Abstract
Seedling establishment is a critical phase in the life of plants when they are the most vulnerable to environment. Growth arrest at post-germinative stage under stress is the major adaptive strategy to help germinating seedlings to survive a spectrum of stressful conditions. ABA signaling is the key pathway to control stress-induced developmental arrest. However, mechanisms controlling the phase transition under abiotic stress are not fully understood. Here, we described miR172b as a new key regulator controlling transition of germinating seedlings from heterotrophic to autotrophic growth under osmotic stress in Arabidopsis. We showed that miR172b and its target SNZ were co-expressed during early seedling development. Expression of miR172b and SNZ was low after radicle emergence and sharply increased at the checkpoint to autotrophic development under normal conditions. Interestingly, activation of miR172b and SNZ was completely abolished by ABA and osmotic stress. miR172b overexpression and snz-1 exhibited increased sensitivity to ABA and osmotic stress during specific post-germinative stage, and resulted in higher expression of ABI3, ABI5 and downstream genes, such as Em6 and RAB18, than wild type under ABA treatment. Our results revealed that miR172b is a critical regulator specifically controlling cotyledon greening during post-germinative growth by directly targeting SNZ under ABA treatment and osmotic stress.
Introduction
Seed germination, post-germinative growth and subsequent seedling establishment are central to successful stand establishment and population maintenance in plants, which are of great importance to agriculture and ecology [1]. The appearance of the radicle represents the end of germination and the beginning of the post-germinative stage, a period that ends when autotrophic growth is established. Over more than 100 years, extensive studies (> 25000 publications) have been made on the physiological and molecular mechanisms of seed germination using various plant species, including Arabidopsis [2]. Many key biological processes and regulators involved in metabolic regulation, hormonal regulation and molecular regulation have been identified [2]. In contrast, control of post-germinative growth and the phase transition from post-germinative growth to seedling establishment remain largely unclear, although they are as important as seed germination for successful plant establishment.
Compelling evidence has shown that the checkpoint from heterotrophic to autotrophic development at the post-germinative stage is genetically controlled, and that this phase transition is also highly vulnerable to various stresses including osmotic stress [2]. In order to prevent the adverse effects of environmental perturbations on the transition from post-germinative stage to subsequent seedling establishment, genetic and molecular mechanisms regulating adaptive response during the phase transition have evolved. It has been shown that ABA signaling plays key roles in both post-germinative growth arrest under stress and subsequent seedling establishment [1]. Mutations of the key components in the ABA signaling pathway, including receptors [3], protein phosphatase 2C (PP2C) family genes (ABI1 and ABI2) [4]–[7], and transcription factors ABI3 and ABI5 [8], result in altered sensitivity to ABA during seed germination and the post-germinative stage [5], [9], [10]. Among these, ABI3 and ABI5 are expressed in a narrow developmental window and control the post-germination developmental arrest checkpoint. For example, the ABI5 level was undetectable on day 1 after stratification, but progressively increased thereafter and reached the highest level on day 4 [1]. Activation of ABI3 and ABI5 is required for maintaining the germinating seedlings at the post-germinative stage (embryonic state) to survive periods of severe drought stress [10].
microRNAs (miRNAs) are small, single-stranded, non-coding RNAs, which down-regulate their target genes at the post-transcriptional level through miRNA cleavage or translational repression [11], [12]. Several miRNAs have been linked with ABA responses and post-germination development. A prominent example in Arabidopsis is miR156, which regulates post-germinative development through modulating the mRNA of its target SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE13 (SPL13) [13]. Expression analysis suggests that this function of miR156 may be achieved through regulation of miR172 and SCHNARCHZAPFEN (SNZ), an AP2-like gene targeted by miR172 in Arabidopsis [14]. miR172 has been proved as a master regulator of the transition from vegetative growth to reproductive development [15], [16]. However, there are no reports to experimentally address the role of miR172 in stress tolerance. Here, we show that miR172b becomes induced at the checkpoint to autotrophic growth and controls the switch to autotrophic development in plants grown under normal conditions. Importantly, we report that miR172b failed to be turned on in the plants treated with ABA and osmotic stress to inhibit greening of cotyledon, and to prevent overly growth arrest from overexpression of ABI3 and ABI5 at post-germination stage. Further, we show that miR172b mediates its function by partially targeting SNZ. Together, these data identify miR172b as a master regulator in stress-induced developmental arrest and adaptation to stress.
Materials and Methods
Plant materials and growth conditions
Seeds of Columbia ecotype (Col-0), 35S: miR172b [16], snz-1 [17], abi5-8 (SALK_013163) [18], snz-1 overexpressing SNZ and snz-1abi5-8 double mutants (Table S5) were surface-sterilized with 50% (v/v) bleach for 5 min and washed at least four times with sterile water. Sterile seeds were plated on MS medium plus 2% sucrose or the medium containing ABA (mixed isomers, Sigma), NaCl and mannitol. Plates were routinely kept for 2 days in the dark at 4°C to break dormancy (stratification) and transferred thereafter to a tissue culture room under constant light at 22°C with a 16/8 h light/dark regime.
Vector construction and Arabidopsis transformation
To generate transgenic plants overexpressing SNZ under the control of cauliflower mosaic virus 35S promoter in snz-1 mutant background, we cloned the coding region of SNZ by polymerase chain reaction (PCR) which was then inserted into the pTF101 vector containing a bar gene. The primers used for the construction are listed in Table S4. Transformation of Arabidopsis was performed using the floral dip method using Agrobacterium tumefaciens strain EHA105. Seeds were harvested and plated on the MS medium containing BASTA to identify transgenic plants. Lines containing single insertions were selected on the basis of the segregation ratio of 3∶1 and homozygous lines were used for the gene expression assay and phenotypic analysis.
Genetic analysis
Homozygous snz-1 was crossed with abi5-8 to generate the snz-1abi5-8 double mutant. Because both mutants contain T-DNA insertions, we performed PCR to identify the homozygous lines containing both mutations in SNZ and ABI5 using the specific primers (LP1 and RP1, or RP1 and T-DNA left-border primer LBb1.3 for abi5-8; LP2 and RP2, or RP2 and LBb1.3 for snz-1). The primers used for genotyping are listed in Table S4.
Germination and seedling development assays under ABA and osmotic stress treatments
For germination assays, at least 60 seeds from 35S: miR172b [16], snz-1 [17] and Col-0 plants were sown onto MS triplicate plates supplemented with 20 gL−1 sucrose, 8 gL−1 agar, and different concentrations of ABA (0, 0.2, 0.4, 0.8, 1 and 1.5 µM), NaCl (0, 50, 75, 100 and 150 mM), Mannitol (0, 100, 200, 300 and 400 mM). After 2-day stratification at 4°C in darkness, the plates were incubated in a growth chamber at 22±2°C under long-day conditions (16 h light/8 h dark). Germination was defined as the emergence of 1 mm or more of the radicle from the seed coat as described previously [19]. The degree of greening is defined as the extent of expanded cotyledons that are capable of turning green after transferred to light. The greening rate was expressed as the percentage of the germinating seedlings turning green.
All experiments were repeated three times with three replicates (n>60) in each. The data reported in the figures are means of the values with standard deviation (SD).
Gene expression analysis
Seeds were kept in darkness at 4°C for 2 days with or without ABA (0.4 µM or 5 µM) and then transferred to constant light at 22°C. Total RNA was extracted from plant tissue using Trizol reagent (Invitrogen) according to the manufacturer's instructions. 2 mg of total RNA was DNase I-treated and single-stranded cDNA was synthesized using oligo (dT) and the RevertAid First Strand cDNA Synthesis Kit (Invitrogen). The sequences of primers that were used for gene expression analysis are listed in Table S3. Quantitative real-time PCR was run on a ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA) using the Platinum SYBR Green qPCR Supermix-UDG (Invitrogen). The expression level was normalized against the geometric mean of GAPC gene, as a reference gene expressed stably. All the experiments were performed three times, each with three replicates. Error bars denote SD.
