ile is present. This interaction takes place via the N-terminal 20 amino acid residues of the Jas degron, and is strongly increased by IP5 (Sheard et al., 2010; Mosblech et al., 2011). IP5 is closely located in the binding pocket of JA-Ile and co-ordinates three arginine residues of COI1 and R206 of the Jas peptide (Sheard et al., 2010). The IP5-free receptor complex is inactive. Previous pull-down experiments revealed that (+)-7-iso-JA-Ile is the most bioactive ligand (Fonseca et al., 2009). This is now substantiated by the crystal structure: most of the ligand is surrounded by COI1 residues, but the keto-group of JA in JA-Ile and the COOH-group of Ile can interact with the Jas domain (Sheard et al., 2010). Initial binding assays with labelled JA-Ile and COI1 protein showed a strong (50-fold) increase in binding and in specificity, if JAZ1 or JAZ6 were used as co-receptor complex component (Katsir et al., 2008b). Site-directed mutagenesis revealed essential amino acid residues for binding of the ligand in the binding pocket established by the COI1–JAZ interaction (Melotto et al., 2008). Although the basic concept of JA-Ile perception is established, there are still several caveats as to the ubiquitination of the JAZs, the exact interaction maps of all the complex members at both low and high JA-lle concentrations and their half-lives.
Recent results show the possibility of the existence of new properties of COI1. Although there is no doubt about the role of COI1 as an F-box protein in JA-dependent signalling via the SCFCOI1 complex, JA-independent signalling by COI1 appeared in analysing a new allele of COI1 involved in regulation of innate immune receptor (NB-LRRs) accumulation (He et al., 2012).
4.4. JA signalling versus OPDA signalling
When the basic concept of JA/JA-Ile perception was established in 2007, a striking exception in binding assays with jasmonate compounds appeared – the JA precursor OPDA was not an active ligand in COI1–JAZ pull-down assays (Thines et al., 2007), although OPDA-specific gene expression had already been reported (Taki et al., 2005). Mechanistic proof came from the crystal structure of the COI1–JAZ co-receptor complex, where OPDA does not fit into the binding pocket for JA-Ile (Sheard et al., 2010). Consequently, there is an increasing number of examples describing an JA/COI1-independent role of OPDA (Wasternack et al., 2013):
Tendril coiling is mainly promoted by OPDA but much less by JA, as previously shown (Stelmach et al., 1998; Blechert et al., 1999).
A distinct set of genes is expressed by OPDA, but only a partial overlap appeared with the expression of JA-induced genes (Taki et al., 2005; Mueller et al., 2008).
Physcomitrella patens is unable to form JA, but accumulates OPDA. The fertility of AOC-knockout lines is decreased, suggesting a requirement for OPDA (Stumpe et al., 2010).
A similar observation was made with developing tomato embryos (Goetz et al., 2012). Here, a preferential and abundant accumulation of OPDA in the seed coat is required for proper embryo development, as shown with tomato mutants defective in OPDA or JA synthesis and JA signalling.
Seed germination is inhibited by JA. However, JA biosynthetic and signalling mutants of Arabidopsis demonstrated that OPDA is the causal compound that inhibits seed germination together with abscisic acid (ABA) in a COI1-independent manner (Dave et al., 2011). According to this scenario, chloroplast-derived OPDA is active in transcriptional activation, but it is not known how the rise in the OPDA is regulated. Here, the above-mentioned esterified OPDA and dinor-OPDA of galactolipids, called arabidopsides, may function as a storage pool of OPDA (Göbel and Feussner, 2009; Mosblech et al., 2009; Dave and Graham, 2012). Moreover, the cytosolic OPDA pool is thought to be regulated via its conjugation with glutathionine (GSH) by GSH transferases and subsequent sequestration in vacuoles (Ohkama-Ohtsu et al., 2011).
OPDA is also thought to have a specific role in the expression of the PHO1;H10 gene, which occurs in several stress responses (Ribot et al., 2008), PHY A signalling and shade avoidance syndrome (SAS) (Robson et al., 2010), hypocotyl growth inhibition (Brüx et al., 2008) and COI1-independent defence signalling via ARF2 (Stotz et al., 2011).
Insect-induced closure of the Venus flytrap, Dionea muscipula, requires OPDA, which affects the secretion of digestive enzymes (Escalante-Pérez et al., 2011).
It is not yet known how OPDA is perceived during OPDA-specific responses. Some of these responses might be explained by the occurrence of an α,β-unsaturated carbonyl group in OPDA, a characteristic of reactive electrophile species (RES) (Farmer and Davoine, 2007).
4.5. Co-repressors interacting with JAZs
Except for the EAR motif of JAZ8 as described above, the JAZ proteins lack a repression motif that is required for direct repression. Consequently, the JAZ proteins are suggested to recruit co-repressors. Indeed, using tandem affinity purification (TAP) NINJA was identified via TAP-tagged JAZ1 and shown to interact with TPL (Pauwels et al., 2010). The hypothetical model for repression of JA-induced gene expression includes the TFs (e.g. MYC2), any JAZ protein and the adaptor NINJA linked to the co-repressor TPL via the EAR motif (Fig. 4). Whereas the JAZ proteins bind to TFs via the Jas domain, the ZIM domain of JAZs mediates homo- and hetero-dimerization as well as binding of NINJA. For JAZ5, JAZ6, JAZ7 and JAZ8 that carry an EAR motif, direct binding to TPL without NINJA is possible (Fig. 4). This is supported by the following data: (1) NINJA over-expressers and knockout lines have a decreased JA response, and (2) the EAR motif of NINJA and its homologue in ABA signalling act specifically in both adaptors (Pauwels et al., 2010). NINJA and TPL were identified as integrators of JA/JA-Ile signalling. Both of them act as co-repressors of JA responses, and link JA and auxin signalling (see section 4.7).
Fig. 4.
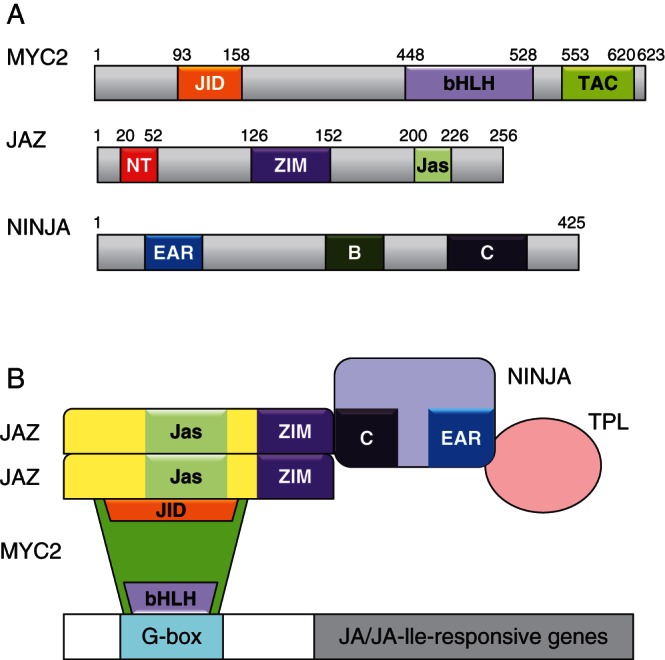
The domain structure of MYC2, Jasmonate ZIM domain proteins (JAZ) and Novel Interactor of JAZ (NINJA) (A) and a hypothetical scheme of interaction between MYC2, JAZ, NINJA and TOPLESS (TPL) (B). Data adapted from Pauwels and Goossens (2011). B, conserved protein domain of NINJA; bHLH, DNA binding domain of MYC2; C, conserved protein domain of NINJA mediating binding to ZIM of JAZ; EAR, ethylene-responsive element binding factor-associated amphiphilic repression motif of NINJA-mediating binding to TPL; Jas, domain of JAZ for binding to COI1, MYC and other TFs; JID, JAZ-interacting domain of MYC2; NT, binding domain of JAZ to other TFs; TAC, domain of MYC2 for homo- and heteromerization; ZIM, domain of JAZ for binding to NINJA and for homo- and heteromerization.
Chromatin modifications performed by histone deacetylases (HDAs) are a basic mechanism underlying the suppression of gene expression, and are involved in Arabidopsis defence responses upon pathogen attack (Berr et al., 2012). HDA6 and HDA19 are known to interfere with JA signalling, thereby affecting pathogen response, senescence and flowering (Zhou et al., 2005; Wu et al., 2008). They are genetically linked to TPL that cannot directly bind to DNA. Possibly, JAZ-mediated repression might finally result from suppression via HDA19 due to its binding to the co-repressor TPL (Fig. 3).
4.6. JAZ targets – TFs mediating JA-specific gene expression
As mentioned above, the plethora of JA signalling is sustained to a remarkable extent by the multiplicity in negative regulation by JAZ proteins and co-repressor activities. The TFs preferentially acting as positive regulators bind to specific elements of the promoters of JA-responsive genes leading to separately acting pathways via singular or combinatorial activities of the TFs. Among them, MYC2 is the most prominent TF, a master switch in JA signalling, because it has been shown to regulate the expression of its most prominent marker gene VEGETATVE STORAGE PROTEIN2 (VSP2). MYCs belong to the bHLH domain-containing TFs, and act as both activator and repressor of distinct JA-responsive gene expressions in Arabidopsis (Lorenzo et al., 2004).
MYC2
This is a prominent member of the MYC-TF-family (Kazan and Manners, 2013). In 2007, it was the only DNA-binding TF known to bind also JAZ family members (Chini et al., 2007). Its central role in numerous signalling pathways such as synthesis of glucosinolates, auxin, tryptophan, ET and JA as well as responses to wounding/insects, oxidative stress, pathogens and ABA-dependent drought stress has already been established (Dombrecht et al., 2007; Kazan and Manners, 2008). It is an activator of JA-induced root growth inhibition, anthocyanin biosynthesis and oxidative stress tolerance, but a repressor in mediating resistance to necrotrophic pathogens and biosynthesis of tryptophan and indol glucosinolates (Lorenzo et al., 2004; Dombrecht et al., 2007). MYC2 activity takes place in a competitive interaction with the ET response factor ETR1 (Lorenzo et al., 2004). The weak phenotype of the myc2/jin1 mutant not only suggested the existence of other MYC-related TFs, but also indicated that JA-responsive gene expression is exclusively controlled by MYC2 (Montiel et al., 2011). MYC2 suppresses the expression of PLETHORA (PLT1 and PLT2) TFs, which are central regulators in auxin-mediated root meristem and root stem cell niche development by directly binding to the promoters (Chen et al., 2011). The PLT1/2 suppression complement the known JA-mediated regulation of auxin biosynthesis in the enzymatic step of anthranilate synthase α1 (ASA1) (Sun et al., 2009), and represents a mechanistic framework for JA-induced root growth inhibition via auxin homeostasis and action. MYC2 is also involved in the circadian clock of JA signalling. Here, TIME FOR COFFEE (TIC) is one of the key components of the circadian clock, which negatively regulates JA signalling (Shin et al., 2012). TIC inhibits MYC2 accumulation, thereby repressing COI1 expression (Shin et al., 2012). There are other examples where MYC2 also acts as a JAZ target: in nicotine biosynthesis (Shoji et al., 2008; Zhang H-B et al., 2012) and in the synthesis of terpenoid indol alkaloids of Catharanthus roseus (Montiel et al., 2011) (see section 5).
Additional TFs are also active downstream of MYC2. The two members of the NAC TF family, ANAC019 and ANAC055, were identified by genetic and biochemical approaches as positive regulators of JA-induced LOX2 and VSP1 expression downstream of COI1 and MYC2 (Bu et al., 2008). In summary, MYC2 is a master regulator in most JA-mediated signalling pathways involved in defence and development in Arabidopsis (Kazan and Manners, 2013): MYC2 mediates (1) antagonistic coordination of two branches in defence responses against herbivores and pathogens, (2) the establishment of induced systemic resistance (ISR) by beneficial soil microbes, (3) effector-mediated suppression of innate immunity in roots, (4) the regulation of cross-talk with SA, ABA, GAs and auxin, (5) a link between JA and other signalling pathways, such as light, phytochromes and circadian clock, and (6) the regulation of development, such as lateral and adventitious root formation, flowering time and SAS.
MYC3 and MYC4
Homologous proteins of MYC2 were picked up independently in three groups by yeast-two-hybrid screening using JAZ as bait (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). MYC3 and MYC4 are closely related to MYC2. Double and triple mutants of the three MYCs and over-expressers of MYC3 and MYC4 showed weak activity of both new MYCs in root growth inhibition as compared with MYC2, but strong involvement in the expression of wound-responsive genes. However, both responses are typical of that mediated by MYC2, thus indicating redundancy. Mutational analysis, however, revealed that the MYC2-regulatory effect was enhanced by MYC3 and MYC4, illustrating another level of modulation in JA signalling by modular and common activities of several TFs. This is supported by the fact that all the three MYC TFs show identical DNA binding specificities and bind preferentially to the G-box (Fernández-Calvo et al., 2011), the cis-element to which most bHLH proteins can potentially bind (Dombrecht et al., 2007).
All the three MYC TFs have two important domains: (1) a JAZ interaction domain (JID) adjacent to the N terminus, which is responsible for JAZ interaction, and (2) a conserved TAC-like domain at the C terminus, which is essential for homo- and hetero-dimerization of MYCs (Cheng et al., 2011; Fernández-Calvo et al., 2011). The JID domain occurring in MYC2, MYC3 and MYC4 is also present in other bHLH TFs, like GL3, EGL3 and TT8, which are known to be involved in anthocyanin formation and trichome initiation, and they have been shown to interact with JAZ1 and JAZ8 (Qi et al., 2011). The WD40/bHLH (GL3, EGL3 and TT8)/MYB (PAP1 and GL1) complex is a regulatory module for anthocyanin and trichome initiation (Qi et al., 2011) (see sections 5.5 and 9.7).
MYB21 and MYB24
Male sterility is the most prominent phenotype of the JA biosynthetic and signalling mutants of Arabidopsis, such as coi1 and opr3 (reviewed by Browse, 2009a, c). In a transcriptome analysis of developing stamens of opr3 plants treated with JA, an up-regulation of TFs occurred, and MYB21 and MYB24 were the TFs identified (Mandaokar et al., 2006). Later, both of them were identified as targets of JAZ1 and JAZ8 by yeast two-hybrid screening (Song et al., 2011), showing that the interactions of both JAZs with MYB21 and MYB24 occur via the N-terminal R2R3 domain (Song et al., 2011). The over-expression of MYB21 in coi1 or opr3 partially rescued stamen filament length for both mutants, but insensitivity to JA in root growth and anthocyanin biosynthesis and susceptibility to Bradysia were not affected (Song et al., 2011). Therefore, MYB21 and MYB24 are more specifically involved in fertility than in other JA-dependent processes.
4.7. Cross-talk between JAZ proteins and other hormone signalling cascades
ET-JA (EIN3/EIL1 and ERF1/ORA59 versus MYCs)
In the JA signalling pathway, there is a parallel branch to the above-mentioned MYC branch, the ETHYLENE RESPONSE FACTOR1 (ERF1), with the marker gene PLANT DEFENSIN1.2 (PDF1.2) (Lorenzo et al., 2004; Pieterse et al., 2012). The synergistic cross-talk between JA and ET is known to occur preferentially for the response to necrotrophic pathogens (Pieterse et al., 2012). Two central TFs of ET signalling, ETHYLENE-INSENSITIVE3 (EIN3) and EIN3-like (EIL1) bind JAZ1, JAZ3 and JAZ9 via the Jas domain of JAZs, resulting in the suppression of EIN3/EIL1 activity (Zhu et al., 2011). This model is the first mechanistic explanation on synergistic cross-talk between ET and JA. Here, as the repressors of JA signalling, JAZs prevent ET signalling by inhibiting the ET-dependent TFs, but in the presence of JA-Ile, where the JAZs are subjected to proteasomal degradation, EIN3/EIL1 becomes free and requires ET for their stabilization as usual.
A second tier of synergistic signalling of JA and ET is conferred by the TFs ORA59 and ERF1 (Pre et al., 2008) that act downstream of EIN3/EIL1 (Leon-Reyes et al., 2009). Here, the synergistic action of JA and ET is mediated by two GCC-boxes, e.g. ORA59 binds to the PDF1.2 promoter (Zarei et al., 2011). The ERF/ORA59 branch is activated upon infection by necrotrophic pathogens leading to the expression of PDF1.2, thus antagonizing the MYC-mediated branch, which is activated by herbivorous insects leading to the expression of VSP2 (Pieterse et al., 2012). Consequently, the defence response against insect attack is expected to be compromised in plants with an activated ERF/ORA59 branch (Fig. 5). Accordingly, an activated MYC-branch of the JA pathway will prevent herbivore-induced stimulation of the ERF branch, and the plants will be less attractive to the herbivores (Verhage et al., 2011).
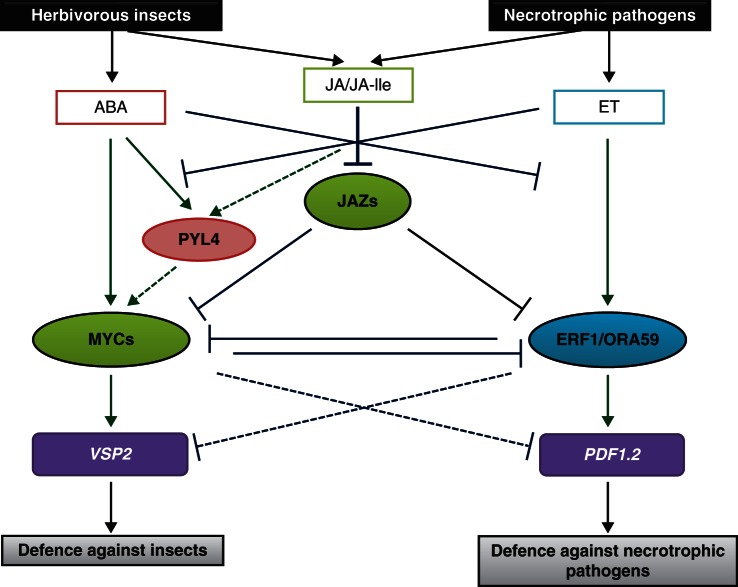
Fig. 5.
The cross-talk between jasmonate (JA), ethylene (ET) and abscisic acid (ABA) triggered in response to herbivorous insects and necrotrophic pathogens (adapted from Pieterse et al., 2012). Attack by herbivorous insects induces JA- and ABA-dependent signalling pathways, whereas infections by necrotrophic pathogens induce JA- and ET-dependent signalling pathways. Both branches are antagonistically regulated. Solid lines, known interactions; dashed lines, hypothetical interactions; green arrows, positive effects; blue inhibition lines, negative effects. Compounds are given in rectangles, transcriptional regulators in circles, regulated genes in purple. ERF1, ethylene response factor 1; ORA59, octadecanoid-responsive Arabidopsis AP2/ERF-domain protein 59; PYL4, PYR1-like protein 4 (ABA receptor); other acronyms are given in Fig. 3.
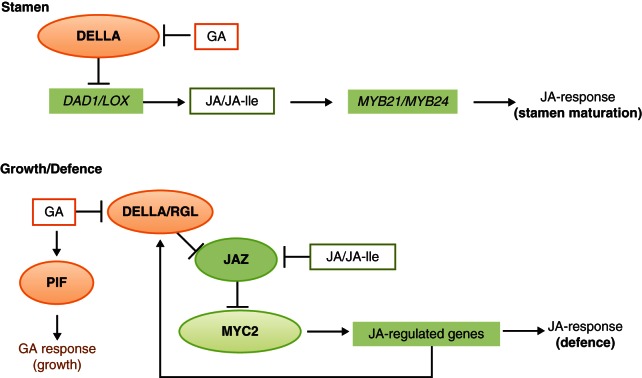
JA-GA (DELLAs versus JAZs)
There are synergistic as well as antagonistic cross-talks between GA and JA depending of the process in which these hormones are involved. For stamen development, both hormones act synergistically (Fig. 6). The DELLA proteins, accumulating upon GA deficiency, prevent JA biosynthesis via the suppression of DAD1 and LOX expression (Cheng et al., 2009; Song et al., 2011). This leads to JA deficiency that causes male sterility by repression of JA-dependent gene expression of the essential TFs MYB21 and MYB24. In the absence of JA/JA-lle, this down-regulation is even attenuated by an inhibition of MYB21 and MYB24 actions through binding of JAZs (Cheng et al., 2009). In contrast, an antagonistic cross-talk between JA and GA occurs in plant growth and defence responses, which are themselves antagonistic because plant defence occurs at the expense of plant growth (Hou et al., 2010; Kazan and Manners, 2012; Yang D-L et al., 2012) (Fig. 6 and see section 9.6).
Fig. 6.
Cross-talk between jasmonic acid (JA)- and gibberellic acid (GA)-signalling pathways in stamen maturation and during growth and defence processes. In stamen, DELLA negatively affects the expression of genes encoding JA biosynthetic enzymes. An increase in GA will result in the removal of DELLA leading to enhanced synthesis of JA/JA-Ile. In turn, this induces the expression of MYB21/24, which is crucial for stamen maturation. In vegetative tissues, the DELLA-TF DELLA RGA-like (RGL) competes with MYC2 for binding to JAZ. With an increasing level of JA/JA-Ile, MYC2 is released and mediates the transcription of not only JA-regulated genes involved in defence but also encoding RGL. An increase in RGL will amplify the defence response by recruiting Jasmonate ZIM domain protein (JAZ) followed by release of MYC2. In contrast, accumulation of GA will lead to degradation of RGL, thereby releasing JAZ to inhibit MYC2. In parallel, GA activates the growth response via phytochrome interacting factor (PIF); other acronyms are given in Fig. 1.
