Abstract
The first complete mitochondrial genome of the lacewing family Osmylidae (Thyridosmylus langii (McLachlan, 1870)) (Neuroptera) was sequenced in this study. The genome is a circular molecule of 16,221 bp containing the typical 37 genes but is arranged in the same order as that of the putative ancestor of hexapod and lacks translocation of trnC as shared by all previously sequenced neuropteran mtDNAs. This reveals that trnC translocation does not represent an organizational synapomorphy in the mitochondrion for the entire Neuroptera clade. Comparative analysis of neuropteran tRNA genes reveals a relatively slow and conserved evolution of the mitochondrion throughout the order. Secondary structure models of the ribosomal RNA genes of T. langii largely agree with those proposed for other insect orders. Nevertheless, domain I of T. langii rrnL is consisted of nine helices rather than eight helices which is typical for neuropteran rrnL. Protein-coding genes have typical mitochondrial start codons, with the exception of COI, which uses the TCG start codon also found in Ithonidae and Chrysopidae. Like other neuropteran insects, the control region is the most AT-rich region and comparatively simple, with little evidence of conserved blocks or long tandem repeats. Considering the issues of base-compositional and branch length heterogeneity, we used a range of phylogenetic approaches to recover neuropteridan relationships and explored the effect of method choice on recovery of monophyly of Neuropterida: ((Neuroptera + Megaloptera) + Raphidioptera). The monophyly of Neuroptera and the more basal position of Osmylidae were also recovered by different datasets and phylogenetic methods.
Introduction
The study of mitochondrial (mt) genome has yielded significant insights into different scientific disciplines including animal health, comparative and evolutionary genomics, molecular evolution, phylogeography, phylogenetics and population genetics [1], [2]. The insect mt genome is a circular, double-stranded molecule of 14–19 kb in length that comprises a set of 37 genes for 22 tRNAs, 2 rRNAs and 13 proteins [3]. More than 270 insect mt genomes have been sequenced, representing 26 of the 30 orders, with most from the mega-diverse orders such as Diptera, Lepidoptera, Hemiptera, Coleoptera and Hymenoptera. Based on a series of analyses, mt genomes may be superior to individual nuclear gene or partial mt genes to reconstruct phylogenetic relationships as they possess a number of evolutionarily interesting and potentially informative features such as length variation, altered tRNA anticodons or secondary structures, atypical start codons, base compositional bias, codon usage, and gene rearrangement [4]–[6].
Neuropterida contains at least 6,500 described valid species divided into three orders: Raphidioptera (with slightly more than 200 species in two families), Megaloptera (with about 300 species in two families) and the extremely heterogeneous Neuroptera (with 6,000 species in 15 families) [7], [8]. Relationships within Neuropterida have been controversial over time. The traditional view is a sister group relationship between Megaloptera and Raphidioptera, which was corroborated by Wheeler et al. (2001) and Wiegmann et al. (2009) [9]–[11]. This view has been challenged recently with some researchers proposing that Raphidioptera are sister to Megaloptera + Neuroptera [8], [12], [13]. Megaloptera and Raphidioptera exhibit the typical insect ancestral gene order [14]–[16]. Conversely, Neuroptera with only six mt genomes fully sequenced to date, all share the common feature of translocation of trnC [15], [17]–[19].
Neuropteran family-level relationships have been controversial. The first quantitative cladistic analysis of neuropteran relationships based on adult and larval morphological characters divided Neuroptera into three suborders: Hemerobiiformia, Myrmeleontiformia and Nevrorthiformia [20]. The former two suborders were proposed as sister groups and comprise most neuropteran species, while Nevrorthiformia includes the single small family Nevrorthidae, supposedly representing the sister group to the rest of the order. The monophyly of the suborder Hemerobiiformia was not recovered with any satisfactory support when originally coined [20] and has been repeatedly shown to be paraphyletic in all subsequent quantitative analyses based on morphological or molecular data [8], [12], [13]. In a detailed analysis of Neuropterida using combined molecular and morphological data by Winterton et al. (2010), Hemerobiiformia were recovered as a paraphyletic grade containing Nevrorthiformia, with Myrmeleontiformia as a monophyletic derived clade [8].
Here, we present the first complete mt genome sequence of Thyridosmylus langii (McLachlan, 1870), a representative of Osmylidae. Osmylidae are considered one of the more basal groups of Neuroptera, exhibiting many plesiomorphic characteristics [8], [11], [12], [19]. Winterton et al. (2010) recovered Osmylidae as sister to Sisyridae and Nevrorthidae in a near-basal clade that originated during the Permian [8]. Characterization of the mt genome of Osmylidae provides an important opportunity to understand the sequence of evolution of the mitochondrion in Neuroptera, especially with reference to gene order, insertion and deletion events and ribosomal secondary structure. We provide analyses of the gene order, nucleotide composition, codon usage, compositional biases, RNA secondary structure, the assessment of base compositional and branch length heterogeneity and evaluate the phylogenetic relationships among the three orders of Neuropterida based on a range of phylogenetic approaches. This work reveals that the trnC translocation, which is absent in Thyridosmylus, occurred within higher Neuroptera and cannot be used as a synapomorphy characteristic for all Neuroptera.
Results and Discussion
Genome organization and structure
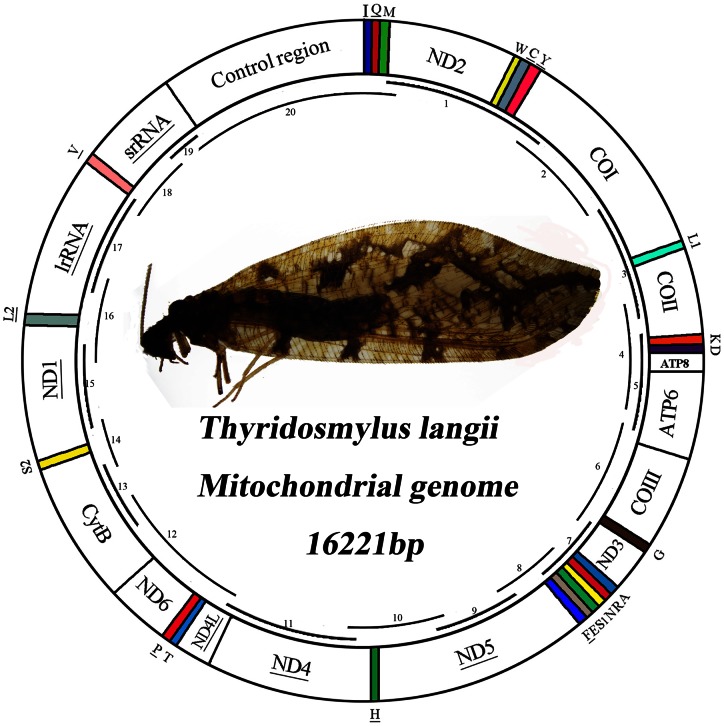
The mt genome of T. langii is a double-stranded circular DNA molecule of 16,221 bp in size and has been deposited in the Genbank under accession number (KC515397). This mt genome contains the entire set of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a control region (also called A+T-rich region) that are typically present in metazoan mt genomes (Fig. 1). The arrangement of mt genes of T. langii is the same as the putative ancestor of insects [21]–[26] and differs with the other six previously sequenced neuropteran mt genomes which share a common synapomorphic feature of transposition of tRNATrp and tRNACys genes. The rearrangement of tRNACys–tRNATrp–tRNATyr (CWY) was considered as a potential synapomorphic character for Neuroptera or some subgroup within the order in the previous study [15]. This opinion is further corroborated in our study that the rearrangement of CWY is just synapomorphic for some subgroup within Neuroptera. The likely explanation for transposition of adjacent genes in Neuroptera involves tandem duplication of the entire region affected by the transposition followed by inactivation and loss of the first copy of the second gene [21].