Electrophoretic mobility shift assays (EMSA)
Recombinant MBP-SNZ fusion protein was expressed and purified using Amylose Resin (NEB)according to the manufacturer's introduction. The binding activity of the protein was analyzed using an oligonucleotide containing 6 copies of RAV1-A motif, 5′-CACCTG(CAACA)6-3′ (wRAV1-A), labeled with biotin at the 3′ end (Invitrogen, USA). An oligonucleotide containing mutated RAV1-A motif, 5′-CACCTG(CGGTA) 6-3′ (mRAV1-A), labeled with biotin at the 3′ end (Invitrogen, USA) was used as a control. The specific probes containing RAV1-A motifs corresponding to the fragment of ABI5 promoter were synthesized and labeled with biotin at the 3′ end as mentioned above. The sequences of probes used for EMSA are listed in Table S4. EMSA was performed using LightShift® Chemiluminescent EMSA Kit according to the manufacturer's protocol. Complementary oligonucleotide pairs were annealed to make double-stranded and biotin-labeled probes (5 pmol) by mixing in a buffer (10 mM Tris and 1 mM EDTA), boiling for 5 min, and cooling slowly overnight. Unlabeled complementary oligonucleotide pairs were also annealed to make double-stranded competitor probes (50 and 250 times the amount of biotin-labeled ABI5 probes). EMSA reaction solutions were prepared by adding the following components in order according to the manufacturer's protocol (LightShift Chemilu minescent EMSA kit): binding buffer, 50 ng poly (dI-dC), 2.5% glycerol, 0.06% NP-40, 5 mM MgCl2, 19 mg BSA, proteins (28 µg), competitor and biotin-labeled probes. Reaction solutions were incubated for 30 min at room temperature. The protein-probes mixture was separated in a 8% poly-acrylamide gel and transferred to a Biodyne B Nylon membrane. Migration of biotin-labeled probes was detected by using streptavidin-horseradish peroxidase conjugates that bind to biotin and chemiluminescent substrate according to the manufacturer's protocol. This experiment was performed three times.
Statistic analysis
All the data were analyzed using software GraphPad Prism 5. The Student's t-test was performed and the statistically significant treatments were marked with ‘*’ (P<0.05), ‘**’ (P<0.01) and ‘***’ (P<0.001).
Results
miR172b controls the transition to autotrophic development and stress tolerance during osmotic stress
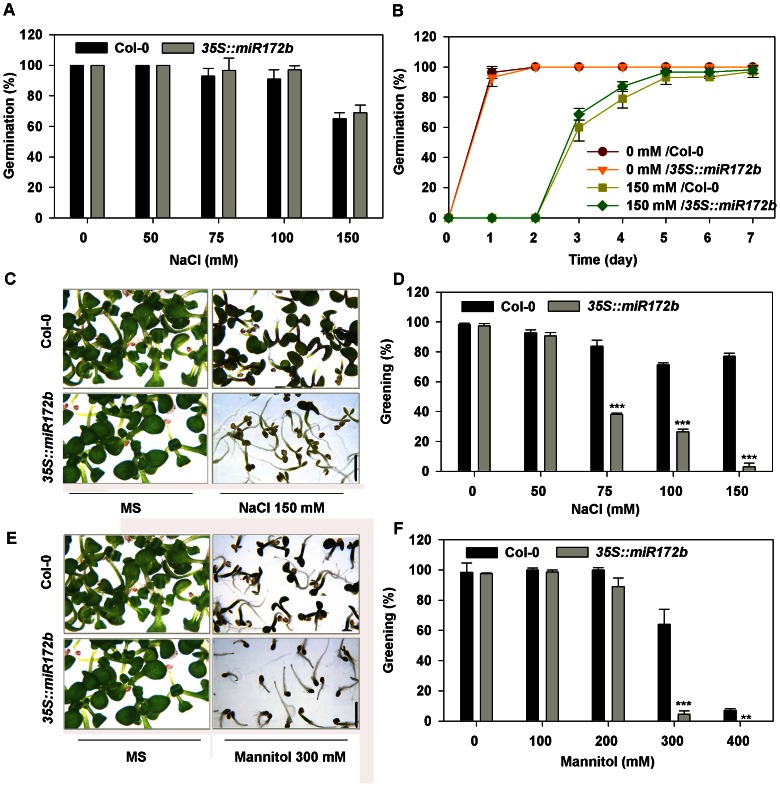
After seed germination, germinating seedlings proceed through a critical developmental stage referred to as the transition from heterotrophic to autotrophic growth that can be tracked by cotyledon greening. Thus, cotyledon greening is thought to be the critical checkpoint in seedling establishment. When plant seeds are germinated under stress conditions, the developmental program to autotrophic growth is interrupted and stress reinstates the embryonic stage [2]. The resulting developmental arrest before autotrophic development is established could allow the germinating seedlings to maintain at quiescent state and survive a period of stress conditions [1]. As it has recently been shown that miR172 mediates post-germinative growth in Arabidopsis grown under normal conditions [13], we sought to determine whether miR172 may have a role in stress response and developmental checkpoint control under osmotic stress. To this end, stress response of plants overexpressing miR172b during early development was examined. No significant differences during seed germination were found between the miR172b overexpressors and the wild type under both normal conditions and salt stress (Figure 1A and B). However, with increasing concentrations of salt stress, miR172b overexpressors displayed a significant reduction in the rate of cotyledon greening compared with that of the wild type (Figure 1C and D). When miR172b overexpressors were exposed to moderate levels of salt (75 to 100 mM NaCl), a more than 50% reduction in greening rate of germinating miR172b overexpression seedlings was observed over a 7 day period. At higher level of NaCl (150 mM NaCl) that allowed about 75% of germinating wild type seedlings to turn green, very few (4% of total) of miR172b overexpressors turned green. No altered salt sensitivity was observed when greened miR172b overexpression seedlings grown under normal conditions were treated with salt stress (Figure S1A, B and E). Salt stress is caused by osmotic effects or ion effect [20]. To investigate whether the hypersensitivity of the miR172b overexpressors to salt is also caused by osmotic stress instead of ion-specific effect, we analyzed the response of 35S: miR172b transgenic plants under mannitol, LiCl and KCl treatments. The results showed that miR172b overexpressors also exhibited increased sensitivity in response to high concentrations of mannitol during early development (Figure 1E and F), but no significant differences were found during seed germination and greening under LiCl and KCl treatments (Figure S3). These data indicate that miR172b regulates general osmotic stress-induced developmental arrest and is specifically required for cotyledon greening, which is an indicator for the successful transition from heterotrophic to autotrophic growth under osmotic stress.
Figure 1. 35S:miR172b transgenic plants are hypersensitive to salt and mannitol at post-germination stages.
