SUMMARY
Long noncoding RNAs (lncRNAs) have been described in cell lines and various whole tissues, but lncRNA analysis of development in vivo is limited. Here, we comprehensively analyze lncRNA expression for the adult mouse subventricular zone neural stem cell lineage. We utilize complementary genome-wide techniques including RNA-seq, RNA CaptureSeq, and ChIP-seq to associate specific lncRNAs with neural cell types, developmental processes, and human disease states. By integrating data from chromatin state maps, custom microarrays, and FACS purification of the subventricular zone lineage, we stringently identify lncRNAs with potential roles in adult neurogenesis. shRNA-mediated knockdown of two such lncRNAs, Six3os and Dlx1as, indicate roles for lncRNAs in the glial-neuronal lineage specification of multipotent adult stem cells. Our data and workflow thus provide a uniquely coherent in vivo lncRNA analysis and form the foundation of a user-friendly online resource for the study of lncRNAs in development and disease.
INTRODUCTION
The mammalian genome encodes thousands of long noncoding RNAs (lncRNAs), and it is becoming increasingly clear that lncRNAs are key regulators of cellular function and development. Loss-of-function studies performed in cell culture indicate that lncRNAs can regulate gene transcription through the targeting and recruitment of chromatin modifying complexes (Guttman et al., 2011; Huarte et al., 2010; Khalil et al., 2009; Tsai et al., 2010). While it is now evident that lncRNAs have important cellular and molecular functions, how they participate in development in vivo is poorly understood.
Emerging studies suggest that lncRNAs play critical roles in central nervous system (CNS) development. For instance, in embryonic stem cells (ESCs), specific lncRNAs repress neuroectodermal differentiation (Guttman et al., 2011), and during in vitro differentiation of ESC-derived neural progenitor cells (ESC-NPCs), lncRNA expression is dynamic (Mercer et al., 2010). In the mouse brain, some lncRNAs are regionally expressed (Mercer et al., 2008), including among the six layers of the adult cortex (Belgard et al., 2011). In vivo functional data is limited, but mice null for the lncRNA Evf2 have abnormal GABAergic interneuron development and function (Bond et al., 2009), and morpholino inhibition of two CNS-specific lncRNAs in Zebrafish affects brain development (Ulitsky et al., 2011).
The subventricular zone (SVZ) of the adult mouse brain represents an ideal system for the study of lncRNAs in vivo. Throughout life, SVZ neural stem cells (SVZ-NSCs) generate large numbers of neuroblasts that migrate to the olfactory bulb (OB) where they differentiate into interneurons (Figure 1A). In addition, SVZ-NSCs are multipotent, capable of generating astrocytes and oligodendrocytes, the other major cell types of the CNS. In contrast to the embryonic brain wherein multipotent precursor cells are inherently transient, continually changing their developmental potential and location over time and with organ morphogenesis, the adult SVZ retains its NSC population in a stable, spatially restricted niche, producing neurons and glia throughout life (Kriegstein and Alvarez-Buylla, 2009). This enduring population of multipotent stem cells and its well-characterized daughter cell lineages make the SVZ a particularly tractable in vivo model for molecular-genetic studies of development. The SVZ has been used to elucidate key principles of neural development including the role of signaling molecules, transcription factors, microRNAs, and chromatin modifiers (Ihrie and Alvarez-Buylla, 2011). We have previously shown that the Mixed-lineage leukemia 1 (Mll1) chromatin modifying factor is required for the SVZ neurogenic lineage (Lim et al., 2009), and recent studies indicate that MLL1 protein can be targeted to specific loci by lncRNAs (Bertani et al., 2011; Wang et al., 2011).
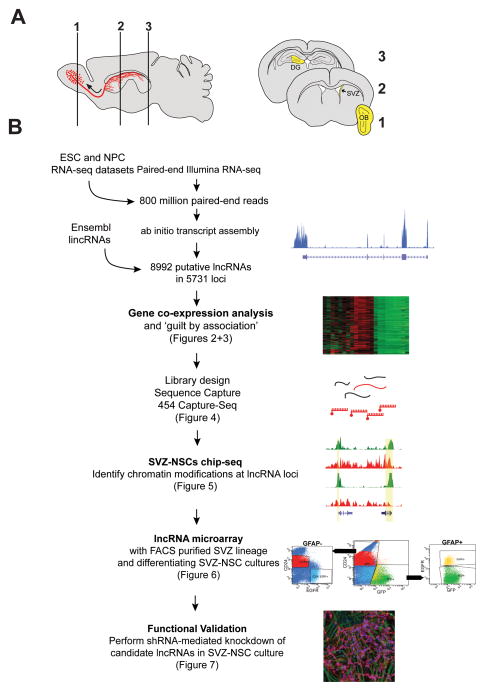
Figure 1. Outline of lncRNA catalog generation, see also Figure S1 and File S1.
(A) Schematic of sagittal section of adult mouse brain. SVZ neural stem cells give rise to migratory neuroblasts (red). These neuroblasts travel along the rostral migratory stream (curved arrow) before terminally differentiating and integrating into olfactory bulb (OB) neuronal circuits. Numbered schematics correspond to coronal brain sections highlighting dissected regions (yellow) used for RNA collection. (B) Workflow for lncRNA catalog construction and characterization.
Here, we leveraged the SVZ-OB system to develop a greater understanding of lncRNA expression and function. First, we used Illumina-based cDNA deep sequencing (RNA-seq) and ab initio reconstruction of the transcriptome to generate a comprehensive lncRNA catalogue inclusive of adult NSCs and their daughter cell lineages. This lncRNA catalogue informed a subsequent RNA Capture-seq approach, which increased the read coverage and read length for our SVZ cell analysis, validating the transcript structure and expression of many of these novel lncRNAs. Gene coexpression analysis identified sets of lncRNAs associated with different neural cells types, cellular processes, and neurologic disease states. In our analysis of genome-wide chromatin state maps, we identified lncRNAs that -- like key developmental genes -- demonstrate chromatin-based changes in a neural lineage-specific manner. Using custom lncRNA microarrays, we found that lncRNAs are dynamically regulated in patterns reminiscent of known neurogenic transcription factors. To define lncRNA expression changes throughout the SVZ neurogenic lineage in vivo, we acutely isolated the major cell types of the SVZ with fluorescent activated cell sorting (FACS) and probed lncRNA expression with our custom microarrays. We integrated these diverse experimental approaches to develop an online resource useful for the identification of lncRNAs with potential roles in SVZ neurogenesis (http://neurosurgery.ucsf.edu/danlimlab/lncRNA/) Furthermore, expression and shRNA-mediated knockdown experiments confirmed functional roles for lncRNAs identified by our integrative approach. Overall, our study demonstrates a generalizable workflow that assimilates genome-wide bioinformatic strategies with experimental manipulations for the identification of lncRNAs that regulate development.