Figure 1. Map of the mt genome of T. langii.
Transfer RNAs are denoted by the color blocks and are labeled according to the IUPACIUB single-letter amino acid codes. Gene name not underlined indicates the direction of transcription from left to right, and with underline indicates right to left. Overlapping lines within the circle denote PCR fragments amplified used for cloning and sequencing.
Genes are encoded by both strands of T. langii mt genome: 23 genes on the majority strand (J-strand) with the rest on the minority strand (N-strand). Eleven pairs of adjacent genes in the mt genome of T. langii overlapped by 1–8 bp and involved a total of 38 bp which make the genome relatively compact (Table S1). Two pairs of overlapping genes are common to all sequenced mt genomes of lacewings: ATP8/ATP6 and ND4/ND4L, and share a near identical seven bp sequence (ATGNTAA), with the exception of the ascalaphids, in which this consensus-overlapped sequence were not found in the ND4/ND4L gene pair.
Transfer RNAs
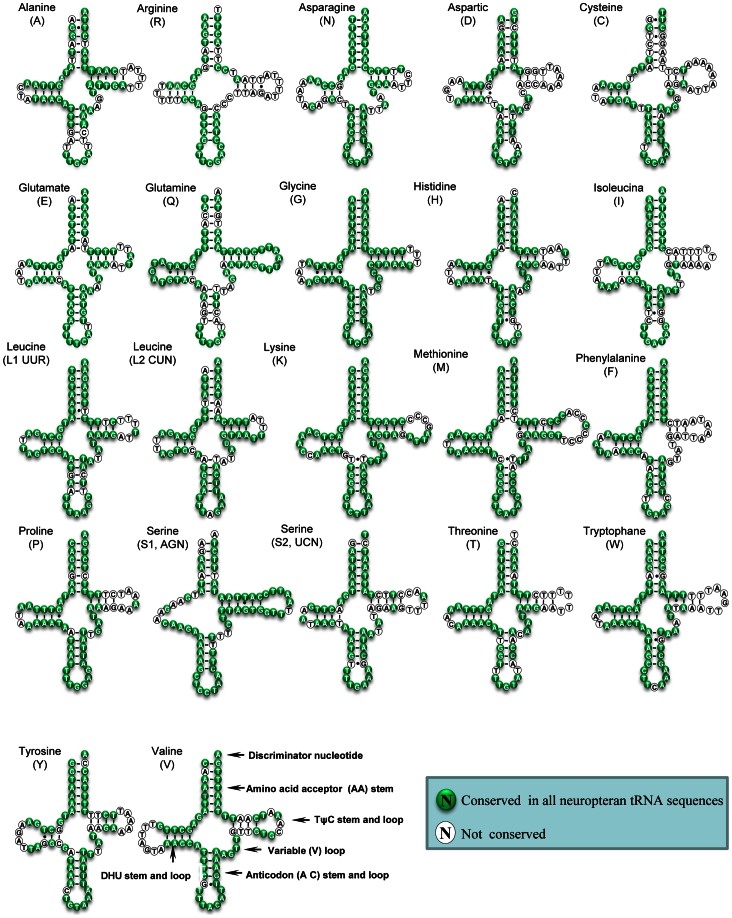
The mt genome of T. langii consists of all 22 tRNA genes, ranging from 64 to 73 bp. Most of them were detected using tRNAscane-SE [27] with two exceptions of the tRNAArg and tRNASer(AGN) genes. Sequences of these two tRNAs were determined through comparison with previously published neuropteran mt genomes [14], [15], [18], [19]. All tRNAs could fold into the typical cloverleaf structure except for tRNASer(AGN), in which its dihydrouridine (DHU) arm simply formed a loop (Fig. 2).
Figure 2. Inferred secondary structure of 22 tRNAs of T. langii mt genome.
22 tRNAs are labeled with the abbreviations of their corresponding amino acids. Dashes (−) indicate Watson-Crick base pairing and dots (•) indicate unmatched base pairing.
Based on the secondary structure, a total of 23 mismatched base pairs are found in the T. langii tRNAs: eighteen of them are G–T pairs, which form a weak bond, located in the AA stem (3 bp), T stem (3 bp), AC stem (5 bp) and the DHU stem (7 bp); the remaining five pairs include T–T mismatches (3 bp) in the AA stem of tRNALeu, the T stem of tRNATrp and the AC stem of tRNALeu, respectively; C–T (1 bp) mismatches in the AC stem of tRNAMet; A–G (1 bp) mismatches in the AA stem of tRNATrp. Mismatches observed in tRNAs are common in arthropod mt genomes and may be corrected through RNA editing processes [28].
Comparative analyses of secondary structures of neuropteridan tRNAs were undertaken based on generating a multiple alignment of tRNAs across exemplars and the percent of identical nucleotides (%INUC) (i.e. conservation) calculated. All tRNAs exhibited high levels of conversation with the average 89%INUC. The pattern of nucleotide conservation was obviously J strand-biased. Indeed, trnG and trnK, which showed the highest levels of nucleotide conservation (%INUC ≥95%), were all located on the J strand; while trnR and trnC showed the lowest levels of nucleotide conservation (%INUC≈80%) on both strands. Among neuropteran tRNAs, nucleotides at the AA and AC stems and loops were conserved: trnG and trnN showed 100%INUC in these two arms; and variations were largely restricted to the TΨC, DHU, and extra loops (Fig. 2). The high levels of nucleotide conservation characterize the low rate of nucleotide substitution and reveal that Neuroptera has in general experienced a slow and conservative evolution.
