SUMMARY
X upregulation in mammals increases levels of expressed X-linked transcripts to compensate for autosomal bi-allelic expression. Here, we present molecular mechanisms that enhance X expression at transcriptional and posttranscriptional levels. Active mouse X-linked promoters are enriched in the initiation form of RNA polymerase II (PolII-S5p) and in specific histone marks including H4K16ac and histone variant H2AZ. The H4K16 acetyltransferase MOF, known to mediate the Drosophila X upregulation, is also enriched on the mammalian X. Depletion of MOF or MSL1 in mouse ES cells causes a specific decrease in PolII-S5p and in expression of a subset of X-linked genes. Analyses of RNA half-life datasets show increased stability of mammalian X-linked transcripts. Both ancestral X-linked genes, defined as those conserved on chicken autosomes, and newly acquired X-linked genes are upregulated by similar mechanisms but to a different extent, suggesting that subsets of genes are distinctly regulated dependent on their evolutionary history.
INTRODUCTION
Many organisms have a single X chromosome in males and two in females. In the heterogametic sex the X and Y chromosomes derived from an ordinary pair of autosomes by progressive degeneration of the Y. Dosage compensation mechanisms evolved to restore a balanced expression both between the X and autosomes and between the sexes (Disteche, 2012). Strategies to achieve this compensation vary among species. In Drosophila, expression of most X-linked genes is increased about two-fold specifically in males (Conrad and Akhtar, 2012). X upregulation also occurs in mammals and in C. elegans but in both sexes (Gupta et al., 2006; Nguyen and Disteche, 2006). Silencing of one X chromosome by X inactivation in mammalian females (Lyon, 1961) and repression of both X chromosomes in C. elegans hermaphrodites (Meyer, 2010) have been adapted to avoid hyper-expression in the homogametic sex. X upregulation is well supported by expression analyses of individual genes and by array-based transcriptome analyses (Adler et al., 1997; Gupta et al., 2006; Nguyen and Disteche, 2006). Although RNA-sequencing analyses did not initially find evidence of X upregulation in mammals and C. elegans (Xiong et al., 2010), re-analyses of RNA-seq data accounting for the skewed gene content of the X chromosome in mammals, C. elegans, and Drosophila re-confirmed that upregulation of expressed X-linked genes occurs in all three species (Deng et al., 2011; Kharchenko et al., 2011; Lin et al., 2011). Interestingly, the mammalian X chromosome is composed of a mosaic of two types of genes based on their evolutionary history: ancestral genes originally present on the autosomes from which the sex chromosomes derived, and acquired genes recently added to the X chromosome (Bellott et al., 2010). Whether these genes are differentially regulated by specific dosage compensation mechanisms is unknown.
X upregulation could be controlled by DNA-sequence-based and/or epigenetic mechanisms to control transcription, RNA stability, and/or translation. In Drosophila, the MSL (male specific lethal) complex directs the acetyltransferase MOF (males absent on the first) to the male X to acetylate histone H4 at lysine 16 (H4K16ac) and enhance transcription (Conrad and Akhtar, 2012). Although increased X expression appears to mainly result from enhanced transcriptional elongation (Larschan et al., 2011), a recent study suggests that initiation may also play a major role (Conrad et al., 2012). In mammals, the existence of epigenetic components involved in X upregulation is supported by our previous analyses of human triploid cells (Deng et al., 2009). In addition, RNA polymerase II phosphorylated at serine 5 (PolII-S5p), which regulates the transition from pre-initiation to early elongation (Buratowski, 2009; Phatnani and Greenleaf, 2006), is higher at the 5’ end of mouse X-linked versus autosomal genes (Deng et al., 2011). A recent study confirmed these findings as well as reported enrichment in other active chromatin marks on the mouse X chromosome (Yildirim et al., 2011).
To identify molecular mechanisms of X upregulation in mammals we determined the genome-wide distributions of the initiation and elongation forms of RNA polymerase II (PolII), PolII-S5p (phosphorylated at serine 5) and PolII-S2p (phosphorylated at serine 2), and of histone modifications in mouse cell lines and tissues. To address the function of the H4K16 acetyltransferase MOF occupancy profiles were established and knockdowns of MOF and MSL1 were done in mouse ES cells. Our results support a conserved role for the MSL complex to enhance transcription initiation and expression of a subset of X-linked genes. Furthermore, we found that the half-life of X-linked transcripts is longer relative to autosomal transcripts. These transcriptional and post-transcriptional mechanisms of dosage compensation appear to regulate ancestral and acquired X-linked genes to various degrees, consistent with differentially regulated subsets of genes.
RESULTS
PolII-S5p but not PolII-S2p is enriched at the promoters of expressed X-linked genes
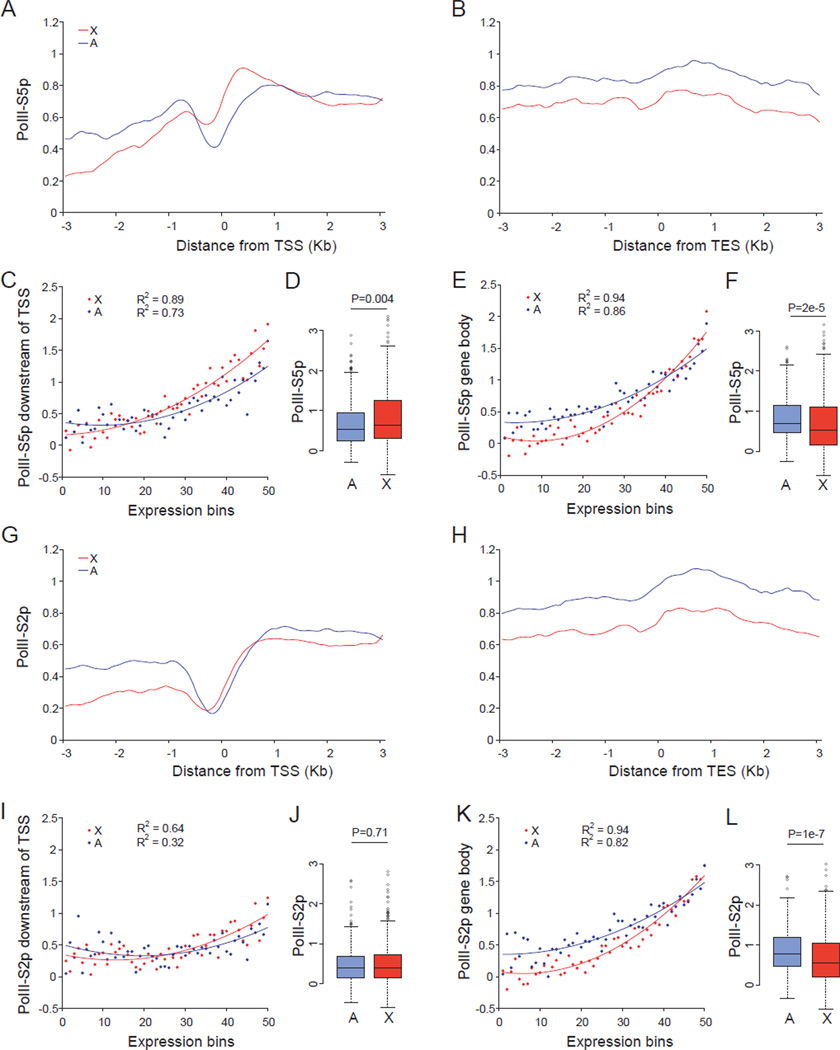
Chromosome-wide occupancy profiles for PolII-S5p and PolII-S2p were established to address the role of transcriptional initiation and elongation in mammalian X upregulation. ChIP-chip analyses (chromatin immunoprecipation combined with arrays) using genome tiling arrays covering mouse chromosomes X and 19 (chr19) were done in undifferentiated female ES cells PGK12.1. In these cells, both X chromosomes are active (Penny et al., 1996), resulting in a 1.3–1.4 X:autosome expression ratio when considering actively expressed genes with ≥1RPKM (reads per kilobase of exon per million mapped reads measured by RNA-seq analysis) (Figure S1A–S1D). Thus, a substantial level of X upregulation is already evident in these ES cells prior to differentiation, in agreement with previous studies (Deng et al., 2011; Lin et al., 2007; Lin et al., 2011). Concordant with X upregulation metagene analyses demonstrated higher PolII-S5p occupancy in a promoter-proximal region downstream of the transcription start site (TSS) of expressed X-linked (387) versus autosomal (355 chr19-linked) genes (Figure 1A). However, no X-specific PolII-S5p enrichment was seen at the gene body and 3' end (Figure 1B).
Figure 1. PolII-S5p but not PolII-S2p occupancy is enhanced at expressed X-linked genes in female ES cells.
(A–L) ChIP-chip analyses using genome tiling arrays to survey X-linked (X) and autosomal (A) genes in undifferentiated female ES cells PGK12.1.