The JAZ proteins have their counterparts in five DELLA proteins of Arabidopsis which are active in GA signalling; GAI/SLY is the homologue of COI1, GID1 is the GA receptor and the DELLAs RGL and RGL1-like (RGL1, RGL2 and RGL3) are the repressors (Schwechheimer, 2012). Interestingly, all these proteins can interact with JAZs via the Jas domain, thereby competing with MYC2 in JAZ binding (Hou et al., 2010; Wild et al., 2012; Yang D-L et al., 2012). In this model GA triggers the degradation of DELLA, thereby allowing JAZ1 to bind to MYC2, which leads to the repression of JA signalling, whereas in the absence of GA, DELLAs exist and bind to JAZs resulting in de-repression of MYC2 (Hou et al., 2010; Wager and Browse, 2012). This was shown in particular for RGL3 (Wild et al., 2012); its expression is induced in a COI1- and MYC2-dependent manner due to direct binding of MYC2 to the RGL3 promoter, and it interacts with JAZ1, again representing a competitive binding for MYC2. Consequently, the rise in JA-Ile will result in an accumulation of RGL3 leading to trapping of JAZ1 and enhancement of the MYC2 activity. In the presence of GA, however, RGL3, a negative regulator in GA signalling, will be subjected to degradation, thereby allowing JAZ1 to inhibit the MYC2 activity and resulting in depression of JA-induced gene expression (Wild et al., 2012). Obviously, there is a flexible balance of both negative regulators DELLAs and JAZs, which sustain the antagonistic behavior of the growth of above-ground plant parts vis-à-vis their defence. Note that cross-talks between plant hormones can differ dramatically between different plant organs. In contrast to this antagonism in above-ground plant parts, there occurs no cross-talk between GA and JA in roots.
Auxin-JA (ARFs versus MYBs)
In roots, the well-known growth inhibition by JA occurs via a cross-talk with auxin. This root growth inhibition does not take place in coi1 and myc2 mutants, but is increased in the jaz10 mutant, indicating the involvement of COI1, MYC2 and JAZ10 (Chen et al., 2011). As MYC2 represses the expression of PLETHORA (see section 4.6), the central regulator of root meristem activity, cell elongation and cell number, this might counteract the auxin–TIR1-AUX/IAA–ARFs signalling cascade, leading to diminished expression of both PLETHORA genes (Chen et al., 2011). Additionally, JA may increase auxin levels by inducing the expression of ASA1 that encodes the first enzyme in auxin biosynthesis (Sun et al., 2009). On the other hand, there is an auxin-induced expression of JAZ1, which might have an integrator function in auxin–JA interaction leading to a regulatory loop in sustaining auxin and JA signalling (Grunewald et al., 2009). Interestingly, the tryptophan-conjugates of JA and IAA are endogenous auxin inhibitors that affect auxin sensitivity in a COI1-independent and/or TIR1-dependent manner (Staswick, 2009), thereby illustrating another mechanism of JA-auxin cross-talk.
In flowers, auxin signalling requires AUXIN RESPONSE FACTOR6 (ARF6) and ARF8, both of which induce the expression of JA biosynthesis genes in filaments (Nagpal et al., 2005). Consequently, arf6-2arf8-3 filaments are characterized by low JA levels. Obviously, petal and stamen growth are determined by a common regulatory network that entails JA-dependent transcription factors MYB21 and MYB24 (see section 4.6) as well as auxin-dependent transcription factors ARF6 and ARF8 (Reeves et al., 2012). In addition, the regulatory effects of ARF6 and ARF8 on JA biosynthesis result in a negative regulation of class1 KNOX genes, which are important negative regulators of optimal flower development (Tabata et al., 2010). It is thus clear that ARF6 and ARF8 function via JA in the progression of floral development.
Brassinosteroid (BR)-JA (DWARF4 versus MYBs)
In contrast to the well-known growth-inhibitory effect of JA, BRs promote the growth of above-ground plant parts. The BR signalling cascade is well described, including the BR receptor BRI1, the BR-associated kinase1 (BAK1) and the transcription factors that are involved in BR-induced expression BES1 and BZR1 (Clouse, 2002). The main phenotype of mutants that are defective in the BR receptor is dwarfism. Interestingly, in a genetic screen on suppressors of coi1, a psc1 mutant was found with partially suppressed coi1-phenotype (Ren et al., 2009). This mutant carries a mutation in DWF4 that encodes a key enzyme in BR biosynthesis, suggesting that BRs might counteract JA signalling. Indeed, psc1 in a background of wild-type COI1 displays JA hypersensitivity, especially in respect to JA-induced inhibition of root growth (Huang et al., 2010). The BR application leads to anthocyanin accumulation, which is a hallmark of JA-induced responses (see section 5.5), whereas JA-induced anthocyanin accumulation is reduced in BR-biosynthetic mutants (Peng et al., 2011). Here, JA-mediated induction of ‘late’ anthocyanin biosynthesis genes was suppressed by reduced BR synthesis (dwf4-102) or disturbed BR perception (bri1-4) via the reduced expression of two MYB genes PAP1 and PAP2 (Peng et al., 2011; Song et al., 2011). In contrast, JA inhibits COI1-dependent DWF4 expression, indicating that DWF4 itself is down-regulated by JA and is located downstream of COI1 in the JA signalling pathway.
JA-ABA (PYL4 versus JA-dependent TFs)
Cross-talk between ABA and JA is not surprising given their common central role in several stress responses (Cutler et al., 2010). ABA was identified as an essential signal in Pythium irregulare-induced defence responses of A. thaliana (Adie et al., 2007). Although contentious, the positive and negative roles of ABA in JA/ET-mediated defence have been described and at least partially linked to callose formation (Ton et al., 2009). Besides its role in plant resistance, there is a role of ABA in JA-mediated wound response (Kazan and Manners, 2008). The recent identification of the direct ABA receptors, the PYR/PYL/RCAR proteins, has allowed the synthesis of a mechanistic view on the cross-talk between ABA and JA. For instance, the tobacco NtPYL4 gene has been shown to encode a functional ABA receptor and is induced by JA (Lackman et al., 2011). A similar link between ABA and JA exists in Arabidopsis, where homologues of NtPYL4, PYL4 and PYL5 are also induced by JA. The pyl4 and pyl5 mutants exhibiting hypersensitivity in JA-mediated biomass reduction recorded a decline in JA-induced anthocyanin accumulation (Lackman et al., 2011). This unequivocally suggests that the ABA–JA cross-talk contributes to maintaining the balance between growth and defence (Fig. 5).
JA–SA (COI1/MYC2 versus NPR1/TGAs)
JA–SA cross-talk has been known for a long time and is the most studied cross-talk among plant hormones. It has been recently reviewed in detail, with reference to its role in plant immunity (Pieterse et al., 2012). Therefore, we will discuss here only key aspects to complement the cross-talks mentioned above. In principle, JA signalling is involved in responses to necrotrophic pathogens and herbivorous insects with key components such as JA biosynthetic enzymes, COI1, JAZs, NINJA, TPL and MYC2 as described above. In response to biotrophic pathogens, however, SA is the central regulator (Vlot et al., 2009; Pieterse et al., 2012). Here, the SA biosynthesis occurs via two parallel pathways – the well-known PAL reaction and the ISOCHORISMATE SYNTHASE (ICS/SID2) reaction (Garcion and Metraux, 2006). As an experimental tool, overexpressing lines of the bacterial NahG gene encoding an SA dehydrogenase have been generated (Mur et al., 1997) significantly compromising the accumulation of SA (Delaney et al., 1994). The central regulator in SA signalling is NONEXPRESSOR OF PR GENES1 (NPR1) that, in the presence of SA, is a transcriptional co-activator for many defence genes. There are NPR1 multimers which monomerize by SA-induced changes of the redox state via thioredoxin followed by the transport of the monomeric forms into the nucleus. Here, they bind as activators to TGA TFs specific for SA-inducible genes, are phosphorylated during transcription initiation and are subsequently ubiquitinylated following binding to a CULLIN3-based ligase (Fu et al., 2012), and are subjected to proteasomal degradation. Later, new nuclear-imported NPR1 monomers can again allow SA-induced gene expression (Spoel et al., 2009). In this model, turnover of the co-activator NPR1 has dual roles in both preventing and stimulating gene expression. In addition to this basic component, several other factors are also known to be involved in the SA signal transduction pathways (Pieterse et al., 2012).
The SA–JA cross-talk was initially observed in the wound response of tomato (Harms et al., 1998). In nature, however, plants are attacked simultaneously and sequentially by a single or several attackers that induce the SA- and/or JA-signalling pathways. The preferential induction of one pathway and its antagonistic interaction with another pathway has been repeatedly demonstrated (Koornneef and Pieterse, 2008; Pieterse et al., 2012) and can be shifted from an antagonistic to synergistic interaction depending on SA and JA concentrations (Wees et al., 2000; Mur et al., 2006). Environmental cues, such abiotic stresses as thermotolerance (Clarke et al., 2009) or shade avoidance (Ballaré, 2011), seem to be involved in maintaining a balance between the SA and JA pathways. The cross-talk between SA and JA has been observed in many Arabidopsis accessions (Koornneef and Pieterse, 2008), and is even transmitted to the next generation (Luna et al., 2012). The adaptability of plants in nature may be attributed to the flexibility of both pathways as conditioned by their individual components as well as interactions.
The putative roles of several new players in the above-mentioned model are as follows (Pieterse et al., 2012):
Mitogen-activated protein (MAP) kinases are clearly involved (Rodriguez et al., 2010a). MPK4 acts as a negative and positive regulator of SA and JA signalling pathways, respectively.
The redox regulators glutaredoxins (GRXs) and thioredoxins (TRXs) that sustain the redox state of proteins in terms of disulfide bridges in a glutathionin-dependent manner represent other regulatory modules, where JA decreases and SA increases the glutathionin pool (Spoel and Loake, 2011).
The above-mentioned NPR1, active as a monomer in the nucleus by binding to TGAs, has a distinct role in the cytoplasm as a multimer (Ramirez et al., 2010) and is regulated by ET (Leon-Reyes et al., 2009).
Some of the clade II TGAs, such as TGA2, TGA5 and TGA6, have been shown to have a positive effect on JA/ET-dependent defence gene expression in the absence of SA (Zander et al., 2010). Obviously, these TGAs counteract the negative effect of MYC2 in the JA/ERF branch (see above). GRXs (called ROXYs) have been postulated to modulate the activities of these TGAs in the JA/ERF branch (Zander et al., 2012; Gatz, 2013). Consequently, the outcome of the JA/ET-induced and SA-suppressed expression of defence genes would be sustained by the simultaneous and sequential action of several key factors, such as the levels of JA, ET and SA, and the expression of MYC2, ROXY19, TGA2, TGA5, ERF/EIN and PDF1.2 (Gatz, 2013).
WRKY TFs, such as WRKY50 and WRKY51, are essential for SA-mediated suppression of JA signalling (Gao et al., 2011), while WRKY62 is a negative regulator of JA signalling acting downstream of cytosolic NPR1 (Mao et al., 2007). Another WRKY TF involved in the cross-talk between SA and JA is WKRY70, which is directly controlled by AtMYB44 that activates the SA-induced defence response and represses the JA branch (Shim et al., 2013).
Subunit 16 of the Mediator complex (MED16) is required for positive regulation of systemic acquired resistance (SAR) via NPR1, but is a negative regulator of resistance to necrotrophic fungal pathogens via the JA/ET pathway. It is thus clear that the SA and JA/ET pathways converge to MED16, which links specific transcriptional regulators with the RNA polymerase II transcription machinery (Zhang X et al., 2012a).
The Ca2+/CaM-binding TF AtSR1 causes down-regulation of SA levels, thereby abolishing its negative impact on basal and induced JA levels upon wounding (Qiu et al., 2012).
The targets of these enumerated effectors of SA–JA signalling are not well understood. In earlier dissection of SA–JA cross-talk, a direct inhibitory effect of SA on JA biosynthesis was assumed due to the inhibitory effect of aspirin (Harms et al., 1998). However, recent mutant analyses have clearly shown that SA antagonizes the JA pathway downstream of JA biosynthesis (Leon-Reyes et al., 2010). Another target for SA-mediated suppression of JA signalling occurs at the transcriptional level. Here, components of the SA signalling, such as TGAs and WRKYs, inhibit the expression of JA-dependent TFs (Pieterse et al., 2012). The numerous components of SA–JA cross-talk are superimposed by other hormones that modulate this cross-talk (Pieterse et al., 2012). This may provide a simpler explanation for the mechanistic basis of the above-mentioned cross-talks such as JA–ET, JA–GA, JA–ABA and JA–BR. Furthermore, evolution of JA–SA cross-talk is a matter of interest because this obviously ancient phylogenetic cross-talk is thought to be of adaptive significance (Thaler et al., 2012).
5. REGULATION OF PLANT SECONDARY METABPOLISM BY JASMONATES
Twenty years ago, it was demonstrated that an endogenous rise in JA levels upon elicitation with a rough yeast elicitor was associated with the induction of alkaloid synthesis in plant cell cultures (Gundlach et al., 1992). Later, the proof of concept for JA-mediated induction of biosynthesis of secondary metabolites came from studies on constitutive activation of the JA signalling pathway in tomato, leading to the constitutive accumulation of caffeoylputrescine (Chen et al., 2006). In 2000, the first TFs involved in JA-dependent terpenoid indole alkaloid (TIA) synthesis in C. roseus were identified (van der Fits and Memelink, 2000). These TFs were called OCTADECANOID-DERIVATIVE RESPONSIVE CATARANTHUS AP2-DOMAIN2 and 3 (ORCA2 and ORCA3) (reviewed by Memelink et al., 2001). Meanwhile, several TF families involved in the synthesis of TIA, nicotine, artemisinin, anthocyanins, camalexin, indol glucosinolates and volatile terpenes have been identified. A common facet underlying JA-mediated transcriptional control of secondary metabolite biosynthesis is involvement of the SCFCOI1 complex, JAZ proteins and MYC2 together with additional components, such as WRKYs, ORCAs, ERFs, MYBs, PAP1 and ZCTs, all of them being active in distinct pathways.
As the involvement of TFs in JA-mediated regulation of secondary metabolite biosynthesis has been recently reviewed (Memelink, 2009; De Geyter et al., 2012), we address here only a few aspects. The examples shown below illustrate that similar and homologous TFs are involved in the JA-dependent biosynthesis of different secondary compounds in different plant species, suggesting an early conserved evolution of the JA signalling network regulating the biosynthesis of secondary metabolites (De Geyter et al., 2012).
5.1. Nicotine
Most of the structural genes encoding enzymes for nicotine biosynthesis are transcriptionally regulated by JA, and depend on a functional COI1–JAZ co-receptor (Shoji et al., 2008). In a genomic screen, bHLH TFs, such as MYC2, were identified to be active in nicotine biosynthesis (Todd et al., 2010; De Boer et al., 2011; Shoji and Hashimoto, 2011). NtMYC2 acts together with AP2/ERFs that occur in the NIC2 locus (Shoji et al., 2010; De Boer et al., 2011). The family of tobacco ERFs has 239 members (Rushton et al., 2008). Seven of them resulting from gene duplication are involved in nicotine biosynthesis, and are clustered in the NIC2 locus. Together, they represent a positive regulatory unit (Shoji et al., 2010). NIC2/ERFs are close homologues of ORCA3 of C. roseus and activate TIA biosynthesis genes via a GCC-box. Here, ORCA/ERF221 and MYC2 act synergistically in binding, and both of them are post-translationally up-regulated by a JA-modulated phosphorylation cascade, where MAPKK1 (JAM1) is active (De Boer et al., 2011). It is interesting that, in two different pathways and two different plant species (nicotine biosynthesis in tobacco and TIA biosynthesis in C. roseus), homologous TFs evolved obviously independently. The widely distributed induction of secondary metabolite biosynthesis by JA may indicate an evolutionary advantage of an established regulatory module.
5.2. Vinblastine
Vinblastine is a TIA and is synthesized in C. roseus cells, where expression of the enzyme-encoding genes is regulated by a cascade of transcription factors including CrMYC2 that regulate the expression of the AP2/ERF domain TFs, such as ORCA2 and ORCA3 (Zhang et al., 2011). CrMYC2 is encoded by an immediate-early JA-responsive gene. CrMYC2 binds in vitro to jasmonate regulatory elements (JREs) in the promoter of ORCA3 leading to the expression of ORCA3. A down-regulation of CrMYC2, however, does not down-regulate the TIA biosynthesis, indicating sufficient basal expression of ORCAs (Zhang et al., 2011). Moreover, at least ORCA3 regulates most but not all steps in TIA synthesis (Suttipanta et al., 2011). The negative regulation of CrMYC2 by JAZ as observed in many JA-dependent pathways remains to be elucidated.
5.3. Artemisinin
Biosynthesis of the antimalarial sesquiterpene lactone artemisinin is positively controlled by two JA-responsive ERFs, namely ERF1 and ERF2 (Yu et al., 2012), which act in a concerted manner with MYC2. WRKY1, a TF in artemisinin biosynthesis, is also assumed to act in such a concerted manner with MYC2 (Ma et al., 2009). Artemisinin is synthesized and stored in glandular secretory trichomes. Global transcript profiling revealed the expression of trichome-specific genes correlating with the expression of genes active in artemisinin biosynthesis (Maes et al., 2011). All these genes are simultaneously activated in a JA-dependent manner.
The artemisinin biosynthetic machinery is confined to specialized cells of glandular trichomes (Olsson et al., 2009). In this context, it is interesting to note another JA-related process in Artemisia annua. The volatile MeJA released from this species has been correlated with the expression of defence genes in a neighbouring tomato plant (Farmer and Ryan, 1990).
5.4. Glucosinolates/camalexin
Glucosinolates are a large group of secondary metabolites involved in plant resistance to insects and pathogens. They are amino acid-derived compounds and can be classified into aliphatic, benzenic and indolic glucosinolates synthesized in numerous steps, most of which are JA-inducible (Sønderby et al., 2010). The main components of the JA/JA-Ile signalling pathway leading to glucosinolates and camalexin have been identified. For camalexin biosynthesis, an SCFCOI1–JAZ-MYC2 branch and an MEK1-MKK3-MPK6-WRKY33 branch have been identified (De Geyter et al., 2012). Camalexin biosynthesis seems to be controlled by ANAC042, a member of the NAC TF family. Expression of ANAC042 is induced by flagellin and depends on ET signalling, but is repressed by the application of MeJA (Saga et al., 2012) indicating a modulation of camalexin formation by JA via ANAC042 (De Geyter et al., 2012). An additional control is given by a member of the DNA-binding one finger (DOF) TF family. Here, the JA/JA-Ile-inducible DOF1.1 is a positive regulator of indole glucosinolates via CYP83B1 expression (Skirycz et al., 2006).
5.5. Anthocyanin
Anthocyanin accumulation represents the most prominent JA/JA-Ile phenotype. TFs, such as PAP1, EGL3, GL3, MYB75 and TT8 being essential components of the WD-repeat/bHLH/MYB transcriptional complexes, are involved in anthocyanin biosynthesis (and trichome development, see section 9.7). Recent biochemical and genetic evidence indicated that these TFs are targets of JAZs, thereby providing a mechanistic framework for JA-induced anthocyanin formation (Qi et al., 2011).
5.6. Benzylisoquinoline alkaloids
This group of compounds comprising about 2500 structures including the most prominent compound morphine is of pharmacological interest. Several of the numerous enzymes in their biosynthesis consisting of different branches (Ziegler and Facchini, 2008) are encoded by JA-inducible genes. These pathways were among the first to be identified, where an endogenous rise in JA upon elicitation was shown to be the reason for alkaloid synthesis (Gundlach et al., 1992; Blechert et al., 1995).
6. JA IN HERBIVORY AND PLANT–INSECT INTERACTIONS
The molecular recognition of pathogens and herbivores by plants exhibits remarkable similarities in the modules which are used for recognition and responses (Erb et al., 2012). For herbivores, at least three different responses can be conceptually distinguished: (1) herbivore-induced immunity (HTI) can appear upon recognition of oviposition-associated compounds, (2) HTI can occur upon perception of herbivore-associated molecular patterns (HAMPs) or damage-associated molecular patterns (DAMPs), and (3) wound-induced resistance (WIR) is generated by mechanical wounding during herbivory. These responses are linked in several tiers of activity. Oral secretions of insects are produced in a species-specific manner with quantitative and qualitative differences among the elicitor compounds, whereas plants respond to these elicitor combinations differentially (Erb et al., 2012).
The elicitation of a wound response in plants appearing upon mechanical wounding or herbivore attack is one of the most prominent examples and extensively studied areas, where JA/JA-Ile is involved as a signal. The data generated in this area of research over the last five years has been thoroughly reviewed (Howe and Jander, 2008; Koo and Howe, 2009; Felton and Tumlinson, 2008; Walling, 2009; Dicke and Baldwin, 2010; Heil and Karban, 2010; Bonaventure et al., 2011b; Sun et al., 2011a; Erb et al., 2012; Meldau et al., 2012). Therefore, only some aspects will be discussed here. Moreover, N. attenuata has been intensively studied in the last decade regarding different aspects of herbivory, JA/JA-Ile biosynthesis and JA/JA-Ile signalling, including field experiments in the desert of Utah (USA). The complexity of JA/JA-Ile biosynthesis and signalling in relation to herbivory as dissected in this species compared with that in other model plants, such as Arabidopsis and tomato, is unique and has been repeatedly reviewed (Kant and Baldwin, 2007; Schwachtje and Baldwin, 2008; Gális et al., 2009; Wu and Baldwin, 2009; Baldwin, 2010; Dicke and Baldwin, 2010; Kessler et al., 2010; Wu and Baldwin, 2010; Bonaventure et al., 2011b; Kessler and Baldwin, 2011; Meldau et al., 2012).
