Abstract
Background
It has been confirmed that tumor necrosis factor-alpha (TNFα), a macrophage-derived pro-inflammatory cytokine, plays an important role in the pathogenesis of psoriasis vulgaris and psoriatic arthritis (PsV&PsA). In contrast, the reported association of TNFα gene promoter region single nucleotide polymorphisms (SNPs) and PsV&PsA has remained controversial. Accordingly, we performed a meta-analysis to provide new evidence that SNPs in the TNFα gene promoter region alter not only the risk of psoriasis vulgaris (PsV) or psoriatic arthritis (PsA) but also of PsV&PsA.
Methods
Interrelated literature dated to October 2012 was acquired from the PubMed, ScienceDirect, and SpringerLink databases. The number of the genotypes and/or alleles for the TNFα promoter in the PsV and PsA and control subjects was obtained. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to calculate the risk of PsV and/or PsA with TNFα promoter SNPs.
Results
A total of 26 papers of 2159 for PsV (2129 normal controls) and 2360 for PsA (2997 normal controls) were included in our meta-analysis. The results showed that the variant genotype and allele of TNFα -308A/G was protective in pooled groups of patients with PsV&PsA (OR = 0.682, 0.750; 95% CI, 0.596-0.779, 0.653-0.861). However, the variant genotypes and alleles of TNFα -238A/G and -857T/C had an increased risk of PsV&PsA (OR = 2.493, 2.228, 1.536, 1.486, 95% CI, 1.777-3.498, 1.628-3.049, 1.336-1.767, 1.309-1.685). Moreover, the meta-analysis revealed a significant association between TNFα -238A/G and -857T/C polymorphism and PsA susceptibility (OR = 2.242, 2.052, 1.419, 1.465; 95% CI, 1.710-2.941, 1.614-2.610, 1.214-1.658, 1.277-1.681). In contrast, the variant genotypes and alleles of TNFα -308A/G proved to be protective against PsV (OR = 0.574, 0.650, 95% CI, 0.478-0.690, 0.556-0.759), whereas TNFα -238A/G was found to have a risk association (OR = 2.636, 2.223, 95% CI, 1.523-4.561, 1.317-3.751).
Conclusions
SNPs in the TNFα gene promoter region alter the risk of PsV and/or PsA.
Introduction
Psoriasis is a common autoimmune disorder that primarily involves the skin (psoriasis vulgaris, PsV). Severe complications, including generalized involvement of the body (erythroderma), extensive pustular lesions, and an associated arthritis known as psoriatic arthritis (PsA), make psoriasis complex and heterogeneous [1]. The worldwide prevalence of PsV varies from 0.91% (United States) to 8.5% (Norway) in adults [2]. However, only 34.4% of patients with PsV also have arthritis, with the onset of skin lesions preceding arthritis in 64.5% of the cases and arthritis appearing after PsV in only 19.35% of cases. In the remaining 16.1% of cases, PsV and PsA began almost simultaneously [3]. Thus, the relationship between the onset of PsV and PsA included in the spondyloarthritis group is closely related, although the exact pathogenesis has yet to be identified. The diseases are multifactorial and result from the interplay between multiple genetic, immunologic, and environmental factors. Under the category of genetic and environmental effects, cytokines, chemokines, adhesion molecules, and growth factors combined with T cells and dendritic cells act in an integrated manner to evolve into unique inflammatory and proliferative processes in psoriasis vulgaris and psoriatic arthritis (PsV & PsA) [1].
Tumor necrosis factor-alpha (TNFα), a macrophage-derived pro-inflammatory cytokine, has been proven to play an important role in the pathogenesis and development of PsV & PsA. By inducing and recruiting multiple cytokines, TNFα activates the inflammatory process of PsV & PsA in the skin, joints, and bowel [4]. The marked clinical response of TNFα blockers in patients has demonstrated the importance of TNFα in both PsV and PsA [5]. The TNFα gene has been localized to the major histocompatibility complex (MHC) region on chromosome 6p21.3, which is 250 kb centromeric from the human leukocyte antigen-B and thought to be a high-priority candidate gene in PsV & PsA [6]. Single nucleotide polymorphisms (SNPs) are common in the TNFα gene, especially in promoter region, and act as markers of disease susceptibility because of their influence on TNFα expression by a cooperative manner [7]. Kaluza et al. found that the psoriasis-associated TNFα promoter allele TNF238.2 showed a significantly decreased transcriptional activity and less tumor necrosis factor alpha production [8]. Although many studies have investigated the relationship between promoter region SNPs in the TNFα gene and the susceptibility to rheumatoid arthritis (RA), ankylosing spondylitis (AS), Crohn’s disease, PsV, and PsA [9]–[13], the association between TNFα promoter SNPs and PsV & PsA risk have produced inconsistent results. Some studies have found a significant association between TNFα promoter SNPs and PsV & PsA, whereas others have failed to confirm these results [14], [15]. Two meta-analyses, one of nine studies, the other of ten, were performed in 2006 and 2007, respectively, to evaluate the association between TNFα promoter SNPs and the susceptibility to PsA and PsV [11], [13]. However, because of the complexity and heterogeneity of disease, the association between PsV and PsA on pathogenesis is still unclear [16]. Thus, it is necessary to pool the results of these studies and systematically analyze the relationship between TNFα promoter SNPs and PsV & PsA risk. Moreover, new studies about the association between TNFα promoter SNPs and the susceptibility to PsV & PsA have recently been published and provide new evidence that was not included in the previous meta-analyses, particularly regarding the susceptibility alleles TNFα -857T/C, TNFα -1031C/T, and TNFα -863A/C [14], [15], [17]–[21]. Therefore, it is essential to collect the most comprehensive evidence and reassess the association of TNFα promoter SNPs with PsV and/or PsA.
Materials and Methods
Search strategy
We searched the PubMed, ScienceDirect, and SpringerLink databases for papers published up to October 2012. An electronic search was performed using combinations of the following search terms without any limits: “tumor necrosis factor-alpha”, “TNFα”, “promoter”, “polymorphisms”, “single nucleotide polymorphism”, “genotype”, “psoriatic arthritis”, “PsA”, and “psoriasis”. Reference lists were checked to identify repeated literature.
Inclusion and exclusion criteria
For selection in the analysis, each study met all of the following criteria: (1) clinical subtypes of psoriasis, including PsV and/or PsA, according to the Moll and Wright or CASPAR criteria or diagnosed by clinical specialists; (2) a case-control or cohort study; (3) the inclusion the distributions of genotypes and/or alleles within the TNFα promoter provided for both the patients and controls; and (4) the reporting of the basic characters of the participants, including ethnicity, age and gender. The exclusion criteria were as follows: (1) a case report, review or descriptive study; (2) clinical subtypes of psoriasis, including erythroderma or extensive pustular lesions; (3) a lack of normal population as controls; (4) duplicate data in the studies; and (5) an inability to obtain the necessary data.
Quality assessment
The quality assessment of the selected studies was performed independently by two authors using the Newcastle-Ottawa Scale (NOS) [22]. Three parameters of quality, selection, comparability, and exposure assessment were assessed for the case-control studies. We defined “low-quality studies” as those with scores of 1–3, “moderate-quality studies” as those with scores of 4–6, and “high-quality studies” as those with scores of 7–9. Any disagreements were resolved by discussion.