(A–B) Metagene analyses of 355 X-linked and 387 chr19-linked expressed genes (≥1RPKM). Average PolII-S5p occupancy (log2 ChIP/input) was plotted 3kb up- and downstream of the TSS (transcriptional start site) (A) and TES (transcriptional end site) (B) in a 500bp sliding window (100bp intervals).
(C–E) Average binding scores of PolII-S5p in a 1kb region downstream of the TSS (C) or in a 3kb region upstream of the TES (3’ gene body) (E) plotted for 50 expression-ranked bins, each containing 10 X-linked and 9 chr19-linked genes. A total of 506 X-linked and 456 chr19-linked genes (>0RPKM) were examined. Regression values (R2) and trend lines are shown.
(D–F) Box plots of PolII-S5p occupancy at the promoter (D) and at the 3’ gene body (F) of expressed X-linked and chr19-linked genes (≥1RPKM). P values from Wilcoxon-test are shown.
(G–L) Occupancy of PolII-S2p at the 5’ or 3’ end of X-linked and chr19-linked genes. Same analyses as described in (A–F).
See also Figure S1.
Average PolII-S5p occupancies within a 1kb region downstream of the TSS and within a 3kb region upstream of the transcription end site (TES) were quantified for 506 X-linked and 456 chr19-linked genes (>0RPKM) grouped in 50 expression-ranked bins. A strong correlation between PolII-S5p occupancy and gene expression was observed for both X-linked and chr19-linked genes (Figure 1C and 1E). Importantly, expressed X-linked promoters (≥1RPKM; Bins 13–50) had significantly higher (27% increase) average PolII-S5p occupancy compared to chr19-linked promoters (Figure 1D). In contrast, the gene body toward the 3’ end did not show an X-specific effect except for a slight increase on highly expressed genes (Bins 40–50) (Figure 1E and 1F). Higher occupancy by PolII-S5p as well as by the general form of RNA PolII (30% increase) at expressed X-linked promoters compared to chr19-linked or to all expressed autosomal promoters was confirmed in a second undifferentiated female ES cell line CD15 (Figures S1E–S1L).
Furthermore, in undifferentiated male ES cells WD44, X -specific higher PolII-S5p occupancy was also observed (Figure S1M and S1N). In contrast, the elongation form of RNA PolII, PolII-S2p, increased towards the 3’ end of expressed genes as expected, but did not show higher occupancy on X-linked genes (Figure 1G–1L).
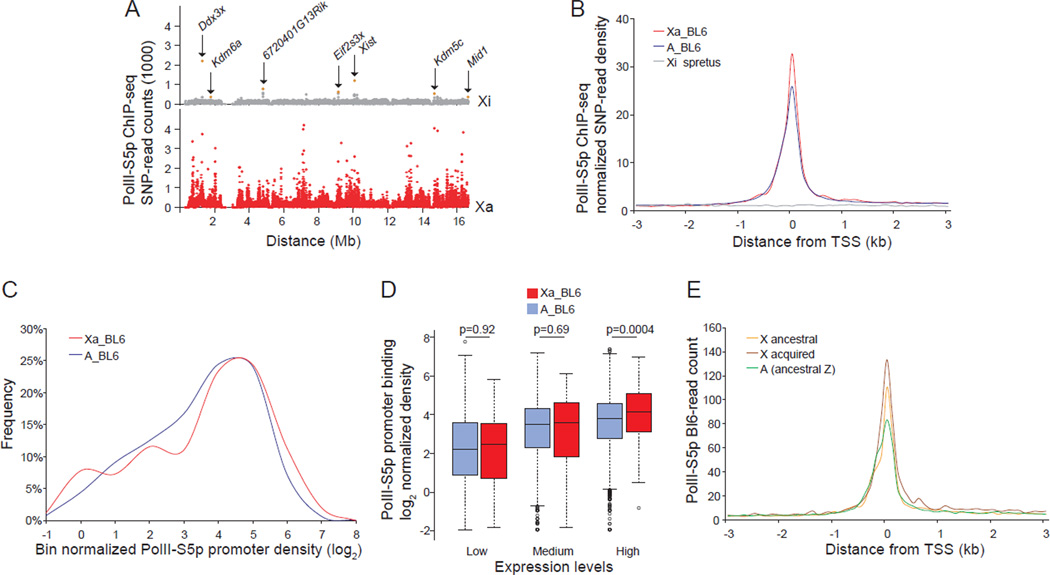
To directly compare the single active X (Xa) to the inactive X (Xi) and to a haploid set of autosomes, we used mouse systems with skewed X inactivation based on crosses between C57BL/6J (BL6) and M. spretus. In these systems, alleles can be differentiated by frequent SNPs between species, and the Xa compared to the haploid set of autosomes from the same species (Lingenfelter et al., 1998; Yang et al., 2010). ChIP-seq (ChIP combined with sequencing) was done using brain from an adult female F1 mouse with a BL6 Xa and a spretus Xi. Uniquely mapped reads containing informative SNPs were then assigned to each haploid chromosome set (BL6 or spretus) to establish allele-specific occupancy profiles. As expected, PolII-S5p occupancy was high on the Xa but very low on the Xi, reflecting gene silencing (Figures 2A, 2B and S2A). Interestingly, the few loci occupied by PolII-S5p on the Xi represent genes known to escape X inactivation that we and others have previously reported (Figure 2A) (Lopes et al., 2011; Reinius et al., 2010; Yang et al., 2010). Metagene analyses showed that PolII-S5p peak levels at promoters of expressed genes on the Xa (≥1RPKM) were 30% higher than those on autosomal genes from the same species (BL6) (Figures 2B and S2A). Control metagene analyses of brain genomic DNA sequencing data (input) showed similar SNP-read counts for the BL6-Xa, spretus-Xi, and BL6-A, indicating absence of sequencing bias (Figure S2C). Thus, the ChIP-seq results substantiate those obtained by ChIP-chip despite apparent differences in the shape of PolII-S5p peaks, which are probably due to differences between the two methodologies (Figures 1A and 2B). An additional analysis in which genes with significant PolII-S5p peaks (FDR ≤1e-5) within a 1kb region up- and downstream of the TSS were selected regardless of their expression levels, confirmed the Xa-specific enhancement in PolII-S5p occupancy (Figure S2B). A noticeable shift in distribution profiles further verified a higher percentage of expressed Xa-linked genes with high PolII-S5p promoter occupancy compared to autosomal genes from the same species haploid set (Figure 2C). Medians of PolII-S5p promoter occupancy were 19%, 8%, and 26% higher, respectively, for genes grouped by low-, medium-, and high-expression on the Xa compared to the haploid autosomal set (p=0.92, 0.69, and 0.0004, respectively, Wilcoxon-test; Figure 2D). An independent allele-specific ChIP-seq analysis performed on mouse fibroblasts (Patski cell line derived from embryonic kidney) that have the opposite X inactivation pattern from the brain sample, i.e. a spretus Xa and a BL6 Xi confirmed high PolII-S5p levels on the Xa (Figure S2D).
Figure 2. Higher PolII-S5p occupancy at expressed X-linked versus autosomal genes by allele-specific ChIP-seq analyses in brain.
(A–E) Allele-specific ChIP-seq in brain of an F1 mouse with the BL6 X chromosome active (Xa), and the spretus X inactive (Xi).
(A) Chromosome-wide allele-specific PolII-S5p occupancy profiles of the Xa and Xi are shown as SNP read counts in 10kb windows. Read counts for genes that escape X inactivation (Ddx3x, Kdm6a, 6720401G13Rik, Eif2s3x, Xist, Kdm5c, Mid1) are indicated (pink dots).
(B) Metagene analysis of PolII-S5p occupancy at the 5' end of expressed genes (≥1RPKM) on the Xa (Xa_BL6) (398 X-linked genes) and on the haploid set of autosomes (A_BL6) (11335 autosomal genes). SNP-read counts plotted 3kb up- and downstream of the TSS in 100bp windows.
(C) Percentage of expressed genes with PolII-S5p promoter occupancy is higher on the Xa (Xa_BL6) than on autosomal genes from the same haploid set (A_BL6) (p =0.007, KS (Kolmogorov–Smirnov)-test). Normalized promoter PolII-S5p density was calculated by averaging SNP-read counts in six 100bp windows around the TSS (peak region) for expressed genes, and normalizing by the median value of genomic DNA-SNP-read counts 3kb up- and downstream of the TSS.
(D) Box plots of normalized allele-specific promoter PolII-S5p density on expressed genes on the Xa compared to those on the haploid set of autosomes in each of the expression groups (low-, medium-, and high-expression). P values from Wilcoxon-test are shown.
(E) Metagene analysis of PolII-S5p allele-specific occupancy at the 5' end of expressed ancestral X-linked genes (282 genes) and acquired X-linked genes (155 genes), compared to ancestral autosomal genes (421 genes).
See also Figure S2.
Taken together, our ChIP-chip and ChIP-seq experiments show enhanced PolII-S5p but not PolII-S2p at the promoter-proximal region of expressed X-linked genes in mouse ES cells, fibroblasts, and brain. Thus, transcription initiation is apparently enhanced to contribute to X upregulation in mammalian cells and tissues.