RESULTS
Cataloging lncRNAs in the adult brain neurogenic zones
Because lncRNAs exhibit tissue-specific expression, previous mouse lncRNA databases were not likely comprehensive for lncRNAs involved in adult neurogenesis. Thus, we identified lncRNAs expressed in the adult brain neurogenic niches by employing an RNA-seq and ab initio transcriptome reconstruction approach. First, we generated cDNA libraries of poly-adenylated RNA extracted from microdissected adult SVZ tissue, which contains NSCs, transit amplifying cells, and young migratory neuroblasts. To include the transcriptome of later stages of neurogenesis and neuronal function, we also generated cDNA libraries from the OB. Furthermore, we generated cDNA libraries from microdissected adult dentate gyrus (DG), the other major adult neurogenic niche, which locally contains all cell types of an entire neuronal lineage. Figure 1A shows a schematic of regions used for the cDNA libraries.
We used Illumina-based sequencing to obtain paired-end reads of these cDNA libraries from the SVZ (229 million reads), OB (248 million reads), and DG (157 million reads). To broaden our lncRNA catalog, we also included RNA-seq data from embryonic stem cells (ESCs) and ESC-derived neural progenitors cells (ESC-NPCs) (Guttman et al., 2010). With this collection of over 800 million paired end reads, we used Cufflinks (Trapnell et al., 2010) to perfom ab initio transcript assembly. This method reconstructed a total of 150,313 multi-exonic transcripts, of which 140,118 (93%) overlapped with known protein-coding genes. Our lncRNA annotation pipeline (see Figure 1B and Experimental Procedures) identified 8992 lncRNAs encoded from 5731 loci (Supplementary File 1). 6876 (76.5%) were novel compared to RefSeq genes, 5044 (56.1%) were novel compared to UCSC known genes, and 3680 (40.9%) were novel compared to all Ensembl genes. Interestingly, 2108 transcripts (23.4%) were uniquely recovered from our SVZ/OB/DG reads.
To substantiate the non-coding nature of our lncRNA candidates, we used the Coding Potential Calculator (Kong et al., 2007) and found that over 80% of these transcripts have essentially no protein coding potential (Figure S1A). Consistent with previous studies, lncRNAs were expressed at lower levels than protein-coding genes (2.49 fold difference; Mann-Whitney U, p<0.0001) (Figure S1B), and their exons were less strongly conserved than protein-coding exons by PhastCons scores (Figure S1C).
The transcriptional start site (TSS) of some lncRNAs is proximal (<10 kb) to the promoters of protein-coding genes (Cabili et al., 2011; Hung et al., 2011), and we found that the TSS of 2265 lncRNAs (25.2%) in our catalog were located within 5 kb of a protein coding gene promoter (Figure S1D). Gene onotology (GO) analysis with the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010) revealed that these protein-coding neighbors are enriched for homeodomain-containing transcription factors, genes expressed in the brain, and genes that are typically repressed by Polycomb Repressive Complex 2 in ESCs (Figure S1E). While some lncRNAs had strongly correlated expression with their protein-coding neighbor, as a group they had no obvious correlation (Figure S1F), indicating that expression of this subset of lncRNA is not likely related to local transcriptional activity of protein coding genes.
To verify that the cDNA libraries of the SVZ and OB together represent a transcriptome enriched for adult neurogenesis, we first analyzed mRNA expression in the RNA-seq data. Differential gene expression identified 1621 genes enriched >2 fold in the SVZ cDNA library as compared to the cDNAs from cells in the adjacent non-neurogenic striatum (76.4 million reads). As the primary site where NSCs and transit amplifying cells proliferate, the SVZ was enriched for gene ontology (GO) terms related to cell cycle and mitosis (Figure S2A and S2B). Neuroblasts migrate through the SVZ and into the OB, and, as expected, transcripts related to this migratory neuroblast stage of neurogenesis were enriched in these regions (Lim et al., 2006). The SVZ/OB expression profile included transcription factors known to play key roles in adult neurogenesis, such as Dlx1, Dlx2, Ascl1, and Pax6 (Hsieh, 2012). Furthermore, in situ hybridization (ISH) data from the Allen Brain Atlas (Lein et al., 2007) confirmed the regional expression of many of these SVZ/OB-enriched genes (Figure S2C and Figure S2D), and the SVZ/OB transcriptional profile (923 genes) was enriched for GO terms related to cell migration, development, and neurogenesis (Figure S2E).
lncRNAs have temporally and spatially unique expression patterns
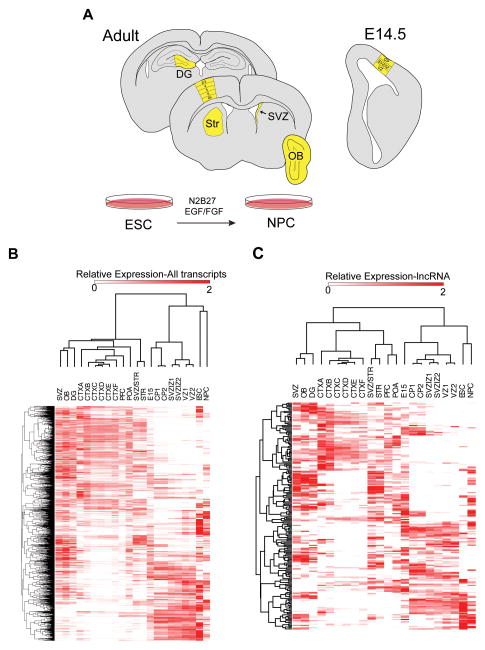
To explore lncRNA expression patterns in multiple adult brain regions and embryonic forebrain development, we analyzed RNA-seq data of the six layers of the adult cortex (Belgard et al., 2011), adult whole prefrontal cortex (PFC), adult preoptic area (POA), whole embryonic day 15 (E15) brain (Gregg et al., 2010), and specific regions of the developing E14.5 cortex (ventricular zone, intermediate zone, and cortical plate) (Ayoub et al., 2011) (Figure 2A and Supplementary File 2). Unsupervised hierarchical clustering of expression profiles revealed region-specific and temporally related expression of both mRNAs and lncRNAs (Figure 2B and 2C). We calculated a specificity score for each transcript (Cabili et al., 2011) and found that the mean score was 0.57 (s.d. 0.21) for lncRNAs, while it was 0.45 (s.d. 0.17) for mRNAs (p-value < 10−325, Wilcoxon Rank Sum Test); thus, lncRNAs exhibit greater brain region and temporal specificity than mRNAs, suggesting that they play important roles in the determination and/or function of specific neural cell types.
Figure 2. mRNAs and lncRNAs have temporally and spatially unique expression patterns, see also Figure S2 and File S2.
(A) Schematic summarizing regions used for this analysis, colored in yellow. (B) Hierarchical clustering results of all transcripts expressed across all samples. (C) Hierarchical clustering results of lncRNAs expressed across all samples. The Pearson correlation coefficient was used as the distance metric. DG=dentate gyrus, STR=striatum, SVZ=subventricular zone, STR/SVZ=mixed dissection including both SVZ and striatal regions, OB= olfactory bulb, CTXA= cortical dissection, layer 2/3, CTXB= cortical dissection, layer 4, CTXC=cortical dissection, layer 5, CTXD=cortical dissection, layer 5, CTXE= cortical dissection, layer 6, CTXF= cortical dissection, layer 6b. POA= preoptic area, PFC= prefrontal cortex, E15= whole embryonic day 15 brain, VZ= ventricular zone of E 14.5 cortex, SVZ/IZ= subventricular zone/ intermediate zone of E14.5 cortex, CP= cortical plate of E14.5 cortex, ESC= cultured embryonic stem cells, NPCs= ESC-derived neural progenitor cells.
lncRNAs are associated with specific neural cell types and neurological disease states
To begin to infer functions for lncRNAs, we investigated the relationship between mRNA and lncRNA transcription by using gene co-expression analysis (GCA) to identify groups of transcripts, or ‘modules,’ whose variation in expression correlate across different brain regions and developmental time points. For mRNAs, module membership distinguishes sets of genes that correspond to specific cell types and biological processes (Oldham et al., 2008), and a similar ‘guilt by association’ approach has been used to assign putative functions to lncRNAs based on their co-expression with protein-coding genes (Guttman et al., 2009).