Local wounding by herbivores leads to a burst in newly synthesized JA. The constitutive occurrence of enzyme proteins involved in JA biosynthesis in all leaf tissues (as in Arabidopsis; Stenzel et al., 2003b) or in vascular bundles (as in tomato; Hause et al., 2000, 2003) may be ascribed to such an immediate rise in JA levels within several minutes (Glauser et al., 2008; Mielke et al., 2011), whereas later (from 15 min onwards) the transcriptional machineries for the expression of LOX, AOS, AOC, OPR3 and JAZs are activated (Stenzel et al., 2003a, b; Chung et al., 2008; Koo and Howe, 2009). In this signalling network compounds of insects' oral secretions, such as volicitin (Bonaventure et al., 2011b), the peptide systemin, H2O2, NO and ET, act as additional positive signals (see Wasternack, 2006; Koo and Howe, 2009).
The involvement of jasmonates in systemic response upon local wounding has been a matter of debate. Primarily, grafting experiments with tomato mutants defective in JA biosynthesis and signalling revealed strong evidence that signalling but no JA biosynthesis is required in systemic leaves (Li et al., 2002). Support for the involvement of JA compounds in systemic response came from observations that showed the occurrence of JA biosynthesis enzymes in mid veins of wounded leaves (Hause et al., 2000, 2003), enrichment of jasmonate compounds in mid veins (Stenzel et al., 2003a; Glauser et al., 2008) as well as their occurrence in phloem exudates of systemic leaves (Truman et al., 2007; Gaupels et al., 2012). However, in Arabidopsis, it has been shown that the occurrence of JA/JA-Ile in systemic leaves requires the presence of intact OPR3 and JAR1, accompanied by a rapid decline in OPDA levels, thus arguing against the transport of any JA compound (Koo et al., 2009). In feeding experiments, labelled JA-Ile could not be recovered in systemic leaves, suggesting that it is not a long-distance signal in N. attenuata (Wang et al., 2008). Less stronger support for this view also came from studies on systemic transport of labelled JA-Ile in tomato (Matsuura et al., 2012). Besides chemical signalling by JA compounds, the involvement of hydraulic and electrical signalling, due to action and variation potentials, in systemic response has been discussed (see Koo and Howe, 2009). More recently, a ‘system potential’ has been proposed for systemic wound signalling that involves stimulation of an H+-ATPase in the plasma membrane concomitant with ion fluxes (Zimmermann et al., 2009). Although volatile MeJA that is released from locally wounded leaves has been proposed to act as a long-distance signal (Heil and Ton, 2008) to evoke the systemic response, this has not yet been experimentally substantiated (Koo and Howe, 2009).
Systemin was the first peptide identified as a signalling compound in wounded tomato leaves (Pearce et al., 1991). Initially, systemin was thought to be a systemic signal involved in long-distance signalling. After two decades of research on wound-induced systemic response, systemin is thought to have a minor role by amplification of systemic wound signalling in a tissue-specific manner (Hind et al., 2010; Sun et al., 2011a). Due to the central role of JA/JA-Ile in SCFCOI1-mediated signalling, one amplification activity of systemin seems to be its positive regulation of the expression of AOC and JA formation upon wounding (Stenzel et al., 2003a). This activation of JA biosynthesis by systemin involves activity of MPK1, MPK2 and MPK3 (Kandoth et al., 2007) (for details see Koo and Howe, 2009; Sun et al., 2011a; Yamaguchi and Huffaker, 2011).
Wounding by herbivores leads to (1) direct defence via synthesis of toxic compounds as well as toxic or antinutritional proteins such as proteinase inhibitors (PIs) and (2) indirect defence by release of volatiles to affect the attraction of carnivores, parasitoids and predators or by altered oviposition of herbivores (Wasternack and Hause, 2002; Howe and Jander, 2008). In this event plants become immunized by the PI expression that affects protein digestion in the herbivore gut. Of the herbivore- and JA-induced proteins, threonine deaminase 1 (TD1) is required for the formation of isoleucine. An interesting adaptive evolution by gene duplication of TD has been demonstrated for tomato (Chen et al., 2007; Gonzales-Vigil et al., 2011). The proteolytic cleavage of the regulatory domain of TD2 may be attributed to plant resistance in response to herbivory.
However, different defence strategies adopted by plants against phloem-feeding insects, such as aphids and whiteflies, must be distinguished (Walling, 2008). These strategies include hormonal signalling by ET, SA and JA affecting pre-entry, entry and colonization by the insects. Recently, the importance of root-derived oxylipins in colonization of the above-ground organs by insects has been elucidated. A phloem sap-consuming green peach aphid of Arabidopsis needs LOX5-derived oxylipins produced within the roots for infestation of the foliage (Nalam et al., 2012). The strategy used by aphids and whiteflies for delivering salivary compounds and proteins is similar to that adopted by phytopathogenic microbes. Here, toxic compounds or effector proteins are released to suppress the host's PAMP-triggered immunity (PTI), but may be perceived by the host plant via different strategies leading to effector-triggered immunity (ETI) that results in various defence responses (Jones and Dangl, 2006; Pieterse et al., 2012).
Finally, plants such as Arabidopsis synchronize defence against herbivores by circadian JA accumulation with circadian insect behavior (Goodspeed et al., 2012). Accumulation of JA and SA is circadian-regulated in different phases, and cabbage loopers (Trichoplusia ni) feed rhythmically on plants grown in light/dark cycles with only moderate tissue damage.
7. JA IN PLANT–PATHOGEN INTERACTIONS
As mentioned above, recognition and response modules can be defined for microbe-, pathogen- and damage-associated molecular patterns (MAMPs, PAMPs and DAMPs) which are similar to that active in herbivory. Pattern recognition receptors (PRRs) that recognize PAMPs develop PTI, which is suppressed by pathogen effectors, whereas the resistance gene products that recognize the effectors lead to ETI (Jones and Dangl, 2006; Erb et al., 2012).
Current results suggest that JA induces resistance against necrotrophic pathogens, some phloem-feeding insects and chewing herbivores, whereas SA induces resistance against biotrophic pathogens and some phloem-feeding insects. This antagonistic SA–JA cross-talk and its evolutionary significance have been discussed recently (Thaler et al., 2012). We have already discussed some molecular aspects of SA–JA cross-talk in section 4.7, while the role of JA in plant interactions with plant–necrotrophic pathogen interactions has been thoroughly reviewed over the last couple of years. However, we refer to a recent and elegant review on hormonal modulation of plant immunity in relation to JA and its cross-talk with SA (Pieterse et al., 2012). Thus, our discussion here will be limited to only some aspects.
7.1. JA, ET and SA in plant–pathogen interactions
The JA signalling cascade via SCFCOI1–JAZ is the backbone representing a link between responses to necrotrophic pathogens and resistance to herbivorous insects (Fig. 5). In both cases, JA is generated. JA acts synergistically with ET upon attack by necrotrophic pathogens, but with ABA during herbivory. ET confers the defence response via the expression of ERF1/ORA59 and PDF1.2, whereas ABA operates via PYL4 and MYCs to elicit the defence response through the expression of VSP2. There are two antagonistic interactions between both pathways: (1) the level of ET and ABA; and (2) at the level of TFs, e.g. MYCs versus ERF/ORA59 (Fig. 5). However, additional antagonistic interactions also originate from the backbone of the JA-mediated signalling cascade that intersects the two pathways. Furthermore, a bioinformatic approach with more than 300 publicly available microarray datasets on the co-expression of ET, JA and SA biosynthesis and signalling components illustrated the signal transduction network intersecting these hormones in plant defence (van Verk et al., 2011). In nature, plants are attacked simultaneously or subsequently by biotrophic or necrotrophic pathogens or by sucking or chewing insects. Consequently, the cross-talk between various signalling components becomes increasingly important. Plants survive in response to interactions between multiple attackers by prioritizing a specific signalling pathway and rewiring the hormone signalling network. Necrotrophic pathogens, such as B. cinerea, have evolved strategies to overcome the host defence system by negotiating the SA–JA cross-talk. Upon infection of tomato, they release β-(1,3)-(1,6) glucan, an exopolysaccharide, that activates the SA–NPR1 pathway, but simultaneously suppresses the JA pathway required for acquiring the host resistance (El Oirdi et al., 2011). Pseudomonas syringae represents an exceptional example of how pathogens hijack hormone-regulated host signalling cascade. This biotrophic pathogen is able to form the plasmid-encoded bacterial toxin coronatine, which is highly active as a molecular mimic of JA-Ile (see sections 2.6 and 4). Biotrophic pathogens such as P. syringae are defended via the SA-NPR1-TGA signalling cascade, but its simultaneous injection of toxins, such as coronatine or bacterial effector proteins into the host, alters the homoeostasis of JA and other hormones such as ABA and auxin, leading to the suppression of host immunity (Pieterse et al., 2012). Recent results show that MYC2-mediated activation of the TFs, such as ANAC019, ANAC055 and ANC972, leads to stomata reopening allowing an entry of P. syringae (Zheng et al., 2012). This provides a mechanistic explanation for coronatine-induced increase in the virulence of P. syringae. Coronatine acting preferentially by suppression of SA signalling (Pieterse et al., 2012) is a multifunctional defence suppressor that also suppresses SA-independent signalling leading to callose deposition and even promotes bacterial growth in a COI1-independent manner (Geng et al., 2012).
Additional oxylipins, such as 9-LOX and α-DOX products, are also involved in conferring resistance against biotrophic pathogens via JA signalling (Vicente et al., 2012). It has been shown that plants produce N-acylamides that confer resistance to necrotrophic pathogens by activating JA biosynthesis and signalling (Méndez-Bravo et al., 2011). Interestingly, arachidonic acid (AA), the counterpart of the JA precursor α-LeA occurring in metazoan species but not in plants, is perceived by plants and acts through an increase in JA levels concomitantly with resistance to necrotrophic pathogens (Savchenko et al., 2010). Obviously, AA is an evolutionarily conserved signalling molecule that acts in plants in response to stress similar to that in animal systems. However, the transport of AA from the pathogen into the plant is unknown.
7.2. Systemic signalling in pathogen defence
SAR has long been known to be induced upon primary infection of a plant. It is an inducible defence mechanism against pathogens established distal to the primarily infected organ. Another type of resistance is the ISR appearing in leaves upon colonization of roots by plant growth-promoting rhizobacteria and during arbuscular mycorrhization (see section 8). Systemic responses occur mainly as priming effects. Similar to systemic signalling upon herbivory (see section 6), mobile signals are involved in linking the local infection with the distal response. Initially, SA was assumed to act as a phloem-mobile systemic signal for inducing SAR. Grafting experiments using transgenic plants in which SA was degraded by a bacterial salicylate hydroxylase (nahG plants), however, argue against the role of SA as a mobile signal in SAR (Mur et al., 1997).
The putative role of JA in SAR is controversial. There are several studies that reported increased JA levels in phloem exudates of systemic leaves, an SAR-induced systemic increase in the expression of JA biosynthesis genes and an attenuation of pathogen-induced SAR in JA biosynthetic and signalling mutants when challenged with an avirulent strain of P. syringae (Truman et al., 2007; Chaturvedi et al., 2008). These all point to a definitive role of JA in SAR. On the other hand, several studies could not detect a role of JA in SAR (Mishina and Zeier, 2007; Attaran et al., 2009). However, note that the dose of SAR-inducing pathogens was remarkably different in the two sets of experiments, without taking into account the impact of the hypersensitive response (HR; Shah, 2009). Possibly, JA is conditionally required to induce SAR depending on whether HR is involved or not (Shah, 2009).
The establishment of SAR is much more complex than previously suggested. At present, methyl salicylate (MeSA) is regarded as a critical mobile signal (Park et al., 2007). Its activity seems to depend on the balance between light-dependent formation of SA and MeSA and interactions with additional compounds, such as azelaic acid (Jung et al., 2009), the abietane diterpenoid dehydroabietanal (DA), the lipid transfer protein DIR1 and pipecolic acid (reviewed by Dempsey and Klessig, 2012). Once these signals are transported to systemic leaves, they may synergistically interact to induce SAR via NPR1.
8. JASMONATES IN SYMBIOTIC INTERACTIONS
Mutualistic symbioses are important in nature and sustainable agriculture, the most important of which are the almost ubiquitously occurring arbuscular mycorrhiza (AM) and root nodule symbiosis (RNS). Whereas the former entails an association with obligate biotrophic fungi of the phylum Glomeromycota (Schüssler et al., 2001), the latter is the interaction of the leguminous roots with nitrogen-fixing bacteria. In these two forms of intracellular (endo)symbioses, the heterotrophically growing microbial partners are accommodated within living root cells (for a review see Oldroyd et al., 2009). The establishment and maintenance of both symbioses require plant resources, such as photosynthetically assimilated carbon. In turn, the respective microbial partners deliver mineral nutrients, mainly phosphate and nitrate in the case of AM and RNS, respectively. Both mutualistic interactions are based on a complex molecular cross-talk between the plant and the microsymbiont (Bonfante and Genre, 2010; Gough and Cullimore, 2011; Geurts et al., 2012). This cross-talk involves recognition of the partners, establishment of mutualistic interactions, regulation of nutrient exchange and maintenance of mutualism. The role of jasmonates in these processes has been intensively studied and reviewed over the last two decades (Ludwig-Müller, 2000; Pozo et al., 2005; Hause et al., 2007; Pozo and Azcon-Aguilar, 2007; van Wees et al., 2008; Hause and Schaarschmidt, 2009; Gutjahr and Paszkowski, 2009; Mortier et al., 2012). Here, we focus on new data showing the involvement of jasmonates in AM and RNS, with a reference to plant interactions with the mutualistic endophyte Piriformospora indica.
8.1. Arbuscular mycorrhiza (AM)
AM are the most common type of mycorrhiza (Smith and Read, 2008). They originated more than 400 million years ago and enabled plants to colonize the land (Brundrett, 2002). Today, this mutualistic interaction is still very common among land plants. About 80 % of plants can interact with the AM fungi (Smith and Read, 2008). Besides supplying mineral nutrients and water, AM can improve the tolerance of plants to certain abiotic and biotic stressors, including drought, salt, heavy metals, and different pathogens and herbivorous insects (García-Garrido and Ocampo, 2002; Pozo and Azcon-Aguilar, 2007; Hartley and Gange, 2009). Therefore, AM are also regarded as inducers of ISR, as evident from plant interactions with plant growth-promoting rhizobacteria (PGPR) (Van der Ent et al., 2009). ISR is induced by non-pathogenic microbes upon interaction with plant roots and confers a broad spectrum of effectiveness in many plant species (Van Wees et al., 2008; Pineda et al., 2010). JA is a central player in mediating ISR, which is depressed in JA biosynthesis and signalling mutants (Van der Ent et al., 2009). Moreover, it has been demonstrated that ISR is based on priming of JA-regulated responses by increasing the sensitivity to JA rather than the production of JA (Pieterse et al., 2002). Similarly to the ISR induced by rhizobacteria, JA might trigger increased local or systemic resistance of the AM plants against pathogens (Pozo and Azcon-Aguilar, 2007; Gutjahr and Paszkowski, 2009; Hause and Schaarschmidt, 2009).
JA seems to be involved in the establishment and maintenance of AM, but the results are partially contradictory. The AM roots exhibit enhanced JA levels accompanied by the expression of JA-induced genes and genes encoding enzymes of JA biosynthesis (Hause et al., 2002; Isayenkov et al., 2005; Lopéz-Ráez et al., 2010). In tomato, however, mycorrhization led also to an accumulation of oxylipins derived from the 9-LOX pathway in colonized parts of the root (Lopéz-Ráez et al., 2010; León-Morcillo et al., 2012). Here, a local induction of expression of LOXA and AOS3 occurred. Both genes are induced by JA (García-Garrido et al., 2010; León-Morcillo et al., 2012), implying that it might control the spread of the fungus via 9-LOX-derived oxylipins. This is supported by the fact that application of JA to mycorrhizal plants results in a diminished mycorrhization rate (Vierheilig, 2004; Herrera-Medina et al., 2008). However, other data show that JA application is associated with an enhanced plant–fungus interaction (Tejeda-Sartorius et al., 2008; Kiers et al., 2010; Landgraf et al., 2012; León-Morcillo et al., 2012). Most probably, these contrasting data accrue from differences in the experimental designs, such as JA concentrations, timing and frequency of JA application, plant organs treated and plant nutritional status. Even the analyses of mycorrhization in mutants and transgenic plants defective in JA biosynthesis or perception did not yield unequivocal results. In comparison with tomato wild-type plants, an increase of mycorrhization occurred in the JA-insensitive mutant jai1, suggesting that JA may control the fungal spread (Herrera-Medina et al., 2008; León-Morcillo et al., 2012). In contrast, JA-deficient mutants spr2 and def1 were characterized by a decrease (Tejeda-Sartorius et al., 2008; León-Morcillo et al., 2012), while the overexpressors of prosystemin (enhanced JA levels) mutants by an increase of mycorrhization (Tejeda-Sartorius et al., 2008 ), supporting the assumption that JA may act as a positive regulator of AM (Fig. 7). Further support for this assumption came from AOC-RNAi Medicago truncatula roots with reduced JA biosynthesis showing a significant decrease in mycorrhization (Isayenkov et al., 2005). Moreover, an endogenous rise in JA levels induced by repeatedly wounding the leaves of M. truncatula led to enhanced mycorrhization (Landgraf et al., 2012).
Fig. 7.
Effects of arbuscular mycorrhization (AM) and root nodule symbiosis (RNS) on endogenous levels of jasmonates as well as the effects of modulated JA levels on AM and RNS in Medicago truncatula. Endogenous jasmonate levels are increased and remain unaffected in roots of AM and RNS plants, respectively. The application of jasmonic acid (JA) or wound-induced rise in JA results in enhanced AM, but no effects on RNS. Similarly, reduced JA levels in transformed ALLENE OXIDE CYCLASE (AOC)-RNAi roots suppress AM, but do not affect RNS.
Fig. 8.
The role of jasmonic acid (JA)/JA-Ile in plant development. JA induces root growth inhibition by stimulating auxin biosynthesis via anthranilate synthase α1 (ASA1) and inhibiting the expression of genes encoding the TFs PLETHORA 1 (PLT1) and PLT2, which ensure the maintenance and activity of stem cells in the root. In tuber formation, jasmonates [JA, tuberonic acid (TA) and TA glucoside (TAG)] might act directly after their rise following activity of LIPOXYGENASE 1 (LOX1). However, most importantly levels of gibberellic acid (GA) are regulated by the combined action of the TFs BEL1-like 5 (BEL5) and POTATO HOMEOBOX 1 (POTH1) at the promoters of GA-20 oxidase-encoding genes. In trichome initialization, JA/JA-Ile act via the COI1 co-receptor complex to activate the trichome-specific TFs MYB75 and GLABRA 3 (GL3), leading to formation of defence proteins as well as terpenoids. Role of jasmonates in flower development is depicted for Arabidopsis thaliana. The TF AGAMOUS activates the phospholipase A1 DAD1, but also auxin induces rise in JA/JA-Ile via the function of the TFs AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8. Jasmonates induce COI1-dependently expression of MYB21 and MYB24, leading to proper stamen development, whereas expression of the TF BIGPETALp (BPEp) restricts petal growth. In senescence, jasmonates act on different levels. On the one hand, chlorophyllase is activated, which leads to chlorophyll breakdown, and RUBISCO activase is inhibited, which switches off photosynthesis. On the other hand, specific TFs, such as WRKY53, WRKY54, WRKY70 and ANAC092/ORE1, are induced, leading to expression of senescence-related genes.
Comparing all data obtained from analyses of mutants and transgenic plants, the positive role of JA in AM can be explained by a systemic signalling to and from the shoot. Jasmonates produced in the roots by AM might result in systemic alterations in the shoot, which in turn might enforce the AM in the root. When JA levels are artificially increased in roots and shoots (JA application, Prosystemin overexpression, wounding), this reinforcement of AM might be increased. In the absence of JA biosynthesis in roots or shoots, the systemic root-to-shoot signals in either direction are missing, resulting in a decline in AM. However, how the shoot supports AM remains to be elucidated, but the role of JA may be attributed to an enhanced allocation of assimilates into the root system, as shown for other wounded or herbivore-affected plants (Babst et al., 2005; Schwachtje et al., 2006; Kaplan et al., 2008). Accordingly, mycorrhization was found to be positively correlated with deregulated carbon (Tejeda-Sartorius et al., 2008). Additionally, JA may influence the action of other hormones (see section 4.7), because ET, ABA, gibberellin and auxin are involved in the regulation of AM (Martín Rodriguez et al., 2010; García-Garrido et al., 2010; de Los Santos et al., 2011; Hanlon and Coenen, 2011; Ortu et al., 2012). A deregulation of the cross-talk between JA and other hormones might occur in JA-insensitive plants, thus resulting in an enhanced mycorrhization.
8.2. Root nodule symbiosis (RNS)
RNS is characterized by the intracellular uptake of nitrogen-fixing bacteria (rhizobia) concomitant with the formation of specialized organs, the root nodules. Specialized Gram-negative bacteria involved in RNS show a narrow host range and exclusively affect legumes. Among them are important agricultural crops such as soybean (Glycine max), common bean (Phaseolus vulgaris) and pea (Pisum sativum) (Kistner and Parniske, 2002). Root nodules provide a suitable microenvironment for nitrogenase activity of nitrogen-fixating bacteria and for protected and controlled development of a high-density bacterial population to maintain the symbiosis.