Specific epigenetic modifications are enriched at the promoters of expressed X-linked genes
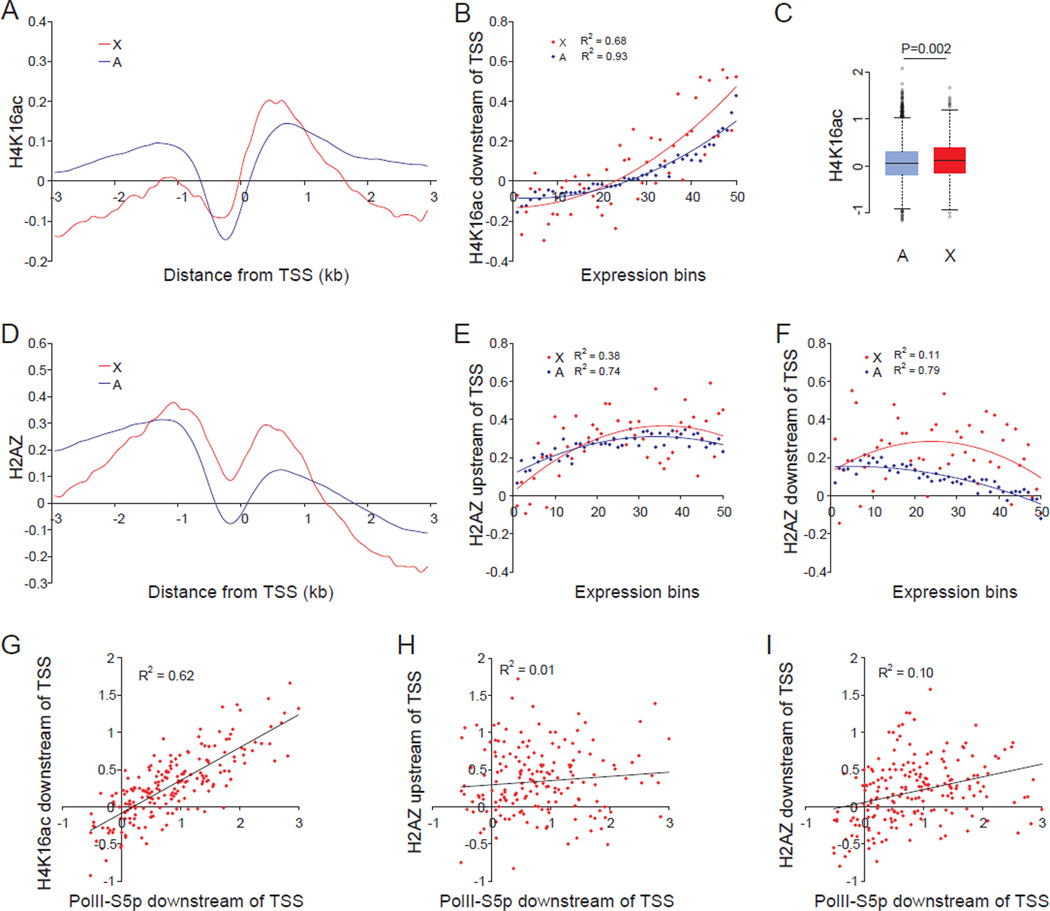
Genome-wide profiles of histone modifications were initially established by ChIP-chip using promoter arrays for mouse undifferentiated female ES cells PGK12.1. Of the three promoter-associated active histone modifications examined (H3K4me3, H3ac, and H4K16ac) only H4K16ac showed a prominent enrichment on expressed X-linked versus autosomal genes (41% average increase in peak values; Figure 3A). H4K16ac levels were well correlated with expression, based on average promoter-binding scores within a 1kb region downstream of the TSS for 506 X-linked and 12755 autosomal genes (>0RPKM) sorted in 50 expression bins. Importantly, binding scores were on average twofold higher for expressed X-linked genes than for autosomal genes (Figures 3B and 3C). H4K16ac enrichment was also well correlated with PolII-S5p (Figure 3G), in agreement with the role of H4K16ac in opening chromatin and activating transcription (Akhtar and Becker, 2000). In contrast, H3K4me3 and H3ac showed little or no X-specific enrichment (Figures S3C and S3D). A control antibody for unmodified pan-H3 demonstrated little difference in nucleosome distribution between X and autosomes (Figure S3A), and when H3 profiles were used for normalization of histone modification profiles no appreciable changes were observed (compare Figures 3A and 3D to S3B and S4E).
Figure 3. Higher levels of H4K16ac and H2AZ at the 5’ end of expressed X-linked versus autosomal genes in female ES cells.
(A–I) ChIP-chip analyses using promoter arrays in undifferentiated female ES cells PGK12.1. Regression values (R2) are shown where appropriate.
(A) Metagene analysis of H4K16ac at the promoter-proximal region of expressed X-linked (X) and autosomal (A) genes. Average enrichment (log2 ChIP/input) was plotted 3kb up- and downstream of the TSS for 387 X-linked and 9800 autosomal expressed genes (≥1RPKM).
(B) Average binding scores of H4K16ac in a 1kb region downstream of the TSS plotted for 50 expression-ranked bins, each containing 10 X-linked and 255 autosomal genes. A total of 506 X-linked and 12755 autosomal genes (>0RPKM) were examined.
(C) Box plots of promoter-binding scores of H4K16ac on expressed X-linked and autosomal genes (≥1RPKM). P value from Wilcoxon-test is shown.
(D) Metagene analysis of H2AZ at the 5’ end of expressed X-linked and autosomal genes. Same analysis as in (A).
(E–F) Average binding scores of H2AZ in a 0.5–1.5kb region upstream of the TSS (E) and a 1kb region downstream of the TSS (F). Same analysis as in (B).
(G–I) PolII-S5p promoter-proximal occupancy at expressed X-linked genes is well correlated with H4K16ac promoter enrichment (G) but not with H2AZ (H–I).
See also Figures S3 and S4.
The X-specific enrichment in H4K16ac at the 5' end of mouse genes suggests that this histone modification may have a conserved role in flies and mammals. However, in Drosophila X-specific enrichment in H4K16ac predominates at the gene body toward the 3’ end, which would facilitate transcription elongation (Gelbart et al., 2009). We did not observe this in mouse cells; rather, the X-specific H4K16ac enrichment was limited to the promoter-proximal region, and did not extend to the 3' end of the gene body, as shown by metagene analyses based on tiling arrays covering mouse chromosomes X and 19. However, H4K16ac levels were higher in genic than intergenic regions and did increased toward the 3’ end of both X-linked and autosomal genes (Figures S3E–S3H). Metagene profiles for H3K36me3, another histone modification involved in active elongation, also showed no X-specific enrichment at the gene body (Figures S3I–S3L). These observations along with high PolII-S5p, but not PolII-S2p, on X-linked genes suggest that transcription initiation rather than elongation may play a major role in mammalian X upregulation.
We also established distribution profiles for H2AZ, a histone variant implicated in different aspects of gene regulation. The acetylated form of H2AZ (H2AZac) is associated with active genes, and the non-acetylated form with repressed genes (Valdes-Mora et al., 2011). Metagene profiles showed that pan-H2AZ levels were higher within two regions at the 5’ end of X-linked versus autosomal genes, a promoter-distal region located 0.5–1.5kb upstream of the TSS (20% increase in peak values) and a promoter-proximal region at 0–1kb downstream of the TSS (two-fold increase in peak values) (Figure 3D). No X-specific H2AZ enrichment was detected at the gene body towards the 3’ end (Figures S4A and S4B). Similar results were obtained for H2AZac, suggesting that profiles obtained for pan-H2AZ represent the acetylated form associated with gene activation (Figures S4C and S4D). The H2AZ enrichment either upstream or downstream of the TSS was not correlated with gene expression levels (Figures 3E and 3F). Similarly, PolII-S5p occupancy was not correlated with H2AZ enrichment at expressed X-linked genes (Figures 3H and 3I), in contrast to what we observed for H4K16ac (Figure 3G). The distinct metagene profiles for H4K16ac and H2AZ suggest that these marks probably play separate roles in regulation of transcription initiation involved in X upregulation.