Ribosomal RNAs
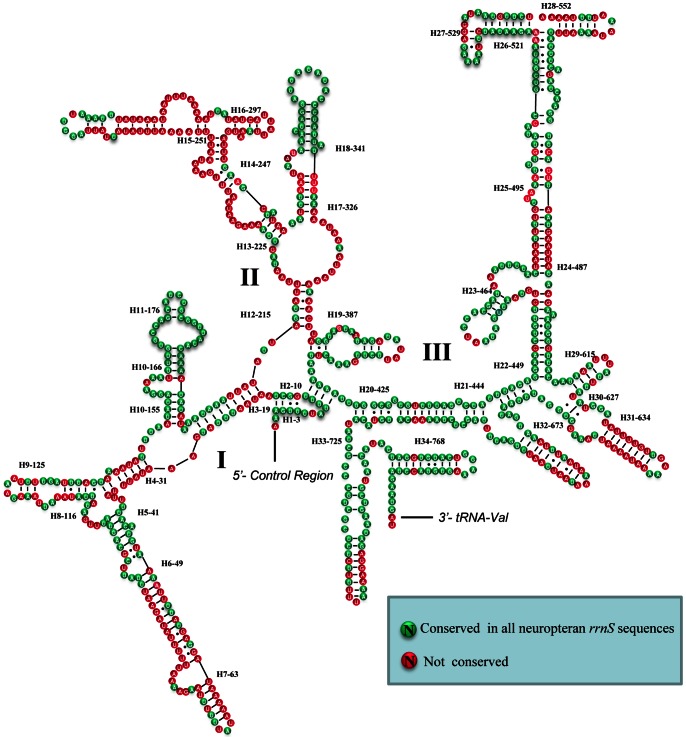
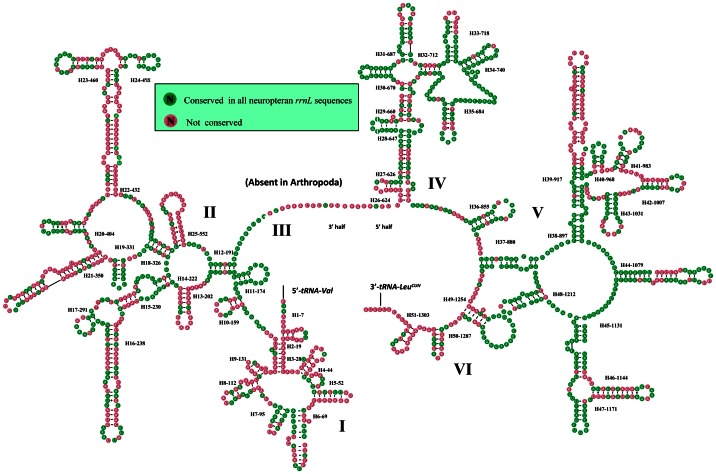
The large and small rRNA subunits (rrnL and rrnS) in T. langii are located at tRNALeu(CUN) – tRNAVal and tRNAVal – control region, respectively (Fig. 1 and Table S1). The length of rrnS and rrnL is determined to be 798 bp and 1,325 bp, respectively. The inferred secondary structure models for rrnS and rrnL are provided in Fig. 3 and Fig. 4, respectively. The secondary structure of both rrnS and rrnL are largely in agreement with the secondary structure models proposed for other endopterygote orders [29]–[31]. The secondary structure of T. langii rrnL consists of six structural domains (domain III is absent in arthropods) and 51 helices (Fig. 4), and the rrnS is comprised of three structural domains and 34 helices (Fig. 3). As domain I in rrnL is highly variable among insects and difficult to predict, most previous studies have no proposed models for this region [29]–[31]. Negrisolo et al. provided the first predicted secondary structure of domain I for rrnL of Libelloides macaronius [18]. Compared with this model, nine helices in domain I of T. langii rrnL were predicted rather than eight helices in L. macaronius.
Figure 3. Secondary structure of T. langii rrnS.
Each helix is numbered progressively from the 5′ to 3′ end together with the first nucleotide belonging the helix itself. Domains are labeled with Roman numbers. Dashes (−) indicate Watson-Crick base pairing and dots (•) indicate unmatched base pairing.
Figure 4. Secondary structure of T. langii rrnL.
Each helix is numbered progressively from the 5′ to 3′ end together with the first nucleotide belonging the helix itself. Domains are labeled with Roman numbers. Dashes (−) indicate Watson-Crick base pairing and dots (•) indicate unmatched base pairing.
The multiple alignment of neuropteran rrnSs spanned 813 positions and contained 319 variable sites including 168 parsimony-informative sites. Nucleotide variability among domains and helices was unevenly distributed. The helices (H11-176, H18-341) exhibit almost 100%INUC, while some others (i.e. H6-49, H7-63, H31-634) display very low %INUC. The multiple alignment of neuropteran rrnLs extended over 1376 positions and contained 583 variable sites, including 321 parsimony-informative sites. Conserved nucleotides were unevenly distributed throughout the rrnL secondary structure, with the highest level of non-varying positions located on domains IV–V and lowest levels observed in domains I–II (Fig. 4).
Protein-coding genes
The mt genome of T. langii includes the canonical 13 PCGs that are also present in all other sequenced lacewings and most animal mt genomes [15], [18], [19]. The orientation of PCGs is identical in the mt genomes of all sequenced lacewings: nine in J-strand and four in N-strand. All but one PCG of T. langii initiate with ATN as the start codon (seven with ATG, four with ATT and one with ATC) (Table S1). The only exception is the COI gene, which uses non-canonical TCG as a start codon. The unusual start condons for the COI gene in Neuroptera is a common feature and include ACG for L. macaronius [18] and Myrmeleon immaculatus [15]; TTA for Ascaloptynx appendiculatus; TCG for Polystoechotes punctatus [17], Chrysoperla nipponensis and Apochrysa matsumurae [19] respectively. It is a widespread phenomenon in Neuroptera and other endopterygote insects that the COI gene often starts with non-standard putative codons [32]–[34].
Nine PCGs of T. langii share the complete termination codons TAA (ND2, ATP8, ATP6, COI, COIII, ND4L and ND6) or TAG (ND1 and ND3), while the remaining four had incomplete termination codons T (ND4, ND5, COII and CytB) (Table S1), which is also common in other lacewings. The truncated stop codons are suggested to be simply a product of the selective pressure to reduce genome size [35], [36] and are presumed to be completed via post-transcriptional polyadenylation [37].
Nucleotide composition and codon usage
The nucleotide composition of the T. langii mt genome was biased toward A and T as in most other insects. The A+T content of the whole genome, PCGs, tRNAs, rRNAs and the control region is 76.7%, 74.3%, 77.4%, 80.4% and 88.5%. The base composition of nucleotide sequences can be described by its skewness: the whole genome and the control region exhibit the negative AT- and GC-skews while the rRNAs show the opposite situation; the PCGs and tRNAs have the negative AT- and positive GC-skews (Table S2).
Analysis of base composition at each codon position of the concatenated 13 PCGs showed that the third codon position (88.9%) was higher in A+T content than the first (68.0%) and second (66.2%) codon positions (Table S2). Nucleotide frequencies in all codon positions differ between the two strands. In the J-strand, the frequencies of nucleotides are T>A>G>C at the first codon position, T>C>A>G at the second codon position and T>A>C>G at the third codon position. In the N-strand, all three codon positions show the common feature (T>A>G>C).
