Abstract
Human mitochondrial mRNAs utilize the universal AUG and the unconventional isoleucine AUA codons for methionine. In contrast to translation in the cytoplasm, human mitochondria use one tRNA, hmtRNAMetCAU, to read AUG and AUA codons at both the peptidyl- (P-), and aminoacyl-(A-) sites of the ribosome. The hmtRNAMetCAU has a unique post-transcriptional modification, 5-formylcytidine, at the wobble position 34 (f5C34), and a cytidine substituting for the invariant uridine at position 33 of the canonical “U-turn” in tRNAs. The structure of the tRNA's anticodon stem and loop domain (hmtASLMetCAU), determined by NMR restrained molecular modeling, revealed how the f5C34 modification facilitates the decoding of AUA at the P- and A-sites. The f5C34 defined a reduced conformational space for the nucleoside, in what appears to have restricted the conformational dynamics of the anticodon bases of the modified hmtASLMetCAU. The hmtASLMetCAU exhibited a “C-turn” conformation that has some characteristics of the U-turn motif. Codon binding studies with both E. coli and bovine mitochondrial ribosomes revealed that the f5C34 facilitates AUA binding in the A-site and suggested that the modification favorably alters the ASL's binding kinetics. Mitochondrial translation by many organisms including humans sometimes initiates with the universal isoleucine codons AUU and AUC. The f5C34 enabled P-site codon binding to these normally isoleucine codons. Thus, the physicochemical properties of this one modification, f5C34, expand codon recognition from the traditional AUG to the non-traditional, synonymous codons AUU and AUC as well as AUA, in the reassignment of universal codons in the mitochondria.
Keywords: 5-formylcytidine, NMR structure, mitochondrial translation, codon expansion, near cognate decoding
Introduction
Mitochondria are membrane-bound organelles with their own genetic information. Through oxidative phosphorylation, the organelle generates the majority of the eukaryotic cell's energy resource, ATP. Most of the approximately 2000 proteins that compose the mitochondrion are encoded by nuclear DNA and are imported. However, the mitochondrial genome encodes thirteen critical components of the electron transfer chain and the ATP synthase. These proteins are the only products of the organelle's protein synthesizing machinery. Other components required for translation, including the ribosomal proteins and the auxiliary initiation, elongation and termination factors, are the products of nuclear genes. The mammalian mitochondrial DNA also encodes 22 tRNAs (mtRNA), one for each of 18 amino acids and two each for serine and leucine.1 Cytoplasmic tRNAs are not imported into the mammalian mitochondria,2 perhaps with the one exception of cytoplasmic glutamine tRNA imported into the isolated mitochondria of cells from individuals with myoclonic epilepsy with ragged-red fiber (MERRF).3
This minimal set of 22 mtRNAs must respond to all of the amino acid codons. In order to facilitate the reading of the amino acid codes in mammalian mitochondria, an unmodified uridine in the “wobble” position 34 of the mtRNA recognizes all four nucleotides at the third position of the synonymous codons, a guanine in position 34 recognizes only pyrimidines, and a modified uridine discriminates between purines and pyrimidines.4 Thus, there are some exceptions to the reading of the universal genetic code. Sequence analysis of mitochondrial genomes from various organisms has revealed that there are many codon reassignments, a unique feature of the non-plant mitochondrial translational systems.4,5 For example, in mammalian mitochondria the universal stop codon UGA codes for tryptophan, and the two universal arginine codons AGA and AGG either terminate translation, or promote frameshifting in the human mitochondrial ribosome.6 Of particular interest is the universal isoleucine codon AUA that is used to specify methionine for initiation and elongation of translation.1,7-10
In the cytoplasm, two tRNAMetCAU decode the single methionine codon AUG.1 One is an initiator tRNA (tRNAiMetCAU for eukaryotes and tRNAfMetCAU for prokaryotes) that decodes AUG in the ribosome's peptidyl- or P-site at the initiation of translation. The other tRNAMetCAU is responsible for recognizing the AUG codons internal to the mRNA (tRNAmMetCAU). In contrast, the single mitochondrial tRNAMetCAU plays a unique role since it supports both the initiation and elongation of protein synthesis by responding to the universal methionine codon AUG in the A- and P-sites.10 The tRNAMetCAU anticodon is the Watson-Crick complement for the methionine codon AUG. This single mtRNAMetCAU also reads the codon AUA which is present in mitochondrial mRNAs as 20% of the initiator codons, and 80% of internal methionine codons. Francis Crick's Wobble Hypothesis explained how a tRNA would be able to read more than one codon, but also predicted an unfavorable pairing of the anticodon CAU to the codon AUA due to the C●A mismatch at the wobble position.11 Since the AUA codon occurs much more frequently than the AUG codon in mammalian mitochondrial genomes,1 it has been assumed that some special mechanism must exist in the mitochondrial translation system to allow the AUA codon to be translated as methionine.
The unique post-transcriptional modification at the human mtRNAMetCAU (hmtRNAMetCAU) wobble position 34, 5-formylcytidine (f5C34) (Fig. 1c),12,13 may play a crucial role in the recognition of both methionine codons, the non-universal AUA and the universal AUG.14,15 The hmtRNAMetCAU shares an important similarity to elongator tRNAMetCAU in that both have a wobble position-34 modification; in Escherichia coli the modification is N4-acetylcytidine, while in humans it is 2′-O-methylcytidine.16 The bacterial and eukaryotic cytoplasmic initiator tRNAMetCAU are unmodified at the wobble position. The f5C modification has been found in the mitochondrial tRNAMetCAU of humans, bovines, and rodents and is considered universal in mammalian mtRNAMetCAU.10,13,17 The modification also is found in the mtRNAMetCAU of nematodes, squids, frogs, chickens, and fruit flies.4,13,18,19 Human mtRNAMetCAU is a hybrid of the mammalian cytoplasmic initiator and elongator tRNAMetCAU species. The hmtRNAMetCAU has the three consecutive G-C pairs at the bottom of the anticodon stem characteristic of initiator tRNAs (Fig. 1). However, this tRNA has a number of unusual features. The dihydrouridine (D)-loop of the human mitochondrial tRNAMetCAU is smaller than that of the canonical cytoplasmic tRNAs and lacks the G residues at 18 and 19 that normally facilitate interactions with the T-loop. The minor loop is small and does not contain the common residue G47. The T-stem has two adjacent pyrimidine:pyrimidine pairs (U-U and U-Ψ). The T-loop lacks the TΨC sequence and contains only 6 nucleotides instead of the normal seven. There is also a mismatch in the acceptor stem.