Data extraction
The following information was extracted from each study: first author, publication year, country of origin, patient ethnicity and demographic characteristics, subtypes of psoriasis (PsV type I, II, and/or PsA), the number of cases and controls, the number of genotypes and/or alleles for the TNFα promoter in the cases and controls, and consistency tests of genotype frequencies with the Hardy-Weinberg equilibrium model (HWE). Two authors analyzed each paper according to the inclusion and exclusion criteria and independently extracted the information. In some studies, only the novel data were extracted because a portion of the data had already been reported. Disagreements were resolved by discussion.
Statistical analysis
The risks of the variant genotypes and alleles compared with the wild-type for six sites of TNFα promoter SNPs were estimated separately. Different subgroups divided by ethnicity or disease subtype were analyzed. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to calculate the risks of PsV and/or PsA with TNFα promoter SNPs. A p value <0.05 was considered statistically significant. In the forest plots, OR>1 represented a risk effect and <1 a protective effect. In our meta-analysis, the heterogeneity among the studies was tested by a χ2-based Q test and I2 statistic [23]. When p<0.10 or I2>50%, the heterogeneity was considered significant. A fixed-effects model was a better choice to calculate the pooled ORs when the heterogeneity was not significant, otherwise a random-effects model was used because of the heterogeneity among the studies. To explore the source of heterogeneity, a meta-regression analysis was performed when the number of studies was more than ten in a group for which heterogeneity existed [24]. A p value <0.05 was considered statistically significant. The size of the heterogeneity source (SH) was assessed by percentage. Both Begg’s and Egger’s tests were performed to check the publication bias [25]; the publication bias was considered significant when p<0.10 in either test. All of the analyses were conducted using Stata software version 11.0 (Stata Corporation, College Station, TX, USA).
Results
Characteristic analysis of the included literature and quality assessment
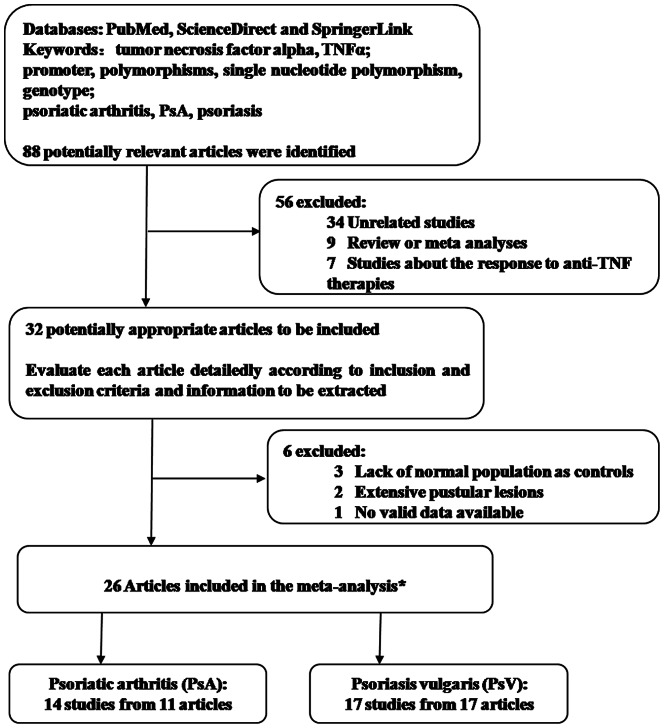
According to the inclusion and exclusion criteria, 26 papers regarding the association between TNFα promoter SNPs and the risk of PsV & PsA, including 14 studies for PsA [13], [17], [20], [21], [26]–[32] and 17 studies for PsV[8], [14], [15], [18], [19], [21], [29], [32]–[42], were included in our analysis (Figure 1). Twenty-five studies (80.6%) were of high quality and six (19.4%) of moderate quality based on the NOS. In our meta-analysis, 2159 PsV (controls, 2129) reports and 2360 PsA reports (controls, 2997) were included to analyze the disease risk from TNFα -308A/G (rs1800629), -238A/G (rs361525), -857T/C (rs1799724), -1031C/T (rs1799964), -863A/C (rs1800630), and -488A/G (rs80267959) polymorphisms. Details about the first author, publication year, country of origin, ethnicity, subtype of psoriasis, number of cases and controls, number of genotypes and/or alleles for the TNFα promoter in the cases and controls, consistency tests of genotype frequencies with HWE, and the NOS score are shown in Tables 1 and 2 for each study selected. The publishing date of all the studies ranged from 1997 to 2012. All of the patients were Caucasian, with the exception of five studies that involved Asian patients. Consistency tests of genotype frequencies with HWE were performed in 8 (47.1%) and 9 (64.3%) studies of PsV and PsA, respectively.
Figure 1. Flow diagram of the selection and nature of the studies.
*including a meta-analysis of PsA with two studies available; both PsV and PsA were involved in the two articles.
Table 1. Characteristics of the individual studies for PsA included in the meta-analysis.
| First author | Year | Country/Ethnicity | Type of psoriasis | SNPs:case/control (n) | HWE | NOS score | |||||
| 308A/G | 238A/G | 857T/C | 1031C/T | 863A/G | 488A/G | ||||||
| Giardina-G | 2011 | Germany/Caucasian | PsA | - | - | 370/560 | - | - | - | Y | 7 |
| Giardina-I | 2011 | Italy/Caucasian | PsA | - | - | 400/400 | - | - | - | Y | 7 |
| Giardina-U | 2011 | UK/Caucasian | PsA | - | - | 125/354 | - | - | - | Y | 7 |
| Popa | 2011 | Romania/Caucasian | PsA | 86/142 | 86/147 | 86/142 | - | - | - | Y | 7 |
| Reich1 | 2007 | Germany/Caucasian | PsA | 361/370 | 368/372 | 370/373 | 367/368 | - | - | Y | 7 |
| Rahman-N | 2005 | Newfoundland/Caucasian | PsA | 225/103 | 228/103 | 220/103 | 220/103 | 231/103 | - | Y | 7 |
| Rahman-T | 2005 | Toronto/Caucasian | PsA | 203/101 | 199/101 | 199/101 | 200/99 | 200/101 | - | Y | 7 |
| Balding | 2003 | Ireland/Caucasian | PsA | 149/390 | - | - | - | - | - | N | 6 |
| Gonzalez-S | 2002 | Spain/Caucasian | PsA | 81/110 | 81/110 | - | - | - | - | N | 8 |
| Al-Heresh | 2002 | Ireland/Caucasian | PsA | 124/101 | 124/101 | - | - | - | 124/101 | N | 8 |
| Hohler-1 | 2002 | Germany/Caucasian | PsA | 87/99 | 87/99 | - | - | - | - | Y | 7 |
| Gonzalez-J | 2001 | Jewish/Caucasoid | PsA | 52/73 | 52/73 | - | - | - | - | N | 8 |
| Hamamoto | 2000 | Japan/Asian | PsA | 20/87 | 20/87 | 20/87 | 20/87 | 20/87 | - | N | 8 |
| Hohler-2 | 1997 | Germany/Caucasian | PsA | 62/99 | 62/99 | - | - | - | - | Y | 7 |
Notes: “PsA”, psoriatic arthritis; “SNPs”, single nucleotide polymorphisms; “HWE”, Hardy–Weinberg equilibrium; “NOS”, Newcastle-Ottawa Scale; “Y”, yes, consistency tests of HWE were performed; “N”, no, consistency tests of HWE were not performed. “-”, no statistics.