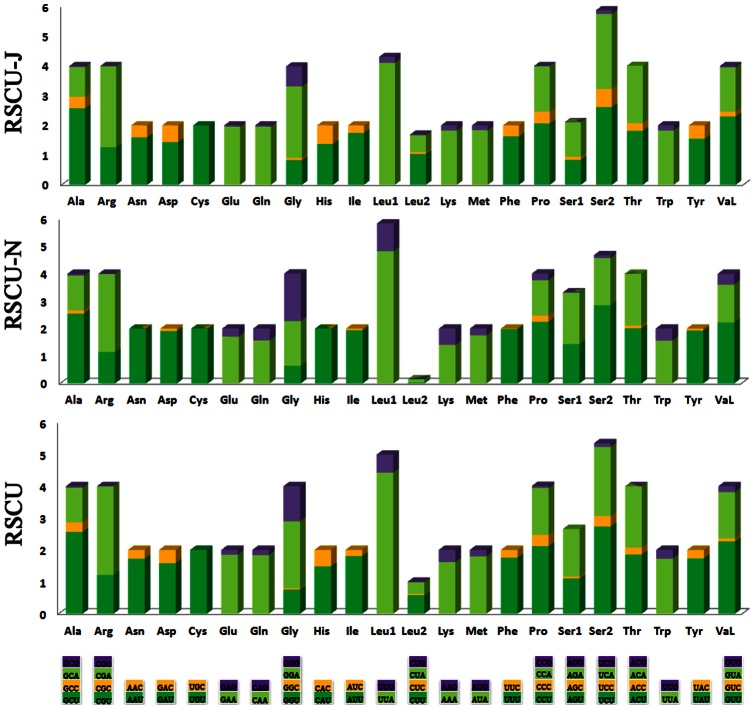
The nucleotide bias was also reflected in the codon usage. Four most frequently used codons, TTA (leucine), ATT (isoleucine), TTT (phenylalanine) and ATA (methionine), were all composed wholly of A and/or T. The two-fold degenerate codon usage also demonstrated bias favoring T/A over C/G at the third codon position on both stands. The four-fold degenerate codon usage presented a definite bias towards thymine at the third codon position of PCGs except Gly (GGN), Arg (CGN) and Ser (AGN), whose most frequently used codons terminated with adenine (Fig. 5).
Figure 5. Relative synonymous codon usage (RSCU) in the (N) and (J) strands of the T. langii mt genome.
Codon families are provided on the x-axis. Along with the different combinations of synonymous codons that code for that amino acid.
Non-coding region
The longest non-coding region (1,358 bp; putative control region) is flanked by rrnS and tRNAIle with 88.5% A+T content, and is the most AT-rich region in the mt genome of T. langii. Unlike hymenopteran [38] or hemipteran [34] insects, this region in the mt genome of sequenced lacewings is comparatively simple, with little evidence of conserved blocks or long tandem repeats. The abundance of A/T is due to the repeated motifs containing mostly/only A/T of different length. The most abundant AT motif occurs 231 times, AAT occurs 91 times, AATT occurs 62 times, AATTT occurs 24 times, AATTTTT occurs 12 times, and TTATATAT occurs six times. The motifs are represented by shorter identical strings as well as longer consensus patterns sharing 80% of identical positions. When an 80% minimum identity was allowed among repeated motifs, two stings were identified in the control region of T. langii, each containing 41 nucleotides, respectively, from base 224 to base 264 and from 428 to 468. Their consensus sequence was ATTT(T/A)AAA(GA/AG)AATAAATTTAAAA(T/C)CGCAGTTTCTCCCTAAA. This trend also occurs in other neuropteran insects [18].
In addition, some short tandem repeat sequences were detected in the control region of T. langii, such as (ATTTT)4, (TAATATAA)2, (AT)4, (AT)6, (AT)7, and (AT)8. These tandem repeat elements can be considered as microsatellites and are useful in the study of geographical structure and phylogenetic relationship of species [25]. Thus, the utility of the AT-rich region as a phylogenetic marker should be most effective at low taxonomic level (family level and below).
Outside the control region, there are 128 bp non-coding sequences in 13 intergenic regions of T. langii, ranging in size from one to 63 bp (Table S1). Three of them spanned more than 15 bp. The longest spacer sequence (63 bp) exists between tRNAIle and tRNAGln which is also present in all part/fully sequenced neuropteran mt genomes, varying from 33 bp to 66 bp; this spacer diverged relatively very fast among different families of the lacewings [18]. The third longest insertion of T. langii is located between ND1 and tRNASer(UCN), which shares the conserved motif (TTAAATNNNNTNAGAT, N = A/T) across Neuroptera. The conservation of this region is also known in many other insects, which is shown to be a binding site for a bidirectional transcription termination factor (DmTTF) [39].
The assessment of base compositional heterogeneity
The total AT% of all included neuropteran species ranged between 74.50% and 79.10% with a mean of 77.63%. The total AT% of megalopteran species ranged between 74.90% and 78.30% with a mean of 76.33%. The AT% of the species from Raphidioptera was 80.30%.
The assessment of base-compositional heterogeneity in the neuropteridan dataset shows that the overall dataset exhibited the complex pattern of compositional bias. To assess the base compositional heterogeneity, we made 55 pair wise comparisons to calculate the disparity index (I D). When we compared all 13 PCGs simultaneously, 48 comparisons had a statistically significant heterogeneous substitution pattern. In general, the I D was low between the taxa with lower ranges of AT% and very high between the taxa with wider ranges of AT%. Eight of eleven neuropteridan species exhibited a similar base composition with a mean I D between four and six in the dataset (PCG123), suggesting the overall dataset exhibited a high level of base compositional heterogeneity (Table 1). The dataset (PCG12) (removal of fast evolving sites) shows a lower level of base compositional heterogeneity than the dataset (PCG123) with a mean I D between one and two in most taxa. The I D values calculated from individual codon partitions suggested that the level of base compositional heterogeneity was the lowest in PCG2, followed by PCG1 and PCG3 (Table 1). Among nucleotide sequences, the ranked order of the datasets from best to worst was: PCG2 > PCG1 > PCG12 > PCG3 > PCG123.
Table 1. Disparity index values calculated from pairwise comparisons among neuropteridan species included in this study.