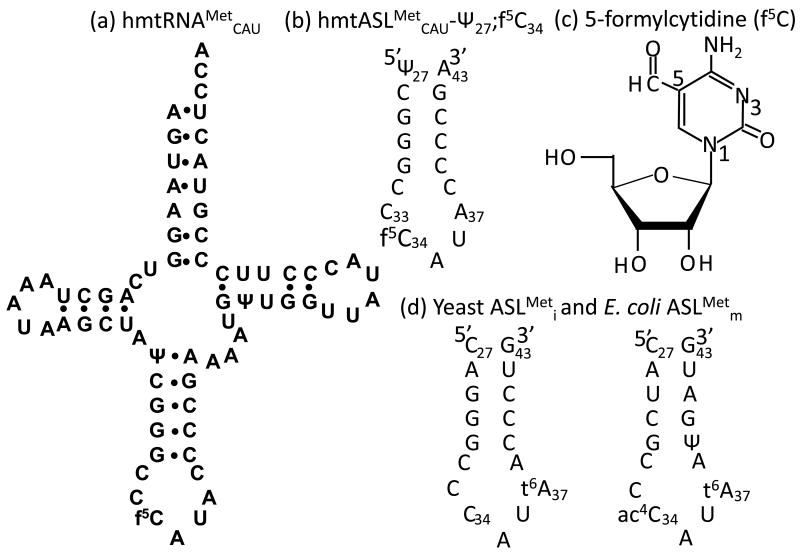
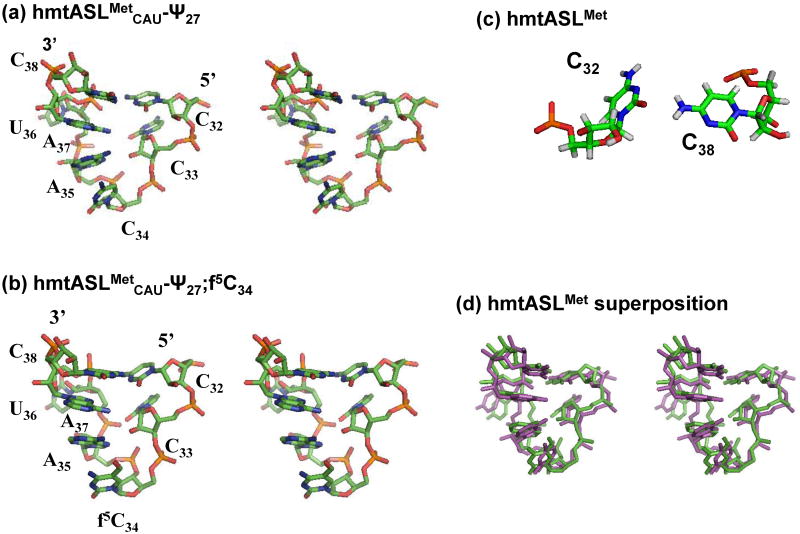
Figure 1.
Primary sequence and secondary structure of the human mtRNAMetCAU and its anticodon stem and loop domain (hmtASLMetCAU-Ψ27;f5C34). Primary sequence and secondary structure of the human (a) mtRNAMetCAU and the (b) hmtASLMetCAU-Ψ27;f5C34. (c) Chemical structure of the 5-formylcytydine modification, f5C34. Numbering of the nucleobase atoms is shown. (d) Nucleoside sequence and secondary structure of the anticodon stem and loop domains of the yeast tRNAMeti (left) and E.coli tRNAMetm (right).