Table 2. Characteristics of the individual studies for PsV included in the meta-analysis.
| First author | Year | Country/Ethnicity | Type of psoriasis | SNPs: case(type I/type II)/control (n) | HWE | NOS score | |||
| 308A/G | 238A/G | 857T/C | 1031C/T | ||||||
| Gallo | 2012 | Spain/Caucasian | PsV with or without PsA | 84/76 | 81/71 | 77/76 | 86/72 | N | 6 |
| Magalhaes | 2010 | Brazil/Caucasian | PsV type I | 69(69/0)/70 | 69(69/0)/70 | - | - | Y | 8 |
| Settin | 2009 | Egypt/Caucasian | PsV type I/type II | 46/98 | - | - | - | N | 7 |
| Nedoszytko | 2007 | Poland/Caucasian | PsV type I/type II | 166/65 | 166(134/32)/65 | - | - | Y | 7 |
| Reich1 | 2007 | Germany/Caucasian | PsV | 368/370 | 373/372 | 374/373 | 370/368 | Y | 7 |
| Baran | 2006 | Poland/Caucasian | PsV type I/type II | 78(54/24)/74 | - | - | - | N | 7 |
| Mossner | 2005 | Germany/Caucasian | PsV with or without PsA | 239/135 | 239/135 | - | - | Y | 6 |
| Long | 2004 | China/Asian | PsV type I/type II | 77/82 | 77(48/29)/82 | - | - | N | 7 |
| Tsunemia | 2003 | Japan/Asian | PsV | 163/96 | 163/96 | - | - | N | 7 |
| Kim | 2003 | Korea/Asian | PsV type I/type II | 103/125 | 103/125 | - | - | N | 6 |
| Chang | 2003 | China/Asian | PsV type I/type II | 105/160 | 105/160 | - | - | N | 7 |
| Reich2 | 2002 | Germany/Caucasian | PsV type I/type II | 231/345 | 231(156/75)/345 | - | - | Y | 7 |
| Hohler1 | 2002 | Germany/Caucasian | PsV type I | 60(60/0)/99 | 60(60/0)/99 | - | - | Y | 7 |
| Craven | 2001 | UK/Caucasian | PsV type I/type II | 81(48/33)/66 | - | - | - | N | 7 |
| Kaluza | 2000 | Germany/Caucasian | PsV type I with or without PsA | 47/43 | 47(47/0)/43 | - | - | N | 6 |
| Reich3 | 1999 | Germany/Caucasian | PsV type I/type II | 151(100/51)/123 | 151(100/51)/123 | - | - | Y | 8 |
| Jacob | 1999 | Germany/Caucasian | PsV type I | 80(80/0)/99 | 83(83/0)/99 | - | - | Y | 6 |
Notes: “PsV”, psoriasis vulgaris; “SNPs”, single nucleotide polymorphisms; “HWE”, Hardy–Weinberg equilibrium; “NOS”, Newcastle-Ottawa Scale; “Y”, yes, consistency tests of HWE were performed; “N”, no, consistency tests of HWE were not performed. “-”, no statistics.
Quantitative synthesis and heterogeneity analysis
1. TNFα -308A/G, -238A/G, -857T/C and -1031C/T polymorphism and susceptibility to PsV & PsA
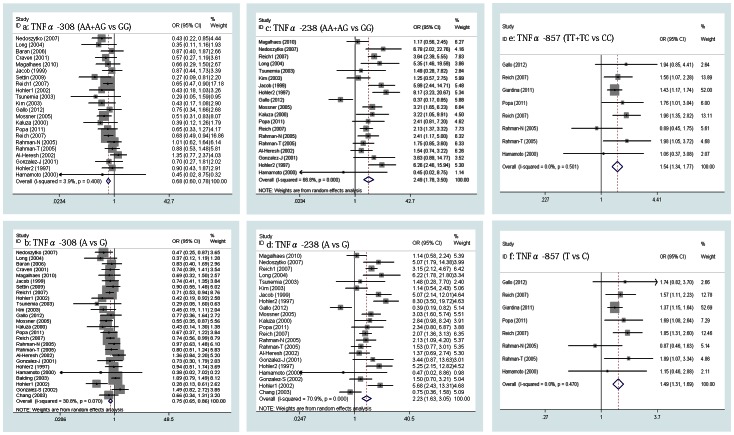
We pooled all the studies involving both PsV and PsA to evaluate the disease risk associated with TNFα -308A/G, -238A/G, -857T/C and -1031C/T polymorphisms. The meta-analysis showed a significant association between the two genotypes of TNFα -308A/G (AA+AG vs GG) and PsV & PsA in a pooled group of 22 studies (OR = 0.682, 95% CI, 0.596-0.779, p = 0.000, Figure 2 a) with the fixed-effects model (Q = 21.85, p = 0.408, I2 = 3.9%), which suggested that the variant genotype of TNFα -308A/G (AA+AG) was protective. For the two alleles of TNFα -308A/G (A vs G), the same protective effect (OR = 0.750, 95% CI, 0.653-0.861, p = 0.000, Figure 2 b) was found with the random-effects model (Q = 36.13, p = 0.070, I2 = 30.8%) because of the heterogeneity among the 26 studies. To explore the heterogeneity source, a meta-regression analysis was performed, and the result indicated that the concomitant variable, the psoriasis subtype, had statistical significance (p = 0.041, SH 56.0%) while the publication year and ethnicity had no statistical significance (p>0.05). A subgroup analysis was performed.
Figure 2. Forrest plot of the relationship between the TNFα promoter SNPs and risk of PsV & PsA.
a–b, Pooled lower risk of PsV & PsA with TNFα -308A/G polymorphism; c-f, Pooled higher risk of PsV & PsA with TNFα -238A/G and TNFα -857T/C polymorphism.
However, the overall meta-analysis involved 19 and 22 studies, which separately indicated that the variant genotype (AA+AG) and allele (A) of TNFα -238A/G exhibited an increased risk of PsV & PsA (OR = 2.493, 2.228, 95% CI 1.777-3.498, 1.628-3.049, p = 0.000, 0.000 [separately], Figure 2 c, d) in the random-effects model (Q = 54.27, 72.07, p = 0.000, 0.000, I2 = 66.8%, 70.9% [separately]). The result of the meta-regression analysis showed that the publication year had statistical significance (p = 0.007, 0.014, SH 53.7%, 35.8% [separately]) while ethnicity and psoriasis subtype had no statistical significance (p>0.05).