The application of JA to roots affects RNS in several ways (for reviews see Ding and Oldroyd, 2009; Gutjahr and Paszkowski, 2009; Hause et al., 2009). On the one hand, JA application to rhizobia causes induction of nod genes (Rosas et al., 1998; Mabood et al., 2006), thereby enhancing the effectiveness of subsequent root nodulation. On the other hand, JA application has been shown to affect the response of M. truncatula to rhizobial Nod factors by inhibiting the Nod factor-induced transcription of ENOD11 and RIP1 and calcium spiking, leading to decreased numbers of nodules (Miwa et al., 2006; Sun et al., 2006). In soybean, JA or OPDA application affects the morphology of nodules by changing the number and size of central and peripheral nodule cells (Costanzo et al., 2012). However, all these pharmacological experiments do not demonstrate an endogenous role of JA in RNS.
Endogenous levels of JA in nodulated roots do not differ from that in non-infected roots (Zdyb et al., 2011). Moreover, no differences in nodule morphology and number were oberserved in transgenic M. truncatula roots with altered JA biosynthesis (Zdyb et al., 2011) (Fig. 7). However, transient transformation of roots resulted in chimeric plants, an observation that could not entirely preclude a possible role of shoot-derived JA in RNS.
Results documenting the role of shoot-derived JA on nodulation are again controversial: (1) JA could act as a negative regulator because MeJA application to Lotus japonicus shoots is known to reduce nodulation (Nakagawa and Kawaguchi, 2006; Seo et al., 2007); (2) JA could act as a positive regulator, as shoot-specific suppression of JA biosynthesis in soybean plants by foliar application of the inhibitor propyl gallate severely reduces nodule number without affecting root growth (Kinkema and Gresshoff, 2008); and (3) JA does not regulate RNS as enhanced JA levels in M. truncatula after wounding and JA application have been shown not to alter nodulation (Landgraf et al., 2012) (Fig. 7). The use of different plant species and growth conditions might have produced such conflicting results. However, under light-limiting conditions, JA seems to have a regulatory function (see section 10). In L. japonicas, nodulation is photomorphogenetically controlled via JA, as demonstrated by the reduced nodule number in phyB mutants (Suzuki et al., 2011). In wild-type plants, PHYB is part of a monitoring system to detect suboptimal light conditions and mediates the initiation of SAS as well as the suppression of nodule development under low R/FR light. Consequently, phyB mutants exhibit a constitutive SAS phenotype under white light. Interestingly, in low R/FR light-grown wild-type and white light-grown phyB plants, transcript levels of JA-induced genes, such as JAR1, are down-regulated, resulting in a decrease of JA-Ile content. Here, two effects – roots do not synthesize JA-Ile due to down-regulation of JAR1 and the translocation of JA-Ile from shoot to root is probably impeded – seem to be involved (Shigeyama et al., 2012). It is thus clear that shoot-derived JA-Ile controls nodule formation in the SAS as a positive regulator.
Another systemic effect in RNS is the ‘autoregulation of nodulation’ (AON). To restrict microbial infections and thereby nodule number, a feedback inhibition occurs, which is controlled by the shoot through CLAVATA1(CLV1)-like receptor kinase (NARK in soybean, HAR1 in L. japonicus and SUNN in M. truncatula) (for reviews see Hause et al., 2009; Reid et al., 2011; Mortier et al., 2012). Mutant plants with a defective receptor kinase have a supernodulating phenotype. Although the nature of the root-derived AON signal is still elusive, CLV3/ESR-related (CLE) peptides binding to the receptor kinase might be involved (Mortier et al., 2012). This signal is transmitted into the formation of an unknown ‘shoot-derived inhibitor’ (SDI) that is transferred back to the roots. Transcript profiling in soybean identified the components of AON acting downstream of NARK in the leaf (Seo et al., 2007; Kinkema and Gresshoff, 2008). Of these, AOC and MYC2 were found to be systemically down-regulated when roots of wild-type, not the nark mutant, were inoculated. Moreover, AON mutants exhibit enhanced levels of JA in leaves (Seo et al., 2007; Kinkema and Gresshoff, 2008). Together with the fact that application of JA biosynthesis inhibitors also reduces nodulation in nark, these data suggest that JA signalling is a positive regulator of RNS and might suppress activity of the SDI (Kinkema and Gresshoff, 2008).
8.3. JA in plant interactions with Piriformospora indica
P. indica has been characterized as a mutualistic, biotrophically living endophyte which colonizes plant roots without causing any disease symptoms (for a review see Qiang et al., 2012). It is a Basidiomycete, belongs to the order Sebacinales (Weiss et al., 2004) and can be cultivated in axenic culture (Lahrmann and Zuccaro, 2012). P. indica is highly effective in root colonization accompanied by immune suppression (Schäfer et al., 2009; Jacobs et al., 2011). It colonizes a broad range of hosts, where it confers significant growth promotion including enhanced seed yield (Franken, 2012). The interaction between plants and P. indica is mutualistic – P. indica enhances the phosphate supply of plants depending on its phosphate transporters (Yadav et al., 2010) and receives carbohydrates from the plant (Schäfer et al., 2009). Additionally, colonization of roots by P. indica induces enhanced resistance against a wide variety of root and leaf pathogens (Molitor and Kogel, 2009). This is similar to ISR and depends on an operative JA pathway as the mutants jar1 and jin1/myc2 are compromised in P. indica-mediated resistance (Stein et al., 2008). In barley, a P. indica-mediated priming leads to enhanced PR and heat-shock gene expression after infection of leaves with powdery mildew (Molitor et al., 2011).
The colonization of roots by P. indica itself is regulated by JA. Mutants, which are impaired in JA biosynthesis or perception, show elevated root immune responses leading to reduced root colonization (Jacobs et al., 2011). This led to the assumption that JA regulates early immune responses by suppression of SA- and glucosinolate-related defence pathways (Jacobs et al., 2011). Indeed, Arabidopsis mutants impaired in SA-associated defence are more susceptible to P. indica. Moreover, there might be a cross-talk with ET, GA and cytokinin (CK). ET biosynthesis, signalling and ET-targeted TFs are required for colonization and the beneficial effects of P. indica in barley and A. thaliana (Camehl et al., 2010; Khatabi et al., 2012). The colonization of barley roots depends on GA as its biosynthesis and perception mutants are significantly less colonized (Schäfer et al., 2009). Moreover, P. indica is able to produce auxin and CK, but not JA or ABA (Sirrenberg et al., 2007; Vadassery et al., 2008), and therefore this mutualistic endophyte might recruit additional plant hormones to manipulate plant defence and development (Lahrmann and Zuccaro, 2012).
9. JASMONATE IN PLANT GROWTH AND DEVELOPMENT
9.1. Seed germination
Inhibition of seed germination was described for JA. However, recent genetic and biochemical evidence has shown that OPDA is the inhibitory compound which acts together with ABA in a COI1-independent manner (Dave et al., 2011) (see section 4.4).
9.2. Root growth inhibition by JA
Growth inhibition and senescence promotion were the first two physiological responses described for JA (Ueda and Kato, 1980; Dathe et al., 1981). Root growth inhibition by JA application has been used in mutant screens since the 1990s. The first mutant insensitive to JA was jar1 (Staswick et al., 1992). Subsequently, JAR1 was cloned as JA-Ile synthase (Staswick and Tiryaki, 2004). Root growth inhibition by JA was also strongly supported by short-root phenotype of mutants with constitutive elevation of JA levels, such as cev1 (Ellis et al., 2002), and reduced sensitivity to JA in coi1 and myc2 mutants (Xie et al., 1998; Lorenzo et al., 2004). For inhibition of root growth, JA requires COI1, as indicated by the JA-unresponsiveness of the coi1 mutant. However, ET and its precursor ACC, which occurs only in the light but not in the dark, are also known to inhibit root growth (Adams and Turner, 2010). The ACC/ET-induced root growth inhibition is light- and COI1-dependent, but JA-independent.
However, JA-induced root growth inhibition needs to be analysed in relation to other factors controlling the complex process of root development (Petricka et al., 2012b; Ubeda-Tomás et al., 2012). Initially, cell- and tissue-specific gene expression maps revealed non-overlapping areas of auxin-, GA- and JA-dependent gene expression (Birnbaum et al., 2003). JA-induced gene expression appeared in outer layers of roots. But such expression data have to be taken into account cautiously, as the cellular proteome map of A. thaliana roots indicated a positive but weak correlation between protein and RNA profiles (Petricka et al., 2012a). Meanwhile, system biology approaches are being used to analyse the complex and hierarchical link between hormonal and mechanic signalling in root growth (Band et al., 2012a). Key players are CK, GA and auxin. However, the cross-talk with other hormones, such as auxin, GA and BR, underlying JA perception and signalling indicates the involvement of JA in root growth. The outcome of root growth is an integration of hormonal and mechanic signalling that affect cell division, membrane traffic, cell-wall loosening and synthesis, turgor, and growth rate (Band et al., 2012b). Many of these processes could be a direct effect of JA or an indirect effect of JA via auxin. The biosynthesis of auxin, the key player in root growth, is affected by JA-induced expression of ASA1 (Sun et al., 2009) (Fig. 8). JA induces auxin redistribution via modulation of endocytosis and an accumulation of PIN2 in the plasma membrane (Sun et al., 2011b). Auxin is also affected by JA-induced MYC2-dependent repression of PLETHORA, the key player in root stem cell niche activity (Chen et al., 2011). Another example of auxin/JA cross-talk is given by the axr1 mutant defective in an SCF-complex component required for auxin signalling (Mockaitis and Estelle, 2008). This mutant shows reduced root growth inhibition by MeJA, indicating that the AXR1-dependent modification of the CULLIN1 subunit of the SCFCOI1 complex is required for JA/JA-Ile signalling (Xu et al., 2002).
Taken together, the JA-induced root growth inhibition seems to occur preferentially via modulation of the effects of auxin in root growth and development (Fig. 8).
9.3. Lateral root formation
Most of the Arabidopsis lateral root mutants are affected in auxin homeostasis, signalling and transport and in PINs (Péret et al., 2009), thereby indicating the dominant role of auxin in lateral root formation (Petricka et al., 2012b). The various possibilities for cross-talk between JA and auxin as described above strongly suggest a role of JA in lateral root formation. The JA-insensitive mutant coi1-16 that produces fewer lateral roots lends further credence to this idea. Furthermore, the high promoter activities of AtAOC3 and AtAOC4 in emerging lateral roots suggest that JA is involved in lateral root formation (Stenzel et al., 2012). It has been shown that lateral root formation is induced by auxin, but is inhibited by the conjugate of JA with tryptophan (Staswick, 2009).
9.4. Adventitious root formation
Adventitious roots are formed naturally or induced by environmental stimuli in aerial organs. Like root growth, adventitious root formation is a complex process regulated by hormones and environmental factors. Auxin is a positive regulator mediated by ARF6 and ARF8, which are targets of miR167 (Gutierrez et al., 2012). Interestingly, downstream of this auxin-induced adventitious root formation, there is a negative COI1- and MYC2-dependent regulation via altered JA/JA-Ile homeostasis. Whereas JAR1, which is the GH3.11 of the GH3 gene family of conjugating enzymes, generates JA-Ile, the other members (GH3.3, GH3.5, GH3.6) conjugate Asp, Met and Trp with JA. The triple-mutant of these genes has fewer adventitious roots and increased expression of JA biosynthesis genes, whereas mutants impaired in JA perception and signalling, such as coi1-16, myc2, myc3, myc4 and jar1 form far more adventitious roots than the wild-type (Gutierrez et al., 2012). These data are in agreement with auxin–JA cross-talk that occurs during adventitious root formation. Here, the positive regulatory effects of ARF6 and ARF8 are increased by the GH3.3, GH3.5, GH3.6 module that attenuates the negative regulatory effect of JA/JA-lle via conjugation of JA to JA-Asp, JA-Met and JA-Trp (Gutierrez et al., 2012). However, this is in opposition to the positive regulatory effects of ARF6 and ARF8 on JA biosynthesis, as recorded in filament elongation during flower development (see section 4.7) (Nagpal et al., 2005; Reeves et al., 2012). Moreover, in leafy cuttings of Petunia hybrida, an increase in JA levels precedes a corresponding increase in auxin levels (Ahkami et al., 2009). However, whether this increase in JA levels is essential for subsequent adventitious rooting remains to be elucidated.
9.5. Tuber formation
For a long time, tuber-inducing activities of jasmonates, particularly 12-OH-JA (TA) and its glucoside (TAG), have been suggested (reviewed by Wasternack and Hause, 2002). StLOX-1 was shown to be involved in tuber yield and tuber formation (Kolomiets et al., 2001), and LOX-derived metabolites such as JA, TA and TAG accumulate at low, tuber-inducing temperature (Nam et al., 2008). There were, however, only correlative data on the endogenous content of jasmonates in stolons and tuber formation. The cloning of a 12-OH-JA sulfotransferase from A. thaliana and tomato (Gidda et al., 2003; J. Heise and C. Wasternack, unpubl. res.) and the occurrence of 12-OH-JA, 12-HSO4-JA and 12-O-Glc-JA in different non-tuber-bearing plant species (Miersch et al., 2008) argue against a specific role of jasmonates in tuber formation. Possibly, tuber-inducing effects might be caused indirectly via cell expansion in stolons accompanied by changes in microtubule orientation (Abe et al., 1990), because JA biosynthesis may occur in developing stolons (Cenzano et al., 2007).
Multiple pathways are involved in tuber formation (Sarkar, 2008) (Fig. 8). Besides hormonal control, tuberization in potato is strictly photoperiod-dependent. Low night temperature and a short-day photoperiod produce a systemic signal in leaves which induces tubers in roots. Tuberization depends on conserved function of the potato orthologue of CONSTANS (CO) and FLOWERING LOCUS T (FT) (Rodríguez-Falcón et al., 2006). Both of them are key players in flower induction. Phytochrome B-mediated photoperiodic control of tuberization is well described (Rodríguez-Falcón et al., 2006). Identification of the BEL5 TF of potato shed new light on the regulation of tuber formation. This TF belongs to the homeobox BEL1 gene family that is involved in different developmental processes. Its mRNA accumulates under short-day conditions in leaves and is transported via the phloem to stolon tips correlating with tuber formation (Banerjee et al., 2006). Additionally, high StBEL5 promoter activity appears in stolons of short-day plants (Chatterjee et al., 2007). StBEL5 mRNA accumulation seems to result from the photoperiod-dependent control via CO and FT and is a long-distance signal with increased mobility mediated by its 3′ untranslated region (Hannapel, 2010). The final step in regulation of tuber formation is an altered GA level (Jackson et al., 1996). StBEL5 binds together with the potato KNOX gene product POTH1 to promoter sequences of the gene encoding the GA-20 oxidase1, which leads to its repression and altered GA level that affect tuber formation and other aspects of vegetative development (Banerjee et al., 2006; Lin et al., 2013) (Fig. 8). Initiation of cell division in stolons by cytokinins is another hormonal control active in tuber formation (Xu et al., 1998).
9.6. Growth versus defense
Besides the above discussed root growth inhibition by JA, growth of above-ground plant parts is also inhibited by JA. Any growth response depends on cell division reflecting the cell cycle activity and on cell expansion mediated by macromolecule formation, ploidy-dependent cell growth, cell-wall elasticity, microtubule organization and turgor pressure (Rymen and Sugimoto, 2012). All these processes are under hormonal control and depend on environmental cues such as biotic and abiotic stresses that are known to suppress the growth of above-ground plant parts. Plant hormones such as auxin, ET and GA have been shown to be involved in such stress-induced growth inhibition (Band et al., 2012a; Murray et al., 2012; Petricka et al., 2012b). JA is the key player in responses to herbivores and mechanical wounding, both of which are known to repress plant growth in A. thaliana (Zhang and Turner, 2008). Endogenously generated JA, but not OPDA, has been shown to repress plant growth by suppressing mitosis in a COI1/JAZ/MYC2-dependent manner. This accords well with a JA-induced reprogramming of the expression of cell cycle-regulated genes in a COI1/JAZ/MYC2-dependent manner (Pauwels et al., 2008). A similar effect was observed when M. truncatula plants were mechano-stimulated by repeatedly touching their leaves, which resulted in an increase of endogenous JA levels concomitant with stunted growth (Tretner et al., 2008). The same experimental setup has been used recently in A. thaliana for analysing touch-induced morphogenesis, which enhances resistance to B. cinerea in a JA-dependent manner (Chehab et al., 2012). Even some aspects of thigmomorphogenesis are ET-dependent; the ET-response mutants show touch-induced thigmomorphogenesis. Mutant analysis revealed a JA-mediated signalling pathway underlying thigmomorphogenesis (Chehab et al., 2012). In contrast to the negative effect of JA and mechano-stimulation on longitudinal growth, a positive effect of mechano-inducible COI1/MYC2/JAZ during secondary growth in cambium formation has been reported (Sehr et al., 2010).
Growth is promoted by GAs that, however, repress defence gene activation (Fig. 6). These antagonistic responses are caused by an imbalanced ratio of GA and JA (see Kazan and Manners, 2012). In the absence of JA, the formation of GA is associated with growth promotion and defence containment, while in the absence of GA, the formation of JA is accompanied by opposing responses. The already described proteins DELLA and PIF (for GA signalling) as well as JAZ and MYC (for JA signalling) are components of this GA–JA cross-talk (see section 4.7), where JA prioritizes defence over growth (Yang D-L et al., 2012). Incidentally, cell elongation and meristem activity required for plant growth are regulated by auxins. Here, JA affects auxin formation and distribution by inducing the expression of ASA1 and by regulating the PINs and PLETHORA, respectively (see sections 4.6 and 4.7).
9.7. Trichome formation
Glandular trichomes are multicellular and often involved in resistance to insects due to formation of terpenoids, flavonoids, alkaloids and defence proteins (Tian et al., 2012). They represent a useful tool for production of secondary metabolites (Tissier, 2012). Genetic evidence for the involvement of JA in glandular trichome formation were obtained by characterizing the tomato homologue of COI1, the central component of JA perception (Li et al., 2004) (Fig. 8). The corresponding tomato mutant jai1 is female sterile, but is impaired in grandular trichome formation, trichome monoterpene content and spider mite resistance. Further support for the link between trichome formation, JA and defence came from the recessive tomato mutant odorless-2 (od-2), which exhibits altered morphology, density and chemical composition of glandular trichomes (Kang et al., 2010). Under natural field conditions, od-2 plants were highly susceptible to Colorado potato beetle larvae and the solanaceous specialist Manduca sexta, indicating that trichome-borne compounds determine host plant selection under natural conditions (Kang et al., 2010; Meldau et al., 2012). Recently, an antagonism between herbivore-induced plant volatiles and trichome formation has been observed in tomato. Using the JA-deficient spr2 mutant and the trichome-free JA-insensitive jai1 mutant, preferential oviposition that was observed on trichome-free JA-insensitive plants indicated a greater impact of trichomes over volatile emission in this tritrophic interaction (Wei et al., 2013). Furthermore, glandular and non-glandular trichomes are involved in defence against herbivores via trichome density and JA-inducible defence compounds, such as PI2, monoterpenes and sesquiterpenes (Tian et al., 2012). It is to be noted here that the cotton fibre represents a special type of single-cell seed trichome. It is well known that its initiation and elongation are under hormonal control including JA. Recently, it has been shown that a member of the class I bHLH TF family of Gossypium barbadense positively regulates JA biosynthesis (Hao et al., 2012). Consequently, the elevated JA level in cotton fibre activates downstream genes involved in Ca2+ signalling and ET biosynthesis. In Arabidopsis, targets of JAZ proteins are TFs such as MYB75, GL3 and EGL3, which are involved in anthocyanin biosynthesis and trichome initiation (Qi et al., 2011) (see section 4.6). JA regulates trichome initiation in a dose-dependent manner via the key TF in trichome formation GL3 and its interaction with JAZ proteins (Yoshida et al., 2009) (Fig. 8).
9.8. Leaf movement
There are several types of leaf movements. Among them are the upward leaf movement (hyponastic growth) and the leaf movement of nyctinastic plants such as Albizzia. Both of them are altered by JA compounds. Hyponastic growth is induced by ET, heat and low light intensity and is stimulated by JA but inhibited by SA (van Zanten et al., 2012).
Nyctinastic leaf movement depends on activity of motor cells (see section 3.6). Here, TAG has a role. Among the enantiomeric forms of TAG, only one mediates activity of motor cells of nyctinastic plants, such as Albizzia and Samanea saman in a COI1-independent manner (Ueda and Nakamura, 2007; Nakamura et al., 2011).
9.9. JA in leaf senescence
Leaf senescence is a complex developmental programme that depends on light/dark conditions, nutrients, biotic and abiotic stresses, and several hormones including JA. Over the last few years, several reviews on leaf senescence in relation to JA have been published (Reinbothe et al., 2009; Guo and Gan, 2012; Zhang and Zhou, 2013). Therefore, only a few aspects will be discussed here. In A. thaliana, comparative large-scale transcript profiling between environmentally and developmentally regulated leaf senescence revealed only limited similarities in early stages, but showed convergence and divergence of gene expression profiles (Guo and Gan, 2012). High-resolution transcript profiling of senescing leaves identified a distinct group of TFs that link metabolic pathways, leaf development and senescence (Breeze et al., 2011). The JA-linked TFs identified to be active in leaf senescence are WRKY53 (Miao and Zentgraf, 2007), WRKY54 and WRKY70 (Besseau et al., 2012), and ANAC092/ORE1 (Balazadeh et al., 2010) (Fig. 8). The F-box protein ORE9 was initially identified from a screen of ABA-, JA- and ET-induced senescence mutants (Woo et al., 2001), but it was found to have different regulatory properties in photomorphogenesis, shoot branching (Stirnberg et al., 2007) and cell death. Leaf senescence is characterized by JA-inducible chlorophyll breakdown. In A. thaliana, between the two key enzymes involved in chlorophyll degradation, the gene encoding the CHLOROPHYLLASE1 is strongly induced by JA (Tsuchiya et al., 1999). Moreover, a mechanistic explanation for the senescence-promoting effects of JA in leaves was provided only recently. It has been shown that Rubisco-activase is down-regulated by JA in a COI1-dependent manner (Shan et al., 2011).