(A) Salt dose–response analysis of Col-0 and 35S:miR172b during seed germination at 5 days after stratification. (B) A germination time course on medium containing 150 mM NaCl. (C) 35S: miR172b plants showed delayed greening under salt treatments; Photographs were taken 7 days after stratification. Bar = 5 mm. (D) Percentage of Col-0 and 35S: miR172b turning green 7 days after stratification under salt. (E) Greening of 35S: miR172b plants was more sensitive to mannitol; photographs were taken 5 days after stratification. Bar = 5 mm. (F) Percentage of Col-0 and 35S: miR172b becoming green at 7 days after stratification under mannitol treatment. All the experiments were performed three times with three replicates. Error bars denote±SD. Student's t test was performed, and the statistically significant treatments are marked with asterisks. (*) P<0.05, (**) P<0.01 and (***) P<0.001.
miR172b mediates ABA induced-developmental arrest during the transition to autotrophic development
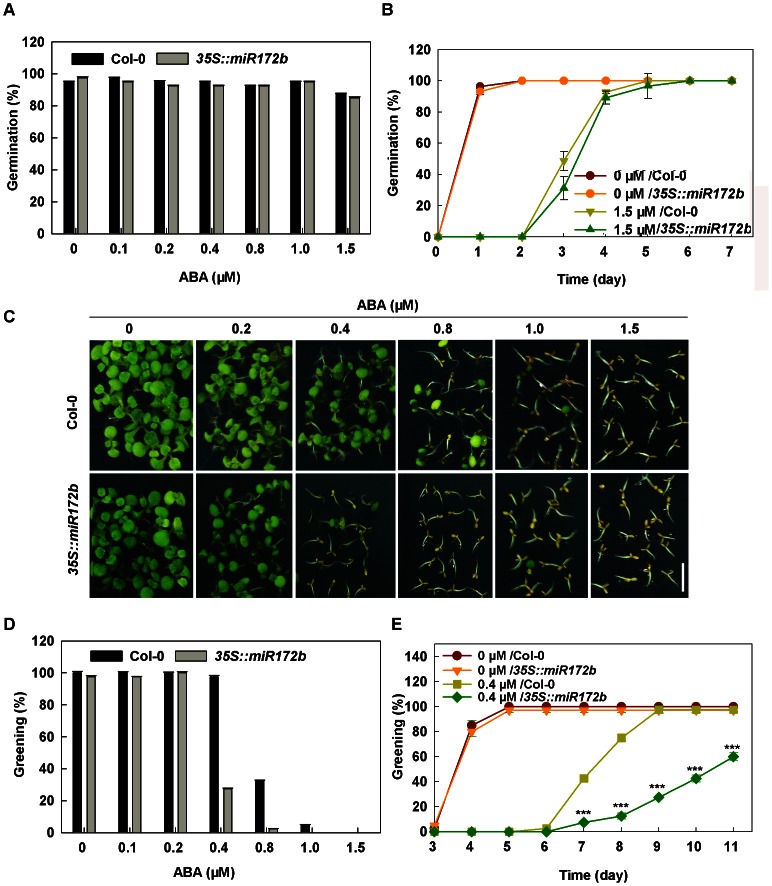
Because ABA is previously shown to play a prominent role in regulation of post-germinative growth arrest under osmotic stress in Arabidopsis [21], [22], we examined the ABA response of miR172b overexpressors during germination and early development stages. We found that miR172b overexpression did not affect germination rate under normal conditions and ABA treatments (Figure 2A and B). Alteration in miR172b expression level also did not influence growth inhibition of the young seedlings caused by exogenous ABA after the transition from heterotrophic to autotrophic development was completed (Figure S1C and D). However, as expected, cotyledon greening of germinating seedlings overexpressing miR172b was dramatically affected when germinated on the medium containing various concentrations of ABA compared with that of the wild type (Figure 2C-E). When exposed to low concentrations of ABA (<0.4 µM), the germinating seedlings overexpressing miR172b displayed delayed greening, although their overall greening rate was the same as that of the wild type at the end of the seven day growing period (Figure 2C and D). In the presence of 0.4 µM ABA, the percentage of germinated seedlings overexpressing miR172b turning green was reduced by more than 70% during the growing period (Figure 2D). The timing for germinated miR172b overexpressors turning green was also dramatically postponed and the greening rate was lower than 30% when the wild type reached 100% on day 9 after stratification (Figure 2E). When germinated on a medium containing ABA concentrations greater than 0.8 µM, cotyledon greening of germinated miR172b overexpressors was completely blocked (Figure 2C and D). However, when the seedlings with yellowish cotyledons on 0.8 µM ABA were transferred onto MS medium, all of them turned green (Figure S2). Together, these results suggest that miR172b may regulate stress-induced developmental arrest and stress tolerance at the post-germinative/seedling establishment stage through the ABA signaling pathway.
Figure 2. 35S: miR172b plants are hypersensitive to ABA in post-germination stages.
(A) ABA dose–response analysis of seed germination for Col-0 and 35S: miR172b at 5 days after stratification. (B) A germination time course on medium containing 1.5 µM ABA. (C) 35S: miR172b plants showed delayed greening under ABA treatments; photographs were taken 9 days after stratification. Bar = 5 mm. (D) ABA dose–response analysis of greening for Col-0 and 35S: miR172b at 9 days after stratification. (E) A greening time course on medium containing 0.4 µM ABA. All the experiments were performed three times with three replicates. Error bars represent±SD. Student's t test was performed, and the statistically significant treatments are marked with asterisks. (*) P<0.05, (**) P<0.01 and (***) P<0.001.
miR172b expression is down-regulated by osmotic stress and ABA
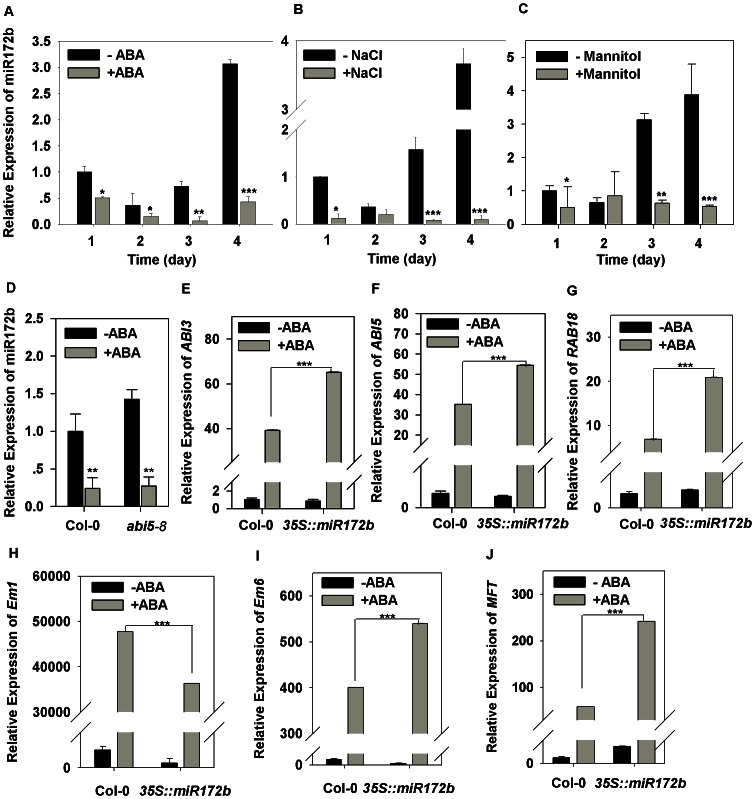
As the levels of miR172 expression are critical for regulating phase transition during plant development, and overexpression of miR172b results in post-germinative developmental arrest under osmotic stress and ABA (Figure 1 and Figure 2), we therefore hypothesized that osmotic stress and ABA may regulate levels of miR172b expression to reprogram the developmental program allowing phase transition changes to occur. To this end, we first analyzed expression of miR172b in response to ABA and osmotic stress induced by NaCl treatment. In the absence of ABA, low levels of miR172b expression were detected in the first two days after stratification, and expression of miR172b decreased and reached its lowest level on day 2 (Figure 3A). miR172b expression then increased starting from day 3 after stratification and a sharp increase in its expression was detected on day 4 (Figure 3A). This observation of a sharp increase of miR172 is consistent with previous reports [16], [23], [24]. Activation of miR172b at the switch point suggests that miR172b plays a pivotal role in regulating the heterotrophic/autotrophic checkpoint during early development.
Figure 3. miR172b regulates post-germinative developmental arrest induced by abiotic stress through the ABA dependent pathway.