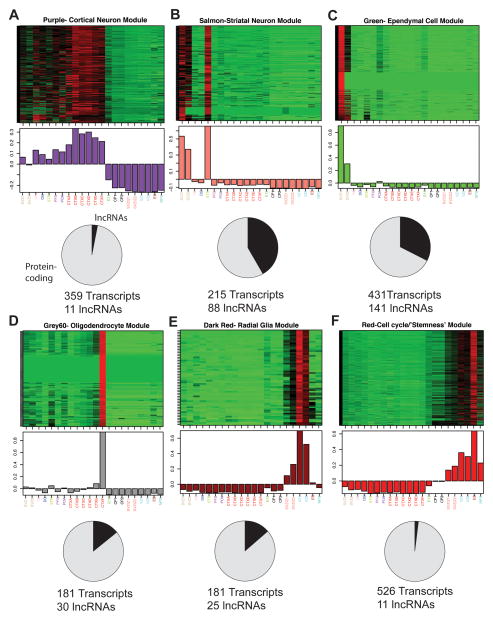
Using RNA-seq data from 22 samples (Figure 2A and Supplementary File 2), we constructed transcript co-expression networks comprised of both mRNAs and lncRNAs. For the 56 modules of co-expressed transcripts, we performed enrichment analysis using gene sets from the Molecular Signatures Database (Subramanian et al., 2005) and other sources (Bult et al., 2008; Cahoy et al., 2008; Thomas et al., 2011; Zhang et al., 2010) to relate modules (described by ‘color’, Figure 3A–F) to specific adult neural cell types including cortical neurons (purple), striatal neurons (salmon), ependymal cells (green), and oligodendrocytes (grey60) (Supplementary File 3).
Figure 3. lncRNAs are associated with specific neural cell types and cellular processes, see also Figure S3 and File S3.
(A–F) Top: heat maps depicting expression levels for six modules of co-expressed transcripts (rows) in 22 samples (columns) representing various brain regions and cell lines. Samples are labeled as in Fig. 2. Red, increased expression; black, neutral expression; green, decreased expression. Middle: barplots of the values of the module eigengenes (Horvath and Dong, 2008), which correspond to the first principal component obtained by singular value decomposition of each module. Modules were characterized by performing enrichment analysis with known gene sets (See Supplemental File 3 and Supplemental Experimental Procedures). Bottom: pie charts indicating the abundance of lncRNAs within each module. Module members are defined as all transcripts that were positively correlated with the module eigengene at P < 2.61e-08 (Supplemental Experimental Procedures).
The dark red module (Figure 3E) was enriched for glial markers but also had a large number of known early neurogenic factors as prominent members (Supplementary File 3). This module was specifically associated with the ventricular zone of the embryonic brain, which contains radial glia, the stem cells of the developing brain and precursors of the adult SVZ-NSCs. We additionally identified a module (red, Figure 3F) specifically associated with the ‘stemness’ transcriptional program and the cell cycle.
Interestingly, some modules were also associated with human disease, notably Huntington’s disease (Thomas et al., 2011), Alzheimer’s disease, convulsive seizures, major depressive disorder, and various cancers (Subramanian et al., 2005; Zhang et al., 2010) (Figure S3A–F). For instance, the striatal neuron module (salmon) correlated with a gene expression set misregulated in Huntington’s disease mouse models, suggesting a potential role for the 88 lncRNAs in this set in this neurodegenerative condition. Taken together, our co-expression analysis provides an important resource as the first annotation of lncRNAs to specific neural cell types in vivo and neurological disease states.
RNA CaptureSeq verifies SVZ lncRNA expression and identifies novel splice isoforms
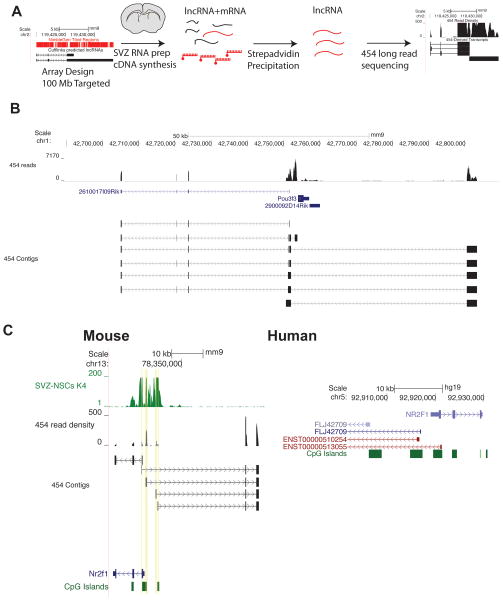
Because many lncRNAs have not been previously annotated and are expressed at low levels, we employed a targeted RNA capture and sequencing strategy (CaptureSeq) to more robustly identify and characterize lncRNAs in the adult SVZ. With RNA CaptureSeq, cDNAs are hybridized to probe libraries tiled against the genomic regions of interest, eluted, and then sequenced (Figure 4A). Through this enrichment, the read coverage of targeted transcripts is dramatically increased (Mercer et al., 2011). Furthermore, by using a 454 GS-FLEX Titanium instrument for sequencing, we obtained longer reads, which improves the delineation of rare splice isoforms.
Figure 4. RNA CaptureSeq validates SVZ lncRNA expression and reveals multiple isoforms and complex locus structures, see also Figure S4.
(A) Schematic of RNA-Capture seq procedure. We used Cufflinlks’ lncRNA assembly to define putative lncRNA loci and designed tiled probe libraries against these loci. The cDNA library was then hybridized to this biotin-labeled probe library, and after purification by streptavidin, the enriched population of lncRNAs was sequenced by 454 (Roche) long-read chemistry. (B) Isotigs assembled at the Pou3f3 locus revealed a distal transcriptional start site for a transcript that can be spliced into known noncoding RNA 2610017I09Rik. (C) CaptureSeq-derived reads correctly assembled known protein-coding gene Nr2f1 and identified 4 distinct TSS’s for a lncRNA transcribed divergently from the Nr2f1 promoter. The syntentic region in human reveals a similar organization of CpG islands and divergent transcriptional start sites for non-coding transcripts. Genes derived from RefSeq are colored purple, genes from Ensembl are red.