Similarly, for the genotypes (TT+TC vs CC) and alleles (T vs C) of TNFα -857T/C, the pooled ORs were found to be 1.536 (95% CI, 1.336-1.767, p = 0.000, Figure 2 e) and 1.486 (95% CI, 1.309-1.685, p = 0.000; Figure 2 f) with the fixed-effects model for each group in eight studies (Q = 6.34, 6.62, p = 0.501, 0.470, I2 = 0.0%, 0.0% [separately]).
Our meta-analysis did not reveal a significant association between the TNFα -1031C/T polymorphism and susceptibility to PsV & PsA. The pooled ORs were 0.894 (95% CI, 0.755-1.059, p = 0.195) for the genotype with the fixed-effects model (Q = 8.27, p = 0.142, I2 = 39.6%) and 0.867 (95% CI, 0.693-1.084, p = 0.211) for the allele with the random-effects model (Q = 10.22, p = 0.069, I2 = 51.1%). A meta-regression analysis was not performed for <10 studies involved in the group in which heterogeneity existed.
2. TNFα -308A/G, -238A/G, -857T/C, -1031C/T, -863A/C, and -488A/G polymorphism and susceptibility to PsA
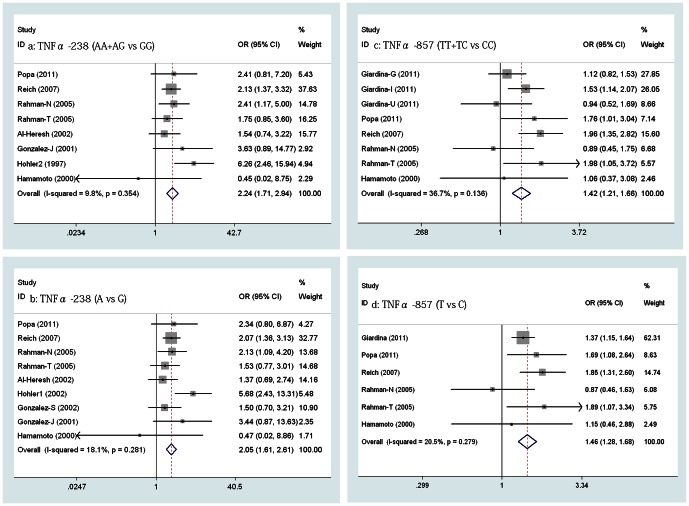
The meta-analysis showed a significant association between the two genotype of TNFα -238A/G (AA+AG vs GG) and PsA in eight pooled studies that included one study involving an Asian group from Japan [31] (OR = 2.242, 95% CI, 1.710-2.941, p = 0.000, Figure 3 a). Similarly, the two alleles of TNFα -238A/G (A vs G) were also correlated with the risk of PsA in the pooled analysis that added another study from Spain [27] (OR = 2.052, 95% CI, 1.614-2.610, p = 0.000; Figure 3 b). Even when the data from Japan was removed, the pooled OR also was statistically significant at 2.284 (95% CI, 1.731-3.004, p = 0.000) and 2.079 (95% CI, 1.632-2.649, p = 0.000 [separately]). All of the ORs stated above were calculated using the fixed-effects model, as there was no significant heterogeneity among the studies in every pooled analysis (Q = 7.76, 9.77, 6.63, 8.82, p = 0.345, 0.281, 0.357, 0.266, I2 = 9.8%, 18.1%, 9.5%, 20.7% [separately]).
Figure 3. Forrest plot of the relationship between TNFα promoter SNPs and risk of PsA.
a–d, Pooled higher risk of PsA with TNFα -238A/G and TNFα -857T/C polymorphism.
The pooled OR from the eight studies including one Asian group [31] was first calculated to be 1.419 (95% CI, 1.214-1.658, p = 0.000) using the fixed-effects model (Q = 11.05, p = 0.136, I2 = 36.7%), which indicated that there was a significant association between the genotypes of TNFα -857T/C (TT+TC vs CC) and PsA (Figure 3 c). Additionally, a statistical significance was found for the two alleles of TNFα -857T/C (T vs C) (OR = 1.465, 95% CI, 1.277-1.681, p = 0.000, Figure 3 d) using the fixed-effects model (Q = 6.29, p = 0.279, I2 = 20.5%). Excluding the Asian group, the pooled ORs were not significantly affected (OR = 1.428, 1.473, 95% CI, 1.220-1.671, 1.281-1.693, p = 0.000, 0.000 [separately]).
Our meta-analysis did not reveal a significant association between TNFα -308A/G, -1031C/T and -863A/C polymorphism and the susceptibility to PsA. Together with the results of heterogeneity and publication bias tests, the values of ORs and 95% CIs for the risk of susceptibility in patients with PsA are shown in Table 3. All of the ORs were calculated using the fixed-effects model, although one pooled OR for the comparison of the TNFα -308A/G allele (A vs G) between PsA and the controls was observed using the random-effects model (Q = 19.20, p = 0.038, I2 = 47.9%). No pooled OR was performed for the TNFα -488A/G polymorphism, as only one study was involved.
Table 3. Summary of odds ratios (95% CI) in the analysis of the relationship between TNF-a promoter SNPs and PsA susceptibility.
| Comparison | No. of studies | Odds Ratio (95% CI) | Test for OR | Test for heterogeneity | Publication bias | ||||
| Z | P | Chi-squared | P | I2 (%) | Begg's Test | Egger's test | |||
| TNFα-308 *(1) | 8 | 0.829(0.681, 1.008)$ | 1.88 | 0.06 | 5.88 | 0.554 | 0.00% | 0.621 | 0.861 |
| TNFα-308* (2) | 7 | 0.831(0.683, 1.012) $ | 1.84 | 0.065 | 5.72 | 0.456 | 0.00% | 0.881 | 0.55 |
| TNFα-308# (1) | 11 | 0.876(0.704, 1.091)& | 1.18 | 0.236 | 19.2 | 0.038 | 47.90% | 0.392 | 0.489 |
| TNFα-308#(2) | 10 | 0.879(0.703,1.100)& | 1.13 | 0.26 | 18.87 | 0.026 | 52.30% | 0.531 | 0.586 |
| TNFα-1031* (1) | 4 | 0.854(0.685, 1.064) $ | 1.4 | 0.16 | 2.35 | 0.504 | 0.00% | 1 | 0.657 |
| TNFα-1031* (2) | 3 | 0.858(0.686, 1.074) $ | 1.33 | 0.183 | 2.29 | 0.319 | 12.50% | 0.602 | 0.264 |
| TNFα-1031# (1) | 4 | 0.860(0.715, 1.034) $ | 1.6 | 0.109 | 2.29 | 0.514 | 0.00% | 1 | 0.686 |
| TNFα-1031# (2) | 3 | 0.863(0.716, 1.040) $ | 1.54 | 0.122 | 2.25 | 0.325 | 11.10% | 0.602 | 0.316 |
| TNFα-863* (1) | 3 | 0.764(0.538, 1.084) $ | 1.51 | 0.132 | 0.26 | 0.877 | 0.00% | 0.602 | 0.733 |
| TNFα-863# (2) | 3 | 0.774(0.569,1.053) $ | 1.63 | 0.103 | 0.47 | 0.789 | 0.00% | 0.602 | 0.587 |
Notes:* The variant genotype; # the variant allele; (1) pooled group, including Asian and Caucasian groups; (2) pooled group including a Caucasian group only; & data from the random-effects model because of significant heterogeneity; $ data from the fixed-effects model.