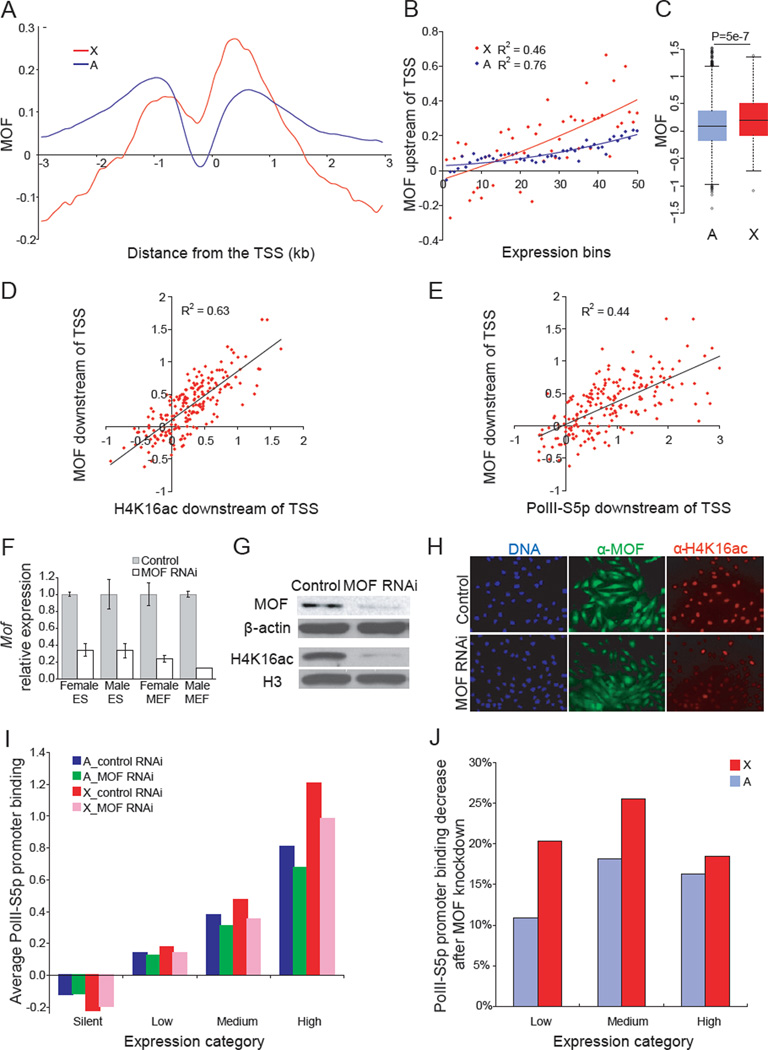
We next determined whether MOF, the enzyme that specifically acetylates H4K16, is involved in mammalian X upregulation. MOF occupancy was specifically enhanced at the promoter-proximal region of expressed X-linked versus autosomal genes in mouse ES cells (Figures 4A, and S5A, and S5B). MOF promoter-binding scores were on average twofold higher for expressed X-linked versus autosomal genes and were correlated with expression (Figures 4B and 4C). As expected, MOF occupancy was also well correlated with H4K16ac enrichment and with PolII-S5p occupancy at expressed X-linked promoters (Figures 4D and 4E). This pattern initially determined in undifferentiated female ES cells PGK12.1 was further confirmed in a second female ES cell line CD15 (data not shown), and in male ES cells (Figure S5C). These findings suggest that MOF and H4K16ac help recruit PolII-S5p for transcription initiation at X-linked promoters.
Figure 4. MOF is enriched at the 5'end of X-linked genes and knockdown reduces accumulation of H4K16ac and PolII-S5p occupancy.
(A–E) ChIP-chip analyses using promoter arrays in undifferentiated female ES cells PGK12.1. Regression values (R2) are shown where appropriate.
(A) Metagene analysis of MOF occupancy at the promoter-proximal region of expressed X-linked (X) and autosomal (A) genes. Same analysis as in Figure 3A.
(B) Average binding scores of MOF in a 1kb region downstream of the TSS plotted for 50 expression-ranked bins. Same analysis as in Figure 3B.
(C) Box plots of promoter-binding scores of MOF on expressed X-linked and autosomal genes (≥1RPKM). P value from Wilcoxon-test is shown.
(D–E) Correlations between MOF promoter-proximal occupancy at expressed X-linked genes with H4K16ac (D) and PolII-S5p promoter enrichment (E).
(F–H) qRT-PCR (F), western blots (G) and immunostaining (H) in control (scramble siRNA) and MOF RNAi treated cells. Mof expression set to 1 in control cells and β-actin used for normalization. Error bars represent standard deviation (SD). β-ACTIN and H3 antibodies serve as controls for the western blot. DNA stained by Hoechst 33342 (blue).
(I) Average PolII-S5p promoter binding scores within 1kb downstream of the TSS for 626 X-linked and 15944 autosomal genes grouped in four expression categories (see Experimental Procedures), compared between control undifferentiated female ES cells PGK12.1 (control RNAi) and MOF knockdown (MOF RNAi).
(J) Percentages of PolII-S5p promoter binding after MOF knockdown for expressed X-linked (X) and autosomal (A) genes in three expression categories.
See also Figure S5.
Both ancestral and acquired genes on the mouse X chromosome have enhanced PolII-S5p, H4K16ac, H2AZ, and MOF
We sorted our data to examine ancestral and acquired X-linked genes separately as they may be differentially regulated. Ancestral mouse X-linked genes represent genes originally present on the autosomes from which the current mouse sex chromosomes derived; they can be identified by comparison to the chicken genome in which orthologs are located within two main blocks of conservation on autosomes 1 and 4 (Kohn et al., 2004). Acquired mouse X-linked genes are those added since the divergence of the mouse sex chromosomes from an ancestral pair of autosomes (Bellott et al., 2010). The acquired category includes many of the multi-copy X-linked genes that have testis-specific expression; these genes being silent in mouse ES cells were excluded from our metagene analyses. We also included a control subset of ancestral mouse autosomal genes (orthologs of chicken Z-linked genes).
Significantly higher PolII-S5p and MOF occupancy as well as H4K16ac and H2AZ enrichment were observed on both ancestral (240 genes) and acquired X-linked genes (147 genes) compared to ancestral autosomal genes (331 genes) (Figures S1O, S4F, S4G and S5D). This is concordant with X:A mean expression ratios of 1.57 and 1.94 for ancestral and acquired genes, respectively, in undifferentiated female ES cells PGK12.1. Acquired X-linked genes appeared to have the highest occupancy/enrichment in active marks, but the promoter-binding scores for each group were not significantly different from each other (Figures S1P, S4H–S4J and S5E). Higher PolII-S5p promoter occupancy in both groups of X-linked genes was confirmed in vivo based on allele-specific ChIP-seq analyses of mouse brain (Figure 2E). In contrast, no or only a slight enrichment was observed for H3K4me3 or H3ac in either group of X-linked genes compared to ancestral autosomal genes (data not shown). Similar results were obtained when all expressed autosomal genes were used for comparison, rather than the subset of orthologs of chicken Z-linked genes (data not shown). We conclude that, regardless of their evolutionary origin, expressed mouse X-linked genes display enhanced levels of PolII-S5p, MOF, H4K16ac, and H2AZ associated with X upregulaton.
MOF knockdown decreases H4K16ac and PolII-S5p occupancy and X-linked gene expression in ES cells
To address the functional role of MOF in X upregulation, mouse cells were transfected with a mixture of three small interfering RNA duplexes, each of which targets a different region of Mof mRNA. Quantitative RT-PCR (qRT-PCR) showed a 70–80% decrease in Mof mRNA, which led to a dramatic decrease in MOF protein detected by western blots and immunostaining (Figures 4F–H). As expected, the levels of H4K16ac were also greatly reduced (Figures 4G–H). While cells grew more slowly after MOF knockdown compared to control cultures as previously reported (Gupta et al., 2008; Thomas et al., 2008), no obvious cell death or abnormal cell morphology were observed (Figure 4H). Interestingly, MOF knockdown in undifferentiated female ES cells PGK12.1 caused a significant reduction not only in H4K16ac but also in PolII-S5p occupancy at the 5’ end of expressed genes, with a more severe effect on medium-expressed X-linked genes (Figures 4I–J, S6A–C). These findings support a role for MOF and H4K16ac in transcription initiation by mediating PolII-S5p recruitment. MOF knockdown did not affect H2AZac enrichment, corroborating the idea that H2AZ acts independent of H4K16ac and MOF (data not shown).