For our RNA CaptureSeq probe library, we tiled across 100 MB of putative lncRNA loci and 30 MB of protein-coding regions as a control. We used this library to capture SVZ cDNA for sequencing (5,882,293 reads, median length of 356 bases per read). As expected, de novo assembly of sequences accurately reconstructed protein-coding transcripts and previously annotated lncRNAs (median identity of 90% for protein-coding RefSeq genes and median identity of 95% for annotated non-coding RefSeq RNA). As an example, Evf1 and Evf2, lncRNAs with roles in neural development, have overlapping genomic structures (Feng et al., 2006), and RNA CaptureSeq identified and distinguished both transcripts in the SVZ (Figure S4A). RNA CaptureSeq also eliminated sequencing bias related to transcript abundance (Figure S4B and S4C), and measured expression values were well correlated between CaptureSeq and conventional RNA-seq strategies (Figure S4D and Supplementary Experimental Procedures).
The enrichment and longer reads provided by RNA CaptureSeq enabled the identification of rare lncRNAs as well as uncommon splice isoforms in the SVZ transcriptome, yielding more than 3,500 novel lncRNAs that could not be detected by the short-read sequencing technology. For example, CaptureSeq identified a lncRNA transcript with an intron overlapping Pou3f3, a known neurogenic transcription factor (Figure 4B). In addition to this discovery of a novel lncRNA locus downstream of Pou3f3, RNA CaptureSeq also identified splice isoforms that includes exons of a previously annotated lncRNA (2620017I09Rik) that lies upstream of the Pou3f3 locus. Some lncRNAs are transcribed from multiple transcriptional start sites (TSS), which can be a challenge for transcript assembly. Adjacent to the locus of neurogenic transcription factor Nr2f1, CaptureSeq identified a series of lncRNAs originating from 4 unique TSS’s. This organization of protein-coding gene and multiple lncRNAs is conserved in humans, hinting at an evolutionarily conserved functional significance (Figure 4C). Thus, RNA CaptureSeq, in addition to providing a genome-wide validation of our SVZ lncRNA analysis, demonstrated previously underappreciated complexity to the structure of lncRNA loci. A complete annotation of Capture-Seq derived transcripts is available at http://neurosurgery.ucsf.edu/danlimlab/lncRNA/.
Correlation between histone modifications and lncRNA expression
Methylation of histone lysine residues is a critical determinant of transcriptional activity (Jenuwein and Allis, 2001). In previous work, lncRNA loci have been identified in part by the presence of H3K4me3 at the TSS (Guttman et al., 2009). For protein-coding genes, H3K4me3 enrichment at the TSS correlates with active transcription whereas H3K27me3 is associated with a repressed state. Genes that are “bivalent” for both H3K4me3 and H3K27me3 are generally silenced but remain transcriptionally “poised” for activation or repression (Bernstein et al., 2006). To investigate whether lncRNA loci exhibit a similar correlation between promoter histone modifications and transcription, we performed ChIP-seq for H3K4me3 and H3K27me3 in SVZ-NSC cultures and included sequencing data from ChIP-seq and RNA-seq studies of mouse ESCs, ESC-NPCs, and mouse embryonic fibroblasts (MEFs) (Mikkelsen et al., 2007).
In SVZ-NSCs, 3671 (40.8%) lncRNAs were marked by either H3K4me3 or H3K27me3 and 928 (10.3%) were bivalent (Figure S5A). As has been described for protein-coding genes, these TSS chromatin modifications correlated strongly with lncRNA expression levels: lncRNAs monovalent for H3K4me3 exhibited higher expression levels than those by marked by only H3K27me3 or bivalent chromatin domains (p<0.0001, Mann-Whitney U, Figure S5B). These data suggest that transcription of both lncRNAs and protein coding genes utilizes similar chromatin-based regulatory mechanisms.
A subset of bivalent lncRNAs in ESCs become resolved to monovalent H3K4me3 in SVZ-NSCs
In ESCs, bivalent domains identify key developmental genes. As ESCs differentiate into lineage-specific cell populations, many of these bivalent genes become activated (monovalent H3K4me3) or repressed (monovalent H3K27me3), reflecting the lineage specification and restriction of developmental potential (Bernstein et al., 2006). Thus, genes that are more likely to play roles in the neural identity of SVZ NSCs would be those that are bivalent in ESCs, activated in SVZ-NSCs, and also repressed (H3K27me3 monovalent or bivalent) in a non-neural cell type (MEFs). We found 302 protein-coding genes that meet these criteria, and analysis revealed that the most statistically significant GO terms for these activated genes pertain to early brain development (Figure S5C). For example, proneural Ascl1, Pou3f3, and Pou3f2 were bivalent in ESCs, H3K4me3-monovalent in SVZ-NSCs, and H3K27me3-repressed in MEFs (Figure S5D).
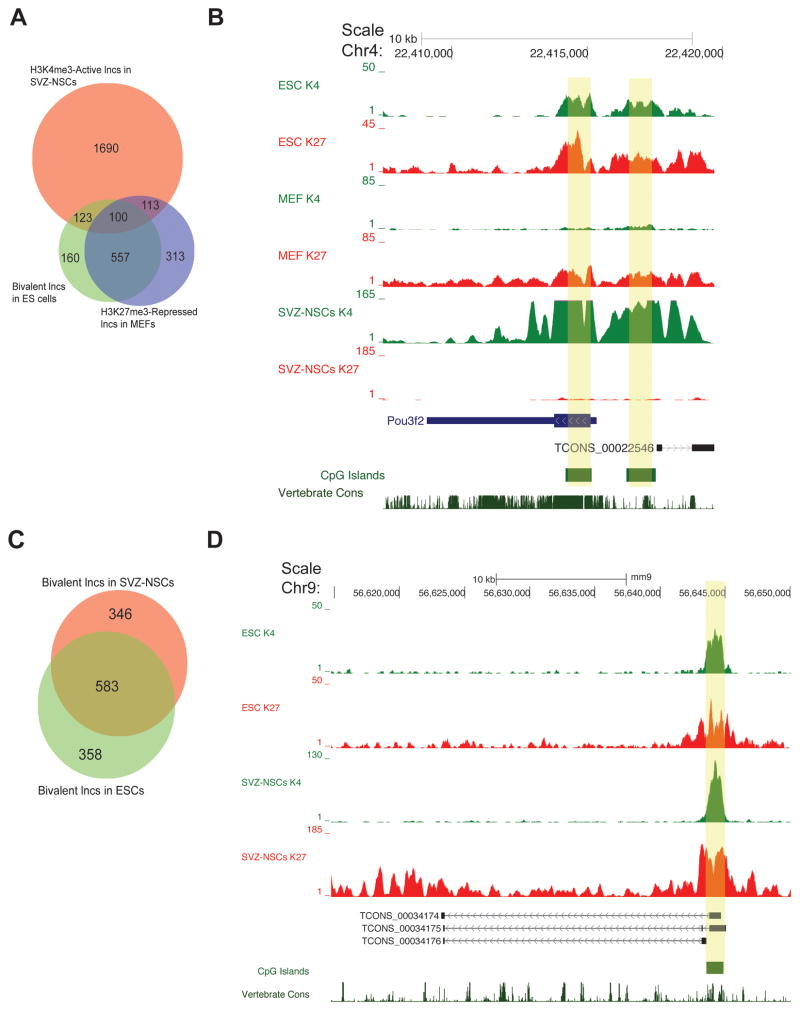
100 lncRNAs have a similar pattern of chromatin-based changes (Figure 5A). Furthermore, 76% of this set of lncRNAs was also monovalent for H3K4me3 in ESC-NPCs, suggesting that these lncRNAs are common to an early neural development transcriptional program. An example is lnc-pou3f2: this novel lncRNA is 2 kb upstream of known neurogenic transcription factor Pou3f2. Both the lnc-pou3f2 and the Pou3f2 loci were bivalent in ESCs, monovalent for H3K4me3 in NSCs, and H3K27me3 monovalent in MEFs (Figure 5B). Given the known relationship between chromatin modifications and the expression of key developmental regulators, we propose that this set of lncRNAs is enriched for those that play roles in early neural commitment in the adult SVZ.