Our studies focus on the folding, structure and codon binding properties of the hmtRNAMetCAU,15,20,21 in particular that of tRNA's anticodon stem and loop domain, hmtASLMetCAU.15 The heptadecamer hmtASLMetCAU has a five base-paired stem consisting of the three contiguous G●C/C●G base pairs (residues 29–31/39–41) that are proximal to the loop and contribute thermodynamic stability to the RNA through stacking (Fig. 1) - a hallmark of initiator tRNAMet that increases its affinity for the P-site.22 As with the mammalian initiator tRNAMet, the hmtASLMetCAU has a cytidine at position-33. The 33rd residue of tRNAs is usually occupied by the invariant U33, responsible for the “U-turn” in the backbone that presents the anticodon for codon binding. Other than in the hmtASLMetCAU, C33 has been found only in the cytoplasmic initiator tRNAMetCAU of insects, plants, and vertebrates.16 In the yeast initiator tRNAiMetCAU, the loop is closed by a non-canonical C32●A38 base-pair, whereas in the hmtRNAMetCAU the loop has only the potential of a non-canonical C32●C38 base pair. Position 37, 3′-adjacent to the anticodons of many of the cytoplasmic tRNAi,fMetCAU and tRNAmMetCAU, have the t6A modification, or a variant,16 whereas the hmtRNAMetCAU has no modification at position 37. Previously, we had reported the procedures for chemical synthesis of the f5C mononucleoside and for its successful, site-selected incorporation into the hmtASLMetCAU.15 In the present paper, we use this chemistry to explore in detail the impact of the modification on the structure and dynamics of the ASL, and the biological ramifications of the formyl modification in the hmtASLMetCAU recognition of non-cognate codons. In our initial report the 5-formyl group appeared to be significant to the binding of the unconventional methionine codon AUA.15 The introduction of the f5C modification lowered the melting temperature and ellipticity, suggesting that this modification can enhance the dynamic of the loop15. Thus, the unique modification such as the f5C can represent a powerful tool in the stabilization of the codon-anticodon interaction. Nevertheless, how the mtRNAMet interacts with the mRNA into the ribosome still remained unclear. Further, ribosome binding experiments using native tRNAMetCAU having the normal modifications and an unmodified transcript of tRNAMetCAU indicate that f5C is important for decoding in the A-site of the bovine mitochondrial ribosome.14 We have sought to understand the role of the 5-formyl group in the efficiency of binding of the hmtRNAMetCAU to the mitochondrial codons AUA or AUG at both the ribosomal P- or A-sites. Here, we report the first high resolution three dimensional structures of the modified and unmodified human mitochondrial anticodon domain of tRNAMetCAU. The conformational dynamic of the hmtASLMetCAU appears to be influenced by the f5C34 modification in the direction of enhanced order in the anticodon loop. In addition, we demonstrate that the wobble position f5C34 of hmtRNAMetCAU is particularly important to the binding of the hmtASLMetCAU to AUA at the A-site. Interestingly, some organisms use the isoleucine codons AUU and AUC in initiation of translation at the P-site.9,18,23-25 The structure and dynamics of the anticodon may be critical to recognition of the unconventional AUA codon at the P- and A-sites of the ribosome, and in expanding its P-site recognition further to AUU and AUC.
Results
In order to determine the effect of f5C34 on codon recognition and its physicochemical contribution to the conformation and dynamics of the hmtASLMetCAU, we have solved the solution structures of the hmtASLMetCAU-Ψ27 and the hmtASLMetCAU-Ψ27;f5C34 by NMR and restrained molecular modeling. In addition, we have analyzed the codon binding of the ASLs and the full length tRNA with E. coli and bovine mitochondrial ribosomes. The heptadecamer hmtASLMetCAU constructs were chemically synthesized with pseudouridine at the 5′-terminal position 27 (Ψ27), and with or without f5C34. The ASLs are designated hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27, respectively (Fig. 1).