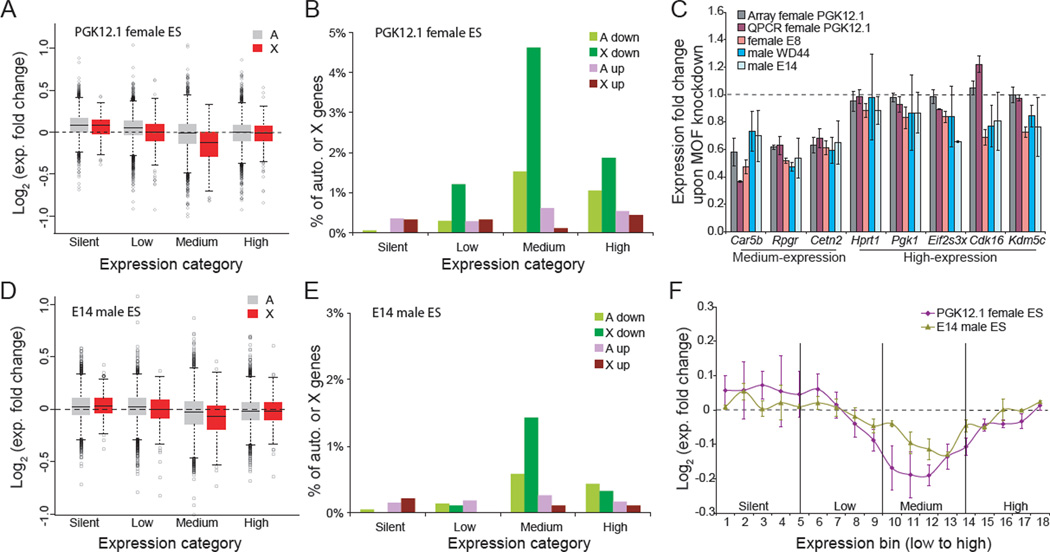
MOF knockdown in female ES cells caused a greater drop in expression of X-linked genes compared to autosomal genes, as measured by expression array analyses. Levels of genes in all expression categories (low, medium, and high) decreased (Figures 5A). However, the strongest effect was observed on medium-expressed X-linked genes whose average expression level decreased by approximately 12% (Figure 5B), in agreement with the significant decrease in H4K16ac (35%) and in PolII-S5p (26%) at these genes (Figures S6C and 4J). Overall, a subset of 70 X-linked genes was apparently MOF-sensitive in female ES cells PGK12.1 (Table S1). The X-specific effect persisted when comparing X-linked genes to genes from a single autosome (chr19) (Figure S6D). Furthermore, a significantly greater proportion of X-linked genes in the medium expression category (19%) were down-regulated compared to other expression categories, or to autosomal genes in any expression category (>1.2 fold down; p <0.05, student t-test; Figure 5B; Table S1). Interestingly, a significantly greater proportion of down-regulated X-linked genes (50 out of 70 genes; p = 2e-6, χ2 test; Figure S6E) were from the ancestral group compared to the acquired group (Figure S6E and Table S1). No such trend was detected for up-regulated ancestral (6 genes) or acquired (5 genes) X-linked genes.
Figure 5. MOF knockdown specifically decreases expression of X-linked genes in female and male ES cells.
(A) Box plots of expression fold changes for X-linked (X) and autosomal (A) genes grouped in four expression categories (see Experimental Procedures) after MOF knockdown in undifferentiated female ES cells PGK12.1. X-linked genes with medium expression levels show the most significant decrease compared to autosomal genes (p <2e-16, one-way ANOVA), or to X-linked genes in other categories (p <6e-10).
(B) Percentages of X-linked (X) and autosomal (A) genes in each expression category with a >1.2-fold expression change (p <0.05, two-tail paired t-test) after MOF knockdown in undifferentiated female ES cells PGK12.1. Genes with decreased expression (down) or with increased expression (up) are indicated.
(C) Quantitative RT-PCR analyses of the effects of MOF knockdown on expression of three medium-expressed (Car5b Rpgr, and Cetn2) and five highly expressed (Hprt1, Pgk1, Eif2s3x, Cdk16 and Kdm5c) X-linked genes in male (WD44 and E14) and female (PGK12.1 and E8) ES cells. The 18S housekeeping gene was used for normalization. Error bars represent SD.
(D) Box plots of expression fold changes for X-linked (X) and autosomal (A) genes grouped in four expression categories after MOF knockdown in undifferentiated male ES cells E14. X-linked genes with medium expression levels show the most significant decrease compared to autosomal genes (p <8e-5, one-way ANOVA), or to X-linked genes in other categories (p <0.0004).
(E) Percentages of X-linked (X) and autosomal (A) genes in each expression category with a >1.2-fold expression change (p <0.05, two-tail paired t-test) after MOF knockdown in undifferentiated male ES cells E14. Same analysis as in Figure 5B.
(F) Expression of X-linked genes sorted in 18 expression–ranked bins (each containing 50 X-linked genes) after MOF knockdown in undifferentiated male ES cells E14 (one active X) and female ES cells PGK12.1 (two active Xs). Average log2 fold change between knockdown and control is shown. Error bars represent SD.
See also Figure S6 and Table S1.
These results were confirmed by qRT-PCR for three medium-expressed and five high-expressed X-linked genes after MOF knockdown in two undifferentiated female ES cell lines (PGK12.1 and E8) (Figures 5C and S6F). Similar results were also obtained for each MOF siRNA duplex separately, ruling out off-target effects (Figure S6G). MOF knockdown did not induce aberrant female ES cell differentiation, which would trigger X-inactivation and confound results due to silencing of one allele. Indeed, expression of ten stem-cell-specific genes showed no obvious changes. In addition, Xist RNA FISH confirmed the absence of an inactive X after knockdown (Figures S6H–I). MOF knockdown in two undifferentiated male ES cell lines (E14 and WD44) also caused a specific reduction in levels of medium-expressed X-linked genes (Figures 5C–F and S6J–L). The extent of this decrease was lesser than in female ES cells (Figure 5F), which could be due either to lower sensitivity of microarray analysis for the detection of expression changes in cells with a single active X chromosome, or to sex-specific differences .
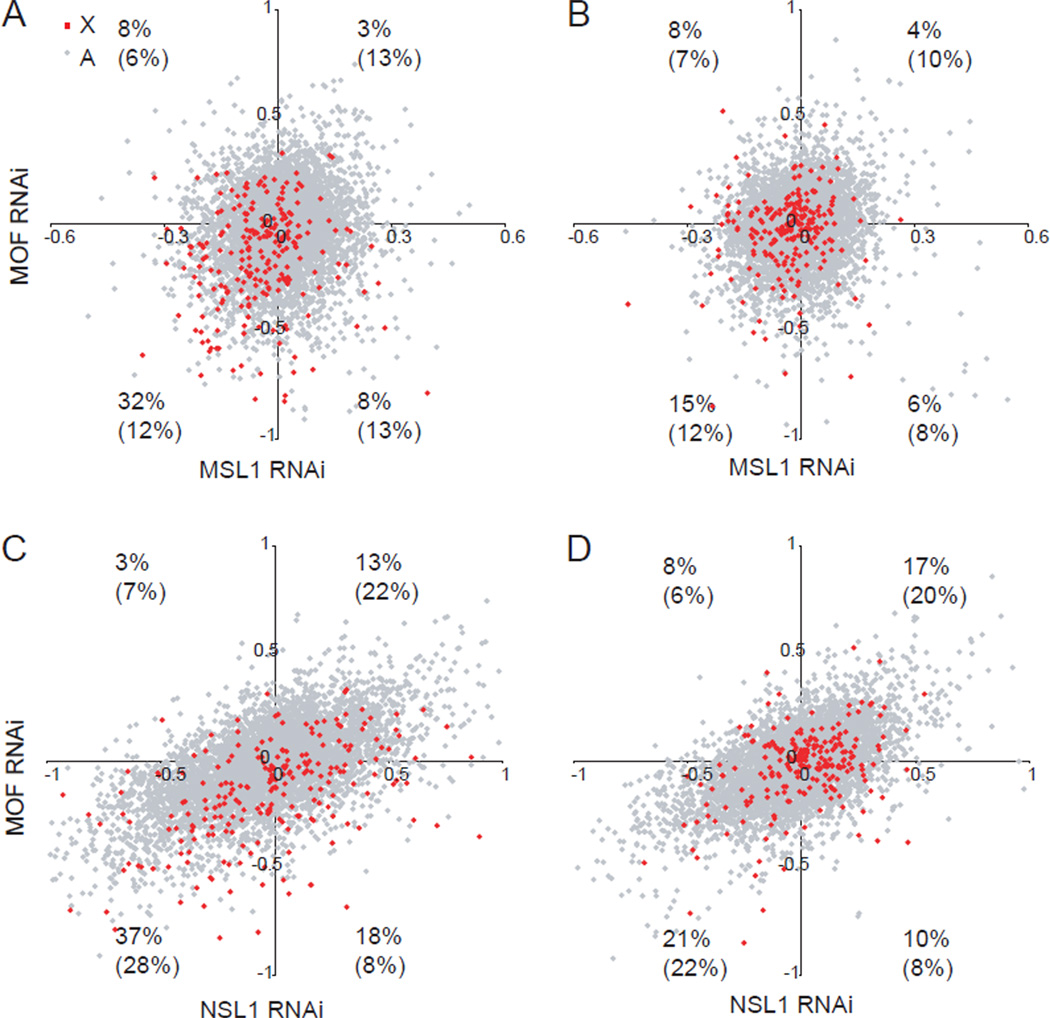
We next examined components of the two known MOF protein complexes, MSL1 (male-specific lethal1) and NSL1 (nonspecific lethal1) (Mendjan et al., 2006; Prestel et al., 2010; Raja et al., 2010). Knockdown of MSL1 but not NSL1 in undifferentiated female ES cells PGK12.1specifically caused a decrease in expression levels of X-linked genes (Figure 5A and Figure S6M–O). Medium-expressed X-linked genes again showed the greatest effect, similar to MOF knockdown. Cells co-transfected with both MOF and MSL1 siRNAs had similar expression changes to MSL1 knockdown alone, indicating that these components probably operate within the same complex but are not additive (Figure S6N). Furthermore, a large subset of medium-expressed X-linked genes (32%) was co-downregulated, while autosomal genes did not show such an effect (12%) (Figures 6A–B; Table S2). In contrast, no prominent X-specific co-downregulation was observed between MOF and NSL1 knockdown (Figures 6C–D). We conclude that key components of the MSL but not NSL complex play a role in upregulation of mammalian X-linked genes in ES cells.