3. TNFα -308A/G, -238A/G, -857T/C and -1031C/T polymorphism and susceptibility to PsV
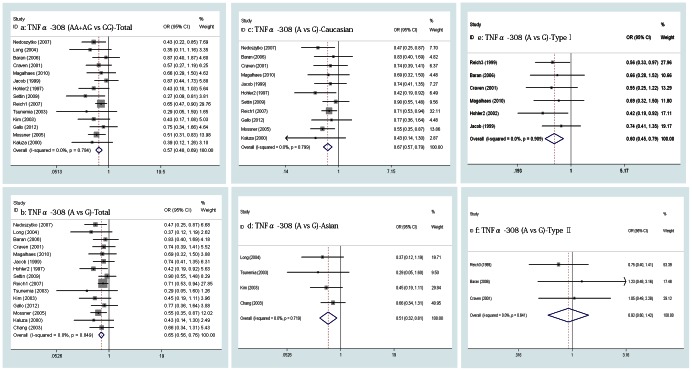
A meta-analysis was performed on all the PsV patients and subgroups divided by ethnicity and subtype of PsV, revealing a significant association between the two genotypes of TNFα -308A/G (AA+AG vs GG) and PsV in 14 pooled studies (OR = 0.574, 95% CI, 0.478-0.690, p = 0.000, Figure 4 a), which suggested that the variant genotypes of TNFα -308A/G (AA+AG) are a protective effect. The same association was revealed in both of the Caucasian and Asian subgroups in eleven and three studies, respectively, but the pooled OR was 0.596 for the Caucasian subgroup (95% CI, 0.492-0.721, p = 0.000) compared with 0.382 for the Asian subgroup (95% CI, 0.196-0.747, p = 0.005). For the two alleles of TNFα -308A/G (A vs G), the same protective effect was found in the pooled groups mentioned above. The OR values were 0.650 (95% CI, 0.556-0.759, p = 0.000), 0.672 (95% CI, 0.569-0.793, p = 0.000) and 0.507 (95% CI, 0.317-0.812, p = 0.005) (Figure 4 b, c, d). The pooled ORs in the type I subgroup (involving six studies) were 0.596 (95% CI, 0.437-0.813, p = 0.001) and 0.597 (95% CI, 0.450-0.791, p = 0.000; Figure 4 e) for the genotypes and the alleles of TNFα -308A/G (separately). However, the results for the genotypes and the alleles of TNFα -308A/G in the type II subgroups involving three studies were not statistically significant in indicating a protective effect (OR = 0.856, 0.923, 95% CI 0.523-1.400, 0.598-1.423, p = 0.535, 0.715 [separately], Figure 4 f). In all of the subgroups, the fixed-effects model was used to calculate the ORs because no heterogeneity was found among the studies (p>0.1, I2<50.0%).
Figure 4. Forrest plot of the relationship between TNFα -308A/G polymorphism and risk of PsV, a–b, Pooled lower risk of PsV with TNFα -308A/G polymorphism; c–d, In Caucasian and Asian subgroups, pooled lower risk of PsV with TNFα -308A/G polymorphism; e, Pooled lower risk of type I PsV with TNFα -308A/G polymorphism; f, no risk of type ΠPsV with TNFα -308A/G polymorphism.
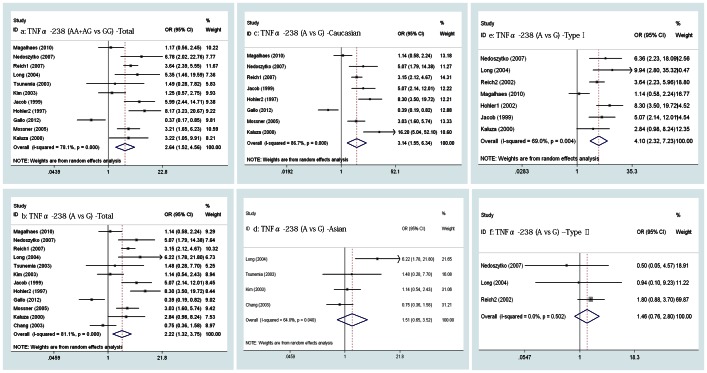
The overall meta-analysis involving 11 and 12 studies separately indicated that the variant genotypes (AA+AG) and allele (A) of TNFα -238A/G had an associated increase risk of PsV (OR = 2.636, 2.223, 95% CI, 1.523-4.561, 1.317-3.751, p = 0.001, 0.003 [separately], Figure 5 a, b) using the random-effects model (Q = 45.76, 58.23, p = 0.000, 0.000, I2 = 78.1%, 81.1% [separately]). To explore the source of heterogeneity, a meta-regression analysis was performed, and the result showed that the concomitant variable of publication year had a statistical significance (p = 0.02, 0.042, SH 54.8%, 37.1% [separately]) while ethnicity, type of PsV and HWE consistency test had no statistical significance (p>0.05). However, we also calculated the pooled ORs in the subgroups divided by ethnicity and type of PsV, and the same risk of the variant genotypes (AA+AG) and allele (A) of TNFα -238A/G was found in all the subgroups with the existence of heterogeneity using the random-effects model, which was not found using the fixed-effects model in subgroups of type II PsV (Figure 5 c, d, e, f).
Figure 5. Forrest plot of the relationship between TNFα -238A/G polymorphism and risk of PsV.
a–b, Pooled higher risk of PsV with TNFα -238A/G polymorphism; c–d, In Caucasian and Asian subgroups, pooled higher risk of PsV with TNFα -238A/G polymorphism; e, Pooled higher risk of type I PsV with TNFα -238A/G polymorphism; f, no risk of type ΠPsV with TNFα -238A/G polymorphism.
No pooled OR was calculated for the TNFα -857T/C and -1031C/T polymorphisms, as only two studies were involved.
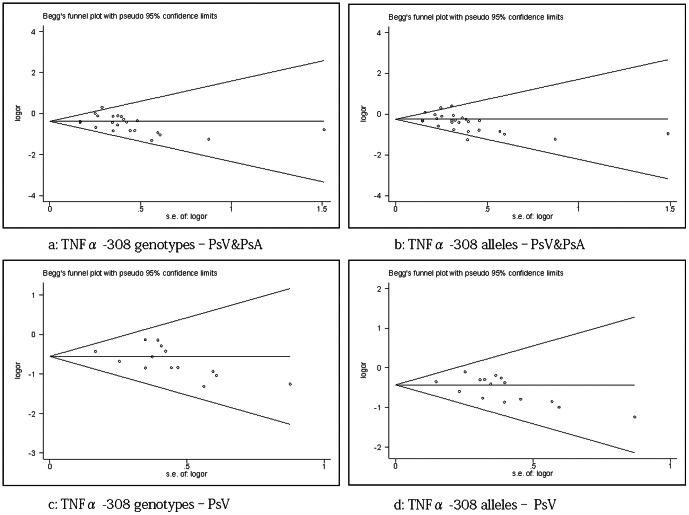
Publication bias
For the genotypes and alleles of TNFα -308A/G, the publication bias showed statistical significance in the pooled groups of PsV & PsA (Begg’s test, p = 0.040, 0.106; Egger’s test, p = 0.021, 0.034 [separately]). Similarly, the same publication bias was found in the pooled groups of PsV (Begg’s test, p = 0.025, 0.054; Egger’s test, p = 0.070, 0.050 [separately]). The funnel plots for publication bias are shown in Figure 6 and suggest the evidence of potential publication bias. In all other groups, neither the Begg’s (p>0.1) nor Egger’s tests (p>0.1) showed statistical significance, which suggested that no publication bias existed (data not shown).