Expression analysis of miR172b under ABA (5 µM) (A), NaCl (100 mM) (B) and mannitol (300 mM) (C) treatment in the first four days after stratification treatment in the first four days after stratification. (D) miR172b expression in wild-type (Col-0) and abi5-8 treated without or with 0.4 µM ABA. Expression analysis of ABI3 (E), ABI5 (F), RAB18 (G), Em1 (H), Em6 (I) and MFT (J) in germinating seeds of the wild type and 35S:miR172b in the third day after stratification with or without 0.4 µM ABA. The expression level was normalized against the reference gene GAPC, and all the experiments were performed three times with three replicates. Error bars denote±SD. Student's t test was performed, and the statistically significant treatments are marked with asterisks. (*) P<0.05, (**) P<0.01 and (***) P<0.001.
When wild type seeds were germinated on the medium containing 5 µM ABA, levels and patterns of miR172b expression were strikingly different (Figure 3A). In the first 2 days after stratification, the levels of miR172b expression were greatly down-regulated by ABA, although miR172b still maintained a downward trend. The decreasing trend in miR172b expression stopped with the lowest expression level on day 3. Most strikingly, we found that on day 4 after stratification, when miR172b was dramatically activated under normal conditions, miR172b expression after ABA treatment still remained at a very low level equal to that on day 2 after stratification under normal conditions. A very similar pattern of miR172b expression was observed of the wild type treated with osmotic stress (Figure 3B and C). These results suggest that down-regulation of miR172b expression by osmotic stress and ABA during early seedling development, in particular the total block of miR172b activation on day 4 after stratification, may contribute to developmental arrest at the post-germinative stage and heterotrophic/autotrophic transition failure under osmotic stress and ABA treatment.
As the expression of miR172b is negatively regulated by osmotic stress and ABA, we sought to determine whether the miR172b promoter contains any cis elements responsive to osmotic stress and/or ABA. Bioinformatic analysis of the miR172b promoter identified several ABA responsive cis elements, such as ABRE, Couple element 3 and cis elements related to stress response (e.g. MYB1, GAREAT and WRKY) in the promoter region of miR172b (Table S1) [25]–[28]. These data support a critical role for miR172b in control of transition to the autotrophic phase and in ABA-mediated stress tolerance during post-germination growth under osmotic stress.
miR172b functions in ABA signaling during post-germinative development
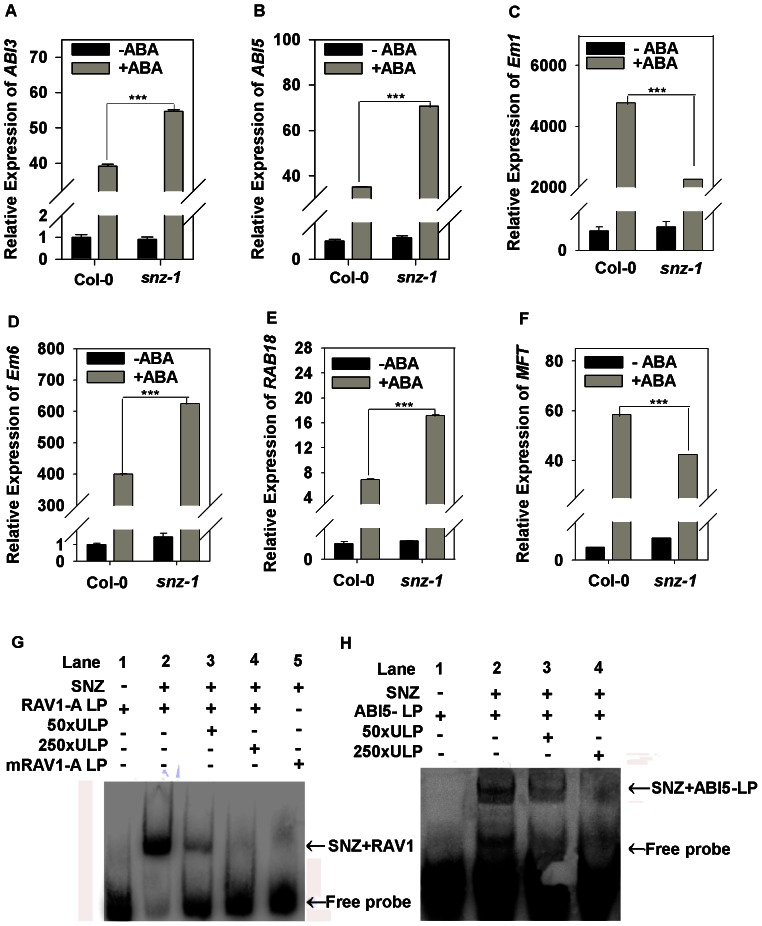
Because ABI3 and ABI5 play a pivotal role in post-germinative arrest checkpoint control [9], [29], we then analyzed the relationship between miR172b and ABI3 and ABI5. We found that the levels and patterns of miR172b expression were not significantly changed in abi5-8 (Figure 3D) [18], a loss of function mutant showing decreased sensitivity to ABA inhibition. Next, we investigated whether the level of miR172b affects transcript level of ABI3 and ABI5 during the early developmental stage. Transcription levels of ABI3 and ABI5 were analyzed in 35S:miR172b transgenic plants in the absence or presence of ABA. In the absence of ABA, ABI3 and ABI5 transcript levels of 35S:miR172b transgenic plants were comparable to those of the wild type (Figure 3E and F). However, the induction levels of both ABI3 and ABI5 were significantly higher in the germinating plants overexpressing miR172b than those of the wild type in the presence of ABA. Expression of ABI3 and ABI5 increased 39-fold and 35-fold in the wild type at 3 day after ABA treatment, in sharp contrast, a 76-fold and 69-fold increase in ABI3 and ABI5 expression occurred by the time of ABA treatment in 35S:miR172b transgenic plants (Figure 3E and F). It has been shown that overexpression of ABI3 and ABI5 greatly increases sensitivity to ABA and osmotic stress [10]. Therefore, it is possible that failure in miR172b activation at the heterotrophic/autotrophic transition point under ABA and osmotic stress may limit ABI3 and ABI5 overexpression and avoid triggering an overreaction of germination seedlings to stress during post-germinative development. To confirm the above hypothesis, we subsequently analyzed the expression of several ABA-responsive genes acting downstream of ABI3 and ABI5 during ABA-induced post-germinative growth arrest in the wild type and 35S:miR172b transgenic line [30], [31]. Among them, LEA genes Em1 and Em6 are directly targeted by ABI5 [30], and RAB18 is regulated by ABI5. As shown in Figure 3G-I, all the genes were up-regulated after ABA treatment in the wild type, and the expression of Em6 and RAB18 was clearly up-regulated in 35S:miR172b compared with wild type control. Previous study has shown that MOTHER OF FT AND TFL1 (MFT) is also directly regulated by ABI3 and ABI5 during seed germination [32]. We found that MFT was induced by ABA at dramatically higher level in 35S:miR172b germinating seedlings (Figure 3J). These results indicate that miR172b may regulate growth arrest of germinated embryos through ABI3/ABI5-dependent signaling pathway.