Contribution of f5C34 to the structure of the hmtRNAMetCAU anticodon: NMR resonance assignments
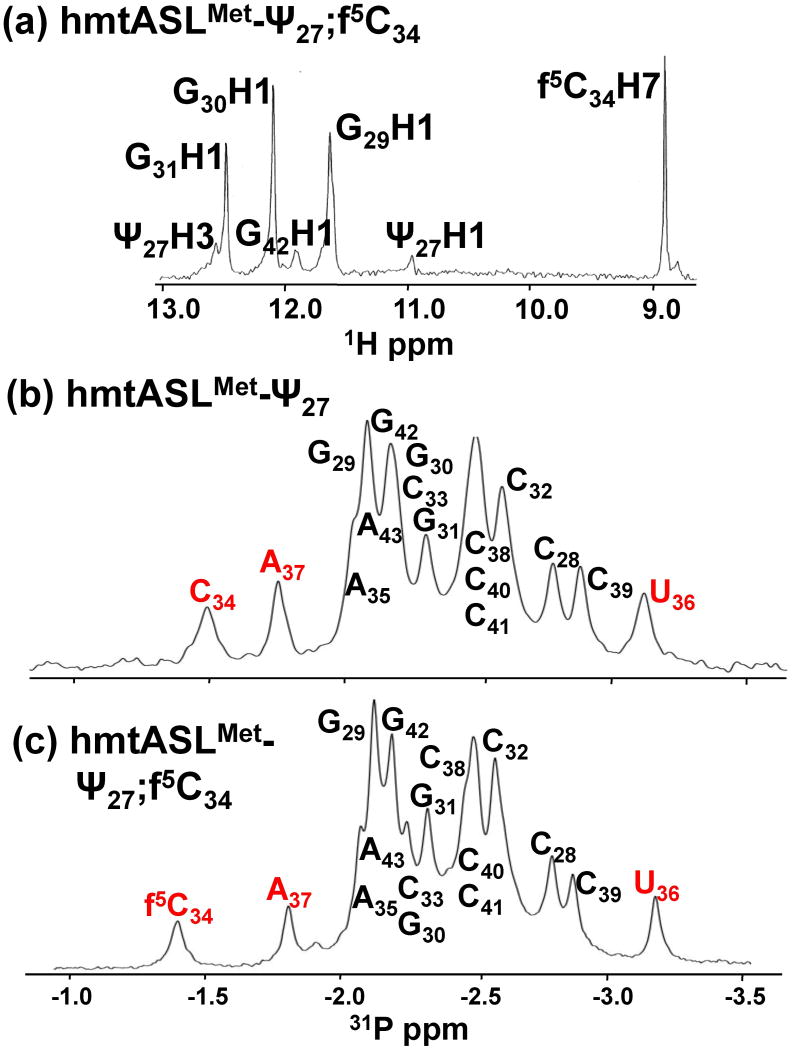
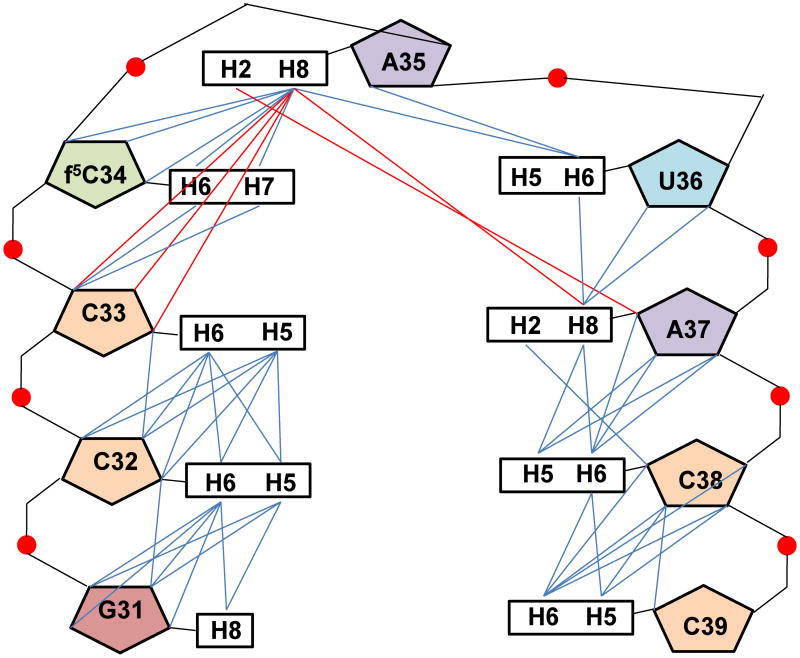
The structural contributions of f5C34 to the hmtASLMetCAU were determined by NMR-restrained molecular modeling. The non-exchangeable resonances were assigned using well established techniques.26 The 1D spectrum recorded exhibited three sharp peaks in the imino region which were assigned to G29, G30 and G31 imino proton resonances, involved in canonical G●C base pairing (Fig. 2a). One other resonance, broader than the previous ones, at 11.0 ppm was assigned to Ψ27H3 (Fig. 2a) and two NOE connectivities were observed between Ψ27H3 and the amino group of A43. The NMR data clearly demonstrated that the stem residues (C28 – G31 and C39 – G42) adopt a classical A-form RNA structure. The sequential pathway (aromatic to H1′ of the 3′ residue) could be followed using the 400ms NOESY spectrum from Ψ27 to C33 and from A37 to G43 (Fig. 3). A break appeared between C33 and f5C34/C34, however, a weak sequential NOE was observed at 400 ms between f5C34/C34 H1′ and A35H8. The two ASLs exhibited non-sequential NOEs between the A35H8 and C33H1′/H2′ protons. No NOE connectivity was observed between A35H8 and U36H1′, nor between U36H6 and A37H1′, even with a long mixing time (400ms). On the 5′ side of the loop, C32 was stacked with G31 and C33, as shown by the inter-aromatic NOEs between the bases (C32H5/G31H8, C32H6/G31H8, C32H6/C33H6 and C32H6/C33H5). Moreover, the chemical shifts of C38 and the C32H5/H6 protons were characteristic of protons experiencing a 5′ purine ring current,26 confirming the stacking of G31/C32, and of A37/C38. The A37H2 resonance was identified based on the natural abundance 1H-13C HSQC spectrum and a strong NOE connectivity was observed between the A37H2 and C38H1′. Non-sequential NOEs were observed between A35H2 and A37H1′, as well as between A35H2 and A37H8 (Fig. 3). The chemical shifts of U36H5 and H6 were not in agreement with the chemical shift of a uridine experiencing a purine ring current, and thus were not supportive of the stacking of A35 and U36. Moreover, weak NOEs were observed between U36H6 and the A35H3′/H2′ protons, as well as very weak NOEs between U36H5 and A35H3′, and H2′. Weak inter-aromatic NOEs were observed between A37H8 and U36H6 and between U36H6 and A35H8. Nevertheless, the intensities of these NOE connectivities were less than the NOE between A35H2 and A37H1′ (Fig. 3). A considerable number of inter-residue NOEs were observed for the hmtASLMetCAU-Ψ27;f5C34 and the hmtASLMetCAU-Ψ27. The inter-nucleoside NOEs of the anticodon loop residues (C32-C38) were particularly important to establishing the contribution of the f5C34. A significant number of these were attributed to the modified nucleoside, and to C33 and A35 (Fig. 4).
Figure 2.
1H and 31P NMR spectra (1D) of the hmtASLMetCAU. (a) 1H NMR spectrum of the hmtASLMetCAU-Ψ27;f5C34. The H2O-exchangeable proton resonances, as well as the formyl-proton (H7) resonance of f5C34 are shown. (b) 31P-NMR spectra (1D) of the hmtASLMetCAU-Ψ27 (top) and the hmtASLMetCAU-Ψ27;f5C34 (bottom). The 31P resonances for f5C34, C34, A37 and U36 are denoted in red.
Figure 3.
The hmtASLMetCAU NOSEY spectra. NOESY spectra of hmtASLMetCAU-Ψ27;f5C34 (a) and hmtASLMetCAU-Ψ27 (b). NOSEY spectra of the hmtASLMetCAU in D2O were recorded at 25° C with a mixing time of 400 ms. The H1′ and H5 to aromatic portion of the spectra are shown. The sequential H1′-aromatic connectivities are indicated, as well as the unusual NOEs connectivities between A35H2 – A37H1′ (blue) and A35H8 – C33H1′ (red).
Figure 4.
Compilation of inter-nucleoside NOE connectivities. The inter-nucleoside NOEs observed in the loop residues 31 to 39 of the hmtASLMetCAU-Ψ27;f5C34 were both sequential and non-sequential. The sequential (i to i+1) inter-nucleosides NOEs are shown in blue and the nonsequential NOEs are shown in red. The hmtASLMetCAU-Ψ27 exhibited comparable NOEs.