| Taxa | PCG123 | PCG12 | PCG1 | PCG2 | PCG3 | |||||
| Sum | Mean | Sum | Mean | Sum | Mean | Sum | Mean | Sum | Mean | |
| Ascaloptynx | 50.08 | 5.01 | 24.60 | 2.46 | 24.85 | 2.49 | 6.26 | 0.63 | 35.90 | 3.60 |
| Apochrysa | 50.76 | 5.01 | 15.05 | 1.51 | 18.63 | 1.86 | 3.58 | 0.36 | 57.43 | 5.74 |
| Chrysoperla | 56.15 | 5.61 | 15.27 | 1.53 | 24.62 | 2.46 | 3.67 | 0.37 | 75.05 | 7.51 |
| Corydalus | 62.00 | 6.20 | 22.43 | 2.24 | 28.27 | 2.83 | 6.79 | 0.68 | 86.54 | 8.65 |
| Ditaxis | 43.72 | 4.37 | 17.51 | 1.75 | 19.15 | 1.92 | 4.27 | 0.43 | 37.73 | 3.77 |
| Libelloides | 95.45 | 9.55 | 47.43 | 4.74 | 35.10 | 3.51 | 3.98 | 0.40 | 103.37 | 10.34 |
| Mongoloraphidia | 65.74 | 6.47 | 31.98 | 3.20 | 50.17 | 5.02 | 26.41 | 2.61 | 29.24 | 2.92 |
| Polystoechotes | 56.27 | 5.62 | 12.97 | 1.30 | 20.78 | 2.08 | 4.46 | 0.45 | 91.73 | 9.17 |
| Protohermes | 41.42 | 4.14 | 15.96 | 1.60 | 18.23 | 1.82 | 6.93 | 0.93 | 38.64 | 3.86 |
| Thyridosmylus | 36.48 | 3.65 | 13.30 | 1.33 | 18.34 | 1.83 | 4.39 | 0.44 | 35.07 | 3.51 |
| Sialis | 27.66 | 2.77 | 11.42 | 1.14 | 12.38 | 1.24 | 3.37 | 0.34 | 26.51 | 2.65 |
‘Sum’ indicates the sum of all I D calculated for a particular taxon, and ‘Mean’ indicates the average of I D for that taxon. Taxa with high I D-values have a significantly different base compositional bias from the rest of the species. Numeric values shown next to PCG (i.e. PCG1) represent codon positions. The values shown here highlight the level of base compositional heterogeneity in our dataset.
Phylogenetic analyses
Base-compositional heterogeneity and branch length heterogeneity are known to be major issues with insect mt genomes in general and neuropteridan/coleopteran mt genomes in particular [8], [40]. According to the test of base-compositional heterogeneity in the neuropteridan dataset herein, and previous studies [40], [41], the datasets of PCG12 and AA are less likely to violate the ‘stationarity’ assumptions, which imply that base composition is constant over all lineages in the dataset [42]. To alleviate the influence of base-compositional heterogeneity and any concerns of possible long-branch attraction, we used a range of phylogenetic approaches to recover the neuropteridan relationships and optimized outgroup selection empirically based on short branches with Trachypachus holmbergi (Coleoptera), Adoxophyes honmai (Lepidoptera) and Drosophila melanogaster (Diptera).
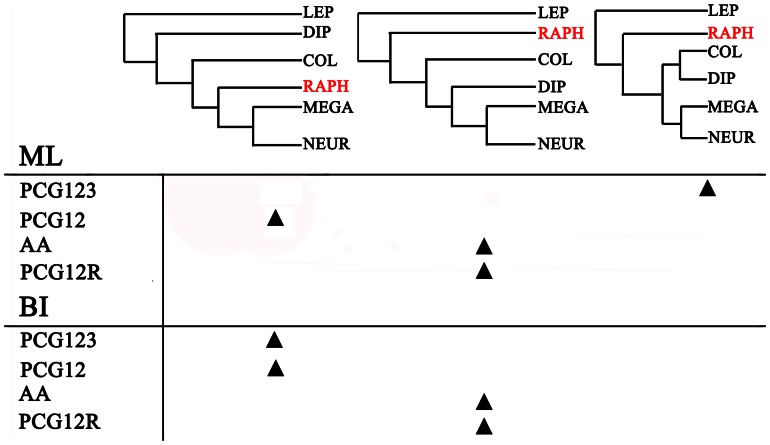
Phylogenetic analyses of the four datasets yielded three different patterns of the relationships between the holometabolan orders (Fig. 6). Examination of the trees recovered from these analyses indicated that each analysis produced identical topologies (Fig. 7), with respect to neuropteran and megalopteran relationships. Bayesian posterior probabilities and bootstrap values of ML of major nodes recovered by various phylogenetic approaches are listed in Table 2. Within Neuropterida, a sister relationship between Megaloptera and Neuroptera was recovered in all analyses of different datasets (node = 9) with high support, which is consistent with the results from the previous mt genome phylogeny of Neuropterida [15], [16]. The largest difference is the uncertain position of Raphidioptera among the different datasets. The datasets of PCG12 (BI and ML) and PCG123 (BI) recovered identical tree topologies which supports a monophyletic Neuropterida: ((Neuroptera + Megaloptera) + Raphidioptera). The datasets (PCG12R and AA) of both BI and ML analyses fail to support a monophyletic Neuropterida as Neuroptera + Megaloptera are recovered as the unlikely sister group to Diptera.
Figure 6. Interordinal relationships inferred by different datasets and analytical methods.
The triangle indicates that the most likely tree recovered for a particular dataset/analysis combination supports one of the three topologies listed. COL: Coleoptera, DIP: Diptera; LEP: Lepidoptera; MEGA: Megaloptera; NEUR: Neuroptera; RAPH: Raphidioptera.
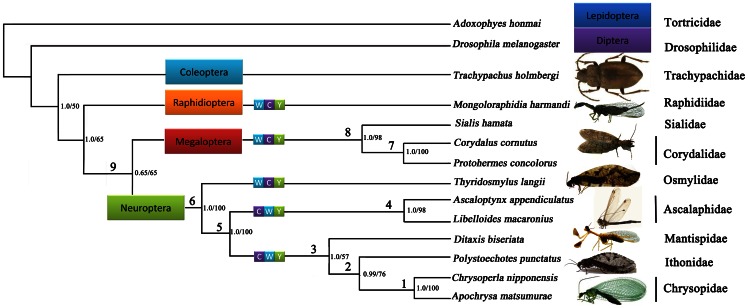
Figure 7. Phylogenetic tree inferred from the sequenced Neuropterida mt genomes.
Phylogenetic analyses are based on the PCG12 dataset. Numbers close to the branching points are ML bootstrap support values (right) and Bayesian posterior probabilities in percentages (left). Numbers above each node relate to values provided in Table 2 for different support statistics for that node recovered by different data combinations.
Table 2. Bayesian posterior probabilities and bootstrap values of ML recovered by various phylogenetic approaches.
| Relationship | PCG123 | PCG12 | AA | PCG12R | ||||
| ML | BI | ML | BI | ML | BI | ML | BI | |
| 1 | 100 | 1.0 | 100 | 1.0 | 100 | 1.0 | 100 | 1.0 |
| 2 | 63 | 0.98 | 76 | 0.99 | 84 | 1.0 | 64 | 0.99 |
| 3 | 94 | 1.0 | 57 | 1.0 | 95 | 1.0 | 72 | 1.0 |
| 4 | 100 | 1.0 | 98 | 1.0 | 100 | 1.0 | 100 | 1.0 |
| 5 | 100 | 1.0 | 100 | 1.0 | 100 | 1.0 | 80 | 1.0 |
| 6 | 100 | 1.0 | 100 | 1.0 | 100 | 1.0 | 100 | 1.0 |
| 7 | 100 | 1.0 | 100 | 1.0 | 100 | 1.0 | 100 | 1.0 |
| 8 | 100 | 1.0 | 98 | 1.0 | 97 | 1.0 | 100 | 1.0 |
| 9 | 86 | 1.0 | 65 | 0.65 | 100 | 1.0 | 97 | 1.0 |
Relationships 1–9 refer to the nodes 1–9 in Figure 7.