Figure 6. A high percentage of medium-expressed X-linked genes is specifically co-downregulated after MOF or MSL1 (but not NSL1) knockdown.
(A–B) Comparison of expression fold changes (log2) between MOF and MSL1 knockdown for X-linked (red) and autosomal (gray) genes in the medium-expression (A) and high-expression (B) categories (representing a total of 227 X-linked and 5316 autosomal genes in each category). The percentages of X-linked and autosomal genes (in parentheses) with greater than 10% change in expression (±0.1 in log2) in both knockdowns are indicated in each quadrant.
(C–D) Comparison of expression fold changes between MOF and NSL1 knockdown. Similar analysis as in (A–B).
See also Table S2.
Longer RNA half-life of X-linked versus autosomal genes contributes to X upregulation
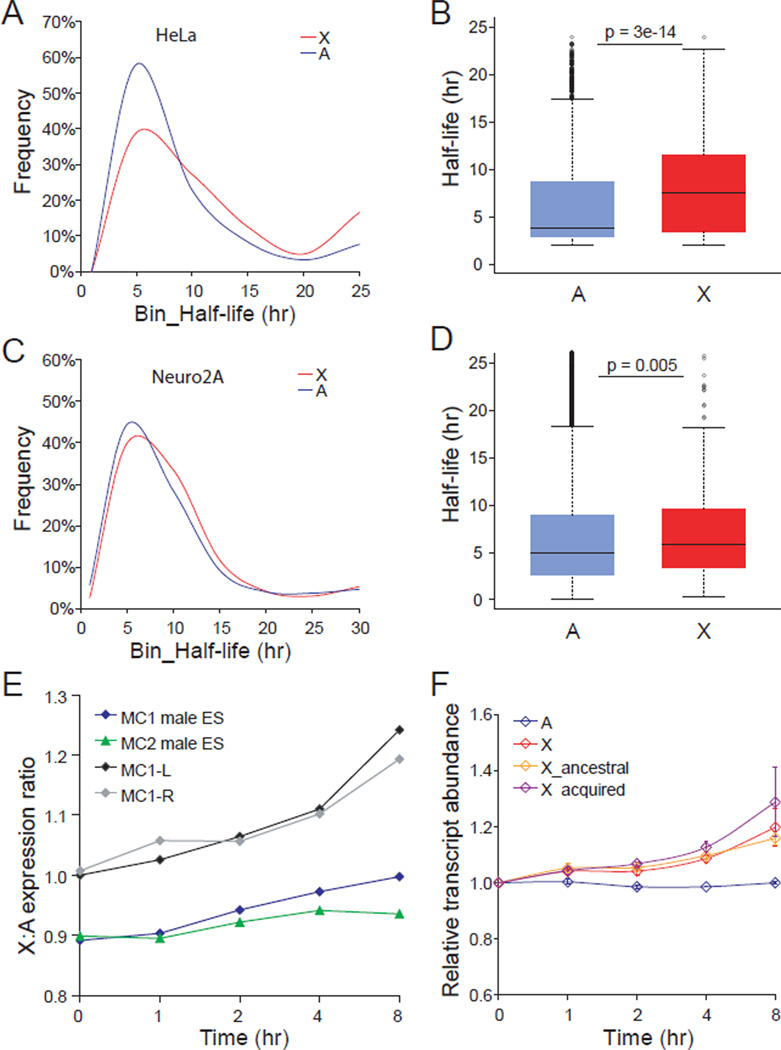
To determine whether posttranscriptional mechanisms were involved in mammalian X upregulation we re-analyzed data from two published studies of genome-wide RNA half-life profiles (Clark et al., 2012; Tani et al., 2012). Distributions of half-life in human HeLa cells and mouse Neuro-2A cells showed a significantly higher percentage of X-linked transcripts with long half-life compared to autosomal transcripts (Figures 7A and 7C). Box plots confirmed an overall longer half-life for X-linked transcripts versus transcripts from all autosomes or from individual autosomes (Figures 7B, 7D, S7A and S7B). There was no significant difference in the half-life of X-linked genes grouped as either ancestral or acquired (using the criteria described above) (Figures S7C–S7F). Additional re-analyses of published microarray data (Sharova et al., 2009) showed that X:autosome expression ratios increased by 10–20% after inhibition of transcription in undifferentiated and differentiated male mouse ES cells, indicating greater stability of X-linked transcripts regardless of their evolutionary origin (Figures 7E and 7F). Interestingly, the 18 MOF-sensitive X-linked genes (representing part of the subset of 70 MOF-sensitive genes, Table S1) for which there was half-life information in the Neuro-2A dataset, had a shorter median half-life (4.2hr), compared to all X-linked (5.8hr) or to all autosomal transcripts (5.0hr). In addition, while 39 of the 70 MOF-sensitive X-linked genes showed enhanced transcript abundance after transcription inhibition in male ES cells, 30 others failed to show any change or even showed a decrease in transcript abundance, suggesting that subsets of X-linked genes may be subject to different types or levels of dosage regulation. Overall, these findings indicate increased stability of X-linked transcripts (or decreased stability of autosomal transcripts), which would contribute to balanced expression between X and autosomes.
Figure 7. Longer half-life of X-linked transcripts compared to autosomal transcripts in mammalian cells.
(A) Distribution of half-life for 327 X-linked and 10252 autosomal transcripts in human HeLa cells shows a higher percentage of X-linked transcripts with long half-life (p=6e-13, KS-test). Published BRIC-seq data were re-analyzed (Tani et al., 2012). Only expressed genes with >1RPKM were included.
(B) Box plots of half-life for human X-linked and autosomal transcripts. P values by Wilcoxon-test are shown.
(C) Distribution of half-life for 266 X-linked and 9131 autosomal transcripts in mouse Neuro-2A cells shows a higher percentage of X-linked transcripts with long half-life (p=0.02, KS-test). Published array data were re-analyzed (Clark et al., 2012).
(D) Box plots of half-life for mouse X-linked and autosomal transcripts. P values by Wilcoxon-test are shown.
(E) X:autosome expression ratios after inhibition of transcription in male mouse ES cells. Reanalysis based on published microarray data on undifferentiated (MC1, MC2) and differentiated (by either LIF withdrawal (MC1-L) or retinoic acid treatment (MC1-R)) mouse ES cells harvested at 0, 1, 2, 4, and 8hr (hour) after addition of the transcription inhibitor actinomycin D (Sharova et al., 2009).
(F) Same analysis as in E for X-linked genes grouped as either ancestral or acquired. The transcript abundance at 0hr was set to 1 and used to calculate the relative transcript abundance of 22166 autosomal (A) genes and of 865 X-linked (X) genes grouped as either ancestral (400 genes) or acquired (465 genes). Means ± SD are shown based on two microarray datasets (MC1-L and MC1-R).
See also Figure S7.
Discussion
Our findings of enhanced chromatin modifications associated with transcription initiation on the mouse X chromosome appear distinct from the proposed mechanism of X upregulation in Drosophila in which enhanced elongation plays a major role (Larschan et al., 2011). However, a recent study reports enhanced recruitment of PolII to X-linked promoters in male Drosophila (Conrad et al., 2012), which is similar to what we observed on mouse X-linked promoters. In their study, Conrad et al. demonstrate that knockdown of MSL2, the key component of the MSL complex that assembles on the male X, affects the recruitment of PolII to X-linked promoters but not to the gene body, suggesting a major role for enhanced transcription initiation. Acetylation of H4K16 mediated by MOF is known to open chromatin structure and activate transcription (Akhtar and Becker, 2000), as well as to enhance the release of paused PolII from promoters (Kapoor-Vazirani et al., 2011; Zippo et al., 2009). Our findings of a good correlation between PolII-S5p occupancy and gene expression support a role in facilitating transcription rather than representing stalling of the RNA polymerase. Enrichment in H4K16ac and MOF was observed specifically at the promoter-proximal region of mammalian X-linked genes, in a region known to be important for efficient transcription initiation and pause release (Appanah et al., 2007; Buratowski, 2009). This may facilitate transcription initiation/re-initiation and PolII pause release, and thus play a conserved role in X upregulation. In male Drosophila, H4K16ac is dominantly enriched on the body of X-linked genes, suggesting a major role in elongation, but it is also observed at the 5’ end of autosomal and X-linked genes in both sexes (Feller et al., 2012; Kind et al., 2008; Prestel et al., 2010; Raja et al., 2010). An important factor that may contribute to differences between flies and mammals is gene length, which is relatively constant in Drosophila but highly variable in mammals (50bp to 2Mb). Transcription levels would be affected by gene length if the elongation rate is a rate-determining step (Conrad and Akhtar, 2012). Given that the process of transcription elongation in mammals is a fast turnover, enhanced initiation rather than elongation would have the advantage of increasing transcript levels despite variable length.