Sugar conformations and backbone geometry
All of the loop residues, with one exception, exhibited small to very small 3JH1′-H2′ coupling constants corresponding to the C3′-endo sugar pucker conformation. The COSY spectra indicated that U36 adopted a C2′-endo sugar pucker conformation. The 31P phosphorous spectra of both ASLs showed that all the resonances are grouped together within ± 0.5 ppm except for three resonances belonging to the residues f5C34/C34, U36 and A37 (Fig. 2b,c). The phosphorous resonance of C34 was downfield by 0.9 ppm from the 31P main cluster, whereas the f5C34 31P resonance was downfield by 1.1ppm (Fig. 2b,c). The 31P resonance of A37 was slightly downfield (∼0.3 ppm), whereas that of U36 was upfield (∼0.3 ppm) from the main cluster of peaks. All of the other resonances remained in the main cluster, indicating that the α and ζ angles were typical of an A-form RNA conformation (Fig. 2b,c).
Structures of the hmtASLMetCAU-Ψ27;f5C34 and the hmtASLMetCAU-Ψ27
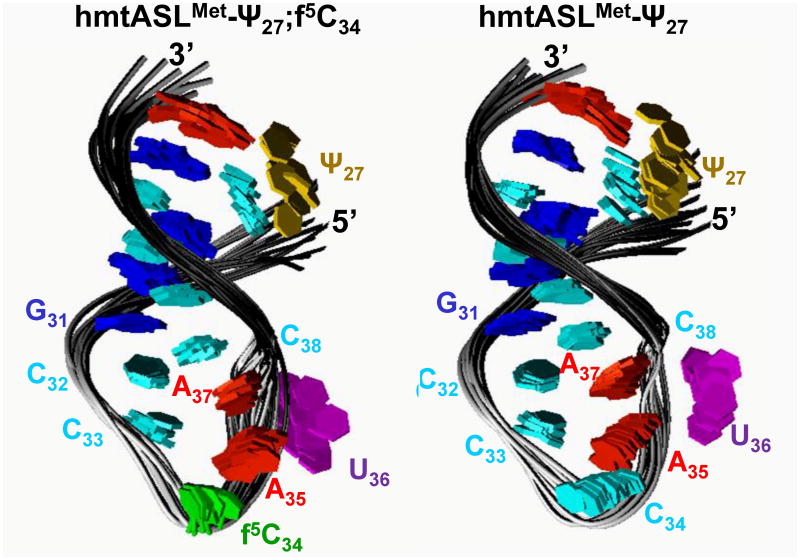
The structures of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were derived from the NMR data using a simulated annealing protocol.26 Ten of the lowest energy structures for the hmtASLMetCAU Ψ27;f5C34 and for the hmtASLMetCAU-Ψ27 were superimposed (Fig. 5). The stems of both ASLs adopt a classical A-form RNA conformation. The ten lowest energy structures for the hmtASLMetCAU-Ψ27;f5C34 and for the hmtASLMetCAU-Ψ27 had overall heavy atom RMSDs relative to the mean structure of 1.71 ± 0.58 Å and 1.51 ± 0.46 Å, respectively (Table 1). The C32●C38, C1′ – C1′ distance was on average 10.19 ± 0.54 Å and 10.39 ± 0.35 Å for the hmtASLMetCAU-Ψ27;f5C34 and for the hmtASLMetCAU-Ψ27, respectively. These values are comparable to the classical A-RNA C1′-C1′ distance (∼10.6 Å). The heavy atom RMSD value observed for the loop residues (C32 to C38) relative to the mean structure were 0.38 ± 0.20 Å and 0.51 ± 0.17 Å, respectively.
Figure 5.
Structures of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27. The ten lowest energy structures for the hmtASLMetCAU-Ψ27;f5C34 (a) and those for the hmtASLMetCAU-Ψ27 (b) are superimposed. To illustrate differences in the degree to which the bases of a family of structures converge, the ribose and phosphodiester bond details are not shown.
Table 1. Structural statistics for hmtASLMetCAU-Ψ27;f5C34 and hmtASL MetCAU-Ψ27.
| hmtASL MetCAU-Ψ27;f5C34 | hmtASL MetCAU-Ψ27 | |
|---|---|---|
| Number of experimental restraints | ||
| Distance restraints | 266 | 275 |
| Intranucleotides | 157 | 161 |
| Internucleotides | 109 | 114 |
| Exchangeable | 13 | 13 |
| Restraints by residue | 15.6 | 16.1 |
| Hydrogen bonding distances restraints | 24 | 24 |
| Dihedral restraints | 82 | 82 |
| Negative NOEs | 26 | 25 |
| Heavy atoms RMSD from mean structure (Å) | ||
| All residues (Y27 - A43) | 1.51 ± 0.58 | 1.71 ± 0.58 |
| Loop residues (C32 – C38) | 0.51 ± 0.17 | 0.38 ± 0.20 |
The two loops (C32 to C38) exhibited the same overall conformation (Fig. 6). The stem was continued from the canonical base pair G31●C39 at the bottom of the stem through the non-canonical C32●C38 mismatch pair (Fig. 6c). Taken together with the stacking of G31/C32 and C39/C38, the C32●C38 non-canonical base pair extended the stem. In the loop, the position of C32 was stabilized by a hydrogen bond between the C32O2 and the C38N4 with acceptor-donor distances of 2.98 ± 0.15 Å and 3.30 ± 0.20 Å for the hmtASLMetCAU-Ψ27;f5C34 and for the hmtASLMetCAU-Ψ27, respectively. Moreover, the structures were stabilized by a probable hydrogen bond between C32N4 and A37O2P with acceptor donor distances of 3.12 ± 0.47 Å and 3.49 ± 0.48 Å for the hmtASLMetCAU-Ψ27;f5C34 and for the hmtASLMetCAU-Ψ27, respectively.
Figure 6.