Among different datasets, PCG12 that exhibits a lower level of base compositional heterogeneity seems most effective at solving the order level relationships. The uncertain position of Raphidioptera that differs in the phylogenetic trees from different datasets may be also influenced by branch length heterogeneity. There exists some variation in the branch lengths within the taxa (Table S3). The families from Megaloptera and Neuroptera show similar uniform branch lengths. The single exemplar of Raphidioptera shows significantly longer branches than that for Megaloptera and Neuroptera. Based on the limited sample of Raphidioptera and the influence of branch length heterogeneity, the position of Raphidioptera is still uncertain in this study. Increased taxon sampling within Raphidioptera, from both families Inocellidae and Raphidiidae, will likely help solve this problem.
The monophyly of Neuroptera is an undoubted hypothesis strongly supported by both larval features [20], [43] and molecular data [11] that is also demonstrated in our study based on different datasets and methods. Comparative analysis of 11 complete mt genomes sequenced to date reveals some important and novel insights into the phylogeny and evolution of mt gene organization of Neuropterida. All nodes among Neuroptera were perfectly supported in both analyses except that subtending Mantispidae and Ithonidae. Due to the limited number of mt genomes sequenced, the family–level relationships within Neuroptera remain still largely unresolved in our analyses, but important patterns are evident. Our data does not support the proposed division of Neuroptera into three suborders (i.e. Nevrorthiformia, Myrmeleontiformia and Hemerobiiformia) by Aspöck et al. (2001) and reflects the results of most subsequent authors where only Myrmeleontiformia are only recovered as monophyletic, with Hemerobiiformia being paraphyletic relative to both Myrmeleontiformia and Nevrorthiformia [8], [12], [13], [44]. The ancestral gene arrangement in Osmylidae presented here represents important additional evidence as to the paraphyly of Hemerobiiformia. In morphological analyses based on characteristics of adults and larvae and on larval and adult head structures [43], [45], Osmylidae were placed in a weakly supported clade Hemerobiiformia. This is contrary to all analyses using molecular data separately [12], [44] or in combination with morphological data [8] that consistently recover Osmylidae, along with Sisyridae and Nevrorthidae near the base of the phylogeny, clearly reinforcing the apparent antiquity of the origin of this family. Osmylidae, together with Nevrorthidae and Sisyridae, also share an apparent synapomorphy of enlarged female external gonocoxites 9 [13] and have a series of sympleisiomorphies characters indicative of the likely early cladogenesis of these families during the Late Permian or Early Triassic [8]. The basal position of Osmylidae within Neuroptera is confirmed in our study with high support nodes recovered by different datasets (node = 6).
As apparently one of the oldest and most primitive groups of Neuroptera [13], Osmylidae also exhibit the plesiomorphic condition of gene order found throughout the rest of Neuropteroidea (i.e. WCY) (Fig. 7). On the basis of the irreversible process of gene rearrangement and using estimates of divergence times by Winterton et al. (2010) [8], then the translocation to the derived gene order in higher Neuroptera may be occurred somewhere around 260 million years ago, after the divergence of the Osmylidae ancestor. Further work is needed to confirm with the more sequenced mitochondrial genomes in Neuroptera to determine the timing of this gene rearrangement more precisely.
Conclusions
This work has added to our current knowledge of gene arrangement of Neuroptera. The first complete mt genome of T. langii (Neuroptera: Osmylidae) presented here displays an ancestral gene organization that lacks translocation of trnC as shared by all previously sequenced neuropteran mtDNAs. This discovery reveals that the trnC translocation is not synapomorphic for the entire neuropteran clade but only for higher Neuroptera. Comparative analyses suggest that gene junctions, tRNAs and rRNAs are comparatively conserved among the neuropteran mt genomes. The mt genomic phylogeny of different datasets and methods herein reconstructed clearly supports the monophyly of Neuroptera, the more basal position of Osmylidae and the sister relationship between Megaloptera and Neuroptera. However, based on the limited taxon sampling and the influence of base compositional and branch length heterogeneity, only the dataset of PCG12 in BI and ML analyses and PCG123 in BI analyses recovered identical tree topologies, which support a monophyletic Neuropterida: ((Neuroptera + Megaloptera) + Raphidioptera). To more fully elucidate phylogenetic relationships within Neuropterida based on comparative analysis of mt genomes, further study is needed based on increased taxon sampling within major groups of Neuropterida, especially representatives of neuropteran families such as Coniopterygidae, Dilaridae, Hemerobiidae, Nevrorthidae and Sisyridae, and well as Inocellidae (Raphidioptera).
Materials and Methods
Sample origin and genomic DNA extraction
Specimens of Thyridosmylus langii were collected from Motuo, Tibet Autonomous Region, China in August 2011. All specimens were preserved in 100% ethanol and stored at −20°C until DNA extraction. Total genomic DNA was extracted from thorax muscle tissue and the leg muscle tissue using the TIANamp Genomic DNA Kit in accordance with the manufacturer's instructions. Voucher specimens (Nos. NEU-0018), preserved in alcohol, are deposited at the Entomological Museum of China Agricultural University (Beijing).
PCR amplification, cloning and sequencing
PCR amplification was performed using a set of insect universal primers [46], [47] and primers specifically designed on the Thyridosmylus langii sequences (Table S4). Initially, 13 fragments were amplified using the universal primers from previous work (Fig. 1). On the basis of these short fragments for secondary PCRs, Seven perfectly matching primers were designed by using Primer Premier 5.0 to bridge the gaps.
Short PCRs were conducted using Qiagen Taq DNA polymerase (Qiagen, Beijing, China) with the following cycling conditions: 5 min at 94°C, followed by 35 cycles of 50 s at 94°C, 30 s at 40–55°C, and 1–2 min at 72°C. The final elongation step was continued for 5 min at 72°C. Long PCRs were performed using NEB Long Taq DNA polymerase (New England Biolabs) under the following cycling conditions: 30 s at 95°C, followed by 45 cycles of 10 s at 95°C, 50 s at 48–55°C, and 3–5 min at 65°C. The final elongation was continued for 10 min at 65°C. These PCR products were detected via electrophoresis in a 1% agarose gel and stained with ethidium bromide.
All PCR Products were sequenced in both forward and reverse directions using the BigDye Terminator Sequencing Kit (Applied Bio Systems) and the ABI 3730XL Genetic Analyzer (PE Applied Biosystems, San Francisco, CA, U.S.A.) with two vector-specific primers and internal primers for primer walking.