SNZ was co-expressed with miR172b in the absence and presence of salt stress and ABA during early development
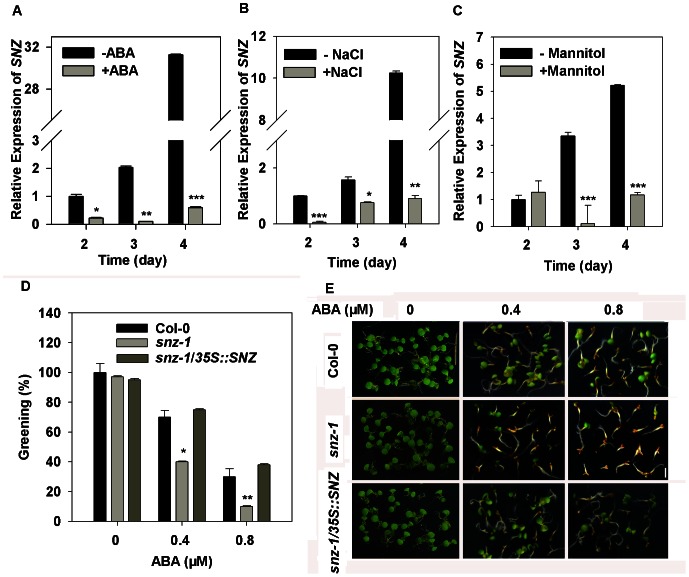
In Arabidopsis, miR172 regulates developmental phase transition through targeting 6 APETALA2-LIKE (AP2-like) transcription factors: AP2, TOE1, TOE2, TOE3, SMZ, SNZ [14], [16], [23], [33]. We presumed that miR172b must regulate stress-induced growth arrest at the post-germinative stage through its target(s). To this end, we first searched the expression profiles of 6 target genes in response to osmotic stress and ABA from the public microarray database (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). We found that among the targets, SNZ and TOE3 were responsive to ABA and osmotic stress/drought in 7 and 18 day-old seedlings (Figure S4A and B). To verify whether SNZ and TOE3 mediate osmotic stress/ABA-induced post-germinative growth arrest, we first analyzed the expression of SNZ and TOE3 in response to ABA during post-germinative development in the wild type. As shown in Figure S5A, expression levels of TOE3 on days 2 and 3 after stratification were low, and an approximately 8-10-fold increase in its expression was detected on day 4 regardless of whether or not they were treated with ABA. In sharp contrast, SNZ showed a quite similar expression pattern to that of miR172b. In the absence of ABA, the level of SNZ expression was the lowest on day 2 after stratification, then increased on day 3 and showed a sharp elevation on day 4 (Figure 4A). However, in the presence of ABA, expression of SNZ was greatly reduced, and remarkably, the sharp elevation of SNZ transcript on day 4 completely disappeared (Figure 4A). A similar pattern in SNZ expression was observed in the germinating seedlings under osmotic stress (Figure 4B and C). In addition, overexpression of miR172b greatly reduced the expression of SNZ under both normal conditions and ABA treatment, although the decreasing trend in its expression remained in response to ABA (Figure S5B). Moreover, promoter analysis revealed several ABA cis elements in the promoter region of SNZ (Table S2) [25], [26], [34]-[36]. Taken together, these results suggest that SNZ may function as a key target to regulate stress/ABA-induced post-germinative growth arrest. As compelling results have shown that miR172 generally co-expresses with its targets and regulates its target genes at a posttranslational level [23], [33], [37], it is conceivable that miR172b may also regulate stress/ABA-induced post-germinative growth arrest and stress tolerance by modulating SNZ at a posttranslational level.
Figure 4. miR172b regulates post-germinative growth arrest by directly targeting SNZ.
Wild-type seeds were germinated on MS with or without treatments, and materials were collected at 2 to 4 days after stratification. (A) SNZ expression in response to ABA (5 µM); (B) SNZ expression in response to NaCl (100 mM); (C) SNZ expression in response to mannitol (300 mM); (D) and (E) snz-1 seedlings showed delayed greening under ABA treatments; Photographs were taken 7 days after stratification. Bar = 4 mm. All the experiments were performed three times with three replicates. Error bars denote±SD. Student's t test was performed, and the statistically significant treatments are marked with asterisks. (*) P<0.05, (**) P<0.01 and (***) P<0.001.The expression level was normalized against the reference gene GAPC. All the experiments were performed three times with three replicates. Error bars denote±SD.
miR172b regulates stress/ABA-induced post-germinative growth arrest by partially targeting SNZ
To further prove whether SNZ is a functional target of miR172b during stress/ABA-induced post-germinative arrest, a loss-of-function mutant snz-1 [17] was analyzed. No significant difference was observed between snz-1 and the wild type during seed germination under normal conditions and ABA treatments (Figure S6B). In the absence of ABA, cotyledon greening and appearance of true leaves of snz-1 germinating seedlings were comparable to the wild type (Figure 4D and E, Figure S6C). However, greening of the mutant seedlings was delayed in comparison with wild type in response to increasing concentrations of ABA (Figure 4D and E), suggesting that loss of function in SNZ affects the developmental arrest induced by ABA at a post-germinative stage. To prove the delayed greening phenotype of snz-1 was due to mutation in the SNZ, we generated transgenic snz-1 plants that express SNZ under control of the cauliflower mosaic virus 35S promoter. We isolated the independent transgenic lines showing similar level of SNZ expression to the wild type and found that SNZ expression in snz-1 plants completely restored their ABA sensitive phenotype (Figure 4D, E, Figure S6A and C). This data confirm the role of SNZ during post-germinative development arrest induced by ABA.
However, when the snz-1 mutant was compared with the 35S:miR172b line, we found that miR172b overexpression plants displayed more severe phenotype in response to ABA (Figure 2E). These data indicate that SNZ may be just one of the genes targeted by miR172b to regulate ABA-dependent developmental arrest during the post-germinative stage. Based on ABA induction of TOE3, we speculate that TOE3 may be another potential target of miR172b in this process.
miR172b-SNZ module functions upstream of ABI5 in regulating stress/ABA-induced post-germinative growth arrest
As overexpression of miR172b affected transcript levels of ABI3 and ABI5 during post-germinative development, we predicted that loss of function in SNZ during early development would result in a similar expression pattern of these genes if SNZ is a direct target of miR172b in the process. To test this, expression of these genes in germinating snz-1 seedlings with and without ABA treatment was analyzed. As expected, ABI3 and ABI5 transcript levels were markedly increased (Figure 5A and B). Notably, the relative increase in ABI3 and ABI5 expression in the snz-1 mutant was lower than that in the miR172b overexpressor (Figure 3E and F, Figure 5A and B). We then examined the expression patterns of ABI5-regulated genes, all the genes we examined were up-regulated by ABA in both of the snz-1 mutant and wild type (Figure 5C-F). Among these genes, Em6 and RAB18 expression was also greatly up-regulated by ABA in snz-1 mutant (Figure 5D and E) compared with wild type, which is consistent with the expression pattern in plants overexpressing miR172b (Figure 3G and I). However, MFT expression was down-regulated in snz-1 mutant compared with the wild type, although it was still able to be induced by ABA (Figure 5F). This result indicates that miR172b may regulate post-germinative growth arrest by targeting SNZ via an ABI3/ABI5 dependent pathway.
Figure 5. miR172b regulates post-germinative growth arrest through modulating the expression of ABI3 and ABI5.
Expression of ABI3 (A), ABI5 (B), Em1 (C), Em6 (D), RAB18 (E) and MFT (F) in the wild type and snz-1 seeds on day 3 after stratification with or without 0.4 µM ABA. The expression level was normalized against the reference gene GAPC. Student's t test was performed, and the statistically significant treatments are marked with asterisks. (*) P<0.05, (**) P<0.01 and (***) P<0.001. (G) SNZ interacts with RAV1 motif. Recombinant MBP-SNZ was used in an EMSA with biotin-labeled oligonucleotide containing 6 copies of the RAV1 motif as the probe; and the probe with the mutated RAV1 (mRAV1) was used as a negative control; (H) SNZ interaction with ABI5 promoter fragment containing RAV1 motif. Unlabeled probes were used as competitors to test the binding specificity. Arrows heads indicate the positions of protein-DNA probe complex and free probes, respectively. All the experiments were performed three times with three replicates, and the representative result is shown. Error bars denote±SD.
SNZ and other target genes of miR172b encode AP2-type transcription factors. We assumed that these transcription factors might bind to the promoters of ABA-responsive genes to regulate ABA signaling and response. To this end, we analyzed a 2-kb upstream region of a putative transcription start code for the ABA-response genes. Interestingly, 7 to 13 RAV1-A (CAACA) cis regulatory elements [38], which are the binding sites of AP2-type transcription factors, in the promoter regions of ABI3, ABI5, and their downstream genes Em1, Em6, RAB18 and MFT were identified (Table S6).