Conformational differences and similarities in the anticodon loops of the f5C34-modified and unmodified hmtASLMetCAU-Ψ27. Stereo view of the average structures of the anticodon loops (residues C32 to A38) for (a) hmtASLMetCAU-Ψ27 and (b) hmtASLMetCAU-Ψ27;f5C34. (c) The anticodon loops contain a C32●C38 intra-loop base pair in both the hmtASLMetCAU-Ψ27;f5C34 and the hmtASLMetCAU-Ψ27. (d) Stereo view of the two average structures superimposed (green, hmtASLMet-Ψ27;f5C34; and purple, the hmtASLMetCAU-Ψ27).
In both ASLs, the unusual C33 residue was stacked with C32. An abrupt turn in the backbone was observed between the residues C33 and f5C34/C34 (Fig. 6a and b) The turn was characterized by a trans α dihedral angle (173 ± 20° for hmtASLMetCAU-Ψ27;f5C and -160.5 ± 8.6° for hmtASLMetCAU-Ψ27). The f5C35/C34 and A35 residues were stacked nicely, consistent with the observation of n/n+2 NOEs between the two residues in the NMR spectra. Both nucleosides adopted the C3′-endo sugar conformation. The average distance between the C332′OH and the A35N7 was 2.88 ± 0.08 Å and 2.90 ± 0.07 Å for the hmtASLMetCAU-Ψ27;f5C34 and for the hmtASLMetCAU-Ψ27, respectively, allowing the formation of a direct hydrogen bond. In both structures, the U36 adopted the C2′-endo sugar pucker conformation (δ dihedral angles of 137.9 ± 4.6° for hmtASLMetCAU-Ψ27;f5C and 143.1 ± 1.6° for hmtASLMetCAU-Ψ27), which is consistent with the NMR data. Thus, the base resides outside the loop and in direct contact with the solvent. The intra-residue NOEs of f5C34 include those between the formyl proton and H6 and H1′. These fix the position of the formyl proton relative to the nucleoside conformation. In addition, there are weak NOEs between the formyl proton and A35H8 and the formyl proton and C33H3′. Importantly, the conformational space occupied by the f5C34 in the ten structures of hmtASLMetCAU-Ψ27;f5C34 was considerably less than that occupied by C34 within the ten structures of hmtASLMetCAU-Ψ27. The limited space that confined the motional dynamics of the modification may be indicative of the modification's contributions to the reading of unconventional codons.
The anticodon wobble position modification, 5-formylcytidine, enhances AUA A-site codon recognition by the hmtASLMetCAU-Ψ27;f5C34
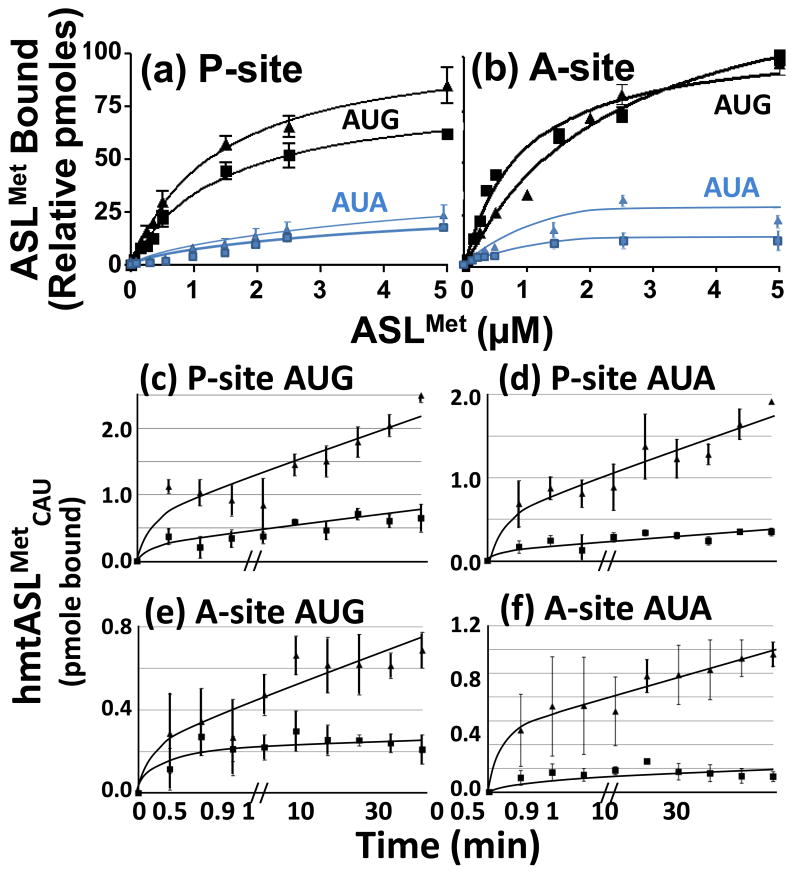
Mitochondrial tRNAMet must recognize both AUG and AUA in the P-site for initiation and in the A-site during polypeptide chain elongation. To study the effect of the f5C34 residue on the codon binding characteristics of the human mitochondrial tRNAMet, we determined the equilibrium binding properties of the hmtASLMetCAU-Ψ27, with and without the f5C34 to the AUG and AUA codons at the P- and A-sites by titrating mRNA-programmed, E. coli 70S ribosomes with increasing amounts of the hmtASLMetCAU (Fig. 7; Table 2). As would be expected,27 the two ASLs bound the P-site codons to a degree (3-4 pmoles bound) exceeding by several fold their binding of the A-site codons (0.5-1.0 pmoles). The 5-formyl modification did not affect the equilibrium binding of hmtASLMetCAU-Ψ27;f5C34 to AUG at the P- site (Table 2). Neither the dissociation constant, nor the extent of binding to AUA, at the P-site appear to be affected greatly by the f5C34 modification (Table 2; Fig. 7a). In binding AUA at the P-site, the hmtASLMetCAU-Ψ27;f5C34 and the hmtASLMetCAU-Ψ27 exhibited steady-state dissociation constants within experimental error of each other, 7.9 ± 5.5 μM and 11.0 ± 4.7 μM, respectively. Binding to AUA in the P-site was weak resulting in larger errors in the determination of the Kds. The most striking observation in this data is that the extent of either hmtASLMetCAU-Ψ27;f5C34 or hmtASLMetCAU-Ψ27 binding at the P-site was substantially lower than observed with the AUG codon. The differences in saturation level between binding to AUA and binding to AUG probably arises from differences in kinetics. First, there appears to be an intrinsically lower ability of the hmtASLMetCAU to capture the formed complex when AUA is present. This is most likely a consequence of the difficulty of forming the f5C34-A base pair and the weakness of this pairing. We suspect that on the E. coli ribosome two pathways to codon binding occur, as has been reviewed previously.28 In one, the ASL binds before the mRNA, but leaves rapidly due to unstable interactions. In the other, the mRNA binds first via the Shine-Delgarno sequence, but also dissociates rapidly in the absence of codon:anticodon interactions. Thus, stable binding of the ASL is only observed when both hmtASLMetCAU and mRNA are present at the same time leading to codon:anticodon interactions. With AUA, the kinetics of the codon:anticodon interaction and/or the stability of the interaction is weaker than that with AUG and the complex cannot form to the same extent. Therefore, the major difference in saturation is that attributed to differences in codon:anticodon interactions. This observation suggests that the use of AUA as the initiation codon may reduce the efficiency of initiation complex formation for specific mRNAs in mammalian mitochondria.