Figure 5. lncRNA loci can be bivalent in stem cell populations, see also Figure S5.
(A) Venn diagram highlighting lncRNAs that were bivalent in ESCs, monovalent H3K4me3 in SVZ-NSCs, and H3K27me3-repressed (monovalent or bivalent) in MEFs. (B) The Pou3f2 promoter and the promoter (yellow boxes) of a nearby lncRNA demonstrated a similar pattern of histone modifications (bivalent in ESCs, repressed in MEFs, and activated in SVZ-NSCs). (C) Venn diagram demonstrating the number of lncRNAs that were bivalent in both ESCs and SVZ-NSCs. (D) A novel lncRNA locus ~50 kb downstream of protein-coding gene Odf3l1. The promoter was bivalent in both SVZ-NSCs and ESCs.
lncRNAs can retain bivalency in an adult stem cell population
Tissue-specific stem cells also retain bivalency at key loci, possibly reflecting retained gene expression plasticity (Cui et al., 2009; Lien et al., 2011). Protein coding genes that are bivalent in ESCs and NSCs were highly enriched for GO terms related to neurogenesis (e.g., neuron differentiation, axonogenesis, Figure S5E). For instance, in adult SVZ NSCs, Dlx1 and Dlx2 are transcription factors required for interneuron development, and these were bivalent in NSCs (Figure S5F). Thus, the identification of lncRNAs that are bivalent in both ESCs and NSCs might enrich for those involved in neuronal differentiation. 583 lncRNAs met these criteria (Figure 5C), such as three splice variants encoded from a novel lncRNA locus located ~50 kb upstream of protein-coding gene Odf3l1 (Figure 5D). We propose that lncRNAs bivalent in SVZ-NSCs are enriched for those that function in neuronal lineage specification.
lncRNAs are dynamically regulated during neuronal differentiation
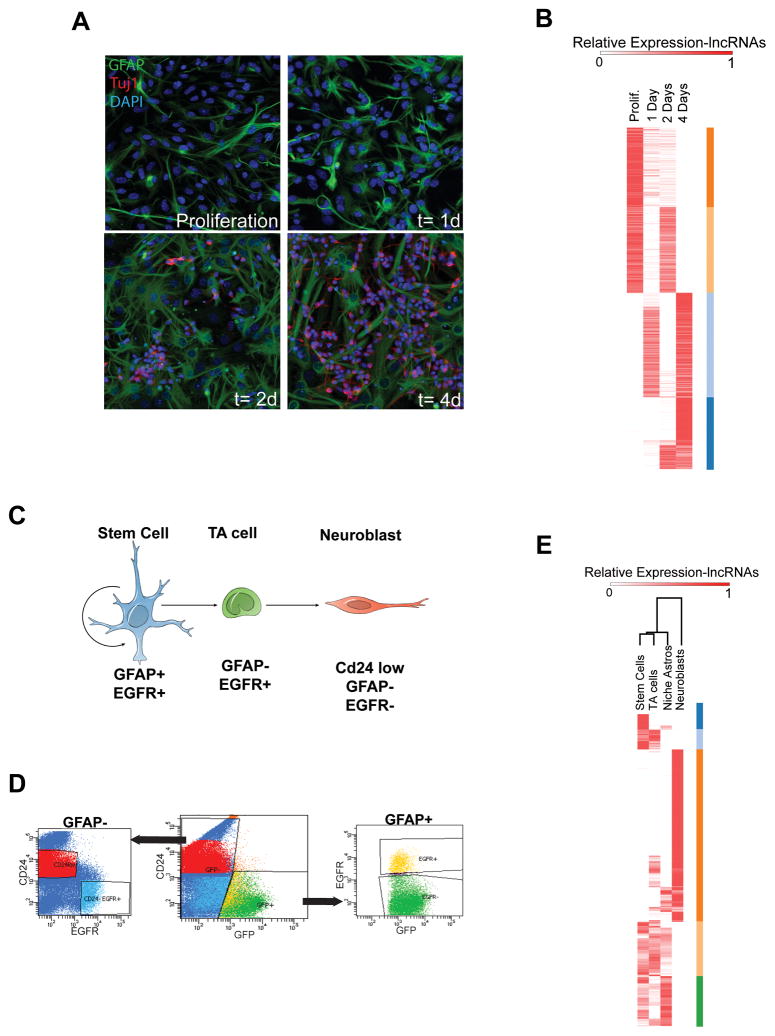
We next sought to define the dynamic changes in lncRNA expression during SVZ neurogenesis. SVZ-NSCs cultured as a monolayer can efficiently recapitulate key aspects of in vivo neurogenesis as assessed by immunocytochemistry (ICC, S6A)(Scheffler et al., 2005). We generated cDNA libraries from SVZ-NSC cultures in self-renewal conditions and after 1,2, and 4 days (1,2, and 4d) of differentiation and hybridized to both custom lncRNA (see Methods) and standard gene expression arrays. Included in the set of upregulated transcripts were genes related to SVZ neurogenesis (e.g., Dlx1/2 and Dlx5/6) (Supplementary File 4). Also as expected, genes expressed at higher levels early in the SVZ lineage (e.g. Egfr and Nestin) were in the set of downregulated transcripts (Supplementary File 4). Like mRNA transcripts, lncRNAs also exhibited similar patterns of induction and repression (Figure 6B and S6B) over this 4-day differentiation timecourse.
Figure 6. Analysis of lncRNA expression in the SVZ lineage in vitro and in vivo, see also Figure S6 and File S4.
(A) Immunocytochemistry (ICC) of SVZ-NSC differentiation in vitro. In proliferation conditions, the culture is composed of neural precursor cells including GFAP+ (green) NSCs. After growth factor withdrawal, cells in these cultures differentiate into Tuj1+ neuroblasts (red, increasing numbers at 2d and 4d). (B) Heat map representing expression of lncRNAs that were changed > 4 fold from proliferation conditions to 4 d of differentiation. Color bars (orange, peach, light blue, dark blue) at the right represent gene clusters resulting from k-means clustering, k=4, Pearson distance metric. (C) Schematic of the SVZ lineage. GFAP+, EGFR+ stem cells (blue) give rise to transit amplifying (TA, green) cells. These TA cells give rise to Cd24+ low migratory neuroblasts (red). (D) FACS plots for isolation of the SVZ lineage. Cells were dissociated from freshly dissected SVZ tissue from the hGFAP-GFP mouse and stained with EGF conjugated to the A667 fluorophore and a CD24 antibody conjugated to PE. (E) Heatmap of lncRNAs differentially expressed throughout the SVZ lineage in vivo. Genes differentially expressed >2 fold between activated NSCs and neuroblasts were k-means clustered using the Pearson correlation metric, k=5. Color bars at the right (dark blue, light blue, orange, peach, green) represent gene clusters resulting from k-means clustering.