To analyze binding activity of SNZ to RAV1-A motif (CAACA) detected in Table S6, we performed electrophoretic mobility shift assays (EMSA) using biotin labeled probe containing RAV1-A motif and MBP-SNZ fusion protein. As shown in Figure 5G, the control lane with only DNA probe showed a single band corresponding to the unbound DNA fragment. However, after adding SNZ protein to the reaction, DNA-SNZ interaction was clearly detected (Figure 5G). By contrast, addition of unlabeled RAV1-A probe resulted in reduced binding activity of MBP-SNZ to the labeled RAV1-A containing oligonucleotide (Figure 5G). To verify the specificity between SNZ protein and RAV1-A motif containing DNA fragment, we replaced the RAV1-A containing probe with a mutated DNA probe (CAACA→CGGTA), and no SNZ-DNA complex was observed. These results indicate that SNZ protein can specifically bind RAV1-A motifs. To determine the physical interactions between SNZ and ABI5 promoters, an EMSA was performed using 31 nt (-1 to -31) promoter fragments of ABI5, which contain the RAV1-A motif, as probes. The results showed that SNZ directly interacted with ABI5 promoter fragment (Figure 5H). By contrast, addition of unlabeled probes inhibited the interactions. The result confirms that SNZ binds to ABI5 promoter, suggesting that SNZ may directly regulate expression of ABI5.
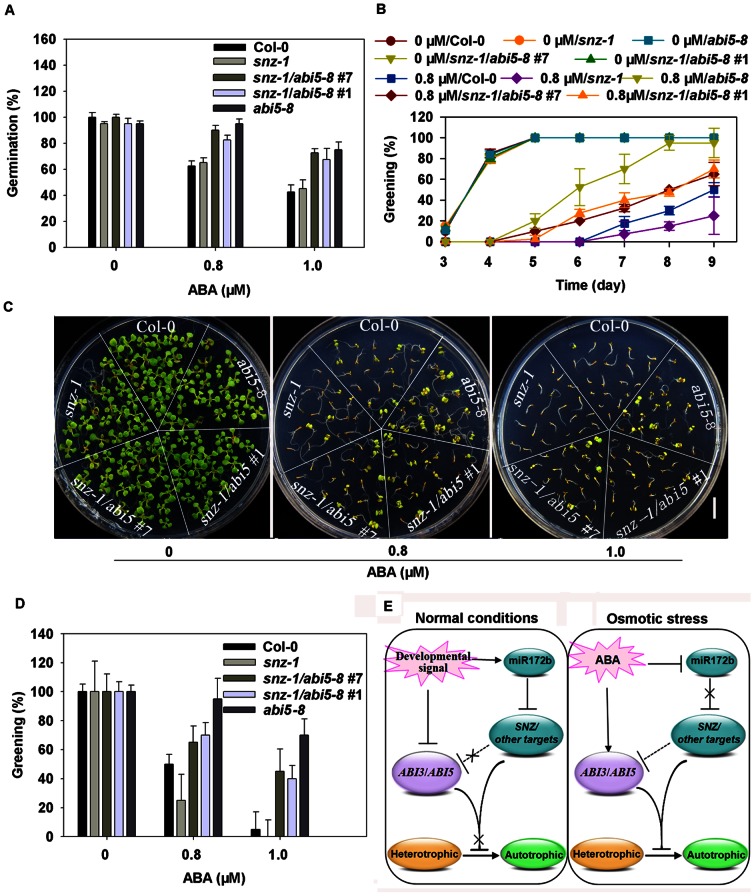
To further prove the genetic relationship between SNZ and ABI5, we performed an epistatic assay for snz-1 and abi5-8. We generated a snz-1abi5-8 double mutant and performed phenotypic analysis. As shown in Figure 6, the snz-1abi5-8 double mutant exhibited ABA insensitive phenotypes similar to the abi5-8 mutant during both germination (Figure 6A) and cotyledon greening stages (Figure 6B, C, D). The result demonstrates that ABI5 functions downstream of SNZ in the ABA signaling pathway and plant response.
Figure 6. Epistatic analysis of relationship between SNZ and ABI5 and a working model for post-germinative growth arrest induced by ABA.
(A) ABA dose–response analysis of Col-0, snz-1, snz-1/abi5-8 #7, snz-1/abi5-8 #1 and abi5-8 during seed germination. (B) A greening time course on medium containing 0.8 µM ABA. (C) abi5-8 mutation is epistatic to snz-1 mutation in ABA-induced inhibition of post-germinative growth; photographs were taken 9 days after stratification. Bar = 10 mm. (D) Greening on medium containing 0, 0.8 and 1.0 µM ABA. All the experiments were performed three times with three replicates. Error bars denote±SD. (E) A hypothetical model for the role of miR172b in controlling phase transition from heterotrophic to autotrophic growth.
Discussion
Plants have two distinct developmental stages: heterotrophic and autotrophic development. Transition from the heterotrophic to autotrophic stage is a key step for plants to become autotrophs and complete their life cycle [39]. As sessile plants have to cope with an adverse environment after germination, it is very important for germinating seedlings to be sensitive to unfavorable conditions and be able to become arrested at the post-germinative growth stage in order to successfully survive a period of stress [40]–[42]. Therefore, transition to the autotrophic phase under various growth conditions must be fine-tuned and strictly regulated. Previous studies have revealed the critical role of the ABA signaling pathway in controlling post-germinative growth arrest and stress tolerance [1], [10], but the molecular mechanism is not fully understood. Here, we identified miR172b as a key molecule that regulates the checkpoint between heterotrophic and autotrophic growth under both normal and stress conditions.
It has been shown that increased expression level of miR172 determines the transition from vegetative growth to reproductive development [23], [33]. In this study, we found that miR172b is dynamically expressed during the post-germination stage, and sharp activation of miR172b at the switch point to the autotrophic stage (Figure 3 A-C) may determine cotyledon greening and subsequent success of the phase transition to phototrophy under normal conditions. Our results that 35S:miR172b showed normal cotyledon greening under normal conditions (Figure 2C-E) support the above hypothesis. The question remaining to be answered is whether miR172b is essential for cotyledon greening under normal condition, and analysis of mutations in miR172b will provide direct evidence. The previous report has shown that the mutated SPL13, a target gene of miR156, led to increased expression of miR172a and miR172b and subsequent delayed appearance of the first pair of true leaves under normal conditions [13], but cotyledon greening and expansion. This observation is also favor of the hypothesis that high levels of miR172b are required for normal cotyledon greening. Seed germination, cotyledon greening and emergence of true leaves are fine-tuned processes regulated by different signaling pathways. It is likely that levels of miR172a and miR172b expression under control of miR156 and its target SPL13 regulate appearance of first pair of true leaves under normal conditions.