Figure 7.
AUG and AUA codon binding properties of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 in the P- and A-sites of the ribosome. (a) AUG and AUA codon binding of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 in the P-site of the ribosome. The E. coli 70S ribosome (5 pmoles), programmed with AUG in the P-site, was titrated with hmtASLMetCAU-Ψ27;f5C34 (▲) and with hmtASLMetCAU-Ψ27 (■), or programmed with AUA and titrated with hmtASLMetCAU-Ψ27;f5C34 ( ) and with hmtASLMetCAU-Ψ27 (
) and with hmtASLMetCAU-Ψ27 ( ). The maximum hmtASLMetCUA bound was between 3-4 pmoles. (b) AUG and AUA codon binding properties of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 in the A-site of the ribosome. The E. coli 70S ribosome (5 pmoles), programmed with AUG in the A-site, was titrated with hmtASLMetCAU-Ψ27;f5C34 (▲) and with hmtASLMetCAU-Ψ27 (■), or programmed with AUA and titrated with hmtASLMetCAU-Ψ27;f5C34 (
). The maximum hmtASLMetCUA bound was between 3-4 pmoles. (b) AUG and AUA codon binding properties of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 in the A-site of the ribosome. The E. coli 70S ribosome (5 pmoles), programmed with AUG in the A-site, was titrated with hmtASLMetCAU-Ψ27;f5C34 (▲) and with hmtASLMetCAU-Ψ27 (■), or programmed with AUA and titrated with hmtASLMetCAU-Ψ27;f5C34 ( ) and with hmtASLMetCAU-Ψ27 (
) and with hmtASLMetCAU-Ψ27 ( ). The maximum hmtASLMetCUA bound was between 0.5-1.0 pmoles. In contrast to equilibrium binding studies, the time-dependent binding of the hmtASLMetCAU-Ψ27;f5C34 (▲) and hmtASLMetCAU-Ψ27 (■) to AUG and AUA were conducted at the single concentration of ASL (0.5 μM), far too low to saturate the ribosomes. The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (c) AUG in the P-site of E. coli 70S ribosome, and to (d) AUA in the P-site). The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (e) AUG in the A-site of E. coli 70S ribosome, and to (f) AUA in the A-site.
). The maximum hmtASLMetCUA bound was between 0.5-1.0 pmoles. In contrast to equilibrium binding studies, the time-dependent binding of the hmtASLMetCAU-Ψ27;f5C34 (▲) and hmtASLMetCAU-Ψ27 (■) to AUG and AUA were conducted at the single concentration of ASL (0.5 μM), far too low to saturate the ribosomes. The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (c) AUG in the P-site of E. coli 70S ribosome, and to (d) AUA in the P-site). The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 were bound to (e) AUG in the A-site of E. coli 70S ribosome, and to (f) AUA in the A-site.
Table 2. Dissociation constants (Kd) of Unmodified and f5C34-Modified hmtASLMetCAU-Ψ27 for the Mitochondrial methionine Codons at the E. coli Ribosomal P or A-sites.
| Dissociation Constants (Kd, μM) | ||||
|---|---|---|---|---|
| hmtASLMetCAU-Ψ27 | P-site | A-site | ||
| AUG | AUA | AUG | AUA | |
| unmodified | 1.4 ± 0.29 | 11.0 ± 4.7 | 0.9 ± 0.1 | 4.5 ± 2.1 |
| f5C34 | 1.3 ± 0.22 | 7.9 ± 5.5 | 2.4 ± 0.4 | 1.6 ± 0.4 |
The f5C34-modified hmtASLMetCAU-Ψ27 bound AUG in the A-site with a higher affinity than that of the hmtASLMetCAU-Ψ27 as indicated by the lower dissociation constant (Table 2; Fig. 7b). At the A-site, the hmtASLMetCAU-Ψ27;f5C34 bound AUA with a Kd of 1.6 ± 0.4 μM (Fig. 7b; Table 2). However, the hmtASLMetCAU-Ψ27 exhibited a a higher Kd of 4.5 ± 2.1 μM. Thus, there appears to be a modification-dependent difference in the hmtASL binding of AUA at the A-site. Again, the largest difference observed in the AUG versus AUA-dependent binding at the A-site is reflected in a lower extent of binding suggesting that the off-rate for codon-anticodon interactions could be more rapid with the AUA codon.