Figure 6. Funnel plot showing publication bias in 4 pooled groups of PsV & PsA and PsV only.
The value of Pr>|z| (continuity corrected) is p<0.10.
Discussion
It is well known that TNFα plays a crucial role in autoimmune and infectious diseases as a proinflammatory cytokine; however, the mechanism of altered PsV & PsA risk owing to SNPs in the TNFα gene promoter region remains unknown. A possible explanation is the abnormal expression of the TNFα gene mediated by variations in the TNFα promoter. This premise was supported by studies noting that the TNFα -857C allele was associated with positive response to drug treatment in PsV patients treated with etanercept (Recombinant Human Tumor Necrosis Factor-α Receptorα:IgG Fc Fusion Protein)[43]. However, the connection between TNFα transcriptional activity or protein production and TNFα promoter SNPs is unresolved, as the reports are inconsistent with some studies revealing no association between them [44]. Therefore, more complex mechanisms than the abnormal expression of the TNFα gene must exist. Another possible explanation is that TNFα promoter SNPs is also associated with the production of other cytokines that play an important role in the pathologic process of the disease. For example, TNFα -308A allele was associated with higher serum interferon-alpha levels in the patients with juvenile dermatomyositis [45]. Furthermore, because of the complexity of biological systems, a single gene polymorphism may result in further polymorphisms or post-transcriptional regulation under different gene-gene or gene-environment interactions. Accordingly, further studies are needed to assess the functional consequences of TNFα promoter SNPs. In the present study, the variant genotypes and alleles of TNFα promoter SNPs were found to contribute to the same risk of disease susceptibility in all groups or subgroups.
It remains controversial whether PsV and PsA are different clinical manifestations of one disease or two different clinical entities. On the one hand, PsV and PsA exist unseparated in patients because of the complex and heterogeneous nature of PsV & PsA [16]. On the other hand, the diversity of skin lesions, including the presence and severity of a clinical rash, does not affect the clinical manifestation of joint disease, which is found in 30% of patients with skin lesions [4]. However, the common pathogenesis, activation of T lymphocytes and proinflammatory cytokines, is now well recognized in patients with PsV and/or PsA [16]. In addition, the curative effect of TNFα blockers in patients with PsV and/or PsA indicates that, to a certain degree, they share a common immune mechanism [5]. Therefore, both PsV and PsA may have a common onset risk with regard to TNFα promoter SNPs. Our meta-analysis results confirmed this hypothesis through the quantitative synthesis of controversial studies. The TNFα -857T/C variant significantly increased the susceptibility to PsV & PsA (OR = 1.536, 95% CI, 1.336-1.767), without heterogeneity among the groups. For TNFα -238A/G, a greater risk (OR = 2.493, 95% CI, 1.777-3.498) was found using the random-effects model, with heterogeneity primarily regarding the year of publication. However, the TNFα -308A/G variant exhibited a protective effect, with heterogeneity primarily in the disease subtype (OR = 0.682, 95% CI, 0.596-0.779). Further subgroup-analysis found the same risk or protective effect in most of the subgroups without heterogeneity. In a word, a variation in the levels of TNFα production attribute to a functional consequence of these TNFα promoter SNPs, which results to a common onset risk in both PsV and PsA.
Although a meta-analysis completed by Rahman et al. revealed that PsA risk was associated with TNFα -238A/G polymorphism (OR = 2.29; 95% CI 1.48-3.55) and not with TNFα -308A/G polymorphism (OR = 0.92, 95% CI, 0.69 -1.23) [13], more recent studies also paid close attention to other SNPs in the TNFα promoter, such as TNFα -857T/C [17]. Additionally, an increasing number of studies have provided new evidence for PsA susceptibility, although the results are controversial. Our meta-analysis gathered the comprehensive evidence and is the first to reveal that TNFα -857T/C polymorphism is associated with PsA susceptibility using the fixed-effects model (OR = 1.419, 95% CI, 1.214-1.658). Lv, K. et al. have observed in vitro that the TNFα -857T/C variant is a stronger transcriptional activator in response to lipopolysaccharide stimulation in the RAW264.7 cell line [46], indicating that the main mechanism of increased PsA risk can be attributed to the TNFα -857T/C polymorphism. Moreover, the results reflecting the association between TNFα -238A/G or TNFα -308A/G polymorphism and PsA susceptibility are remarkably consistent compared with the previous meta-analysis, although the pooled ORs differ because more studies and the fixed-effects model were used in our meta-analysis.
Furthermore, similar to a previous meta-analysis performed by Chunying Li et al. [11], our meta-analysis results showed that the TNFα -238A/G and TNFα -308A/G polymorphisms separately had the same risks and protective effects with regard to PsV susceptibility. However, in the subgroup-analysis, the results differ because recent studies have provided new evidence for the role of TNFα promoter SNPs in PsV susceptibility [15]. In the Asian subgroup, the TNFα -238A/G variant increased PsV risk by 1.912 (95% CI 1.047-3.491) using the fixed-effects model compared with the value of 1.46 (95% CI 0.66-3.19) revealed previously. However, no association was found between PsV risk and TNFα -238A/G or TNFα -308A/G polymorphism (OR = 1.569, 0.856, 95% CI 1.11-8.71, 0.523-1.400) using the fixed-effects model for the type II PsV subgroup compared to the previous Chunying Li et al. meta-analysis (OR = 3.11, 0.57, 95% CI, 0.800-3.078, 0.37-0.88). The inconsistency in the meta-analysis results indicates that the alterative risk of PsV result from TNFα promoter SNPs may be more complex because of the presence of stratification factors, such as ethnic admixture and the subtype or severity of disease.
Our meta-analysis has several limitations. First, heterogeneity was observed in some groups; although a subgroup analysis was performed, heterogeneity also existed with unclear sources in some subgroups. This finding indicated that the genetic background, combined with linkage disequilibrium patterns, epigenetic modifications, and clinical heterogeneity, made the relationship between the risk of disease susceptibility and TNFα promoter SNPs more complicated. Second, on analyses the risk of psoriasis with TNFα promoter SNPs, patients only with PsV and/or PsA were involved, but not with erythroderma or pustular lesions. Besides, only Caucasian and Asian patients were involved in the subgroup analysis; therefore, the assessment of disease risk is not applicable to other disease subtypes and ethnicities. Third, apart from the limited sample size or quality control of genotyping in some of studies, some subgroup analyses only involved three studies; this small sample size indicated insensitivity with regard to exploring the real association. Finally, as some negative-association studies are often unpublished, it seems unavoidable that publication bias may influence the meta-analysis results. Accordingly, caution should be exercised when interpreting the results of this meta-analysis.