In vivo SVZ lineage analysis of lncRNA expression
The adult SVZ contains three major cell types that represent a developmental continuum: (1) activated NSCs, which express glial fibrillary acidic protein (GFAP) and the epidermal growth factor receptor (EGFR), (2) transit amplifying cells, which are EGFR-positive but GFAP-negative, and (3) migratory neuroblasts, which are CD24-positive (Figure 6C). In addition, the SVZ contains GFAP+ cells that do not express EGFR, and these have been termed ‘niche’ astrocytes (Pastrana et al., 2009). We used these cell-specific characteristics to perform fluorescent-activated cell sorting (FACS) to acutely isolate cell populations representing each stage of this neurogenic lineage and the niche astrocytes (Figure 6D). cDNA libraries for each SVZ cell type were generated and hybridized to our custom lncRNA and standard gene expression microarrays. Expression levels of both protein-coding genes and lncRNAs were visualized by heat maps organized by k-means clustering (transcripts) and unsupervised hierarcical clustering (cell types) (Figure 6E and S6C).
To confirm the separation of SVZ cells, we examined differential mRNA expression. 12,812 protein-coding probesets were differentially expressed (>2-fold) in a comparison between activated SVZ-NSCs and migratory neuroblasts (Supplementary file 4). As SVZ-NSCs become activated and differentiate into transit-amplifying cells, Dlx1/2 and Ascl1 become upregulated (Doetsch et al., 2002; Kim et al., 2007) and this was reflected in our transcriptional profiles (Supplementary file 4). As expected, the transcriptome of migratory neuroblasts was enriched for Dlx1/2 downstream targets, including Dlx5 and Arx, as well as markers of young neurons, including Tubb3 (Supplementary File 4). Thus, these transcriptomes represent distinct stages of the SVZ neurogenesis and also distinguish niche astrocytes from NSCs.
Similar to the cell culture data (Figure 6B), we found sets of lncRNAs that showed transient increases in TA cells, repression throughout differentiation, and significant induction in the terminally differentiated neuroblast population (Figure 6E). By integrating our chromatin state maps with this microarray expression data, we were able to begin to define a ‘lncRNA signature’ for each stage of neurogenesis in vivo (Supplementary File 5).
Identification of lncRNAs with roles in adult SVZ neurogenesis
To facilitate the identification of lncRNAs with potential roles in SVZ neurogenesis, we constructed an online resource that allows the user to easily filter the lncRNA catalogue for multiple variables including locus chromatin-state, expression in FACS-isolated SVZ cells, and regulation during in vitro neurogenesis (http://neurosurgery.ucsf.edu/danlimlab/lncRNA/). Using this resource, we filtered for those lncRNAs that were bivalent in ESCs and H3K27me3-repressed in MEFs. Of this set, which includes lnc-pou3f2, 100 lncRNAs were monovalent for H3K4me3 in SVZ-NSCs (Figure 5A and 5B), which would predict expression in the adult SVZ; indeed, in situ hybridization (ISH, Figure S7A and S7B) revealed lnc-pou3f2 expression in the SVZ, and, as predicted by the FACS microarray data, this transcript was not detected in the OB (Figure S7B).
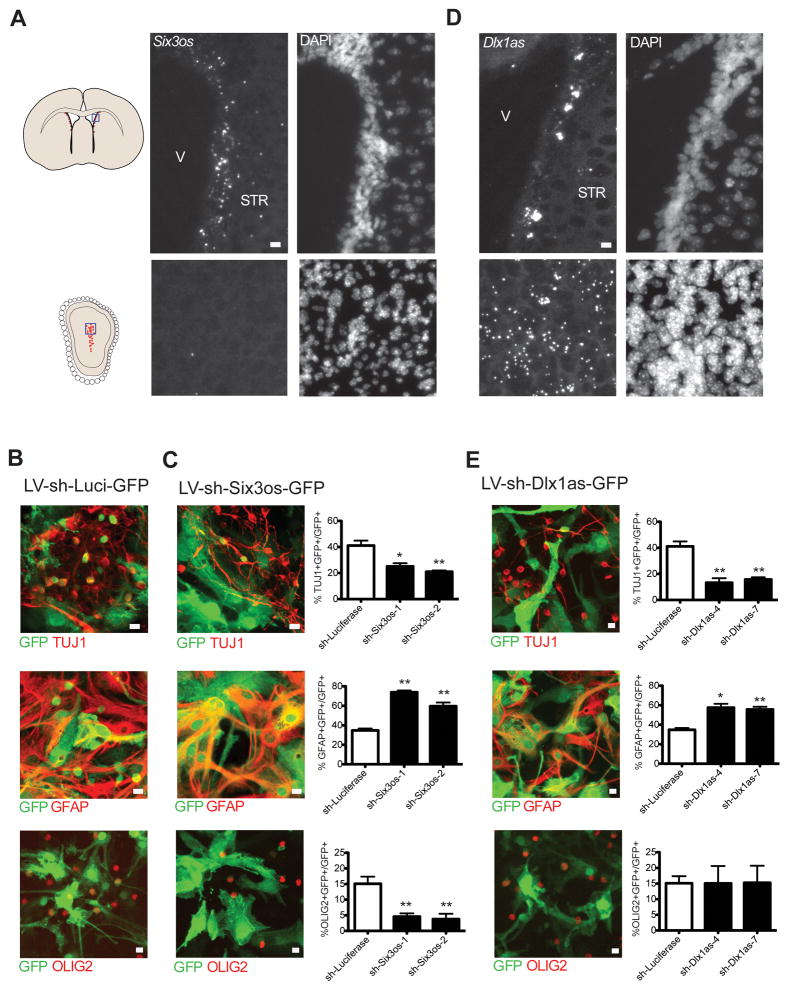
Like lnc-pou3f2, lncRNA Six3os was also monovalent for H3K4me3 in SVZ-NSCs and downregulated in neuroblasts. Consistent with these observations, Six3os transcripts were detected in the SVZ but not the OB core (Figure 7A). To further investigate the role of Six3os in SVZ NSCs, we used lentiviruses to separately introduce two different short hairpin RNA (shRNA) sequences to knockdown Six3os (LV-sh-Six3os-GFP) in monolayers of SVZ-NSCs. After confirming Six3os knockdown in proliferating NSCs (Figure S7C), we assessed neuronal and glial lineages from LV-sh-Six3os-GFP infected cells in comparison to controls infected with LV-sh-luc-GFP. After 7 d of differentiation, there were 2-fold fewer Tuj1-positive cells and 2.5-fold fewer cells expressing OLIG2, a marker of the oligodendrocyte lineage (Zhou et al., 2002). These decreases were accompanied by an increase in the number of GFAP+ cells (Figure 7B and 7C).
Figure 7. Functional validation of lncRNA candidates, see also Figure S7 and File S5.