Sequence analysis and inferences of secondary structures
Original sequence files were manually checked and assembled using the software BioEdit 7.0.5.3 [48], Chromas 1.62 and DNAMAN 6.0. The tRNAs were identified by tRNAscan-SE Search Server v.1.21 [27] with default setting or recognized manually by comparison with published neuropteran mt sequences (i.e. Polystoechotes punctatus (Ithonidae), Libelloides macaronius (Ascalaphidae) and Chrysoperla nipponensis (Chrysopidae)) [15], [18], [19]. Secondary structures of the small and large ribosomal RNAs were inferred using alignment to the models predicted for L. macaronius, which is largely in agreement with those proposed for other endopterygote orders (i.e. Coleoptera, Diptera, Hymenoptera, and Lepidoptera) [4], [18], [30], [49]. PCGs were identified using Clustal X [50] by similarity of published neuropteran mt gene amino acid sequences. Translation of PCG open reading frames were further analyzed by MEGA version 4.0 [51] for the codon usage under the condition of the invertebrate mtDNA genetic code. Sequence motifs in the AT-rich region were identified using the Spectral Repeat Finder program [52]. Strand asymmetry was calculated using the formulae: AT skew = [A−T]/[A+T] and GC skew = [G−C]/[G+C] for the strand encoding the majority of the PCGs [53].
Assessment of the degree of heterogeneity
To measure the variation in evolutionary pattern, we calculated the disparity index (I D) [54] for all 13 PCGs together and pairwise. We also tested the homogeneity of substitution pattern (I D test) using a Monte–Carlo method with 1000 replicates as implemented in MEGA 4.0 [51]. We calculated the probability of rejecting the null hypothesis that sequences have evolved with the same pattern of substitution at α<0.01. All positions containing gaps and missing data were removed from the data set (complete deletion option). Also to assess the degree of branch length heterogeneity among Neuropterida, we extracted branch length estimates from the most likely tree, after Maximum-likelihood and Bayesian analysis of the datasets. Branch lengths were conveniently displayed on this tree, using FigTree (v. 1). We then manually calculated the branch length for each taxon, from the tip to the basal node of the tree.
Phylogenetic analysis
Phylogenetic analysis was carried out based on the 13 complete mt genomes of neuropteridan and endopterygote outgroups from Genbank (Table S5) in addition to the one presented here for Osmylidae. Within Endopterygota, the closest living relatives of Neuropterida are the beetles [11], [15], so a representative of Coleoptera and two species from Diptera and Lepidoptera respectively were selected for outgroup comparison. All PCGs were aligned at the amino acid level using the default settings in Mega version 4.0 [51]. The alignments were back-translated into the corresponding nucleotide sequences. The stop codons of PCGs were excluded when aligned. Two rRNAs were respectively aligned in Mega version 4.0 [51] as nucleotides. Ambiguously aligned regions of PCGs and rRNAs were checked by eye.
To overcome the base compositional heterogeneity and branch length heterogeneity and explore the effect of method choice on recovery of monophyly of Neuropterida, four datasets were used for phylogenetic analyses: 1) a concatenated nucleotide sequence alignment of PCGs (PCG123); 2) a concatenated nucleotide sequence alignment of the first and the second codon positions of PCGs (PCG12); 3) a concatenated nucleotide sequence alignment of the first and the second codon positions of PCGs and two rRNA genes (PCG12R); and 4) a concatenated amino acid sequence alignment of PCGs (AA). We analyzed these datasets in both Bayesian inference (BI) and Maximum-likelihood (ML) frameworks.
Partitioned ML and Bayesian analyses were run with PCG123, PCG12, PCG12R, and AA matrix, using PHYML online web server [55] and MrBayes Version 3.1.1 [56]. The best-fit model for the amino acid sequence alignment was determined with ProtTest [57] and the jModelTest 0.1.1 [58] was used for the nucleotide sequence of each gene, according to the Akaike Information Criterion (AIC). GTR+I+G model for nucleotide sequence and MtREV model for amino acid sequence were used to optimize the topology. For Bayesian analyses, two simultaneous runs of 5 to 10 million generations were conducted for the matrix and trees were sampled every 1,000 generations, with the first 25% discarded as burn-in. Stationarity was considered to be reached when the average standard deviation of split frequencies was below 0.01. In ML analysis, the parameters were estimated during analysis and the node support values were assessed by bootstrap resampling (BP) [59] calculated with 1000 replicates.
Supporting Information
Organization of Thyridosmylus langii mt genome.
(DOCX)
Nucleotide composition of the Thyridosmylus langii mt genome.
(DOC)
Comparision of branch lengths among Neuropterida insects.
(DOCX)
Primer sequences used in this study.
(DOC)
Mt genomes used in comparative analyses.
(DOCX)
Acknowledgments
We thank Dr. Qingbin Zhan (China Agricultural University) for his help in the collection of the specimens, Dr. Yongjie Wang (Capital Normal University, China) for the identification of the specimens, Dr Ding Yang, Dr Wanzhi Cai, Mr. Aimin Shi and Dr. Fan Song (China Agricultural University) for technical assistance and advice on experiments.
Funding Statement
This study was supported by grants from the National Natural Foundation of China (No. 31071962, No. 201103022, and No. 31272352). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Salvato P, Simonato M, Battisti A, Negrisolo E (2008) The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC genomics 9: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, et al. (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18: 489–522. [Google Scholar]
- 3. Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sheffield NC, Song H, Cameron SL, Whiting MF (2008) A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol 25: 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyce TM, Zwick ME, Aquadro CF (1989) Mitochondrial DNA in the bark weevils: size, structure and heteroplasmy. Genetics 123: 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia W, Higgs PG (2008) Codon usage in mitochondrial genomes: distinguishing context-dependent mutation from translational selection. Mol Biol Evol 25: 339–351. [DOI] [PubMed] [Google Scholar]
- 7. Winterton SL, Makarkin VN (2010) Phylogeny of moth lacewings and giant lacewings (Neuroptera: Ithonidae, Polystoechotidae) using DNA sequence data, morphology, and fossils. Ann Entomol Soc Am 103: 511–522. [Google Scholar]
- 8. Winterton SL, Hardy NB, Wiegmann BM (2010) On wings of lace: phylogeny and Bayesian divergence time estimates of Neuropterida (Insecta) based on morphological and molecular data. Syst Entomol 35: 349–378. [Google Scholar]
- 9. Kristensen NP (1999) Phylogeny of endopteryygote insects, the most successful lineage of living organisms. Eur J Entomol 96: 237–254. [Google Scholar]
- 10. Wheeler WC, Whiting M, Wheeler QD, Carpenter JM (2001) The phylogeny of the extant hexapod orders. Cladistics 17: 113–169. [DOI] [PubMed] [Google Scholar]
- 11. Wiegmann BM, Trautwein MD, Kim JW, Cassel BK, Bertone MA, et al. (2009) Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC biology 7: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haring E, Aspöck U (2004) Phylogeny of the Neuropterida: a first molecular approach. Syst Entomol 29: 415–430. [Google Scholar]
- 13. Aspöck U, Aspöck H (2008) Phylogenetic relevance of the genital sclerites of Neuropterida (Insecta: Holometabola). Syst Entomol 33: 97–127. [Google Scholar]