9.10. Gravitropism
Gravitropism is a well-studied, morphogenic response, in which intra- and intercellular communication by auxin takes place. Traditionally, the Cholodny–Went hypothesis is used to explain the asymmetric growth as a consequence of auxin redistribution. Regarding the repeatedly discussed cross-talk between auxin and JA (see sections 4.7 and 9.2), it is not surprising that JA has a role in gravitropism (Gutjahr et al., 2005). Using rice coleoptiles, the Cholodny–Went hypothesis was found to be true. In addition to an auxin gradient, gradients of JA and auxin responsiveness were found to be involved in gravitropism (Gutjahr et al., 2005). A mechanistic framework might be given by an interaction between auxin and JA signalling pathways. This became evident by identification of tryptophan conjugates of indolyl-3-acetic acid and JA as endogenous inhibitors of the gravitropic response, one of the most prominent auxin responses (Staswick, 2009). In rice a gravitropism-related gene, LAZY1, was identified that is required for gravity responses in leaf lamina, but not in roots (Yoshihara and Ino, 2007). The function of its gene product remains unknown.
9.11. JA in development of reproductive organs of dicotyledonous plants
The most diagnostic phenotype of Arabidopsis mutants impaired in JA biosynthesis and perception, such as coi1, opr3, dde1, dde2, dad1, aos and fad3-2fad7-2fad8 (Browse, 2009c; Wilson et al., 2011), is male sterility (Browse, 2009a, c). Three characteristic phenotypes were identified: (1) insufficient filament elongation, (2) non-viable pollen and (3) delayed anther dehiscence. In mutants impaired in JA biosynthesis, fertility can be restored by JA treatment when applied in stages 11 and 12 of floral development, but not by OPDA (Mandaokar et al., 2006). Transcript profiling of JA-treated stamens of opr3 plants allowed detection of stamen- and JA-specific mRNAs preferentially regulating genes involved in metabolic pathways required for the synthesis of terpenoid volatiles, wax and pollen constituents (Mandaokar et al., 2006) (Fig. 8). Most interestingly, new TFs required for stamen development were identified in the stamen transcriptome of opr3 plants. Among them were MYB21 and MYB24 (Mandaokar et al., 2006). Subsequent genetic analysis identified MYB108, which, together with MYB24, is involved in JA-regulated stamen and pollen developments (Mandaokar and Browse, 2009). MYB21 and MYB24 were further identified as targets of JAZ repressors (Song et al., 2011) (see section 4.6). In the coi1 mutant, the overexpression of MYB21 could partially restore the delayed anther dehiscence, but not JA insensitivity in terms of root growth inhibition, anthocyanin formation and susceptibility to necrotrophic pathogens (Song et al., 2011). These data suggest a dominant role of MYB21 in stamen and pollen development.
The essential role of JA in stamen development is also obvious by DAD1, an Arabidopsis PLA1 involved in JA formation of flowers. DAD1 is expressed in flowers, and dad1 shows a phenotype similar to coi1 (Ishiguro et al., 2001). This gene is a target of the central TF AGAMOUS (Ito et al., 2007) (see section 2.1). Of all the JA cross-talks involving other hormones, the JA–auxin cross-talk is the most important in flower development. It has been unequivocally demonstrated that ARF6 and ARF8 regulate JA biosynthesis in anther filaments (Nagpal et al., 2005; Reeves et al., 2012) (see section 4.7).
In contrast to the male sterile phenotype of coi1, the homologous tomato mutant jai1 impaired in the tomato homologue of COI1 is female sterile, suggesting that JA signalling plays distinct roles in flower development in Arabidopsis and tomato (Li et al., 2004). Recently, embryo development in tomato has been shown to be OPDA-specific, but JA-independent (Goetz et al., 2012), implying the difference in flower development vis-à-vis fertility between Arabidopsis and tomato.
Beside its role in stamen development in Arabidopsis, JA has a role in petal growth (Brioudes et al., 2009). The final stages of petal growth are largely dependent on cell proliferation and/or cell expansion. The bHLH TF BIGPETALp (BPEp), expression of which is controlled by JA, limits petal size by controlling cell expansion. Consequently, the opr3 bpe-1 mutants are characterized by a larger petal size that can be restored by JA treatment (Brioudes et al., 2009).
9.12. JA in development of reproductive organs of monocotyledonous plants
JA has a central role in sex determination of maize (Acosta et al., 2009; Browse, 2009b). In maize, sex organs are located on the same plant in the male tassel at the top and the female ear(s). Originating from a bisexual floral meristem, the pistil primordia are aborted undergoing a tasselseed-mediated cell death (Acosta et al., 2009). There are two tasselseed genes in maize, namely ts1 and ts2. Whereas the ts2 gene encodes a short-chain dehydrogenase/reductase with broad substrate specificity, the ts1 gene has been identified recently by positional cloning and encodes a plastid-targeted 13-LOX (Acosta et al., 2009). The homozygous ts1 mutant is characterized by a loss of 13-LOX activity and lower JA levels in inflorescences, but the mutant phenotype could be rescued by JA application. TS1 and TS2 are both required for sex determination. Possibly, TS2 plays a role similar to TS1 in JA biosynthesis by regulating β-oxidation steps of the carboxylic acid side chain of OPDA (Acosta et al., 2009; Browse, 2009b).
Cytoplasmic male sterility (CMS), a maternally inherited phenomenon leading to pollen abortion, is associated with JA biosynthesis. In rice, proteins of mitochondrial complexes together with a sex-determining TASSELSEED2-like protein were found to be affected in a CMS line YuetaiB, leading to aberrant changes in JA biosynthesis during microspore development (Liu et al., 2012).
10. JA IN LIGHT SIGNALLING
The amount and quality of light sensed by plants is important in different developmental programmes such as skotomorphogenesis, photomorphogenesis and SAS. The molecular mechanism of light signalling and essential components of light perception and responses have been intensively studied over the last two decades (reviewed by Chory, 2010; Kami et al., 2010; Lau and Deng, 2010). The involvement of plant hormones, such as auxins, cytokinins, GA, ET and BRs, in light-dependent regulation of developmental processes has been intensively studied (Wolters and Jürgens, 2009; Chory, 2010). However, the involvement of JA in light signalling has been described only over the last few years. The newly discovered key players in JA perception and signalling (JA receptor, JAZ proteins and MYC/MYB TFs) have provided new mechanistic insights into how JA and light are integrated in growth and development and how competition between growth and defence occurs. This interplay has been most authoritatively reviewed (Lau and Deng, 2010; Kazan and Manners, 2011). To avoid an overlap, we will discuss here only to some important aspects.
The mutant myc2 impaired in the key TFs of JA signalling pathways shows high sensitivity to SAS or FR light, whereas phy A mutant exhibits reduced JA-regulated growth inhibition (Robson et al., 2010). The wound and shade avoidance responses are integrated via JAZ1.
The mutant hy1-101, which is affected in a heme-oxygenase required for phytochrome chromophore biosynthesis, has an overproduction of JA concomitant with increased expression of JA-responsive genes (Zhai et al., 2007).
HY5, a bZIP TF, is a positive regulator of photomorphogenesis and a key regulator of light signalling (Lau and Deng, 2010). It binds to the LOX3 promoter (Lee et al., 2007) involved in JA biosynthesis (Caldelari et al., 2011) (see section 2.2).
Light responses are organ- and tissue-specific. Roots of phytochrome chromophore-deficient mutants, such as hy1-1 and hy2-1, have reduced sensitivity to JA similar to the JA-insensitive mutants jar1 and myc2 (Costigan et al., 2011), suggesting a photoregulation of root elongation via an acquisition of JA sensitivity.
The CSN is clearly linked to JA biosynthesis and JA-dependent defence responses. Tomato plants with a silenced CSN subunit are less resistant to herbivorous insects and necrotrophic pathogens (Hind et al., 2011).
The positive regulator of PHY B-mediated SAS is PHYTOCHROME AND FLOWERING TIME1 (PFT1), which is an important regulator of JA signalling. PFT1 encodes the conserved MED25 subunit of the mediator complex active in JA signalling (see section 4.2). Possibly, the light signalling component PFT1/MED25 negotiates the activation of TFs, such as MYC2 and ERF1 and their binding to the RNA polymerase II apparatus (Kidd et al., 2009; Çevik et al., 2012). This integrates a variety of interdependent environmental stimuli, such as flowering time control via CONSTANS, light quality control and JA-dependent defence responses (Iñigo et al., 2012).
Light stress affects JA biosynthesis through plastid proteins called fibrillins (Youssef et al., 2010; Kazan and Manners, 2011). Additionally, anthocyanin accumulation, the most prominent JA/JA-Ile-induced phenotype, is elevated by light stress.
When plants are treated with JA and UV-B light, there occurs an overlap in gene expression resulting in stronger defence even under field conditions (Ballare et al., 2012).
R/FR light ratio causes weaker resistance to necrotrophic pathogens and decreased JA responses in a COI1–JAZ10-dependent but SA-independent manner (Cerrudo et al., 2012).
In rice, light regulates JA/JA-lle biosynthesis via PHY-dependent up-regulation of OsAOS1 and OsJAR1 (reviewed by Svyatyna and Riemann, 2012).
The secretion of extrafloral nectar in lima bean, a JA-dependent indirect defence mechanism, is controlled by light via the formation of JA-Ile irrespective of R/FR light ratio (Radhika et al., 2010).
Hyponastic growth (upward leaf movement), a component of SAS in A. thaliana, is an ET- and heat-controlled process, and is modulated by both JA and SA (van Zanten et al., 2012).
11. CONCLUDING REMARKS
In the last decade, a vast amount of data has been accumulated by transcriptomic, proteomic, lipidomic and metabolomic studies. Many of them were performed to elucidate hormone action including that of jasmonates in a developmental or stress-related context. Such analyses will continue to expand by analytical and bioinformatic improvements, which will allow analysis of spatial and temporal hormone-induced changes at the level of organs, tissues and even specific cell types. Many new components or key players in hormone signalling are expected to be identified by these ‘omics’ studies.
To date, at the protein level, signalling components are analysed by interaction studies, such as yeast two-hybrid and three-hybrid screening and BiFC analyses in vivo. Technical improvements in combination with specialized genetic tools will allow us to address the complexity inherent in the plant hormone cross-talk. Another emerging issue will be to optimize sensitive methods for recording hormone homeostasis and detecting active and inactive hormones sustained by their metabolic conversion.
In the jasmonate field the following aspects will become of interest in the near future.
The assembly of the SCFCOI1–JAZ co-receptor complex and stability of its components.
A mechanistic insight into gene activation and repression by studying protein interactions between JA/JA-lle signalling components involving JAZs and TFs.
Epigenetic regulation of JA/JA-Ile signalling components.
Mechanistic insights into the cross-talks between JA/JA-Ile and other hormones such as auxin, ABA or GA in a process-related context.
Fine-mapping of JA, JA-Ile and OPDA levels as well as related metabolites at cells, tissue and organ levels in relation to external stimuli and developmental phases.
Chemical and genetic screens to pick up new components involved in JA/JA-Ile formation and signalling.
Translational and post-translational control mechanisms including protein phosphorylation that affect JA/JA-lle-dependent processes.
However, a system biology approach will help understand many of these data and elucidate how hormone-dependent processes evolved during adaptation of plants to their environments.
ACKNOWLEDGMENTS
The authors work was supported by the Deutsche Forschungsgemeinschaft (SFB 363 project C5 and SFB 648 project C2 to C.W. and projects HA2655/7-3 and HA2655/12-1 to B.H.) and the Region HANA for Biotechnological and Agricultural Research, Czech Republic (grant no. ED0007/01/01 to C.W.). We thank two anonymous reviewers for excellent suggestions and language editing of the manuscript.
NOTE ADDED IN PROOF
Recently it was shown that COI1 is stabilized by SCFCOI1 and degraded via the 26S proteasome (Yan et al., 2013, The Plant Cell 25: 486–498).
The SA suppression on JA biosynthesis and signalling was localized downstream of SCFCOI1–JAZ and includes the TF ORA59 (Van der Does et al., 2013, The Plant Cell 25: 744–761).
LITERATURE CITED
- Abe M, Shiboaka H, Yamane H, Takahashi N. Cell cycle-dependent disruption of microtubules by methyl jasmonate in tobacco BY-2 cells. Protoplasma. 1990;156:1–8. [Google Scholar]
- Acosta IF, Farmer EE. Jasmonates. The Arabidopsis Book. 2010;8 doi: 10.1199/tab.0129. e0129. http://dx.doi.org/10.1199/tab.0129 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta IF, Laparra H, Romero SP, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- Adams E, Turner J. COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. Journal of Experimental Botany. 2010;61:4373–4386. doi: 10.1093/jxb/erq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie BAT, Perez-Perez J, Perez-Perez MM, et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. The Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahkami AH, Lischewski S, Haensch K-T, et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: involvement of wound response and primary metabolism. New Phytologist. 2009;181:613–625. doi: 10.1111/j.1469-8137.2008.02704.x. [DOI] [PubMed] [Google Scholar]
- Andreou A, Brodhun F, Feussner I. Biosynthesis of oxylipins in non-mammals. Progress in Lipid Research. 2009;48:148–170. doi: 10.1016/j.plipres.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Attaran E, Zeier TE, Griebel T, Zeier J. Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. The Plant Cell. 2009;21:954–971. doi: 10.1105/tpc.108.063164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst BA, Ferrieri RA, Gray DW, et al. Jasmonic acid induces rapid changes in carbon transport and partitioning in Populus. New Phytologist. 2005;167:63–72. doi: 10.1111/j.1469-8137.2005.01388.x. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. The Plant Journal. 2010;62:250–264. doi: 10.1111/j.1365-313X.2010.04151.x. [DOI] [PubMed] [Google Scholar]
- Baldwin IT. Plant volatiles. Current Biology. 2010;20:R392–R397. doi: 10.1016/j.cub.2010.02.052. [DOI] [PubMed] [Google Scholar]
- Ballaré CL. Jasmonate-induced defenses: a tale of intelligence, collaborators and rascals. Trends in Plant Science. 2011;16:249–257. doi: 10.1016/j.tplants.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Mazza CA, Austin AT, Pierik R. Canopy light and plant health. Plant Physiology. 2012;160:145–155. doi: 10.1104/pp.112.200733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Fozard JA, Godin C, et al. Multiscale systems analysis of root growth and development: Modeling beyond the network and cellular scales. The Plant Cell. 2012a;24:3892–3906. doi: 10.1105/tpc.112.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Úbeda-Tomás S, Dyson RJ, et al. Growth-induced hormone dilution can explain the dynamics of plant root cell elongation. Proceedings of the National Academy of Sciences of the USA. 2012b;109:7577–7582. doi: 10.1073/pnas.1113632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Chatterjee M, Yu Y, Suh S-G, Miller WA, Hannapel DJ. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. The Plant Cell. 2006;18:3443–3457. doi: 10.1105/tpc.106.042473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannenberg G, Martínez M, Hamberg M, Castresana C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids. 2009;44:85–95. doi: 10.1007/s11745-008-3245-7. [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman R, Mullet J. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Ménard R, Heitz T, Shen W-H. Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cellular Microbiology. 2012;14:829–839. doi: 10.1111/j.1462-5822.2012.01785.x. [DOI] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany. 2012;63:2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhl D, Hörtensteiner S, Martinoia E, et al. The fou2 mutation in the major vacuolar cation channel TPC1 confers tolerance to inhibitory luminal calcium. The Plant Journal. 2009;58:715–723. doi: 10.1111/j.1365-313X.2009.03820.x. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha D, Wang J, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Blechert S, Brodschelm W, Hölder S, et al. The octadecanoid pathway: Signal molecules for the regulation of secondary pathways. Proceedings of the National Academy of Sciences of the USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert S, Bockelmann C, Füßlein M, et al. Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta. 1999;207:470–479. [Google Scholar]
- Bonaventure G, Gfeller A, Proebsting WM, et al. A gain-of-function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. The Plant Journal. 2007a;49:889–898. doi: 10.1111/j.1365-313X.2006.03002.x. [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Gfeller A, Rodríguez VM, Armand F, Farmer EE. The fou2 gain-of-function allele and the wild-type allele of Two Pore Channel 1 contribute to different extents or by different mechanisms to defense gene expression in Arabidopsis. Plant and Cell Physiology. 2007b;48:1775–1789. doi: 10.1093/pcp/pcm151. [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Schuck S, Baldwin IT. Revealing complexity and specificity in the activation of lipase-mediated oxylipin biosynthesis: a specific role of the Nicotiana attenuata GLA1 lipase in the activation of jasmonic acid biosynthesis in leaves and roots. Plant, Cell & Environment. 2011a;34:1507–1520. doi: 10.1111/j.1365-3040.2011.02348.x. [DOI] [PubMed] [Google Scholar]
- Bonaventure G, VanDoorn A, Baldwin IT. Herbivore-associated elicitors: FAC signaling and metabolism. Trends in Plant Science. 2011b;16:294–299. doi: 10.1016/j.tplants.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A. Mechanisms underlying beneficial plant – fungus interactions in mycorrhizal symbiosis. Nature Communications. 2010;1 doi: 10.1038/ncomms1046. 48. http://dx.doi.org/10.1038/ncomms1046 . [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. The Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breithaupt C, Strassner J, Breitinger U, et al. X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE. Structure. 2001;9:419–429. doi: 10.1016/s0969-2126(01)00602-5. [DOI] [PubMed] [Google Scholar]
- Breithaupt C, Kurzbauer R, Lilie H, et al. Crystal structure of 12-oxophytodienoate reductase 3 from tomato: Self-inhibition by dimerization. Proceedings of the National Academy of Sciences of the USA. 2006;103:14337–14342. doi: 10.1073/pnas.0606603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brioudes F, Joly C, Szécsi J, et al. Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. The Plant Journal. 2009;60:1070–1080. doi: 10.1111/j.1365-313X.2009.04023.x. [DOI] [PubMed] [Google Scholar]
- Brodhun F, Feussner I. Oxylipins in fungi. FEBS Journal. 2011;278:1047–1063. doi: 10.1111/j.1742-4658.2011.08027.x. [DOI] [PubMed] [Google Scholar]
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annual Review in Plant Biology. 2009a;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- Browse J. Jasmonate: preventing the maize tassel from getting in touch with his feminine side. Science Signalling. 2009b;2 doi: 10.1126/scisignal.259pe9. pe9. [DOI] [PubMed] [Google Scholar]
- Browse J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry. 2009c;70:1539–1546. doi: 10.1016/j.phytochem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Brundrett MC. Coevolution of roots and mycorrhizas of land plants. New Phytologist. 2002;154:275–304. doi: 10.1046/j.1469-8137.2002.00397.x. [DOI] [PubMed] [Google Scholar]
- Brüx A, Liu T-Y, Krebs M, et al. Reduced V-ATPase activity in the trans-golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. The Plant Cell. 2008;20:1088–1100. doi: 10.1105/tpc.108.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li C-B, et al. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Research. 2008;18:756–767. doi: 10.1038/cr.2008.53. [DOI] [PubMed] [Google Scholar]
- Caldelari D, Wang G, Farmer E, Dong X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Molecular Biology. 2011;75:25–33. doi: 10.1007/s11103-010-9701-9. [DOI] [PubMed] [Google Scholar]
- Camehl I, Sherameti I, Venus Y, et al. Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytologist. 2010;185:1062–1073. doi: 10.1111/j.1469-8137.2009.03149.x. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Banerjee AK, Hannapel DJ. A BELL1-like gene of potato is light activated and wound inducible. Plant Physiology. 2007;145:1435–1443. doi: 10.1104/pp.107.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenzano A, Abdala G, Hause B. Cytochemical immuno-localization of allene oxide cyclase, a jasmonic acid biosynthetic enzyme, in developing potato stolons. Journal of Plant Physiology. 2007;164:1449–1456. doi: 10.1016/j.jplph.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, et al. Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiology. 2012;158:2042–2052. doi: 10.1104/pp.112.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çevik V, Kidd BN, Zhang P, et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiology. 2012;160:541–555. doi: 10.1104/pp.112.202697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, et al. Plastid ω3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. The Plant Journal. 2008;54:106–117. doi: 10.1111/j.1365-313X.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- Chauvin A, Caldelari D, Wolfender J-L, Farmer EE. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytologist. 2013;197:566–575. doi: 10.1111/nph.12029. [DOI] [PubMed] [Google Scholar]
- Chehab EW, Kim S, Savchenko T, Kliebenstein D, Dehesh K, Braam J. Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiology. 2011;156:770–778. doi: 10.1104/pp.111.174169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Yao C, Henderson Z, Kim S, Braam J. Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Current Biology. 2012;22:701–706. doi: 10.1016/j.cub.2012.02.061. [DOI] [PubMed] [Google Scholar]
- Chen H, Jones AD, Howe GA. Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Letters. 2006;580:2540–2546. doi: 10.1016/j.febslet.2006.03.070. [DOI] [PubMed] [Google Scholar]
- Chen H, Gonzales-Vigil E, Wilkerson CG, Howe GA. Stability of plant defense proteins in the gut of insect herbivores. Plant Physiology. 2007;143:1954–1967. doi: 10.1104/pp.106.095588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. The Plant Cell. 2012;24:2898–2916. doi: 10.1105/tpc.112.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, et al. The basic helix–loop–helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. The Plant Cell. 2011;23:3335–3352. doi: 10.1105/tpc.111.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, et al. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000440. e1000440. http://dx.doi.org/10.1371/journal.pgen.1000440 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Sun L, Qi T, et al. The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Molecular Plant. 2011;4:279–288. doi: 10.1093/mp/ssq073. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Chini A, Boter M, Solano R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS Journal. 2009a;276:4682–4692. doi: 10.1111/j.1742-4658.2009.07194.x. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. The Plant Journal. 2009b;59:77–87. doi: 10.1111/j.1365-313X.2009.03852.x. [DOI] [PubMed] [Google Scholar]