When plants are subjected to osmotic stress, osmotic stress-induced modification of miR172b expression, in particular the failure of sharp activation at the switch to the autotrophic mode (Figure 6 E), may reinstate the embryonic program and metabolism, thus preventing damage and death in response to osmotic stress. Under osmotic stress, activation of the ABA signaling pathway is crucial for post-germination growth arrest by induction of ABI3 and ABI5, with maximum expression level on day 4 after stratification. Activation of ABI3 and ABI5 and their level control are necessary for survival of germinating seedlings under stress and regrowth after stress removal. Indeed, overexpression of ABI3 and ABI5 results in phenotypes displaying more severe arrest in response to osmotic stress or ABA [10]. Inhibition of miR172b activation on day 4 after stratification by ABA may function as a positive regulator to control the level of ABI3 and ABI5 under osmotic stress, thus protecting germinating young seedlings from excessive stress. Indeed, overexpression of miR172b results in higher levels of ABI3 and ABI5 and hypersensitivity in response to ABA and stress treatments (Figure 1C and E, Figure 2C, Figure 3E and F). Most importantly, our results demonstrate that SNZ has a sequence-specific DNA-binding activity and can bind to the ABI5 promoter region harboring RAV1 motif (Figure 5H). Further genetic evidence confirmed that SNZ functions upstream of ABI5 and negatively regulates ABI5 and the ABA signaling (Figure 5B-F, Figure 6A-D). The data that overexpression of miR172b results in substantial increases in expression levels of Em6, MFT and RAB18, downstream components of ABI3 and ABI5 also support the above hypothesis (Figure 3G-J) [30], [32], [43]. Our findings suggested that SNZ is involved in transcriptional regulation of ABI5 in ABA mediated plant response to osmotic stress during post-germinative development and seedling establishment. Previously, it has been shown that MFT represses ABI5 transcription during seed germination[32]. Since MFT protein does not have any DNA binding domain, it has been proposed that it may regulate ABI5 expression through other transcription factors[32]. Here we demonstrate a transcription factor SNZ physically binds the ABI5 promoter to negatively regulate its expression. It remains to be determined whether SNZ is involved in guiding MFT to the ABI5 promoter and corporately modulating ABI5 transcription level.
Post-germinative development is a complex biological process, and successful phase transition from heterotrophic to autotrophic growth involves the complex interplay of the developmental and stress response signaling pathways and precise regulation at multiple levels. miRNA has been considered as fine-tuned regulators of the expression levels of their targets and the related biological processes. It is apparent that miR172b-SNZ is an important regulatory module in regulation of ABI5 expression and subsequent post-germinative development of plants in response to abiotic stress. However, we do not exclude the possibility that miR172b may achieve its regulatory function by targeting multiple genes, because loss of function in SNZ alone exhibited less severe phenotypes compared with miR172b overexpressing plants in response to ABA (Figure 2C-E, Figure 4D, E).
Our studies reveal that miR172b may have evolved to fine-tune adaptive regulation of the heterotrophic to autotrophic checkpoint and developmental remodeling in response to adverse environmental conditions in Arabidopsis. However, many questions remain unknown. How is the ABA signal perceived and transmitted to miR172b? How do the multiple target genes of miR172b fine-tuning the post-germinative development and adaptation? Does SNZ function as a MFT interacting partner to repress ABI5 expression? Answering these questions in future research will provide novel insight into the molecular mechanisms underlying the miR172b-SNZ mediated ABA signal transduction during post-germinative development stage in response to abiotic stress.
Supporting Information
Phenotypic analysis of young seedlings of 35S::miR172b and Col-0 in response to ABA and NaCl. The seeds of 35S::miR172b and Col-0 were germinated on MS medium for 5 days and then transferred to MS medium containing NaCl (0, 50, 100 and 150 mM) or ABA (0, 50 and 100 µM). The photographs were taken 7 days after transfer to NaCl (A), and the primary root length was estimated (B). Photographs were taken 5 days after transfer to ABA (C), and the primary root length was estimated (D). (E) Identification of overexpression of miR172b in 35S:miR172b transgenic lines. Bar = 250 µm. All the experiments were performed three times with three replicates. Bars represent mean±SD.
(TIF)
Post-geminative developmental arrest of 35S:: miR172b on ABA was restored after removing ABA. Seeds of 35::miR172b and Col-0 were germinated on MS medium containing 0.8 µM ABA, the seedlings which turned green were counted at 12 days after stratification (A); the arrested seedlings were transferred onto MS medium (B) or MS plus 0.8 µM ABA (C) for another 3 days, and the number of seedlings turning green were recorded. Bar = 5mm. All the experiments were performed three times with three replicates. Bars represent mean±SD.
(TIF)
Phenotype of 35::miR172b transgenic plants and snz-1 under LiCl and KCl treatment. (A) KCl and LiCl dose–response analysis of seed germination for Col-0, 35S:: miR172b and snz-1 at 3 days after stratification. (B) and (C) Greening of 35:: miR172b transgenic plants and snz-1 was not affected under KCl and LiCl treatment. Photographs were taken 7 days after stratification. Bar = 5 mm. All the experiments were performed three times with three replicates. Error bars denote±SD.
(TIF)
SNZ and TOE3 were up-regulated by ABA and osmotic stress/drought. Plant materials from 7 and 18 day old wild-type Col-0 was treated by ABA and osmotic stress, respectively. (A) and (B) Expression of SNZ (A) and TOE3 (B) in response to ABA and osmotic stress/drought; The colors from yellow to red indicate the increased absolute signal values of SNZ and TOE3 expression retrieved from microarray data. (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi).
(TIF)
Expression of SNZ and TOE3 in response to ABA. (A) Expression analysis of TOE3 in response to ABA. Wild-type seeds were germinated on MS with or without 5 µM, and materials were collected at 2 to 4 days after stratification. (B) SNZ expression in wild-type (Col-0) and 35S:: miR172b plants on day 3 and 4 after stratification with or without 0.4 µM ABA. Student's t test was performed, and the statistically significant treatments are marked with asterisks. (*) P<0.05, (**) P<0.01 and (***) P<0.001. All the experiments were performed three times with three replicates. Error bars denote±SD.
(TIF)
SNZ regulates post-germinative developmental arrest induced by abiotic stress through the ABA dependent pathway. SNZ cDNA under control of 35S promoter was expressed in snz-1 mutant, and the transgenic lines were characterized and used for phenotypic analysis. (A) Transcript abundance of SNZ in wild-type, snz-1 and 35S:: SNZ/snz-1 transgenic lines was monitored using RT-PCR. Shown are RT-PCR products amplified after 30 cycles, and the transcript expression levels were normalized against the reference gene GAPC. The red boxes indicate the line which was used in the phenotypic analysis. (B) ABA dose–response analysis of seed germination for Col-0 and snz-1 at 5 days after stratification. (C) A greening time course on medium containing 0.4 µM ABA. All the experiments were performed three times with three replicates. Error bars denote±SD.
(TIF)
Genotyping of snz-1 / abi5-8 . (A) Schematics showing the location of the T-DNA insertion in abi5-8(SALK_013163) and snz-1(SALK_030031). Black boxes indicate exons, green box indicate 5′UTR, yellow box indicates 3′UTR, and the horizontal lines indicate introns. (B) Confirmation of the homologous T-DNA insertion in snz-1/abi5-8 by PCR. Col-0 was used as positive control. The red boxes indicate the lines with the homologous T-DNA insertions both snz-1 and abi5-8 mutations, which were used in the phenotypic analysis.
(TIF)
cis-acting regulatory elements analysis of promoter sequence of miR172b by PLACE and PROMO.
(DOC)
cis-acting regulatory elements analysis of the SNZ promoter sequence by PLACE and AtcisDB.
(DOC)
Primers pairs used for real-time RT-PCR (Sequence 5′→3′).
(DOC)
Primers pairs used for construction and genotyping, probes used for EMSA (Sequence 5′→3′).
(DOC)
Information of the plant materials used in this work.
(DOC)
cis-acting regulatory elements RAV1 analysis of promoter sequence of ABA response genes by PLACE and AtcisDB.
(DOC)
Acknowledgments
We thank Professor Markus Schmid (Department of Molecular Biology, Max Planck Institute for Developmental Biology, Germany) for kindly providing the snz-1 mutant, Professor R. Scott Poethig (Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA) for kindly providing the 35S:miR172b transgenic line.