- 14. Hua J, Li M, Dong P, Xie Q, Bu W (2009) The mitochondrial genome of Protohermes concolorus Yang et Yang 1988 (Insecta: Megaloptera: Corydalidae). Mol Biol Rep 36: 1757–1765. [DOI] [PubMed] [Google Scholar]
- 15. Cameron SL, Sullivan J, Song H, Miller KB, Whiting MF (2009) A mitochondrial genome phylogeny of the Neuropterida (lace-wings, alderflies and snakeflies) and their relationship to the other holometabolous insect orders. Zool Scr 38: 575–590. [Google Scholar]
- 16. Wang Y, Liu X, Winterton SL, Yang D (2012) The first mitochondrial genome for the fishfly subfamily Chauliodinae and implications for the higher phylogeny of Megaloptera. PLoS ONE 7: e47302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beckenbach ATBA, Stewart JBSJ (2008) Insect mitochondrial genomics 3: the complete mitochondrial genome sequences of representatives from two neuropteroid orders: a dobsonfly (order Megaloptera) and a giant lacewing and an owlfly (order Neuroptera). Genome 52: 31–38. [DOI] [PubMed] [Google Scholar]
- 18. Negrisolo E, Babbucci M, Patarnello T (2011) The mitochondrial genome of the ascalaphid owlfly Libelloides macaronius and comparative evolutionary mitochondriomics of neuropterid insects. BMC genomics 12: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haruyama N, Mochizuki A, Sato Y, Naka H, Nomura M (2011) Complete mitochondrial genomes of two green lacewings, Chrysoperla nipponensis (Okamoto, 1914) and Apochrysa matsumurae Okamoto, 1912 (Neuroptera: Chrysopidae). Mol Biol Rep 38: 3367–3373. [DOI] [PubMed] [Google Scholar]
- 20. Aspöck U, Plant JD, Nemeschkal HL (2001) Cladistic analysis of Neuroptera and their systematic position within Neuropterida (Insecta: Holometabola: Neuropterida: Neuroptera). Syst Entomol 26: 73–86. [Google Scholar]
- 21. Mueller RL, Boore JL (2005) Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol Biol Evol 22: 2104–2112. [DOI] [PubMed] [Google Scholar]
- 22. Li H, Gao J, Liu H, Liu H, Liang A, et al. (2011) The architecture and complete sequence of mitochondrial genome of an assassin bug Agriosphodrus dohrni (Hemiptera: Reduviidae). Int J Biol Sci 7: 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Erler S, Ferenz HJ, Moritz RFA, Kaatz HH (2010) Analysis of the mitochondrial genome of Schistocerca gregaria (Orthoptera: Acrididae). Biol J Linn Soc 99: 296–305. [Google Scholar]
- 24. Yu DJ, Xu L, Nardi F, Li JG, Zhang RJ (2007) The complete nucleotide sequence of the mitochondrial genome of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Gene 396: 66–74. [DOI] [PubMed] [Google Scholar]
- 25. Li H, Liu H, Shi A, Štys P, Zhou X, et al. (2012) The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp.(Hemiptera: Enicocephalidae). PloS one 7: e29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Liu H, Song F, Shi A, Zhou X, et al. (2012) Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera). PloS one 7: e45925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lavrov DV, Brown WM, Boore JL (2000) A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus . Proc Natl Acad Sci 97: 13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cameron SL, Whiting MF (2008) The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 408: 112–123. [DOI] [PubMed] [Google Scholar]
- 30. Gillespie JJ, Johnston JS, Cannone JJ, Gutell RR (2006) Characteristics of the nuclear (18S, 5.8 S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): structure, organization, and retrotransposable elements. Insect Mol Biol 15: 657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cannone J, Subramanian S, Schnare M, Collett J, D'Souza L, et al. (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao JL, Zhang YY, Luo AR, Jiang GF, Cameron SL, et al. (2011) The complete mitochondrial genome of Spilonota lechriaspis Meyrick (Lepidoptera: Tortricidae). Mol Biol Rep 38: 3757–3764. [DOI] [PubMed] [Google Scholar]
- 33. Caterino MS, Sperling FAH (1999) Papilio phylogeny based on mitochondrial Cytochrome Oxidase I and II genes. Mol Phylogenet Evol 11: 122–137. [DOI] [PubMed] [Google Scholar]
- 34. Li H, Liu H, Cao L, Shi A, Yang H, et al. (2012) The complete mitochondrial genome of the damsel bug Alloeorhynchus bakeri (Hemiptera: Nabidae). Int J Biol Sci 8: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stanton DJ, Daehler LL, Moritz CC, Brown WM (1994) Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics 137: 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rand DM (1993) Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol 37: 281–295. [DOI] [PubMed] [Google Scholar]
- 37. Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- 38. Wei S, Shi M, Sharkey MJ, Van Achterberg C, Chen X (2010) Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC genomics 11: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberti M, Bruni F, Polosa PL, Gadaleta MN, Cantatore P (2006) The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res 34: 2109–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song H, Sheffield NC, Cameron SL, Miller KB, Whiting MF (2010) When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst Entomol 35: 429–448. [Google Scholar]
- 41. Sheffield NC, Song H, Cameron SL, Whiting MF (2009) Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst Biol 58: 381–394. [DOI] [PubMed] [Google Scholar]
- 42. Galtier N, Gouy M (1995) Inferring phylogenies from DNA sequences of unequal base compositions. Proc Natl Acad Sci U S A 92: 11317–11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beutel RG, Friedrich F, Aspöck U (2010) The larval head of Nevrorthidae and the phylogeny of Neuroptera (Insecta). Zool J Linn Soc-Lond 158: 533–562. [Google Scholar]
- 44. Winterton SL (2003) Molecular phylogeny of Neuropterida with emphasis on the lacewings (Neuroptera). Entomol Abh 61: 158–160. [Google Scholar]
- 45. Beutel RG, Zimmermann D, Krauß M, Randolf S, Wipfler B (2010) Head morphology of Osmylus fulvicephalus (Osmylidae, Neuroptera) and its phylogenetic implications. Org Divers Evol 10: 311–329. [Google Scholar]
- 46. Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT (2006) Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst 37: 545–579. [Google Scholar]
- 47. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Ent Soc Am 87: 651–701. [Google Scholar]
- 48. TA H (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows. 95/98/NT: 95–98. [Google Scholar]
- 49. Beard CB, Hamm D, Collins FH (1993) The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol 2: 103–124. [DOI] [PubMed] [Google Scholar]
- 50. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 52. Sharma D, Issac B, Raghava G, Ramaswamy R (2004) Spectral Repeat Finder (SRF): identification of repetitive sequences using Fourier transformation. Bioinformatics 20: 1405–1412. [DOI] [PubMed] [Google Scholar]
- 53. Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41: 353–358. [DOI] [PubMed] [Google Scholar]
- 54. Kumar S, Gadagkar SR (2001) Disparity index: a simple statistic to measure and test the homogeneity of substitution patterns between molecular sequences. Genetics 158: 1321–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 56. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 57. Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 58. Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 59.Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution: 783–791. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Organization of Thyridosmylus langii mt genome.
(DOCX)
Nucleotide composition of the Thyridosmylus langii mt genome.
(DOC)
Comparision of branch lengths among Neuropterida insects.
(DOCX)
Primer sequences used in this study.
(DOC)
Mt genomes used in comparative analyses.
(DOCX)