- Chory J. Light signal transduction: an infinite spectrum of possibilities. The Plant Journal. 2010;61:982–991. doi: 10.1111/j.1365-313X.2009.04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. The Plant Cell. 2009;21:131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiology. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Niu Y, Browse J, Howe GA. Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry. 2009;70:1547–1559. doi: 10.1016/j.phytochem.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Cooke TF, DePew CL, et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. The Plant Journal. 2010;63:613–622. doi: 10.1111/j.1365-313X.2010.04265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SM, Cristescu SM, Miersch O, Harren FJM, Wasternack C, Mur LAJ. Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytologist. 2009;182:175–187. doi: 10.1111/j.1469-8137.2008.02735.x. [DOI] [PubMed] [Google Scholar]
- Clouse SD. Brassinosteroid signal transduction: clarifying the pathway from ligand perception to gene expression. Molecular Cell. 2002;10:973–982. doi: 10.1016/s1097-2765(02)00744-x. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Andrade A, del Carmen Tordable M, Cassán F, Abdala G. Production and function of jasmonates in nodulated roots of soybean plants inoculated with Bradyrhizobium japonicum. Archives of Microbiology. 2012;194:837–845. doi: 10.1007/s00203-012-0817-y. [DOI] [PubMed] [Google Scholar]
- Costigan SE, Warnasooriya SN, Humphries BA, Montgomery BL. Root-localized phytochrome chromophore synthesis is required for photoregulation of root elongation and impacts root sensitivity to jasmonic acid in Arabidopsis. Plant Physiology. 2011;157:1138–1150. doi: 10.1104/pp.111.184689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid. Annual Review of Plant Biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. emergence of a core signaling network. [DOI] [PubMed] [Google Scholar]
- Dabrowska P, Boland W. iso-OPDA: An early precursor of cis-jasmone in plants? ChemBioChem. 2007;8:2281–2285. doi: 10.1002/cbic.200700464. [DOI] [PubMed] [Google Scholar]
- Dabrowska P, Freitak D, Vogel H, Heckel DG, Boland W. The phytohormone precursor OPDA is isomerized in the insect gut by a single, specific glutathione transferase. Proceedings of the National Academy of Sciences of the USA. 2009;106:16304–16309. doi: 10.1073/pnas.0906942106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiology. 2012;159:1511–1523. doi: 10.1104/pp.112.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe W, Rönsch H, Preiss A, Schade W, Sembdner G, Schreiber K. Endogenous plant hormones of the broad bean, Vicia faba L. (–)-Jasmonic acid, a plant growth inhibitor in pericarp. Planta. 1981;155:530–535. doi: 10.1007/BF00385537. [DOI] [PubMed] [Google Scholar]
- Dave A, Graham IA. Oxylipin signalling: a distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA) Frontiers in Plant Science. 2012;3 doi: 10.3389/fpls.2012.00042. 42. http://dx.doi.org/10.3389/fpls.2012.00042 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A, Hernández ML, He Z, et al. 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. The Plant Cell. 2011;23:583–599. doi: 10.1105/tpc.110.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer K, Tilleman S, Pauwels L, et al. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix–loop–helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. The Plant Journal. 2011;66:1053–1065. doi: 10.1111/j.1365-313X.2011.04566.x. [DOI] [PubMed] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends in Plant Science. 2012;17:349–359. doi: 10.1016/j.tplants.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Delaney T, Ukness S, Vernooij B, et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Demianski AJ, Chung KM, Kunkel BN. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Molecular Plant Pathology. 2012;13:46–57. doi: 10.1111/j.1364-3703.2011.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DMA, Klessig DF. SOS – too many signals for systemic acquired resistance? Trends in Plant Science. 2012;17:538–545. doi: 10.1016/j.tplants.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Dicke M, Baldwin IT. The evolutionary context for herbivore-induced plant volatiles: beyond the ‘cry for help. Trends in Plant Science. 2010;15:167–175. doi: 10.1016/j.tplants.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Ding Y, Oldroyd GED. Positioning the nodule, the hormone dictum. Plant Signaling & Behavior. 2009;4:89–93. doi: 10.4161/psb.4.2.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. The Plant Cell. 2007;19:2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, et al. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature. 2009;457:1154–1158. doi: 10.1038/nature07612. [DOI] [PubMed] [Google Scholar]
- El Oirdi M, El Rahman TA, Rigano L, et al. Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. The Plant Cell. 2011;23:2405–2421. doi: 10.1105/tpc.111.083394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D, Stingl N, Kubigsteltig II, et al. DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiology. 2010;153:114–127. doi: 10.1104/pp.110.155093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. The Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA. Role of phytohormones in insect-specific plant reactions. Trends in Plant Science. 2012;17:250–259. doi: 10.1016/j.tplants.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Pérez M, Krol E, Stange A, et al. A special pair of phytohormones controls excitability, slow closure, and external stomach formation in the Venus flytrap. Proceedings of the National Academy of Sciences of the USA. 2011;108:15492–15497. doi: 10.1073/pnas.1112535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmaki T, Sanmartin M, Jimenez P, et al. Differential distribution of the lipoxygenase pathway enzymes within potato chloroplasts. Journal of Experimental Botany. 2007;58:555–568. doi: 10.1093/jxb/erl230. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Davoine C. Reactive electrophile species. Current Opinion in Plant Biology. 2007;10:380–386. doi: 10.1016/j.pbi.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proceedings of the National Academy of Sciences of the USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Tumlinson JH. Plant-insect dialogs: complex interactions at the plant-insect interface. Current Opinion in Plant Biology. 2008;11:457–463. doi: 10.1016/j.pbi.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C. The lipoxygenase pathway. Annual Review in Plant Biology. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, et al. (+)-7-iso-jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nature Chemical Biology. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- Franken P. The plant strengthening root endophyte Piriformospora indica: potential application and the biology behind. Applied Microbiology and Biotechnology. 2012;96:1455–1464. doi: 10.1007/s00253-012-4506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire MA, Tourneur C, Granier F, et al. Plant lipoxygenase 2 is a translation initiation factor-4E-binding protein. Plant Molecular Biology. 2000;44:129–140. doi: 10.1023/a:1006494628892. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gális I, Gaquerel E, Pandey SP, Baldwin IT. Molecular mechanisms underlying plant memory in JA-mediated defence responses. Plant Cell & Environment. 2009;32:617–627. doi: 10.1111/j.1365-3040.2008.01862.x. [DOI] [PubMed] [Google Scholar]
- Gao Q-M, Venugopal S, Navarre D, Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiology. 2011;155:464–476. doi: 10.1104/pp.110.166876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Garrido J, Ocampo J. Regulation of the plant defence response in arbuscular mycorrhizal symbiosis. Journal of Experimental Botany. 2002;53:1377–1386. [PubMed] [Google Scholar]
- García-Garrido JM, León-Morcillo RJ, Rodríguez JÁM, Bote JAO. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Molecular Plant–Microbe Interactions. 2010;23:651–664. doi: 10.1094/MPMI-23-5-0651. [DOI] [PubMed] [Google Scholar]
- Garcion C, Metraux J-P. Salicylic acid. In: Hedden P, Thomas S, editors. Plant hormone signaling. Harpenden: Blackwell Publishing; 2006. pp. 229–256. [Google Scholar]
- Gatz C. From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Molecular Plant–Microbe Interactions. 2013;26:151–159. doi: 10.1094/MPMI-04-12-0078-IA. [DOI] [PubMed] [Google Scholar]
- Gaupels F, Sarioglu H, Beckmann M, et al. Deciphering systemic wound responses of the pumpkin extrafascicular phloem by metabolomics and stable isotope-coded protein labeling. Plant Physiology. 2012;160:2285–2299. doi: 10.1104/pp.112.205336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Cheng J, Gangadharan A, Mackey D. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. The Plant Cell. 2012;24:4763–4774. doi: 10.1105/tpc.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R, Lillo A, Bisseling T. Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Current Opinion in Plant Biology. 2012;15:438–443. doi: 10.1016/j.pbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Gfeller A, Baerenfaller K, Loscos J, Chételat A, Baginsky S, Farmer EE. Jasmonate controls polypeptide patterning in undamaged tissue in wounded Arabidopsis leaves. Plant Physiology. 2011;156:1797–1807. doi: 10.1104/pp.111.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda S, Miersch O, Levitin A, Schmidt J, Wasternack C, Varin L. Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. Journal of Biological Chemistry. 2003;278:17895–17900. doi: 10.1074/jbc.M211943200. [DOI] [PubMed] [Google Scholar]
- Glauser G, Grata E, Dubugnon L, Rudaz S, Farmer EE, Wolfender J-L. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. Journal of Biological Chemistry. 2008;283:16400–16407. doi: 10.1074/jbc.M801760200. [DOI] [PubMed] [Google Scholar]
- Glauser G, Dubugnon L, Mousavi SAR, Rudaz S, Wolfender J-L, Farmer EE. Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. Journal of Biological Chemistry. 2009;284:34506–34513. doi: 10.1074/jbc.M109.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel C, Feussner I. Methods for the analysis of oxylipins in plants. Phytochemistry. 2009;70:1485–1503. doi: 10.1016/j.phytochem.2009.07.040. [DOI] [PubMed] [Google Scholar]
- Goetz S, Hellwege A, Stenzel I, et al. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiology. 2012;158:1715–1727. doi: 10.1104/pp.111.192658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Vigil E, Bianchetti CM, Phillips GN, Howe GA. Adaptive evolution of threonine deaminase in plant defense against insect herbivores. Proceedings of the National Academy of Sciences of the USA. 2011;108:5897–5902. doi: 10.1073/pnas.1016157108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proceedings of the National Academy of Sciences of the USA. 2012;109:4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C, Cullimore J. Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Molecular Plant–Microbe Interactions. 2011;24:867–878. doi: 10.1094/MPMI-01-11-0019. [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JDG. Hormone (dis)harmony moulds plant health and disease. Science. 2009;324:750–752. doi: 10.1126/science.1173771. [DOI] [PubMed] [Google Scholar]
- Grunewald W, Vanholme B, Pauwels L, et al. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Reports. 2009;10:923–928. doi: 10.1038/embor.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach H, Müller M, Kutchan T, Zenk M. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proceedings of the National Academy of Sciences of the USA. 1992;89:2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S-S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant, Cell & Environment. 2012;35:644–655. doi: 10.1111/j.1365-3040.2011.02442.x. [DOI] [PubMed] [Google Scholar]
- Guranowski A, Miersch O, Staswick PE, Suza W, Wasternack C. Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1) FEBS Letters. 2007;581:815–820. doi: 10.1016/j.febslet.2007.01.049. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. The Plant Cell. 2012;24:2515–2527. doi: 10.1105/tpc.112.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Paszkowski U. Weights in the balance: Jasmonic acid and salicylic acid signaling in root-biotroph interactions. Molecular Plant–Microbe Interactions. 2009;22:763–772. doi: 10.1094/MPMI-22-7-0763. [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Riemann M, Müller A, Düchting P, Weiler E, Nick P. Cholodny–Went revisited: A role for jasmonate in gravitropism of rice coleoptiles. Planta. 2005;222:575–585. doi: 10.1007/s00425-005-0001-6. [DOI] [PubMed] [Google Scholar]
- Hanlon MT, Coenen C. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytologist. 2011;189:701–709. doi: 10.1111/j.1469-8137.2010.03567.x. [DOI] [PubMed] [Google Scholar]
- Hannapel DJ. A model system of development regulated by the long-distance transport of mRNA. Journal of Integrative Plant Biology. 2010;52:40–52. doi: 10.1111/j.1744-7909.2010.00911.x. [DOI] [PubMed] [Google Scholar]
- Hao J, Tu L, Hu H, et al. GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. Journal of Experimental Botany. 2012;63:6267–6281. doi: 10.1093/jxb/ers278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms K, Ramirez I, Pena-Cortés H. Inhibition of wound-induced accumulation of allene oxide synthase transcripts in flax leaves by aspirin and salicylic acid. Plant Physiology. 1998;118:1057–1065. doi: 10.1104/pp.118.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SE, Gange AC. Impacts of plant symbiotic fungi on insect herbivores: Mutualism in a multitrophic context. Annual Review of Entomology. 2009;54:323–342. doi: 10.1146/annurev.ento.54.110807.090614. [DOI] [PubMed] [Google Scholar]
- Hause B, Schaarschmidt S. The role of jasmonates in mutualistic symbioses between plants and soil-born microorganisms. Phytochemistry. 2009;70:1589–1599. doi: 10.1016/j.phytochem.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Hause B, Stenzel I, Miersch O, et al. Tissue-specific oxylipin signature of tomato flowers – allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. The Plant Journal. 2000;24:113–126. doi: 10.1046/j.1365-313x.2000.00861.x. [DOI] [PubMed] [Google Scholar]
- Hause B, Maier W, Miersch O, Kramell R, Strack D. Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiology. 2002;130:1213–1220. doi: 10.1104/pp.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Hause G, Kutter C, Miersch O, Wasternack C. Enzymes of jasmonate biosynthesis occur in tomato sieve elements. Plant and Cell Physiology. 2003;44:643–648. doi: 10.1093/pcp/pcg072. [DOI] [PubMed] [Google Scholar]
- Hause B, Wasternack C, Strack D. Jasmonates in stress responses and development. Phytochemistry. 2009;70:1483–1484. doi: 10.1016/j.phytochem.2009.07.004. [DOI] [PubMed] [Google Scholar]
- He Y, Chung E-H, Hubert DA, Tornero P, Dangl JL. Specific missense alleles of the Arabidopsis jasmonic acid co-receptor COI1 regulate innate immune receptor accumulation and function. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1003018. e1003018. http://dx.doi.org/10.1371/journal.pgen.1003018 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Karban R. Explaining evolution of plant communication by airborne signals. Trends in Ecology & Evolution. 2010;25:137–144. doi: 10.1016/j.tree.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Heil M, Ton J. Long-distance signalling in plant defence. Trends in Plant Science. 2008;13:264–272. doi: 10.1016/j.tplants.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Heitz T, Widemann E, Lugan R, et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. Journal of Chemical Biology. 2012;287:6296–6306. doi: 10.1074/jbc.M111.316364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Medina M, Tamayo M, Vierheilig H, Ocampo J, García-Garrido J. The jasmonic acid signalling pathway restricts the development of the arbuscular mycorrhizal association in tomato. Journal of Plant Growth Regulation. 2008;27:221–230. [Google Scholar]
- Hind S, Malinowski R, Yalamanchili R, Stratmann JW. Tissue-type specific systemin perception and the elusive systemin receptor. Plant Signaling & Behavior. 2010;5:42–44. doi: 10.4161/psb.5.1.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind SR, Pulliam SE, Veronese P, et al. The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. The Plant Journal. 2011;65:480–491. doi: 10.1111/j.1365-313X.2010.04437.x. [DOI] [PubMed] [Google Scholar]
- Hofmann E, Zerbe P, Schaller F. The crystal structure of Arabidopsis thaliana allene oxide cyclase: Insights into the oxylipin cyclization reaction. The Plant Cell. 2006;18:3201–3217. doi: 10.1105/tpc.106.043984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Developmental Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Howe G, Jander G. Plant immunity to insect herbivores. Annual Review in Plant Biology. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Huang Y, Han C, Peng W, et al. Brassinosteroid negatively regulates jasmonate inhibition of root growth in Arabidopsis. Plant Signaling & Behavior. 2010;5:140–142. doi: 10.4161/psb.5.2.10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Choi S, Hwang H-J, et al. Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Developmental Cell. 2008;14:183–192. doi: 10.1016/j.devcel.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdán PD. PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. The Plant Journal. 2012;69:601–612. doi: 10.1111/j.1365-313X.2011.04815.x. [DOI] [PubMed] [Google Scholar]
- Isayenkov S, Mrosk C, Stenzel I, Strack D, Hause B. Suppression of allene oxide cyclase in hairy roots of Medicago truncatula reduces jasmonate levels and the degree of mycorrhization with Glomus intraradices. Plant Physiology. 2005;139:1401–1410. doi: 10.1104/pp.105.069054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kwai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation. The Plant Cell. 2001;13:2191–2209. doi: 10.1105/tpc.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ng K-H, Lim T-S, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. The Plant Cell. 2007;19:3516–3529. doi: 10.1105/tpc.107.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD, Heyer A, Dietze J, Prat S. Phytochrome B mediates the photoperiodic control of tuber formation in potato. The Plant Journal. 1996;9:159–166. [Google Scholar]
- Jacobs S, Zechmann B, Molitor A, et al. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiology. 2011;156:726–740. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. Priming in systemic plant immunity. Science. 2009;324:89–91. doi: 10.1126/science.1170025. [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. In: Marja CPT, editor. Current Topics in Developmental Biology. New York: Academic Press; 2010. pp. 29–66. [DOI] [PubMed] [Google Scholar]
- Kandel S, Sauveplane V, Compagnon V, et al. Characterization of a methyl jasmonate and wounding-responsive cytochrome P450 of Arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. FEBS Journal. 2007;274:5116–5127. doi: 10.1111/j.1742-4658.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, et al. Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proceedings of the National Academy of Sciences of the USA. 2007;104:12205–12210. doi: 10.1073/pnas.0700344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-H, Liu G, Shi F, Jones AD, Beaudry RM, Howe GA. The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiology. 2010;154:262–272. doi: 10.1104/pp.110.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Kim HB, Lee H, et al. Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fungal infection. Molecules and Cells. 2006;21:418–427. [PubMed] [Google Scholar]
- Kant MR, Baldwin IT. The ecogenetics and ecogenomics of plant–herbivore interactions: rapid progress on a slippery road. Current Opinion in Genetics & Development. 2007;17:519–524. doi: 10.1016/j.gde.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Kaplan I, Halitschke R, Kessler A, Rehill BJ, Sardanelli S, Denno RF. Physiological integration of roots and shoots in plant defense strategies links above- and belowground herbivory. Ecology Letters. 2008;11:841–851. doi: 10.1111/j.1461-0248.2008.01200.x. [DOI] [PubMed] [Google Scholar]
- Katsir L, Chung HS, Koo AJ, Howe GA. Jasmonate signaling: a conserved mechanism of hormone sensing. Current Opinion in Plant Biology. 2008a;11:428–435. doi: 10.1016/j.pbi.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proceedings of the National Academy of Sciences of the USA. 2008b;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. Jasmonate signaling: toward an integrated view. Plant Physiology. 2008;146:1459–1468. doi: 10.1104/pp.107.115717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. The interplay between light and jasmonate signalling during defence and development. Journal of Experimental Botany. 2011;62:4087–4100. doi: 10.1093/jxb/err142. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. JAZ repressors and the orchestration of phytohormone crosstalk. Trends in Plant Science. 2012;17:22–31. doi: 10.1016/j.tplants.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. MYC2: the Master in Action. Molecular Plant. 2013 doi: 10.1093/mp/sss128. in press. http://dx.doi.org/10.1093/mp/sss128 . [DOI] [PubMed] [Google Scholar]
- Kelley DR, Estelle M. Ubiquitin-mediated control of plant hormone signaling. Plant Physiology. 2012;160:47–55. doi: 10.1104/pp.112.200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D, Baldwin IT. Back to the past for pollination biology. Current Opinion in Plant Biology. 2011;14:429–434. doi: 10.1016/j.pbi.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Kessler D, Diezel C, Baldwin IT. Changing pollinators as a means of escaping herbivores. Current Biology. 2010;20:237–242. doi: 10.1016/j.cub.2009.11.071. [DOI] [PubMed] [Google Scholar]
- Khatabi B, Molitor A, Lindermayr C, et al. Ethylene supports colonization of plant roots by the mutualistic fungus Piriformospora indica. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0035502. e35502. http://dx.doi.org/10.1371/journal.pone.0035502 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, et al. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. The Plant Cell. 2009;21:2237–2252. doi: 10.1105/tpc.109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, Adler LS, Grman EL, Van Der Heijden MGA. Manipulating the jasmonate response: How do methyl jasmonate additions mediate characteristics of aboveground and belowground mutualisms? Functional Ecology. 2010;24:434–443. [Google Scholar]
- Kinkema M, Gresshoff PM. Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Molecular Plant–Microbe Interactions. 2008;21:1337–1348. doi: 10.1094/MPMI-21-10-1337. [DOI] [PubMed] [Google Scholar]
- Kistner C, Parniske M. Evolution of signal transduction in intracellular symbiosis. Trends in Plant Science. 2002;7:511–518. doi: 10.1016/s1360-1385(02)02356-7. [DOI] [PubMed] [Google Scholar]
- Kitaoka N, Matsubara T, Sato M, et al. Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant and Cell Physiology. 2011;52:1757–1765. doi: 10.1093/pcp/pcr110. [DOI] [PubMed] [Google Scholar]
- Knopf RR, Feder A, Mayer K, et al. Rhomboid proteins in the chloroplast envelope affect the level of allene oxide synthase in Arabidopsis thaliana. The Plant Journal. 2012;72:559–571. doi: 10.1111/j.1365-313X.2012.05090.x. [DOI] [PubMed] [Google Scholar]
- Koch T, Bandemer K, Boland W. Biosynthesis of cis-jasmone: a pathway for the inactivation and the disposal of the plant stress hormone jasmonic acid to the gas phase? Helvetica et Chimica Acta. 1997;80:838–850. [Google Scholar]
- Kolomiets M, Hannapel D, Chen H, Tymeson M, Gladon R. Lipoxygenase is involved in the control of potato tuber development. The Plant Cell. 2001;13:613–626. doi: 10.1105/tpc.13.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink E. Chemical and genetic exploration of jasmonate biosynthesis and signaling paths. Planta. 2012;236:1351–1366. doi: 10.1007/s00425-012-1705-z. [DOI] [PubMed] [Google Scholar]
- Koo AJK, Howe GA. The wound hormone jasmonate. Phytochemistry. 2009;70:1571–1580. doi: 10.1016/j.phytochem.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ, Howe GA. Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Frontiers in Plant Science. 2012;3 doi: 10.3389/fpls.2012.00019. 19. http://dx.doi.org/10.3389/fpls.2012.00019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Gao X, Jones AD, Howe GA. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. The Plant Journal. 2009;59:974–986. doi: 10.1111/j.1365-313X.2009.03924.x. [DOI] [PubMed] [Google Scholar]