Conclusions
This meta-analysis confirms the previous results that TNFα -238A/G and TNFα -308A/G polymorphisms are associated with PsV or PsA susceptibility. Furthermore, this meta-analysis shows that the TNFα -857T/C variant increases the risk of PsA significantly, whereas TNFα -857T/C and TNFα -238A/G polymorphisms are risk effects in the pooled analysis of PsV & PsA. In contrast, the TNFα -308A/G polymorphism is a protective effect for PsV & PsA. No significant association between TNFα-308A/G, -1031C/T, or -863A/C polymorphism and PsA susceptibility was revealed by this meta-analysis. Thus, further studies are needed to elucidate the precise contribution of those SNPs to the pathogenesis of PsV& PsA by using proteome-wide analysis of SNPs (PWAS) [47]. Additionally, prospective studies on the response to the TNFα blockers in patients with PsV& PsA to their TNFα promoter SNPs are benefit for properly assessing the impact and value of TNFα blockers in patients.
Supporting Information
PRISMA 2009 Checklist.
(DOC)
Funding Statement
This work was supported by the Natural Science Foundation of China (NO. 81173456) and the Natural Science Foundation of Guangdong Province (NO. S2012010009164). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Raychaudhuri SP (2012) A Cutting Edge Overview: Psoriatic Disease. Clin Rev Allergy Immunol. [DOI] [PubMed]
- 2.Parisi R, Symmons DP, Griffiths CE, Ashcroft DM (2012) Global Epidemiology of Psoriasis: A Systematic Review of Incidence and Prevalence. J Invest Dermatol. [DOI] [PubMed]
- 3. Scarpa R, Oriente P, Pucino A, Torella M, Vignone L, et al. (1984) Psoriatic arthritis in psoriatic patients. Br J Rheumatol 23: 246–250. [DOI] [PubMed] [Google Scholar]
- 4. Scarpa R, Ayala F, Caporaso N, Olivieri I (2006) Psoriasis, psoriatic arthritis, or psoriatic disease? J Rheumatol 33: 210–212. [PubMed] [Google Scholar]
- 5. Mease P (2004) TNFalpha therapy in psoriatic arthritis and psoriasis. Ann Rheum Dis 63: 755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kane D, FitzGerald O (2004) Tumor necrosis factor-alpha in psoriasis and psoriatic arthritis: a clinical, genetic, and histopathologic perspective. Curr Rheumatol Rep 6: 292–298. [DOI] [PubMed] [Google Scholar]
- 7. Cui G, Wang H, Li R, Zhang L, Li Z, et al. (2012) Polymorphism of tumor necrosis factor alpha (TNF-alpha) gene promoter, circulating TNF-alpha level, and cardiovascular risk factor for ischemic stroke. J Neuroinflammation 9: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaluza W, Reuss E, Grossmann S, Hug R, Schopf RE, et al. (2000) Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism. J Invest Dermatol 114: 1180–1183. [DOI] [PubMed] [Google Scholar]
- 9. Lee YH, Ji JD, Song GG (2007) Tumor necrosis factor-alpha promoter -308 A/G polymorphism and rheumatoid arthritis susceptibility: a metaanalysis. J Rheumatol 34: 43–49. [PubMed] [Google Scholar]
- 10. Li B, Wang P, Li H (2010) The association between TNF-alpha promoter polymorphisms and ankylosing spondylitis: a meta-analysis. Clin Rheumatol 29: 983–990. [DOI] [PubMed] [Google Scholar]
- 11. Li C, Wang G, Gao Y, Liu L, Gao T (2007) TNF-alpha gene promoter -238G>A and -308G>A polymorphisms alter risk of psoriasis vulgaris: a meta-analysis. J Invest Dermatol 127: 1886–1892. [DOI] [PubMed] [Google Scholar]
- 12. Xie C, Liu XF, Yang MS (2012) A meta-analysis on the association between three promoter variants of TNF-alpha and Crohn's disease. Mol Biol Rep 39: 1575–1583. [DOI] [PubMed] [Google Scholar]
- 13. Rahman P, Siannis F, Butt C, Farewell V, Peddle L, et al. (2006) TNFalpha polymorphisms and risk of psoriatic arthritis. Ann Rheum Dis 65: 919–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gallo E, Cabaleiro T, Roman M, Abad-Santos F, Dauden E (2012) [Study of genetic polymorphisms in the tumor necrosis factor alpha promoter region in Spanish patients with psoriasis]. Actas Dermosifiliogr 103: 301–307. [DOI] [PubMed] [Google Scholar]
- 15. Nedoszytko B, Szczerkowska-Dobosz A, Zablotna M, Glen J, Rebala K, et al. (2007) Associations of promoter region polymorphisms in the tumour necrosis factor-alpha gene and early-onset psoriasis vulgaris in a northern Polish population. Br J Dermatol 157: 165–167. [DOI] [PubMed] [Google Scholar]
- 16. Ruiz DG, Azevedo MN, Santos OL (2012) Psoriatic arthritis: a clinical entity distinct from psoriasis? Rev Bras Reumatol 52: 630–638. [PubMed] [Google Scholar]
- 17. Giardina E, Huffmeier U, Ravindran J, Behrens F, Lepre T, et al. (2011) Tumor necrosis factor promoter polymorphism TNF*-857 is a risk allele for psoriatic arthritis independent of the PSORS1 locus. Arthritis Rheum 63: 3801–3806. [DOI] [PubMed] [Google Scholar]
- 18. Magalhaes RF, Biral AC, Pancoto JA, Donadi EA, Mendes CJ, et al. (2010) Human leukocyte antigen (HLA) and single nucleotide polymorphisms (SNPs) tumor necrosis factor (TNF)-alpha -238 and -308 as genetic markers of susceptibility to psoriasis and severity of the disease in a long-term follow-up Brazilian study. Int J Dermatol 49: 1133–1140. [DOI] [PubMed] [Google Scholar]
- 19. Settin A, Hassan H, El-Baz R, Hassan T (2009) Association of cytokine gene polymorphisms with psoriasis in cases from the Nile Delta of Egypt. Acta Dermatovenerol Alp Panonica Adriat 18: 105–112. [PubMed] [Google Scholar]
- 20. Popa OM, Bojinca M, Bojinca V, Dutescu MI, Meirosu M, et al. (2011) A pilot study of the association of tumor necrosis factor alpha polymorphisms with psoriatic arthritis in the romanian population. Int J Mol Sci 12: 5052–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reich K, Huffmeier U, Konig IR, Lascorz J, Lohmann J, et al. (2007) TNF polymorphisms in psoriasis: association of psoriatic arthritis with the promoter polymorphism TNF*-857 independent of the PSORS1 risk allele. Arthritis Rheum 56: 2056–2064. [DOI] [PubMed] [Google Scholar]
- 22.Wells GA, Shea B, O'Connell D, Peterson J, Welch V, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. ottawa hospital research institute website. Available: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 2013 Apr 22.