By ChIP-seq we measured a 20–30% greater level of PolII-S5p at the 5’ end of expressed X-linked genes compared to autosomal genes, with a higher level in brain than in fibroblasts, probably reflecting high X expression in brain (Nguyen and Disteche, 2006). Our results are comparable to those reported in a recent allele-specific ChIP-seq study in which cloned mouse fibroblasts showed a similar Xa-specific enrichment in PolII-S5p (31%) (Yildirim et al., 2011). However, while we did not observed an X-specific enrichment in PolII-S2p and H3K36me3 by ChIP-chip in mouse ES cells, this study reported some enrichment in PolII-S2p (20% increase) and H3K36me3 (9%) along the body of X-linked genes (Yildirim et al., 2011). These differences may be related to the use of different cell types/tissues and/or methodologies. For example, a high turnover of PolII in ES cells could explain the absence of detectable enrichment at the gene body in our study. Our results on H3K36me3 are similar to those obtained in Drosophila, where no X-specific enrichment has been detected, even though H3K36me3 may facilitate MSL binding to the male X (Larschan et al., 2007). We found that H2AZ was increased at the 5’ end of X-linked genes but that it probably plays a separate role from H4K16ac. H2AZ often associates with chromatin remodelers, for example ATP-dependent ISWI (Goldman et al., 2010). Studies in Drosophila suggest that H4K16ac directly counteracts the chromatin compaction mediated by ISWI (Corona et al., 2002). In mammals, H2AZ could potentially constrain X upregulation by counteracting H4K16ac to fine-tune gene expression levels and achieve doubling.
In mouse ES cells, MOF and MSL1 knockdown but not NSL1 knockdown significantly diminished the expression of medium-expressed X-linked genes via lower levels of PolII-S5p. The MSL complex may facilitate mammalian X upregulation in early development, which is supported by high MOF and MSL expression specifically in early mouse embryos (Zeng et al., 2004). Additional studies will be necessary to determine whether MOF is still important for X upregulation later in development and in adult tissues. Our studies also suggest that MOF may regulate only certain subsets of X-linked genes. We speculate that specific subsets such as those with medium expression could be more dosage sensitive and respond more extensively than other genes to changes in key dosage regulators such as MOF or other components of the dosage compensation machinery in early development. This is consistent with our previous study of human triploid cells in which expression of a subset of dosage sensitive X-linked genes is apparently adjusted to achieve balance with the autosomes (Deng et al., 2009). Interestingly, expression levels of the mouse X-linked orthologs to these human dosage-sensitive genes were significantly decreased (by 9%) after MOF knockdown, suggesting that this same subset of genes may be more sensitive to changes in MOF and H4K16ac. The existence of a subset of dosage sensitive genes on the mammalian X chromosome is supported by analyses of X-linked and autosomal genes involved in large protein complexes that require maintenance of stoichiometry (Pessia et al., 2012). Our re-analysis of published data shows that in Drosophila downregulation of gene expression after knockdown of components of the MSL complex including MOF is also most pronounced for medium-expressed genes (data not shown) (Hamada et al., 2005; Kind et al., 2008). Yet, the extent of gene expression changes observed after MOF knockdown in mouse ES cells is lesser than that reported in Drosophila, perhaps partly due to incomplete knockdown in mouse cells. Unfortunately, the lethality of MOF null mutations (Gupta et al., 2008; Thomas et al., 2008) and the dramatic changes in chromatin structure and expression of thousands of genes (including the core ES essential genes) observed in inducible MOF knockout ES cells (Li et al., 2012), preclude the study of X-specific effects resulting from complete loss of MOF.
In addition to transcriptional regulation posttranscriptional regulatory mechanisms such as RNA transportation and stability may also be involved in X upregulation (Disteche, 2012). Nonsense-mediated mRNA decay (NMD) has been previously implicated in mammalian X upregulation by demonstrating that inhibition of the NMD pathway decreases the X:A expression ratio in human cells (Yin et al., 2009). Our results based on re-analyses of published measurements of RNA amounts following inhibition of transcription (Clark et al., 2012; Sharova et al., 2009; Tani et al., 2012) are in agreement with a longer half-life for X-linked genes compared to autosomal genes. While the half-life data we re-analyzed were obtained in transformed human and mouse cell lines we also confirmed greater stability of X-linked transcripts in mouse ES cells. Thus, X upregulation in mammals apparently results from a combination of mechanisms that involve transcriptional and posttranscriptional regulation. Transcriptional regulation may involve targeting of factors to the active X and potential sequestration in a special nuclear compartment as suggested in Drosophila (Conrad and Akhtar, 2012), while regulation of RNA stability would suggest transcript sequence differences at the 5' and 3' end UTR. In Drosophila, doubling of gene expression on the male X chromosome apparently involves both a basal buffering mechanism in response to aneuploidy and a feed-forward mechanism targeted to the X via the MSL complex (Zhang et al., 2010). Reverse dosage effects on autosomes have also been proposed (Birchler et al., 2011). In mammals, enhanced initiation of transcription via MOF and H4K16ac may be a conserved feed-forward upregulation mechanism, with increased RNA stability further contributing to X upregulation.
Ancestral X-linked genes have persisted on the mammalian X chromosome even though the sex chromosomes have diverged for more than 130 million years (Lahn and Page, 1999). During that time hundreds of genes have also been acquired (Bellott et al., 2010). For ancestral X-linked genes, dosage compensation may have immediately followed loss of the Y-linked copy, while for acquired X-linked genes dosage regulation may result in part from position effects due to relocation. We found that ancestral X-linked genes may be more sensitive to changes in MOF compared to newly acquired X-linked genes, suggesting that ancestral X-linked genes are the primary targets of this dosage compensation machinery. Two recent studies based on expression analyses using RNA-seq have compared expression of ancestral X-linked genes to their orthologs in chicken and concluded that there is little or no upregulation of ancestral X-linked genes in eutherian mammals (Julien et al., 2012; Lin et al., 2012). Our conclusions differ as we did find higher levels of PolII-S5p, H4K14ac, and H2AZ, and longer half-life for both ancestral and acquired expressed X-linked genes compared to autosomal genes. A potential problem with the RNA-seq studies is the inclusion of non-expressed genes, which are abundant on the mammalian X chromosome and lower median expression values (Deng et al., 2011). In addition, the RNA-seq data normalization needed to compare expression levels of hundreds of genes across distant species is complicated because the chicken genome is not well annotated compared to the mouse or human genomes. A major difference with our study is that we examined expressed X-linked genes, an appropriate gene set when considering mechanisms of dosage compensation. It should be noted that in Drosophila, the X upregulation machinery preferentially assembles at expressed genes (Alekseyenko et al., 2006; Gilfillan et al., 2006), and that enhanced PolII occupancy at the 5' end of genes is observed only on expressed genes, similar to what we report (Conrad et al., 2012).
In conclusion, mammalian X upregulation is apparently mediated by both transcriptional and posttranscriptional mechanisms. Transcriptional effects appear to predominantly operate by epigenetic modifications at the 5’ end of X-linked genes. Posttranscriptional effects on RNA stability may result from epigenetic and/or sequence-specific characteristics. Our results further suggest that subsets of X-linked genes may be differentially regulated by these mechanisms and that the timing of acquisition of genes on the X chromosome may play an important role in regulatory mechanisms.
Experimental Procedures
ChIP-chip analysis
ChIP-chip was performed using fixed chromatin from mouse cell lines and brain as previously described (Yang et al., 2010). Details about the cell lines and brain sample and about antibodies used for ChIP are described in Supplemental Experimental Procedures. After chromatin labeling hybridization was done using two types of arrays: NimbleGen mouse 2.1M tiling arrays (set10; mm8 design) that cover the entire X chromosome, half of chr18, and the entire chr19; and (2) NimbleGen 2.1M mouse promoter tiling arrays (mm8 and mm9 design: tiling regions covering 8kb upstream −3kb downstream of ~24,000 and ~24,507 promoters, respectively). GFF ratios (log2 ChIP/input) and peak files were generated using Nimblescan software with default values. Average enrichment profiles at the 5’ or 3’ ends of genes were calculated using a 500bp sliding window (100bp interval) by metagene analysis (Mito et al., 2005). Only unique RefSeq genes including 626 X-linked and 15944 autosTSS were compared. Normalization of metagenomal genes were analyzed. For the genome tiling array data, 456 chr19-linked and 506 X-linked genes (>0RPKM) were grouped into 50 expression bins based on expression levels from RNA-seq. For the promoter tiling array data, 12755 (12004 for mm9) autosomal genes and 506 (478 for mm9) X-linked genes (>0RPKM) were grouped into 50 expression bins.