- Koo AJK, Cooke TF, Howe GA. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proceedings of the National Academy of Sciences of the USA. 2011;108:9298–9303. doi: 10.1073/pnas.1103542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ. Cross talk in defense signaling. Plant Physiology. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubigsteltig I, Laudert D, Weiler E. Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta. 1999;208:463–471. doi: 10.1007/s004250050583. [DOI] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. The Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K, Walcher C, Nemhauser J. Cross-regulatory mechanisms in hormone signaling. Plant Molecular Biology. 2009;69:375–381. doi: 10.1007/s11103-008-9389-2. [DOI] [PubMed] [Google Scholar]
- Lackman P, González-Guzmán M, Tilleman S, et al. Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proceedings of the National Academy of Sciences of the USA. 2011;108:5891–5896. doi: 10.1073/pnas.1103010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahrmann U, Zuccaro A. Opprimo ergo sum—Evasion and suppression in the root endophytic fungus Piriformospora indica. Molecular Plant–Microbe Interactions. 2012;25:727–737. doi: 10.1094/MPMI-11-11-0291. [DOI] [PubMed] [Google Scholar]
- Landgraf R, Schaarschmidt S, Hause B. Repeated leaf wounding alters the colonization of Medicago truncatula roots by beneficial and pathogenic microorganisms. Plant, Cell & Environment. 2012;35:1344–1357. doi: 10.1111/j.1365-3040.2012.02495.x. [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. Plant hormone signaling lightens up: integrators of light and hormones. Current Opinion in Plant Biology. 2010;13:571–577. doi: 10.1016/j.pbi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Lee B-R, Huseby S, Koprivova A, et al. Effects of fou8/fry1 mutation on sulfur metabolism: Is decreased internal sulfate the trigger of sulfate starvation response? PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0039425. e39425. http://dx.doi.org/10.1371/journal.pone.0039425 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Morcillo RJ, Ángel J, Martín-Rodríguez, Vierheilig H, Ocampo JA, García-Garrido JM. Late activation of the 9-oxylipin pathway during arbuscular mycorrhiza formation in tomato and its regulation by jasmonate signalling. Journal of Experimental Botany. 2012;63:3545–3558. doi: 10.1093/jxb/ers010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, De Lange ES, et al. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiology. 2009;149:1797–1809. doi: 10.1104/pp.108.133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Van der Does D, De Lange E, et al. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta. 2010;232:1423–1432. doi: 10.1007/s00425-010-1265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li C, Lee G, Howe G. Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proceedings of the National Academy of Sciences of the USA. 2002;99:6416–6421. doi: 10.1073/pnas.072072599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, McCaig B, Wingerd B, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. The Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Sharma P, Gonzalez DH, Viola IL, Hannapel DJ. The impact of the long-distance transport of a BEL1-like mRNA on development. Plant Physiology. 2013;161:760–772. doi: 10.1104/pp.112.209429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Tian H, Huang Y-Q, et al. Alterations of mitochondrial protein assembly and jasmonic acid biosynthesis pathway in Honglian (HL)-type cytoplasmic male sterility rice. Journal of Biological Chemistry. 2012;287:40051–40060. doi: 10.1074/jbc.M112.382549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopéz-Ráez JA, Verhage A, Fernandez I, et al. Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. Journal of Experimental Botany. 2010;61:2589–2601. doi: 10.1093/jxb/erq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Los Santos RT, Vierheilig H, Ocampo JA, Garrido JMG. Altered pattern of arbuscular mycorrhizal formation in tomato ethylene mutants. Plant Signaling & Behavior. 2011;6:755–8. doi: 10.4161/psb.6.5.15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Bruce TJA, Roberts MR, Flors V, Ton J. Next-generation systemic acquired resistance. Plant Physiology. 2012;158:844–853. doi: 10.1104/pp.111.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Pu G, Lei C, et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant and Cell Physiology. 2009;50:2146–2161. doi: 10.1093/pcp/pcp149. [DOI] [PubMed] [Google Scholar]
- Mabood F, Souleimanov A, Khan W, Smith DL. Jasmonates induce Nod factor production by Bradyrhizobium japonicum. Plant Physiology and Biochemistry. 2006;44:759–765. doi: 10.1016/j.plaphy.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Maes L, Van Nieuwerburgh FCW, Zhang Y, et al. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytologist. 2011;189:176–189. doi: 10.1111/j.1469-8137.2010.03466.x. [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Browse J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiology. 2009;149:851–862. doi: 10.1104/pp.108.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, et al. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. The Plant Journal. 2006;46:984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- Mao P, Duan M, Wei C, Li Y. WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant and Cell Physiology. 2007;48:833–842. doi: 10.1093/pcp/pcm058. [DOI] [PubMed] [Google Scholar]
- Martín Rodriguez JÁ, León Morcillo R, Vierheilig H, Antonio Ocampo J, Ludwig-Müller J, García Garrido JM. Mycorrhization of the notabilis and sitiens tomato mutants in relation to abscisic acid and ethylene contents. Journal of Plant Physiology. 2010;167:606–613. doi: 10.1016/j.jplph.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Matsui K, Minami A, Hornung E, et al. Biosynthesis of fatty acid derived aldehydes is induced upon mechanical wounding and its products show fungicidal activities in cucumber. Phytochemistry. 2006;67:649–657. doi: 10.1016/j.phytochem.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Matsuura H, Takeishi S, Kiatoka N, Sato C, Sueda K, Masuta C, Nabeta K. Transportation of de novo synthesized jasmonoyl isoleucine in tomato. Phytochemistry. 2012;83:25–33. doi: 10.1016/j.phytochem.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Matthes M, Bruce T, Ton J, Verrier P, Pickett J, Napier J. The transcriptome of cis-jasmone-induced resistance in Arabidopsis thaliana and its role in indirect defence. Planta. 2010;232:1163–1180. doi: 10.1007/s00425-010-1244-4. [DOI] [PubMed] [Google Scholar]
- Meldau S, Erb M, Baldwin IT. Defence on demand: mechanisms behind optimal defence patterns. Annals of Botany. 2012;110:1503–1514. doi: 10.1093/aob/mcs212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Mecey C, Niu Y, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. The Plant Journal. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J. Regulation of gene expression by jasmonate hormones. Phytochemistry. 2009;70:1560–1570. doi: 10.1016/j.phytochem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Memelink J, Verpoorte R, Kijne J. ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends in Plant Science. 2001;6:212–219. doi: 10.1016/s1360-1385(01)01924-0. [DOI] [PubMed] [Google Scholar]
- Méndez-Bravo A, Calderón-Vázquez C, Ibarra-Laclette E, et al. Alkamides activate jasmonic acid biosynthesis and signaling pathways and confer resistance to Botrytis cinerea in Arabidopsis thaliana. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0027251. e27251. http://dx.doi.org/10.1371/journal.pone.0027251 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. The Plant Cell. 2007;19:819–830. doi: 10.1105/tpc.106.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke K, Forner S, Kramell R, Conrad U, Hause B. Cell-specific visualization of jasmonates in wounded tomato and Arabidopsis leaves using jasmonate-specific antibodies. New Phytologist. 2011;190:1069–1080. doi: 10.1111/j.1469-8137.2010.03638.x. [DOI] [PubMed] [Google Scholar]
- Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C. Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytologist. 2008;177:114–127. doi: 10.1111/j.1469-8137.2007.02252.x. [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. The Plant Journal. 2007;50:500–513. doi: 10.1111/j.1365-313X.2007.03067.x. [DOI] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GED, Allan Downie J. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. The Plant Journal. 2006;48:883–894. doi: 10.1111/j.1365-313X.2006.02926.x. [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. Auxin receptors and plant development: A new signaling paradigm. Annual Review of Cell and Developmental Biology. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- Molitor A, Kogel K-H. Induced resistance triggered by Piriformospora indica. Plant Signaling & Behavior. 2009;4:215–216. doi: 10.4161/psb.4.3.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor A, Zajic D, Voll LM, et al. Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica–mediated systemic induced resistance to powdery mildew. Molecular Plant–Microbe Interactions. 2011;24:1427–1439. doi: 10.1094/MPMI-06-11-0177. [DOI] [PubMed] [Google Scholar]
- Montiel G, Zarei A, Körbes AP, Memelink J. The jasmonate-responsive element from the ORCA3 promoter from Catharanthus roseus is active in Arabidopsis and is controlled by the transcription factor AtMYC2. Plant and Cell Physiology. 2011;52:578–587. doi: 10.1093/pcp/pcr016. [DOI] [PubMed] [Google Scholar]
- Mortier V, Holsters M, Goormachtig S. Never too many? How legumes control nodule numbers. Plant, Cell & Environment. 2012;35:245–258. doi: 10.1111/j.1365-3040.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- Mosblech A, Feussner I, Heilmann I. Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiology and Biochemistry. 2009;47:511–517. doi: 10.1016/j.plaphy.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Mosblech A, Thurow C, Gatz C, Feussner I, Heilmann I. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. The Plant Journal. 2011;65:949–957. doi: 10.1111/j.1365-313X.2011.04480.x. [DOI] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, et al. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. The Plant Cell. 2008;20:768–785. doi: 10.1105/tpc.107.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, et al. Disruption of adenosine-5'-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. The Plant Cell. 2009;21:910–927. doi: 10.1105/tpc.109.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtarova LS, Mukhitova FK, Gogolev YV, Grechkin AN. Hydroperoxide lyase cascade in pea seedlings: Non-volatile oxylipins and their age and stress dependent alterations. Phytochemistry. 2011;72:356–364. doi: 10.1016/j.phytochem.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Bi Y-M, Darby RM, Firek S, Draper J. Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. The Plant Journal. 1997;12:1113–1126. doi: 10.1046/j.1365-313x.1997.12051113.x. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiology. 2006;140:249–262. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JAH, Jones A, Godin C, Traas J. Systems analysis of shoot apical meristem growth and development: Integrating hormonal and mechanical signaling. The Plant Cell. 2012 doi: 10.1105/tpc.112.102194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kawaguchi M. Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant and Cell Physiology. 2006;47:176–180. doi: 10.1093/pcp/pci222. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Mithöfer A, Kombrink E, et al. 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiology. 2011;155:1226–1236. doi: 10.1104/pp.110.168617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam VJ, Keeretaweep J, Sarowar S, Shah J. Root-derived oxylipins promote green peach aphid performance on Arabidopsis foliage. The Plant Cell. 2012;24:1643–1653. doi: 10.1105/tpc.111.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K-H, Kong F, Matsuura H, Takahashi K, Nabeta K, Yoshihara T. Temperature regulates tuber-inducing lipoxygenase-derived metabolites in potato (Solanum tuberosum) Journal of Plant Physiology. 2008;165:233–238. doi: 10.1016/j.jplph.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Nelson D, Werck-Reichhart D. A P450-centric view of plant evolution. The Plant Journal. 2011;66:194–211. doi: 10.1111/j.1365-313X.2011.04529.x. [DOI] [PubMed] [Google Scholar]
- Neumann P, Brodhun F, Sauer K, et al. Crystal structures of Physcomitrella patens AOC1 and AOC2: insights into the enzyme mechanism and differences in substrate specificity. Plant Physiology. 2012;160:1251–1266. doi: 10.1104/pp.112.205138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AK, Fahlberg P, Ellerström M, Andersson MX. Oxo-phytodienoic acid (OPDA) is formed on fatty acids esterified to galactolipids after tissue disruption in Arabidopsis thaliana. FEBS Letters. 2012;586:2483–2487. doi: 10.1016/j.febslet.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. Journal of Experimental Botany. 2011;62:2143–2154. doi: 10.1093/jxb/erq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Sasaki-Sekimoto Y, Oikawa A, et al. 12-Oxo-phytodienoic acid-glutathione conjugate is transported into the vacuole in Arabidopsis. Plant and Cell Physiology. 2011;52:205–209. doi: 10.1093/pcp/pcq181. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Harrison MJ, Paszkowski U. Reprogramming plant cells for endosymbiosis. Science. 2009;324:753–754. doi: 10.1126/science.1171644. [DOI] [PubMed] [Google Scholar]
- Olsson ME, Olofsson LM, Lindahl A-L, Lundgren A, Brodelius M, Brodelius PE. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry. 2009;70:1123–1128. doi: 10.1016/j.phytochem.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Ortu G, Balestrini R, Pereira PA, Becker JD, Küster H, Bonfante P. Plant genes related to gibberellin biosynthesis and signaling are differentially regulated during the early stages of AM fungal interactions. Molecular Plant. 2012;5:951–954. doi: 10.1093/mp/sss027. [DOI] [PubMed] [Google Scholar]
- Park S-W, Kaimoyo E, Kumar D, Mosher S, Klessig DF. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science. 2007;318:113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Goossens A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. The Plant Cell. 2011;23:3089–3100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Inzé D, Goossens A. Jasmonate-inducible gene: what does it mean? Trends in Plant Science. 2009;14:87–91. doi: 10.1016/j.tplants.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, et al. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proceedings of the National Academy of Sciences of the USA. 2008;105:1380–1385. doi: 10.1073/pnas.0711203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan C. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–897. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- Peng Z, Han C, Yuan L, Zhang K, Huang H, Ren C. Brassinosteroid enhances jasmonate-induced anthocyanin accumulation in Arabidopsis seedlings. Journal of Integrative Plant Biology. 2011;53:632–640. doi: 10.1111/j.1744-7909.2011.01042.x. [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, et al. Arabidopsis lateral root development: an emerging story. Trends in Plant Science. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, et al. The protein expression landscape of the Arabidopsis root. Proceedings of the National Academy of Sciences of the USA. 2012a;109:6811–6818. doi: 10.1073/pnas.1202546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis root development. Annual Review of Plant Biology. 2012b;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C, Wees Sv, Ton J, Pelt JV, Loon LV. Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biology. 2002;4:535–544. [Google Scholar]
- Pieterse CMJ, van der Does D, Zamioudis C, Leon-Reyes A, van Wees SCM. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- Pineda A, Zheng S-J, van Loon JJA, Pieterse CMJ, Dicke M. Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends in Plant Science. 2010;15:507–514. doi: 10.1016/j.tplants.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Pinot F, Beisson F. Cytochrome P450 metabolizing fatty acids in plants: characterization and physiological roles. FEBS Journal. 2011;278:195–205. doi: 10.1111/j.1742-4658.2010.07948.x. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Azcon-Aguilar C. Unraveling mycorrhiza-induced resistance. Current Opinion in Plant Biology. 2007;10:393–398. doi: 10.1016/j.pbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Pre M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, et al. The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. The Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang X, Weiss M, Kogel K-H, Schäfer P. Piriformospora indica—a mutualistic basidiomycete with an exceptionally large plant host range. Molecular Plant Pathology. 2012;13:508–518. doi: 10.1111/j.1364-3703.2011.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Xi J, Du L, Suttle J, Poovaiah BW. Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Molecular Biology. 2012;79:89–99. doi: 10.1007/s11103-012-9896-z. [DOI] [PubMed] [Google Scholar]
- Radhika V, Kost C, Mithöfer A, Boland W. Regulation of extrafloral nectar secretion by jasmonates in lima bean is light dependent. Proceedings of the National Academy of Sciences of the USA. 2010;107:17228–17233. doi: 10.1073/pnas.1009007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez V, Van der Ent S, Garcia-Andrade J, Coego A, Pieterse C, Vera P. OCP3 is an important modulator of NPR1-mediated jasmonic acid-dependent induced defenses in Arabidopsis. BMC Plant Biology. 2010;10 doi: 10.1186/1471-2229-10-199. 199. http://dx.doi.org/10.1186/1471-2229-10-199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, et al. A regulatory network for coordinated flower maturation. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1002506. e1002506. http://dx.doi.org/10.1371/journal.pgen.1002506 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Hayashi S, Lin Y-H, Gresshoff PM. Molecular mechanisms controlling legume autoregulation of nodulation. Annals of Botany. 2011;108:789–795. doi: 10.1093/aob/mcr205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C, Springer A, Samol I, Reinbothe S. Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS Journal. 2009;276:4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x. [DOI] [PubMed] [Google Scholar]
- Ren C, Han C, Peng W, et al. A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiology. 2009;151:1412–1420. doi: 10.1104/pp.109.140202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C, Zimmerli C, Farmer EE, Reymond P, Poirier Y. Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiology. 2008;147:696–706. doi: 10.1104/pp.108.119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Annual Review of Phytopathology. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, et al. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. The Plant Cell. 2010;22:1143–1160. doi: 10.1105/tpc.109.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Falcón M, Bou J, Prat S. Seasonal control of tuberization in potato: Conserved elements with the flowering response. Annual Review of Plant Biology. 2006;57:151–180. doi: 10.1146/annurev.arplant.57.032905.105224. [DOI] [PubMed] [Google Scholar]
- Rodriguez MCS, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annual Review of Plant Biology. 2010a;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Rodriguez VM, Chetelat A, Majcherczyk P, Farmer EE. Chloroplastic phosphoadenosine phosphosulfate metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiology. 2010b;152:1335–1345. doi: 10.1104/pp.109.150474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas S, Soria R, Correa N, Abdala G. Jasmonic acid stimulates the expression of nod genes in Rhizobium. Plant Molecular Biology. 1998;38:1161–1168. doi: 10.1023/a:1006064807870. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Bokowiec MT, Han S, et al. Tobacco transcription factors: Novel insights into transcriptional regulation in the Solanaceae. Plant Physiology. 2008;147:280–295. doi: 10.1104/pp.107.114041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Sugimoto K. Tuning growth to the environmental demands. Current Opinion in Plant Biology. 2012;15:683–690. doi: 10.1016/j.pbi.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Saga H, Ogawa T, Kai K, et al. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Molecular Plant–Microbe Interactions. 2012;25:684–696. doi: 10.1094/MPMI-09-11-0244. [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. The Plant Journal. 2010;61:1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D. The signal transduction pathways controlling in planta tuberization in potato: an emerging synthesis. Plant Cell Reports. 2008;27:1–8. doi: 10.1007/s00299-007-0457-x. [DOI] [PubMed] [Google Scholar]
- Savchenko T, Walley JW, Chehab EW, et al. Arachidonic acid: An evolutionarily conserved signaling molecule modulates plant stress signaling networks. The Plant Cell. 2010;22:3193–3205. doi: 10.1105/tpc.110.073858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko T, Pearse IS, Ignatia L, Karban R, Dehesh K. Insect herbivores selectively suppress the HPL branch of the oxylipin pathway in host plants. The Plant Journal. 2013;73:653–662. doi: 10.1111/tpj.12064. [DOI] [PubMed] [Google Scholar]
- Schäfer P, Pfiffi S, Voll LM, et al. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. The Plant Journal. 2009;59:461–474. doi: 10.1111/j.1365-313X.2009.03887.x. [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. Enzymes in jasmonate biosynthesis – Structure, function, regulation. Phytochemistry. 2009;70:1532–1538. doi: 10.1016/j.phytochem.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Scherer GFE, Ryu SB, Wang X, Matos AR, Heitz T. Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends in Plant Science. 2010;15:693–700. doi: 10.1016/j.tplants.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology. 2008;6 doi: 10.1371/journal.pbio.0060230. e230. http://dx.doi.org/10.1371/journal.pbio.0060230 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B, Dabrowska P, Boland W. Rapid enzymatic isomerization of 12-oxophytodienoic acid in the gut of Lepidopteran larvae. ChemBioChem. 2007;8:208–216. doi: 10.1002/cbic.200600379. [DOI] [PubMed] [Google Scholar]
- Schuman M, Barthel K, Baldwin I. Herbivory-induced volatiles function as defences increasing fitness of the native plant Nicotiana attenuata in nature. eLife. 2012;1 doi: 10.7554/eLife.00007. e00007. http://dx.doi.org/10.7554/eLife.00007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüssler A, Schwarzott D, Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycological Research. 2001;105:1413–1421. [Google Scholar]
- Schwachtje J, Baldwin IT. Why does herbivore attack reconfigure primary metabolism? Plant Physiology. 2008;146:845–851. doi: 10.1104/pp.107.112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proceedings of the National Academy of Sciences of the USA. 2006;103:12935–12940. doi: 10.1073/pnas.0602316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C. Gibberellin signalling in plants (the extended version) Frontiers in Plant Science. 2012;2 doi: 10.3389/fpls.2011.00107. 107. http://dx.doi.org/10.3389/fpls.2011.00107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehr EM, Agusti J, Lehner R, Farmer EE, Schwarz M, Greb T. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. The Plant Journal. 2010;63:811–822. doi: 10.1111/j.1365-313X.2010.04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann MA, Stingl NE, Lautenschlaeger JK, Krischke M, Mueller MJ, Berger S. Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiology. 2010;152:1940–1950. doi: 10.1104/pp.110.153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Song J, Cheong J-J, et al. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proceedings of the National Academy of Sciences of the USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Li J, Lee S-Y, et al. The hypernodulating nts mutation induces jasmonate synthetic pathway in soybean leaves. Molecules and Cells. 2007;24:185–193. [PubMed] [Google Scholar]