(A) In situ hybridization (ISH) for Six3os using branched DNA probes. Positive signal is revealed by Fast Red alkaline phosphatase substrates, which appears as highly fluorescent, punctate deposits (left panels); DAPI nuclear counterstain is shown at the right. Blue box in SVZ and OB coronal schematics at left indicate regions shown at right. Scale bars= 10 μm. V=ventricle, STR= striatum. (B) Control (LV-sh-Luci-GFP) lentiviral infections in SVZ-NSC cultures after 7 days of differentiation. Top, immunocytochemistry (ICC) for Tuj1 (red) and GFP (green), Middle, ICC for GFAP (red) and GFP (green), Bottom, ICC for OLIG2 (red) and GFP (green). (C) Analysis of Six3os knockdown in SVZ-NSCs after 7 days of differentiation. Two different constructs were used (sh-Six3os-1, sh-Six3os-2). Top, immunocytochemistry (ICC) for Tuj1 (red) and GFP (green), Middle, ICC for GFAP (red) and GFP (green), Bottom, ICC for OLIG2 (red) and GFP (green) after infection with control vector expressing shRNAs targeting Six3os (LV-sh-Six3os-GFP). Quantification of data is presented at right. Scale bars= 10 μm. Error bars= SEM, 5–6 replicates for control group, 2–3 per experimental group. *p<.05, **p<0.01, compared to sh-Luci, two-tailed t-test. (D) ISH with branched DNA probes for Dlx1as in the SVZ (top) and OB (bottom). Scale bars= 10 μm. V=ventricle, STR= striatum. (E) Analysis of Dlx1as knockdown after 7 days of differentiation. Two unique targeting sequences were used (sh-Dlx1as-4, sh-Dlx1as-7). Top, immunocytochemistry (ICC) for Tuj1 (red) and GFP (green), Middle, ICC for GFAP (red) and GFP (green), Bottom, ICC for OLIG2 (red) and GFP (green). Quantification of of data is presented at right. Scale bars= 10 μm. Error bars= SEM, 5–6 replicates for control group, 3 per experimental group. *p<.05, **p<0.01, compared to sh-Luci, two-tailed t-test.
The Dlx1/2 bigene cluster encodes lncRNA Dlx1as (Liu et al., 1997; Dinger et al., 2008), and this locus was also bivalent in ESCs and H3K27me3 monovalent in MEFs. We used SVZ-NSC monolayer cultures to further investigate the chromatin state of the Dlx1as TSS. In self-renewal conditions, Dlx1as was bivalent, and after 30 hours (h) of differentiation, H3K27me3 decreased, correlating with the start of Dlx1as upregulation (Figure S7D and S7E). Interestingly, we also found enrichement of the H3K27me3-specific demethylase JMJD3 (Agger et al., 2007) at the Dlx1as TSS during differentiation (Figure S7F), suggesting that this chromatin-modifying factor plays a role in the activation of this lncRNA. Consistent with the transcriptional upregulation of Dlx1as during SVZ neurogenesis in vitro, we observed robust Dlx1as expression in SVZ regions with migratory neuroblasts and the OB core (Figure 7D). We designed two knockdown constructs targeting the splice junction of Dlx1as, and verified that these constructs target Dlx1as and not full-length Dlx1 transcript (Figure S7G). Knockdown of Dlx1as caused a decrease in expression of Dlx1 and Dlx2 after two days of differentiation compared to control (Figure S7H), suggesting this lncRNA can regulate expression of its protein-coding gene neighbors. After 7 d of differentiation, we found a nearly 3-fold decrease in Tuj1+ neuroblasts, and a ~60% increase in the number of GFAP+ astrocytes. In contrast to knockdown of Six3os, the production of OLIG2+ cells was unaffected by Dlx1as knockdown (Figure 7B and 7E).
DISCUSSION
We performed an in-depth analysis of lncRNA expression of adult SVZ-OB neurogenesis, an excellent in vivo model system for the study of multipotent stem cells and neural development. Our use of two high-throughput sequencing-based approaches for the study of the lncRNA transcriptome (RNA-seq and RNA CaptureSeq) provided complementary datasets that together allowed the identification of thousands of novel lncRNAs, confirmation of rare transcripts, and resolution of previously unappreciated complexity of lncRNA loci.
Like the loci of genes encoding key developmental transcription factors, a subset of lncRNA loci showed changes of chromatin state during lineage specification. By integrating these chromatin state maps with data from custom microarrays and FACS purification of the SVZ lineage, our online resource (http://neurosurgery.ucsf.edu/danlimlab/lncRNA/) and files (Supplementary File 5) facilitate the identification lncRNAs with potential roles in adult NSCs as well as neural development. Interestingly, we found that Dlx1as is required selectively for the SVZ neuronal lineage, whereas Six3os appears to play a role in both neuronal and oligodendrocyte differentiation. These data indicate that lncRNAs can play key roles in the glial-neuronal lineage specification of multipotent adult stem cells.
2,265 lncRNAs had proximal protein-coding gene neighbors (Figure S1D), and this gene set was enriched for homeodomain-containing genes. For instance, Six3os is proximal to the Six3 homeobox gene, and Dlx1as is encoded from the Dlx1/2 bigene cluster (Liu et al., 1997; Dinger et al., 2008). Interestingly, Dlx1as knockdown caused a decrease in Dlx1/2 expression (Figure S7H), suggesting that this lncRNA plays a role in neuronal differentiation by regulating expression of its homeobox gene neighbors. In developing retina, knockdown of Six3os results in deficits in lineage specification through modulation of SIX3 activity (Rapicavoli et al., 2011); it will be interesting to investigate whether the defect in SVZ neurogenesis with Six3os knockdown (Figure 7C) similarly involves a change in SIX3 activity. Taken together, our genome wide analysis and functional data further support the notion that lncRNAs and homeobox-gene neighbors function cooperatively (Rapicavoli and Blackshaw, 2009).
A recent model of lncRNA action suggests that lineage-specific lncRNAs become activated during differentiation and guide histone modifications that create cell-type specific transcriptional programs (Guttman et al., 2011). MLL1 is a trithorax group (trxG) chromatin-modifying factor that is enriched at Dlx2 during SVZ NSC differentiation and is required for proper Dlx2 expression (Lim et al., 2009); however, how MLL1 is targeted to Dlx2 is not known. Interestingly, in mouse ESCs, lncRNA Mistral directly binds MLL1 and recruits it to Hoxa6 and Hoxa7 (Bertani et al., 2011) and lncRNA HOTTIP recruits MLL1 through an interaction with WDR5 to distal HOXA genes in human fibroblasts (Wang et al., 2011). Our work provides a useful resource for the identification of such lncRNAs. For instance, lncRNAs that immunoprecipitate with chromatin modifiers could be identified by hybridization to the lncRNA microarray and then filtered online by multiple other criteria (e.g., enrichment in neuroblasts, upregulation during neurogenesis, bivalency in ESCs, repression in MEFs).
Our analysis of chromatin state maps and transcript expression suggest that histone modifications correlate with lncRNA expression in a manner similar to that of protein coding genes. Some lncRNA loci were bivalent in both ESCs and SVZ-NSCs, and many of these lncRNA loci became transcriptionally active in SVZ neuroblasts, supporting their candidacy as key determinates of neurogenesis. In SVZ-NSC monolayer cultures, Dlx1as was bivalent and H3K27me3 repression decreased during neuronal differentiation (Figure S7D), correlating with the upregulation of Dlx1as transcription (Figure S7E). Interestingly, we also found enrichment of the H3K27me3-specific demethylase JMJD3 at the Dlx1as locus (Figure S7F), suggesting that active removal of repressive histone modifications plays a role in the expression of lncRNAs. Overall, our data raise the possibility that lncRNA loci, like protein coding genes, are targeted by chromatin modifying factors that have critical roles in development.