- 23. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 24. Thompson SG, Higgins JP (2002) How should meta-regression analyses be undertaken and interpreted? Stat Med 21: 1559–1573. [DOI] [PubMed] [Google Scholar]
- 25. Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Balding J, Kane D, Livingstone W, Mynett-Johnson L, Bresnihan B, et al. (2003) Cytokine gene polymorphisms: association with psoriatic arthritis susceptibility and severity. Arthritis Rheum 48: 1408–1413. [DOI] [PubMed] [Google Scholar]
- 27. Gonzalez S, Martinez-Borra J, Lopez-Vazquez A, Garcia-Fernandez S, Torre-Alonso JC, et al. (2002) MICA rather than MICB, TNFA, or HLA-DRB1 is associated with susceptibility to psoriatic arthritis. J Rheumatol 29: 973–978. [PubMed] [Google Scholar]
- 28. Al-Heresh AM, Proctor J, Jones SM, Dixey J, Cox B, et al. (2002) Tumour necrosis factor-alpha polymorphism and the HLA-Cw*0602 allele in psoriatic arthritis. Rheumatology (Oxford) 41: 525–530. [DOI] [PubMed] [Google Scholar]
- 29. Hohler T, Grossmann S, Stradmann-Bellinghausen B, Kaluza W, Reuss E, et al. (2002) Differential association of polymorphisms in the TNFalpha region with psoriatic arthritis but not psoriasis. Ann Rheum Dis 61: 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gonzalez S, Brautbar C, Martinez-Borra J, Lopez-Vazquez A, Segal R, et al. (2001) Polymorphism in MICA rather than HLA-B/C genes is associated with psoriatic arthritis in the Jewish population. Hum Immunol 62: 632–638. [DOI] [PubMed] [Google Scholar]
- 31. Hamamoto Y, Tateno H, Ishida T, Muto M (2000) Lack of association between promoter polymorphism of the tumor necrosis factor-alpha gene and psoriatic arthritis in Japanese patients. J Invest Dermatol 115: 1162–1164. [DOI] [PubMed] [Google Scholar]
- 32. Hohler T, Kruger A, Schneider PM, Schopf RE, Knop J, et al. (1997) A TNF-alpha promoter polymorphism is associated with juvenile onset psoriasis and psoriatic arthritis. J Invest Dermatol 109: 562–565. [DOI] [PubMed] [Google Scholar]
- 33. Baran W, Szepietowski JC, Mazur G, Baran E (2006) A - 308 promoter polymorphism of tumor necrosis factor alpha gene does not associate with the susceptibility to psoriasis vulgaris. No difference either between psoriasis type I and type II patients. Acta Dermatovenerol Alp Panonica Adriat 15: 113–118. [PubMed] [Google Scholar]
- 34. Mossner R, Kingo K, Kleensang A, Kruger U, Konig IR, et al. (2005) Association of TNF -238 and -308 promoter polymorphisms with psoriasis vulgaris and psoriatic arthritis but not with pustulosis palmoplantaris. J Invest Dermatol 124: 282–284. [DOI] [PubMed] [Google Scholar]
- 35. Long F, Sun C, Deng D, Zhou X, Li XP, et al. (2004) TNF-238A is associated with juvenile onset psoriasis in patients of Han population in Southwest China. J Dermatol Sci 36: 109–111. [DOI] [PubMed] [Google Scholar]
- 36. Tsunemi Y, Nishibu A, Saeki H, Oyama N, Nakamura K, et al. (2003) Lack of association between the promoter polymorphisms at positions -308 and -238 of the tumor necrosis factor alpha gene and psoriasis vulgaris in Japanese patients. Dermatology 207: 371–374. [DOI] [PubMed] [Google Scholar]
- 37. Kim TG, Pyo CW, Hur SS, Kim YK, Hwang HY, et al. (2003) Polymorphisms of tumor necrosis factor (TNF) alpha and beta genes in Korean patients with psoriasis. Arch Dermatol Res 295: 8–13. [DOI] [PubMed] [Google Scholar]
- 38. Chang YT, Tsai SF, Lee DD, Shiao YM, Huang CY, et al. (2003) A study of candidate genes for psoriasis near HLA-C in Chinese patients with psoriasis. Br J Dermatol 148: 418–423. [DOI] [PubMed] [Google Scholar]
- 39. Reich K, Mossner R, Konig IR, Westphal G, Ziegler A, et al. (2002) Promoter polymorphisms of the genes encoding tumor necrosis factor-alpha and interleukin-1beta are associated with different subtypes of psoriasis characterized by early and late disease onset. J Invest Dermatol 118: 155–163. [DOI] [PubMed] [Google Scholar]
- 40. Craven NM, Jackson CW, Kirby B, Perrey C, Pravica V, et al. (2001) Cytokine gene polymorphisms in psoriasis. Br J Dermatol 144: 849–853. [DOI] [PubMed] [Google Scholar]
- 41. Reich K, Westphal G, Schulz T, Muller M, Zipprich S, et al. (1999) Combined analysis of polymorphisms of the tumor necrosis factor-alpha and interleukin-10 promoter regions and polymorphic xenobiotic metabolizing enzymes in psoriasis. J Invest Dermatol 113: 214–220. [DOI] [PubMed] [Google Scholar]
- 42. Jacob N, Ruschendorf F, Schmitt-Egenolf M, Hennies HC, Friedl G, et al. (1999) Promoter polymorphism at -238 of the tumor necrosis factor alpha gene is not associated with early onset psoriasis when tested by the transmission disequilibrium test. J Invest Dermatol 112: 514–516. [DOI] [PubMed] [Google Scholar]
- 43. Vasilopoulos Y, Manolika M, Zafiriou E, Sarafidou T, Bagiatis V, et al. (2012) Pharmacogenetic analysis of TNF, TNFRSF1A, and TNFRSF1B gene polymorphisms and prediction of response to anti-TNF therapy in psoriasis patients in the Greek population. Mol Diagn Ther 16: 29–34. [DOI] [PubMed] [Google Scholar]
- 44. Mekinian A, Tamouza R, Pavy S, Gestermann N, Ittah M, et al. (2011) Functional study of TNF-alpha promoter polymorphisms: literature review and meta-analysis. Eur Cytokine Netw 22: 88–102. [DOI] [PubMed] [Google Scholar]
- 45. Niewold TB, Kariuki SN, Morgan GA, Shrestha S, Pachman LM (2009) Elevated serum interferon-alpha activity in juvenile dermatomyositis: associations with disease activity at diagnosis and after thirty-six months of therapy. Arthritis Rheum 60: 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lv K, Chen R, Cai Q, Fang M, Sun S (2006) Effects of a single nucleotide polymorphism on the expression of human tumor necrosis factor-alpha. Scand J Immunol 64: 164–169. [DOI] [PubMed] [Google Scholar]
- 47. Butter F, Davison L, Viturawong T, Scheibe M, Vermeulen M, et al. (2012) Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet 8: e1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA 2009 Checklist.
(DOC)