- Seto Y, Hamada S, Matsuura H, et al. Purification and cDNA cloning of a wound inducible glucosyltransferase active toward 12-hydroxy jasmonic acid. Phytochemistry. 2009;70:370–379. doi: 10.1016/j.phytochem.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Shah J. Plants under attack: systemic signals in defence. Current Opinion in Plant Biology. 2009;12:459–464. doi: 10.1016/j.pbi.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Shan X, Wang J, Chua L, Jiang D, Peng W, Xie D. The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiology. 2011;155:751–764. doi: 10.1104/pp.110.166595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Yan J, Xie D. Comparison of phytohormone signaling mechanisms. Current Opinion in Plant Biology. 2012;15:84–91. doi: 10.1016/j.pbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeyama T, Tominaga A, Arima S, et al. Additional cause for reduced JA-Ile in the root of a Lotus japonicus phyB mutant. Plant Signaling & Behavior. 2012;7(746 – 748) doi: 10.4161/psb.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, Jung C, Lee S, et al. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. The Plant Journal. 2013;73:483–495. doi: 10.1111/tpj.12051. [DOI] [PubMed] [Google Scholar]
- Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, Davis SJ. TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. The Plant Cell. 2012;24:2470–2482. doi: 10.1105/tpc.111.095430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Hashimoto T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant and Cell Physiology. 2011;52:1117–1130. doi: 10.1093/pcp/pcr063. [DOI] [PubMed] [Google Scholar]
- Shoji T, Ogawa T, Hashimoto T. Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant and Cell Physiology. 2008;49:1003–1012. doi: 10.1093/pcp/pcn077. [DOI] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. The Plant Cell. 2010;22:3390–3409. doi: 10.1105/tpc.110.078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C, Figueroa P, DePew CL, et al. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. The Plant Cell. 2012;24:536–550. doi: 10.1105/tpc.111.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg A, Gobel C, Grond S, et al. Physiologia Plantarum. Piriformospora indica affects plant growth by auxin production. 2007;131:581–589. doi: 10.1111/j.1399-3054.2007.00983.x. [DOI] [PubMed] [Google Scholar]
- Skirycz A, Reichelt M, Burow M, et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. The Plant Journal. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. London: Academic Press; 2008. [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates – gene discovery and beyond. Trends in Plant Science. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, et al. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. The Plant Cell. 2011;23:1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Loake GJ. Redox-based protein modifications: the missing link in plant immune signalling. Current Opinion in Plant Biology. 2011;14:358–364. doi: 10.1016/j.pbi.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiology. 2009;150:1310–1321. doi: 10.1104/pp.109.138529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P, Su W, Howell S. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences of the USA. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E, Molitor A, Kogel K-H, Waller F. Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant and Cell Physiology. 2008;49:1747–1751. doi: 10.1093/pcp/pcn147. [DOI] [PubMed] [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Laudert D, Andert L, Weiler EW. Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry. 1998;47:539–546. doi: 10.1016/s0031-9422(97)00547-5. [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Maucher H, et al. Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato – amplification in wound signaling. The Plant Journal. 2003a;33:577–589. doi: 10.1046/j.1365-313x.2003.01647.x. [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, et al. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Molecular Biology. 2003b;51:895–911. doi: 10.1023/a:1023049319723. [DOI] [PubMed] [Google Scholar]
- Stenzel I, Otto M, Delker C, et al. ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: tissue- and organ-specific promoter activities and in vivo heteromerization. Journal of Experimental Botany. 2012;63:6125–6138. doi: 10.1093/jxb/ers261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer E. Plant defense in the absence of jasmonic acid: The role of cyclopentanones. Proceedings of the National Academy of Sciences of the USA. 2001;98:12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Leyser HMO. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. The Plant Journal. 2007;50:80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- Stitz M, Gase K, Baldwin IT, Gaquerel E. Ectopic expression of AtJMT in Nicotiana attenuata: Creating a metabolic sink has tissue-specific consequences for the jasmonate metabolic network and silences downstream gene expression. Plant Physiology. 2011;157:341–354. doi: 10.1104/pp.111.178582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz HU, Jikumaru Y, Shimada Y, et al. Jasmonate-dependent and COI1-independent defense responses against Sclerotinia sclerotiorum in Arabidopsis thaliana: Auxin is part of COI1-independent defense signaling. Plant and Cell Physiology. 2011;52:1941–1956. doi: 10.1093/pcp/pcr127. [DOI] [PubMed] [Google Scholar]
- Stumpe M, Göbel C, Faltin B, et al. The moss Physcomitrella patens contains cyclopentenones but no jasmonates: mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytologist. 2010;188:740–749. doi: 10.1111/j.1469-8137.2010.03406.x. [DOI] [PubMed] [Google Scholar]
- Sun J, Cardoza V, Mitchell DM, Bright L, Oldroyd G, Harris JM. Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. The Plant Journal. 2006;46:961–970. doi: 10.1111/j.1365-313X.2006.02751.x. [DOI] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. The Plant Cell. 2009;21:1495–1511. doi: 10.1105/tpc.108.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J-Q, Jiang H-L, Li C-Y. Systemin/jasmonate-mediated systemic defense signaling in tomato. Molecular Plant. 2011a;4:607–615. doi: 10.1093/mp/ssr008. [DOI] [PubMed] [Google Scholar]
- Sun J, Chen Q, Qi L, et al. Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytologist. 2011b;191:360–375. doi: 10.1111/j.1469-8137.2011.03713.x. [DOI] [PubMed] [Google Scholar]
- Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiology. 2011;157:2081–2093. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suza W, Rowe M, Hamberg M, Staswick P. A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-L-isoleucine in wounded leaves. Planta. 2010;231:717–728. doi: 10.1007/s00425-009-1080-6. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Suriyagoda L, Shigeyama T, et al. Lotus japonicus nodulation is photomorphogenetically controlled by sensing the red/far red (R/FR) ratio through jasmonic acid (JA) signaling. Proceedings of the National Academy of Sciences of the USA. 2011;108:16837–16842. doi: 10.1073/pnas.1105892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svyatyna K, Riemann M. Light-dependent regulation of the jasmonate pathway. Protoplasma. 2012;249:137–145. doi: 10.1007/s00709-012-0409-3. [DOI] [PubMed] [Google Scholar]
- Swiatek A, Dongen WV, Esmans EL, Onckelen HV. Metabolic fate of jasmonates in tobacco bright yellow-2 cells. Plant Physiology. 2004;135:161–172. doi: 10.1104/pp.104.040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, et al. Arabidopsis AUXIN RESPONSE FACTOR6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant and Cell Physiology. 2010;51:164–175. doi: 10.1093/pcp/pcp176. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, et al. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. The Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiology. 2005;139:1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T, Sobajima H, Okada K, et al. Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta. 2008;227:517–526. doi: 10.1007/s00425-007-0635-7. [DOI] [PubMed] [Google Scholar]
- Tejeda-Sartorius M, Martinez de la Vega O, Delano-Frier JP. Jasmonic acid influences mycorrhizal colonization in tomato plants by modifying the expression of genes involved in carbohydrate partitioning. Physiol. Plant. 2008;133:339–353. doi: 10.1111/j.1399-3054.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK. Evolution of jasmonate and salicylate signal crosstalk. Trends in Plant Science. 2012;17:260–270. doi: 10.1016/j.tplants.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCFC°I1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Tian D, Tooker J, Peiffer M, Chung S, Felton G. Role of trichomes in defense against herbivores: comparison of herbivore response to woolly and hairless trichome mutants in tomato (Solanum lycopersicum) Planta. 2012;236:1053–1066. doi: 10.1007/s00425-012-1651-9. [DOI] [PubMed] [Google Scholar]
- Tissier A. Glandular trichomes: what comes after expressed sequence tags? The Plant Journal. 2012;70:51–68. doi: 10.1111/j.1365-313X.2012.04913.x. [DOI] [PubMed] [Google Scholar]
- Todd AT, Liu E, Polvi SL, Pammett RT, Page JE. A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. The Plant Journal. 2010;62:589–600. doi: 10.1111/j.1365-313X.2010.04186.x. [DOI] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends in Plant Science. 2009;14:310–317. doi: 10.1016/j.tplants.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Tong X, Qi J, Zhu X, et al. The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. The Plant Journal. 2012;71:763–775. doi: 10.1111/j.1365-313X.2012.05027.x. [DOI] [PubMed] [Google Scholar]
- Tretner C, Huth U, Hause B. Mechanostimulation of Medicago truncatula leads to enhanced levels of jasmonic acid. Journal of Experimental Botany. 2008;59:2847–2856. doi: 10.1093/jxb/ern145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proceedings of the National Academy of Sciences of the USA. 2007;104:1075–1080. doi: 10.1073/pnas.0605423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, et al. Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proceedings of the National Academy of Sciences of the USA. 1999;96:15262–15367. doi: 10.1073/pnas.96.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Beemster GTS, Bennett MJ. Hormonal regulation of root growth: integrating local activities into global behaviour. Trends in Plant Science. 2012;17:326–331. doi: 10.1016/j.tplants.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.) Plant Physiology. 1980;66:246–249. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Nakamura Y. Chemical basis of plant leaf movement. Plant and Cell Physiology. 2007;48:900–907. doi: 10.1093/pcp/pcm060. [DOI] [PubMed] [Google Scholar]
- Ueda M, Yang G, Ishimaru Y, et al. Hybrid stereoisomers of a compact molecular probe based on a jasmonic acid glucoside: Syntheses and biological evaluations. Bioorganic & Medicinal Chemistry. 2012;20:5832–5843. doi: 10.1016/j.bmc.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Vadassery J, Ritter C, Venus Y, et al. The role of auxins and cytokinins in the mutualistic interaction between Arabidopsis and Piriformospora indica. Molecular Plant–Microbe Interactions. 2008;21:1371–1383. doi: 10.1094/MPMI-21-10-1371. [DOI] [PubMed] [Google Scholar]
- Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithöfer A. CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiology. 2012;159:1159–1175. doi: 10.1104/pp.112.198150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S, Van Wees SCM, Pieterse CMJ. Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry. 2009;70:1581–1588. doi: 10.1016/j.phytochem.2009.06.009. [DOI] [PubMed] [Google Scholar]
- van Verk M, Bol J, Linthorst H. Prospecting for genes involved in transcriptional regulation of plant defenses, a bioinformatics approach. BMC Plant Biology. 2011;11 doi: 10.1186/1471-2229-11-88. 88. http://dx.doi.org/10.1186/1471-2229-11-88 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees SC, Van der Ent S, Pieterse CM. Plant immune responses triggered by beneficial microbes. Current Opinion in Plant Biology. 2008;11:443–448. doi: 10.1016/j.pbi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. The tify family previously known as ZIM. Trends in Plant Science. 2007;12:239–244. doi: 10.1016/j.tplants.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Vellosillo T, Martinez M, Lopez MA, et al. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. The Plant Cell. 2007;19:831–846. doi: 10.1105/tpc.106.046052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage A, Vlaardingerbroek I, Raaijmakers C, et al. Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Frontiers in Plant Science. 2011;2 doi: 10.3389/fpls.2011.00047. 47. http://dx.doi.org/10.3389/fpls.2011.00047 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J, Cascón T, Vicedo B, García-Agustín P, Hamberg M, Castresana C. Role of 9-lipoxygenase and α-dioxygenase oxylipin pathways as modulators of local and systemic defense. Molecular Plant. 2012;5:914–928. doi: 10.1093/mp/ssr105. [DOI] [PubMed] [Google Scholar]
- Vierheilig H. Regulatory mechanisms during the plant-arbuscular mycorrhizal fungus interaction. Canadian Journal of Botany. 2004;82:1166–1176. [Google Scholar]
- Vlot AC, Dempsey DMA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Wager A, Browse J. Social Network: JAZ protein interactions expand our knowledge of jasmonate signaling. Frontiers in Plant Science. 2012;3 doi: 10.3389/fpls.2012.00041. 41. http://dx.doi.org/10.3389/fpls.2012.00041 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling L. Adaptive defense responses to pathogens and insects. In: Loon LV, editor. Plant innate immunity. Amsterdam: Elsevier; 2009. [Google Scholar]
- Walling LL. Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiology. 2008;146:859–866. doi: 10.1104/pp.107.113142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Allmann S, Wu J, Baldwin IT. Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiology. 2008;146:904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. Oxylipins: biosynthesis, signal transduction and action. In: Hedden P, Thomas S, editors. Plant Hormone Signaling. Harpenden: Blackwell Publishing; 2006. pp. 185–228. [Google Scholar]
- Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Annals of Botany. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. Jasmonates and octadecanoids – signals in plant stress response and development. In: Moldave K, editor. Progress in nucleic acid research and molecular biology. New York: Academic Press; 2002. pp. 165–222. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Kombrink E. Jasmonates: Structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chemical Biology. 2010;5:63–77. doi: 10.1021/cb900269u. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Forner S, Strnad M, Hause B. Jasmonates in flower and seed development. Biochimie. 2013;95:79–85. doi: 10.1016/j.biochi.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Wees S, Swart E, Pelt J, Loon L, Pieterse C. Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA. 2000;97:8711–8716. doi: 10.1073/pnas.130425197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Yan L, Ren QIN, Li C, Ge F, Kang LE. Antagonism between herbivore-induced plant volatiles and trichomes affects tritrophic interactions. Plant, Cell & Environment. 2013;36:315–327. doi: 10.1111/j.1365-3040.2012.02575.x. [DOI] [PubMed] [Google Scholar]
- Weiss M, Selosse M, Rexer K, Urban A, Oberwinkler F. Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycological Research. 2004;108:1003–1010. doi: 10.1017/s0953756204000772. [DOI] [PubMed] [Google Scholar]
- Westfall CS, Zubieta C, Herrmann J, Kapp U, Nanao MH, Jez JM. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science. 2012;336:1708–1711. doi: 10.1126/science.1221863. [DOI] [PubMed] [Google Scholar]
- Wild M, Davière J-M, Cheminant S, et al. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. The Plant Cell. 2012;24:3307–3319. doi: 10.1105/tpc.112.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ZA, Song J, Taylor B, Yang C. The final split: the regulation of anther dehiscence. Journal of Experimental Botany. 2011;62:1633–1649. doi: 10.1093/jxb/err014. [DOI] [PubMed] [Google Scholar]
- Withers J, Yao J, Mecey C, Howe GA, Melotto M, He SY. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proceedings of the National Academy of Sciences of the USA. 2012;109:20148–20153. doi: 10.1073/pnas.1210054109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldemariam MG, Onkokesung N, Baldwin IT, Galis I. Jasmonoyl-l-isoleucine hydrolase 1 (JIH1) regulates jasmonoyl-l-isoleucine levels and attenuates plant defenses against herbivores. The Plant Journal. 2012;72:758–767. doi: 10.1111/j.1365-313X.2012.05117.x. [DOI] [PubMed] [Google Scholar]
- Wolters H, Jürgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nature Reviews Genetics. 2009;10:305–317. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
- Woo H, Chung K, Park J-H, et al. The Plant Cell. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Annals of Botany. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. Herbivory-induced signalling in plants: perception and action. Plant, Cell & Environment. 2009;32:1161–1174. doi: 10.1111/j.1365-3040.2009.01943.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. New insights into plant responses to the attack from insect herbivores. Annual Review of Genetics. 2010;44:1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. The Plant Cell. 2007;19:1096–1122. doi: 10.1105/tpc.106.049353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Zhang L, Zhou C, Yu C-W, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. Journal of Experimental Botany. 2008;59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- Wu Q, Wu J, Sun H, Zhang D, Yu D. Sequence and expression divergence of the AOC gene family in soybean: Insights into functional diversity for stress responses. Biotechnology Letters. 2011;33:1351–1359. doi: 10.1007/s10529-011-0585-9. [DOI] [PubMed] [Google Scholar]
- Xie D-X, Feys B, James S, Nieto-Rostro M, Turner J. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, et al. The SCF-coi1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. The Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Vreugdenhil D, van Lammeren AAM. Cell division and cell enlargement during potato tuber formation. Journal of Expimental Botany. 1998;49:573–582. [Google Scholar]
- Yadav V, Kumar M, Deep DK, et al. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. Journal of Biological Chemistry. 2010;285:26532–26544. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yamaguchi Y, Huffaker A. Endogenous peptide elicitors in higher plants. Current Opinion in Plant Biology. 2011;14:351–357. doi: 10.1016/j.pbi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chetelat A, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. The Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Christensen S, Isakeit T, et al. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. The Plant Cell. 2012;24:1420–1436. doi: 10.1105/tpc.111.094151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-H, Hettenhausen C, Baldwin IT, Wu J. Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiology. 2012;159:1591–1607. doi: 10.1104/pp.112.199018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-L, Yao J, Mei C-S, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences of the USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Devaiah SP, Pan X, Isaac G, Welti R, Wang X. AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. Journal of Chemical Biology. 2007;282:18116–18128. doi: 10.1074/jbc.M700405200. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Sano R, Wada T, Takabayashi J, Okada K. Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development. 2009;136:1039–1048. doi: 10.1242/dev.030585. [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Greulich F. Biosynthesis of jasmonoids and their function. In: Barton D, Nakanishi K, Meth-Cohn O, editors. Comprehensive natural product chemistry. Oxford: Elsevier; 1998. pp. 117–138. [Google Scholar]
- Yoshihara T, Iino M. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant and Cell Physiology. 2007;48:678–688. doi: 10.1093/pcp/pcm042. [DOI] [PubMed] [Google Scholar]
- Youssef A, Laizet Yh, Block MA, et al. Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. The Plant Journal. 2010;61:436–445. doi: 10.1111/j.1365-313X.2009.04067.x. [DOI] [PubMed] [Google Scholar]
- Yu Z-X, Li J-X, Yang C-Q, Hu W-L, Wang L-J, Chen X-Y. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Molecular Plant. 2012;5:353–365. doi: 10.1093/mp/ssr087. [DOI] [PubMed] [Google Scholar]
- Zander M, Camera SL, Lamotte O, Métraux J-P, Gatz C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. The Plant Journal. 2010;61:200–210. doi: 10.1111/j.1365-313X.2009.04044.x. [DOI] [PubMed] [Google Scholar]
- Zander M, Chen S, Imkampe J, Thurow C, Gatz C. Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Molecular Plant. 2012;5:831–840. doi: 10.1093/mp/ssr113. [DOI] [PubMed] [Google Scholar]
- van Zanten M, Ritsema T, Polko J, et al. Modulation of ethylene- and heat-controlled hyponastic leaf movement in Arabidopsis thaliana by the plant defence hormones jasmonate and salicylate. Planta. 2012;235:677–685. doi: 10.1007/s00425-011-1528-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A, Körbes A, Younessi P, Montiel G, Champion A, Memelink J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Molecular Biology. 2011;75:321–331. doi: 10.1007/s11103-010-9728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdyb A, Demchenko K, Heumann J, et al. Jasmonate biosynthesis in legume and actinorhizal nodules. New Phytologist. 2011;189:568–579. doi: 10.1111/j.1469-8137.2010.03504.x. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Li C-B, Zheng W, et al. Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis. Plant and Cell Physiology. 2007;48:1061–1071. doi: 10.1093/pcp/pcm076. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou C. Signal transduction in leaf senescence. Plant Molecular Biology. 2013 doi: 10.1007/s11103-012-9980-4. in press. http://dx.doi.org/10.1007/s11103-012-9980-4 . [DOI] [PubMed] [Google Scholar]
- Zhang H, Hedhili S, Montiel G, et al. The basic helix–loop–helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. The Plant Journal. 2011;67:61–71. doi: 10.1111/j.1365-313X.2011.04575.x. [DOI] [PubMed] [Google Scholar]
- Zhang H-B, Bokowiec MT, Rushton PJ, Han S-C, Timko MP. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Molecular Plant. 2012;5:73–84. doi: 10.1093/mp/ssr056. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang C, Zhang Y, Sun Y, Mou Z. The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. The Plant Cell. 2012a;24:4294–4309. doi: 10.1105/tpc.112.103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu Q, Ren J, et al. Two novel RING-type ubiquitin ligases, RGLG3 and RGLG4, are essential for jasmonate-mediated responses in Arabidopsis. Plant Physiology. 2012b;160:808–822. doi: 10.1104/pp.112.203422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0003699. e3699. http://dx.doi.org/ 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-Y, Spivey Natalie W, et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host & Microbe. 2012;11:587–596. doi: 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. The Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Fu C, et al. Identification and characterization of petiolule-like pulvinus mutants with abolished nyctinastic leaf movement in the model legume Medicago truncatula. New Phytologist. 2012;196:92–100. doi: 10.1111/j.1469-8137.2012.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2011;108:12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Facchini PJ. Alkaloid biosynthesis: Metabolism and trafficking. Annual Review of Plant Biology. 2008;59:735–769. doi: 10.1146/annurev.arplant.59.032607.092730. [DOI] [PubMed] [Google Scholar]
- Zimmermann MR, Maischak H, Mithofer A, Boland W, Felle HH. System potentials, a novel electrical long-distance apoplastic signal in plants, induced by wounding. Plant Physiology. 2009;149:1593–1600. doi: 10.1104/pp.108.133884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller M, Stingl N, Krischke M, et al. Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: Biogenesis of pimelic and azelaic acid. Plant Physiology. 2012;160:365–378. doi: 10.1104/pp.112.202846. [DOI] [PMC free article] [PubMed] [Google Scholar]