Funding Statement
This work was supported by grants from the National Program on Key Basic Research Project (2009CB118305), NSFC (31230050), Main Direction Program of Knowledge Innovation of Chinese Academy of Sciences (KSCX2-EW-J-5), and National Transgenic Key Project of the Ministry of Agriculture of China (2011ZX08009-003-002). This work was also supported by Youth Innovation Promotion Association, CAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, et al. (2012) Seed germination and vigor. Annu Rev Plant Biol 63: 507–533. [DOI] [PubMed] [Google Scholar]
- 3. Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, et al. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, et al. (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, et al. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A 106: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soon FF, Ng LM, Zhou XE, West GM, Kovach A, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, et al. (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci RES 20: 55–67. [Google Scholar]
- 9. Koornneef M, Reuling G, Karssen CM (1984) The Isolation and Characterization of Abscisic-Acid Insensitive Mutants of Arabidopsis-Thaliana. Physiol PlantarumLANTARUM 61: 377–383. [Google Scholar]
- 10. Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328. [DOI] [PubMed] [Google Scholar]
- 11. He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531. [DOI] [PubMed] [Google Scholar]
- 12. Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524. [DOI] [PubMed] [Google Scholar]
- 13. Martin RC, Asahina M, Liu PP, Kristof JR, Coppersmith JL, et al. (2010) The microRNA156 and microRNA172 gene regulation cascades at post-germinative stages in Arabidopsis. Seed Sci RES 20: 79–87. [Google Scholar]
- 14. Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, et al. (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- 15. Lauter N, Kampani A, Carlson S, Goebel M, Moose SP (2005) microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci U S A 102: 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu G, Park MY, Conway SR, Wang JW, Weigel D, et al. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mathieu J, Yant LJ, Murdter F, Kuttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci U S A 109: 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu C, Hills MJ (2002) Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid, sugars, and osmotic stress during germination and seedling development. Plant Physiol 129: 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sumer A, Zorb C, Yan F, Schubert S (2004) Evidence of sodium toxicity for the vegetative growth of maize (Zea mays L.) during the first phase of salt stress. J Appl Bot Food Qual 78: 135–139. [Google Scholar]
- 21. Wang ZY, Xiong L, Li W, Zhu JK, Zhu J (2011) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23: 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129. [DOI] [PubMed] [Google Scholar]
- 25. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prestridge DS (1991) Signal Scan - a Computer-Program That Scans DNA-Sequences for Eukaryotic Transcriptional Elements. Computer Applications in the Biosciences 7: 203–206. [DOI] [PubMed] [Google Scholar]
- 27. Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, et al. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18: 333–334. [DOI] [PubMed] [Google Scholar]
- 28. Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, et al. (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31: 3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Finkelstein R, Gampala SS, Lynch TJ, Thomas TL, Rock CD (2005) Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol Biol 59: 253–267. [DOI] [PubMed] [Google Scholar]
- 30. Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koornneef M, et al. (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383. [DOI] [PubMed] [Google Scholar]
- 31. Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635. [DOI] [PubMed] [Google Scholar]
- 32. Xi W, Liu C, Hou X, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22: 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, et al. (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, et al. (2006) AGRIS and AtRegNet. A platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, et al. (2003) AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen M, Thelen JJ (2010) The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. Plant Cell 22: 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401. [DOI] [PubMed] [Google Scholar]
- 41. Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14 Suppl: S15–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bies-Etheve N, da Silva Conceicao A, Giraudat J, Koornneef M, Leon-Kloosterziel K, et al. (1999) Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol Biol 40: 1045–1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotypic analysis of young seedlings of 35S::miR172b and Col-0 in response to ABA and NaCl. The seeds of 35S::miR172b and Col-0 were germinated on MS medium for 5 days and then transferred to MS medium containing NaCl (0, 50, 100 and 150 mM) or ABA (0, 50 and 100 µM). The photographs were taken 7 days after transfer to NaCl (A), and the primary root length was estimated (B). Photographs were taken 5 days after transfer to ABA (C), and the primary root length was estimated (D). (E) Identification of overexpression of miR172b in 35S:miR172b transgenic lines. Bar = 250 µm. All the experiments were performed three times with three replicates. Bars represent mean±SD.
(TIF)
Post-geminative developmental arrest of 35S:: miR172b on ABA was restored after removing ABA. Seeds of 35::miR172b and Col-0 were germinated on MS medium containing 0.8 µM ABA, the seedlings which turned green were counted at 12 days after stratification (A); the arrested seedlings were transferred onto MS medium (B) or MS plus 0.8 µM ABA (C) for another 3 days, and the number of seedlings turning green were recorded. Bar = 5mm. All the experiments were performed three times with three replicates. Bars represent mean±SD.
(TIF)
Phenotype of 35::miR172b transgenic plants and snz-1 under LiCl and KCl treatment. (A) KCl and LiCl dose–response analysis of seed germination for Col-0, 35S:: miR172b and snz-1 at 3 days after stratification. (B) and (C) Greening of 35:: miR172b transgenic plants and snz-1 was not affected under KCl and LiCl treatment. Photographs were taken 7 days after stratification. Bar = 5 mm. All the experiments were performed three times with three replicates. Error bars denote±SD.
(TIF)
SNZ and TOE3 were up-regulated by ABA and osmotic stress/drought. Plant materials from 7 and 18 day old wild-type Col-0 was treated by ABA and osmotic stress, respectively. (A) and (B) Expression of SNZ (A) and TOE3 (B) in response to ABA and osmotic stress/drought; The colors from yellow to red indicate the increased absolute signal values of SNZ and TOE3 expression retrieved from microarray data. (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi).
(TIF)
Expression of SNZ and TOE3 in response to ABA. (A) Expression analysis of TOE3 in response to ABA. Wild-type seeds were germinated on MS with or without 5 µM, and materials were collected at 2 to 4 days after stratification. (B) SNZ expression in wild-type (Col-0) and 35S:: miR172b plants on day 3 and 4 after stratification with or without 0.4 µM ABA. Student's t test was performed, and the statistically significant treatments are marked with asterisks. (*) P<0.05, (**) P<0.01 and (***) P<0.001. All the experiments were performed three times with three replicates. Error bars denote±SD.
(TIF)
SNZ regulates post-germinative developmental arrest induced by abiotic stress through the ABA dependent pathway. SNZ cDNA under control of 35S promoter was expressed in snz-1 mutant, and the transgenic lines were characterized and used for phenotypic analysis. (A) Transcript abundance of SNZ in wild-type, snz-1 and 35S:: SNZ/snz-1 transgenic lines was monitored using RT-PCR. Shown are RT-PCR products amplified after 30 cycles, and the transcript expression levels were normalized against the reference gene GAPC. The red boxes indicate the line which was used in the phenotypic analysis. (B) ABA dose–response analysis of seed germination for Col-0 and snz-1 at 5 days after stratification. (C) A greening time course on medium containing 0.4 µM ABA. All the experiments were performed three times with three replicates. Error bars denote±SD.
(TIF)
Genotyping of snz-1 / abi5-8 . (A) Schematics showing the location of the T-DNA insertion in abi5-8(SALK_013163) and snz-1(SALK_030031). Black boxes indicate exons, green box indicate 5′UTR, yellow box indicates 3′UTR, and the horizontal lines indicate introns. (B) Confirmation of the homologous T-DNA insertion in snz-1/abi5-8 by PCR. Col-0 was used as positive control. The red boxes indicate the lines with the homologous T-DNA insertions both snz-1 and abi5-8 mutations, which were used in the phenotypic analysis.
(TIF)
cis-acting regulatory elements analysis of promoter sequence of miR172b by PLACE and PROMO.
(DOC)
cis-acting regulatory elements analysis of the SNZ promoter sequence by PLACE and AtcisDB.
(DOC)
Primers pairs used for real-time RT-PCR (Sequence 5′→3′).
(DOC)
Primers pairs used for construction and genotyping, probes used for EMSA (Sequence 5′→3′).
(DOC)
Information of the plant materials used in this work.
(DOC)
cis-acting regulatory elements RAV1 analysis of promoter sequence of ABA response genes by PLACE and AtcisDB.
(DOC)