ChIP-seq analysis
Sequencing libraries were prepared each from four pooled independent PolII-S5p ChIP samples from Patski cells or from an Xist mutant F1 brain using the Illumina seq sample prep kit. Genomic DNA from the same batch of nuclei from F1 brain was used for control library preparation. These libraries were sequenced on Illumina Genome Analyzers, yielding 36bp single-end reads for ChIP-seq of Patski cells and 100bp single-end reads for ChIP-seq and gDNA-seq of F1 brain.
A pseudo-spretus genome was assembled by replacing available SNPs between C57BL/6J and M. spretus (Sanger Center) into the BL6 reference genome. Reads were aligned to the BL6 reference sequence (mm9, UCSC) (Waterston et al., 2002) and to the pseudo-spretus genome separately using BWA (Burrows-Wheeler Aligner) (Li and Durbin, 2009). Only high-quality uniquely-mapped reads were assigned to each haploid genome (See Supplemental Experimental Procedures). The SNP-associated read counts were used to assess average allelic enrichment at expressed genes (≥1RPKM). PolII-S5p peak regions were selected using uniquely mapped reads by both CisGenome (FDR=1e-5 as the cutoff) and Model-based Analysis of ChIP-Seq (p=1e-5). 10325 autosomal and 326 X-linked genes containing significant PolII-S5p peaks within a 2kb region of the TSS were compared. Normalization of metagene ChIP-seq results was done using genomic gDNA-seq data (See Supplemental Experimental Procedures).
Classification of ancestral and acquired genes
Mouse ancestral X-linked genes were identified as those corresponding to human X-linked genes that have orthologs on chicken chromosomes 1 and 4 based on a previous study (Bellott et al., 2010). The rest of mouse X-linked genes were grouped as acquired. A set of mouse autosomal genes corresponding to human genes on chromosomes 5 and 9, which match to orthologs on chicken chromosome Z, were grouped as control mouse ancestral genes.
Expression array and qRT-PCR analyses
Affymetrix 430 2.0 and 1.0 ST Gene mouse arrays were used for expression analyses in undifferentiated PGK12.1 ES cells and in MOF RNAi and control RNAi treated ES cells (See Supplemental Experimental Procedures). X:A expression ratios were calculated from the mean expression values of X-linked and autosomal genes. For analysis of individual genes, expression was normalized to the mean autosomal expression value of the array. 21253 autosomal and 908 X-linked genes were grouped into four expression categories (‘silent’, ‘low’, ‘medium’, and ‘high’), each containing 5313 autosomal and 227 X-linked genes. These four categories were also applied to classify 626 X-linked and 15944 autosomal genes for metagene analyses and calculations of binding changes. Two-tail student t-tests were used to test the significance of expression changes of individual genes. One-way ANOVA was used to test whether X-linked expression fold changes differed between the four expression categories and from autosomal gene expression changes. Quantitative RT-PCR was performed using a SYBR green master mix (Roche) after reverse transcription of 1µg of total RNA. Detailed RT-PCR protocols and primer information are described in Supplemental Experimental Procedures.
RNAi knockdown
A mixture of three Stealth Select™ siRNA duplexes (Invitrogen) was selected to target different parts of the Mof mRNA. One Silencer Select™ siRNA duplex (Invitrogen) was selected to target Msl1 mRNA and a mixture of two Silencer Select™ siRNA duplexes to target Nsl1 (1700081L11Rik) mRNA. RNAi knockdown was performed using gene-specific siRNAs and negative control siRNAs with Invitrogen Lipofectamine RNAiMax (See Supplemental Experimental Procedures).
Supplementary Material
Highlights.
-
➢
PolII-S5p, H4K16ac, and H2AZ high at ancestral and acquired X-linked gene promoters
-
➢
MOF knockdown in ES cells lowers H4K16ac and PolII-S5p specially at X-linked genes
-
➢
MOF/MSL1 knockdown in ES cells lowers expression of a subset of X-linked genes
-
➢
RNA stability is enhanced at ancestral and acquired X-linked genes
ACKNOWLEDGEMENTS
We thank C. Ware for the WD44 cell line, N. Brockdorff for the PGK12.1 cell line, J. Gribnau for the cell lines E8 and E14 and T. Sado for the Xistm mice. We thank R. Beyer for help with statistical analyses. This work was supported by NIH grants GM046833 and GM079537 (C.M.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The accession number for microarray and sequencing data reported in this article is GSE30761.
The authors have declared no competing financial interests.
References
- Adler DA, Rugarli EI, Lingenfelter PA, Tsuchiya K, Poslinski D, Liggitt HD, Chapman VM, Elliott RW, Ballabio A, Disteche CM. Evidence of evolutionary up-regulation of the single active X chromosome in mammals based on Clc4 expression levels in Mus spretus and Mus musculus. Proc Natl Acad Sci U S A. 1997;94:9244–9248. doi: 10.1073/pnas.94.17.9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appanah R, Dickerson DR, Goyal P, Groudine M, Lorincz MC. An unmethylated 3' promoter-proximal region is required for efficient transcription initiation. PLoS Genet. 2007;3:e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–616. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J, Sun L, Fernandez H, Donohue R, Xie W, Sanyal A. Re-evaluation of the function of the male specific lethal complex in Drosophila. J Genet Genomics. 2011;38:327–332. doi: 10.1016/j.jgg.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2012;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- Conrad T, Cavalli FM, Vaquerizas JM, Luscombe NM, Akhtar A. Drosophila Dosage Compensation Involves Enhanced Pol II Recruitment to Male X-Linked Promoters. Science. 2012 doi: 10.1126/science.1221428. [DOI] [PubMed] [Google Scholar]
- Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–247. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Nguyen DK, Hansen RS, Van Dyke DL, Gartler SM, Disteche CM. Dosage regulation of the active X chromosome in human triploid cells. PLoS Genet. 2009;5:e1000751. doi: 10.1371/journal.pgen.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. Dosage Compensation of the Sex Chromosomes. Annu Rev Genet. 2012 doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller C, Prestel M, Hartmann H, Straub T, Soding J, Becker PB. The MOF-containing NSL complex associates globally with housekeeping genes, but activates only a defined subset. Nucleic Acids Res. 2012;40:1509–1522. doi: 10.1093/nar/gkr869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol. 2009;16:825–832. doi: 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JA, Garlick JD, Kingston RE. Chromatin remodeling by imitation switch (ISWI) class ATP-dependent remodelers is stimulated by histone variant H2A.Z. J Biol Chem. 2010;285:4645–4651. doi: 10.1074/jbc.M109.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schutz F, Daish t, Grutzner F, Kaessmann H. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol. 2011;31:1594–1609. doi: 10.1128/MCB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Xi R, Park PJ. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet. 2011;43:1167–1169. doi: 10.1038/ng.991. [DOI] [PubMed] [Google Scholar]
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Kohn M, Kehrer-Sawatzki H, Vogel W, Graves JA, Hameister H. Wide genome comparisons reveal the origins of the human X chromosome. Trends Genet. 2004;20:598–603. doi: 10.1016/j.tig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:877–879. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Xing K, Zhang J, He X. Expression reduction in mammalian X chromosome evolution refutes Ohno's hypothesis of dosage compensation. Proc Natl Acad Sci U S A. 2012;109:11752–11757. doi: 10.1073/pnas.1201816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O'Neill LP, Turner BM. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Halsall JA, Antczak P, O'Neill LP, Falciani F, Turner BM. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno's hypothesis. Nat Genet. 2011;43:1169–1170. doi: 10.1038/ng.992. [DOI] [PubMed] [Google Scholar]
- Lingenfelter PA, Adler DA, Poslinski D, Thomas S, Elliott RW, Chapman VM, Disteche CM. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- Lopes AM, Arnold-Croop SE, Amorim A, Carrel L. Clustered transcripts that escape X inactivation at mouse XqD. Mamm Genome. 2011;22:572–582. doi: 10.1007/s00335-011-9350-6. [DOI] [PubMed] [Google Scholar]
- Lyon M. Gene action in the X-chromosome of the mouse (Mus musculus L) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Prestel M, Feller C, Straub T, Mitlohner H, Becker PB. The activation potential of MOF is constrained for dosage compensation. Mol Cell. 2010;38:815–826. doi: 10.1016/j.molcel.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, Kadlec J, Fraterman S, Luscombe NM, Akhtar A. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell. 2010;38:827–841. doi: 10.1016/j.molcel.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Reinius B, Shi C, Hengshuo L, Sandhu KS, Radomska KJ, Rosen GD, Lu L, Kullander K, Williams RW, Jazin E. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics. 2010;11:614. doi: 10.1186/1471-2164-11-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 2011 doi: 10.1101/gr.118919.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Chen X, Chen Z, Wang X, Shi S, Wang X, Zhang J, He X. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Deng W, Zheng H, Zhang Z, Hu L, Kong X. Evidence that the nonsense-mediated mRNA decay pathway participates in X chromosome dosage compensation in mammals. Biochem Biophys Res Commun. 2009;383:378–382. doi: 10.1016/j.bbrc.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.