While this study attempted to be as comprehensive as possible, it is possible that some lncRNAs important for SVZ neurogenesis were not identified. The initial sequencing experiments were performed on microdissected tissues that contain several cell types. Even at our sequencing depth, transcripts that are expressed at low copy number in a small number of cells might not be detected. Despite this potential shortcoming, we were still able to identify thousands of novel lncRNA transcripts. Furthermore, our initial catalog proved sufficient for our primary objective, which was to integrate complementary data analysis strategies and experimental methods to identify lncRNA expression patterns coherent to an in vivo experimental model system.
The role of lncRNAs in development and disease is in the early states of investigation, and our analysis of the SVZ lineage provides a resource for the movement of this research into in vivo studies. More broadly, this work presents a generalizable workflow for the identification and categorization of novel transcripts, both coding and noncoding.
Experimental Procedures
Brain Dissection for RNA extraction
The brain from adult (>P60) male C57/B6 mice was removed from the skull and placed in ice-cold L15 media and a 0.5 mm thick coronal slab was obtained. The lateral SVZ and striatum were then microdissected, avoiding contamination from the corpus callosum. FACS of SVZ cells was performed as described (Pastrana, et al. 2009). DG was microdissected in ice-cold L15 media from 300 μm thick coronal sections obtained with a Vibratome.
High-Throughput sequencing
Illumina libraries were prepared using standard protocols. Sequencing was performed with a Genome Analyzer IIx or Hi-Seq instrument (Illumina). CaptureSeq followed by 454 sequencing (GS-FLX Titanium, Roche) was performed essentially as described (Mercer et al., 2011).
Sequencing Data Processing
Sequencing analysis was performed on the Galaxy platform (Goecks et al., 2010), using TopHat (Trapnell et al., 2009) and Bowtie (Langmead et al., 2009) for read alignment. Supplementary file 2 describes all novel and previously published datasets used in this work. Cufflinks and Cuffdiff (Trapnell et al., 2010) were used for transcript reconstruction, quantification, and differential expression analysis. MACS was used to call Chip-Seq peaks (Zhang et al., 2008). For longer 454 reads, BLAT (Kent, 2002) was used to map the 454 reads to the transcriptome, and Newbler (Roche) was used to construct contigs.
lncRNA Catalog Assembly
.gtf files from Cufflinks were filtered using the UCSC genome browser to remove all single-exonic transcripts as well as any transcripts overlapping RefSeq genes, known protein-coding genes from other species using the ‘Other Ref seq’ track and the ‘XenoRefGene’ table. These filtered .gtf files were used as input for CuffCompare, and the complete Ensembl gene annotation .gtf file (Downloaded from: www.ensembl.org/info/data/ftp on 5/23/2011) was used as reference. We kept any transcript with the CuffCompare designation “I,” which marks all novel intergenic sequences. To these sequences, we added annotated lincRNAs and processed_transcripts from Ensembl. Finally, we excluded all genes known to RefSeq, giving the 8992 transcripts in Supplementary File 1, a.bed file containing our putative lncRNAs. This file was merged with the Illumina iGenomes UCSC mm9 gtf annotation of all RefSeq genes (http://cufflinks.cbcb.umd.edu/igenomes.html on 1/5/2012), generating a final comprehensive annotation of non-coding and coding transcripts, which was then used for all subsequent analysis of RNA-seq data.
CaptureSeq library and Expression Arrays
Capture-Seq and Expression Array designs are available upon request. For the Capture-Seq probe set, probes were designed to tile across ~100 MB of putative noncoding loci and ~30 MB protein-coding control loci, for a total of 6287 genomic targets. For expression array probe selection, a FASTA file was generated representing all putative lncRNAs. A description of probe selection strategies for both sequence-capture and expression arrays can be found in Technical Note: Roche Nimblegen Probe Design Fundamentals.
In situ hybridization
In situs were performed using the QuantiGene ViewRNA ISH tissue Assay (Affymetrix). Probes were designed based on lncRNA sequences determined by Cufflinks. Images were taken on a DMI4000B fluorescent microscope (Leica).
Supplementary Material
Highlights.
RNA-seq, RNA Capture-Seq of SVZ-NSCs identifies novel lncRNAs and splice variants.
LncRNA loci have dynamic chromatin signatures, including bivalency in SVZ-NSCs.
Online resource facilitates discovery of lncRNAs involved in adult neurogenesis.
LncRNAs can regulate glial-neuronal lineage specification of SVZ-NSCs.
Acknowledgments
This project was supported by NIH DP2-OD006505-01, VA 1I01 BX000252-01, and the Sontag Foundation to D.A.L, R01CA163336 and the Sontag Foundation to J.S.S., the UCSF Program for Breakthrough Biomedical Research, which is funded in part by the Sandler Foundation, to M.C.O, NIH GMS K12-GM081266 to G.G.-R., a training grant from the California Institute for Regenerative Medicine (CIRM) to A.D.R., MSTP training grant 2T32GM007618-34 to A.D.R. and R.N.D, and facilities and resources provided by the San Francisco Veterans Affairs Medical Center.
Footnotes
ACCESSION NUMBERS
All data are deposited in NCBI GEO under accession number XXXXX
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsibler J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, García-Moreno F, Molnár Z, Margulies EH, Ponting CP. A Transcriptomic Atlas of Mouse Neocortical Layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The Noncoding RNA Mistral Activates Hoxa6 and Hoxa7 Expression and Stem Cell Differentiation by Recruiting MLL1 to Chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Mouse Brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comp Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 2012;26:1010–1021. doi: 10.1101/gad.187336.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–686. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Leung CT, Reed RR, Johnson JE. In vivo analysis of Ascl1 defined progenitors reveals distinct developmental dynamics during adult neurogenesis and gliogenesis. J Neurosci. 2007;27:12764–12774. doi: 10.1523/JNEUROSCI.3178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev of Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E. Genome-wide Maps of Histone Modifications Unwind In Vivo Chromatin States of the Hair Follicle Lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Suárez-Fariñas M, Naef F, Hacker CR, Menn B, Takebayashi H, Magnasco M, Patil N, Alvarez-Buylla A. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JL. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2011;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn. 2009;238:2103–2114. doi: 10.1002/dvdy.21844. [DOI] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler B, Walton NM, Lin DD, Goetz AK, Enikolopov G, Roper SN, Steindler DA. Phenotypic and functional characterization of adult brain neuropoiesis. Proc Natl Acad Sci. 2005;102:9353–9358. doi: 10.1073/pnas.0503965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA, Coppola G, Tang B, Kuhn A, Kim S, Geschwind DH, Brown TB, Luthi-Carter R, Ehrlich ME. In vivo cell-autonomous transcriptional abnormalities revealed in mice expressing mutant huntingtin in striatal but not cortical neurons. Hum Mol Genet. 2011;20:1049–1060. doi: 10.1093/hmg/ddq548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved Function of lincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, De S, Garner JR, Smith K, Wang SA, Becker KG. Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med Genom. 2010;3:1. doi: 10.1186/1755-8794-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.