Kinetics of the f5C34-modified and unmodified hmtASLMetCAU-Ψ27 binding AUG and AUA at the P- and A-site
The unmodified CAU anticodon of tRNAMet is a canonical match for the codon AUG. Therefore, we did not expect the 5-formyl modification of hmtASLMetCAU-Ψ27;f5C34 to have a significant effect on the binding of the ASL to AUG. As expected, steady-state binding assays of hmtASLMetCAU-Ψ27;f5C34 or the hmtASLMetCAU-Ψ27 to AUG at the P-site resulted in similar Kds. However, there were differences in the levels of ASL concentration at which saturation was attained for the binding of AUG at the P-site, and also for AUA at the A-site (Fig. 7a,b). Thus, we explored the possibility that the two ASLs had significant differences in the kinetics of their binding. A significant difference in the kinetics of binding became apparent at a concentration of ASL (0.5 μM) far too low to saturate the ribosomes (Fig. 7c-d). The amount of hmtASLMetCAU-Ψ27;f5C34 bound to AUG in the P-site was five-fold greater than that of the hmtASLMetCAU-Ψ27 after only 30 seconds (Fig. 7). While the hmtASLMetCAU-Ψ27 maintained a steady amount of weak binding over time, the amount of hmtASLMetCAU-Ψ27;f5C34 bound to AUG continued to increase, reaching a ten-fold greater amount bound than that of the hmtASLMetCAU-Ψ27 at the final time point of 30 min. These results were repeated for the binding of the two ASLs to AUG at the A-site, and AUA at both the P- and A-sites (Fig. 7). Though we were deterred in determining the rate constants for lack of sufficient modified hmtASLMetCAU, the time courses clearly indicate a significant difference in rates of association that are dependent on the presence of f5C34. The binding appears to be biphasic. We suspect that the 70S preparation has some portion of 30S subunits present. The subunits would form complexes rapidly accounting for an initial rapid phase in the kinetics. The slower phase may represent binding to 70S particles which probably takes longer either because access to the P-site is more restricted or because there is a dissociation step required in providing 30S subunits prior to the mRNA binding step. The equilibrium binding constants of the hmtASLMetCAU-Ψ27 to the P-site codons AUG and AUA are not affected by the presence of f5C34 (Table 2), though there seems to be an effect on the extent of binding (Fig. 7). The similarities of the Kds taken together with the results of the time courses at low ASL concentration indicate that the modification affects both on- and off- rates. In comparison, the equilibrium binding of the hmtASLMetCAU-Ψ27 to the A-site codons AUG and AUA is enhanced by the presence of the f5C34, and the respective time courses reflect this result. This suggests that in the absence of f5C, the off-rate may predominate.
Codon binding of hmtASLMetCAU-Ψ27;f5C34 and the hmtASLMetCAU-Ψ27 on the bovine 55S mitochondrial ribosome
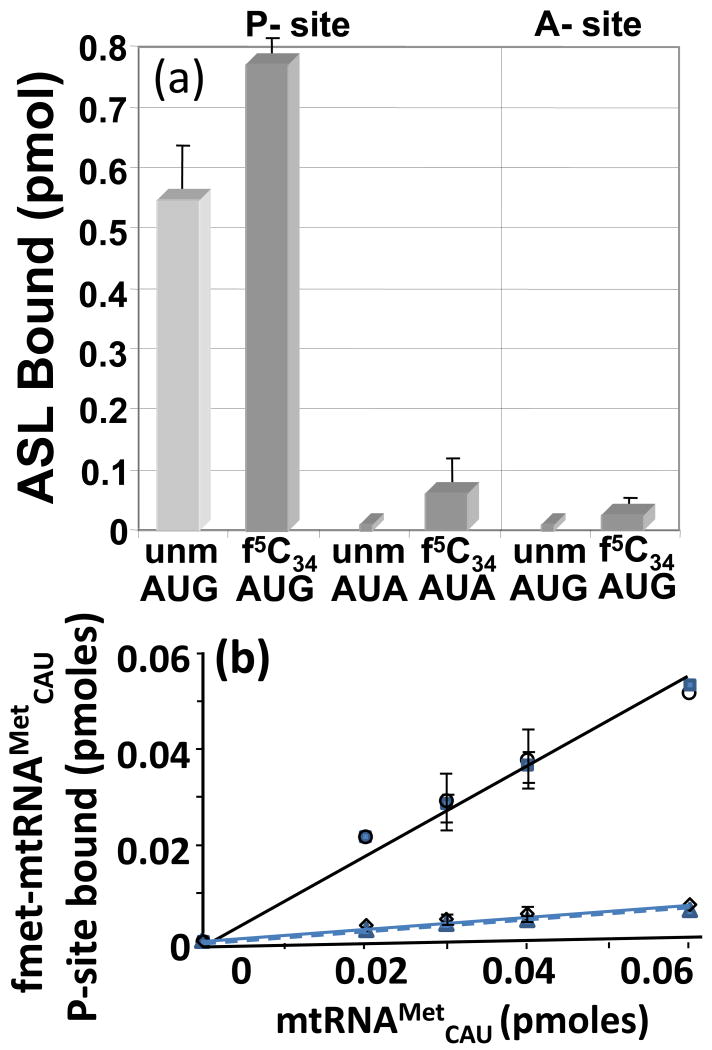
Thus far, the codon binding experiments we have described for hmtASLMetCAU were conducted with the model E. coli 70S ribosomes. However, previous studies of translation in vitro using bovine mitochondrial 55S ribosomes succeeded in demonstrating low levels of incorporation of methionine into protein using the native bovine mtRNAMetCAU, unmodified transcripts and mitochondrial ribosomes programmed with poly(AUA).14 Therefore, we thought it appropriate to attempt the binding of hmtASLMetCAU to programmed, purified bovine mitochondrial 55S ribosomes. The bovine mitochondrial 55S ribosomes were programmed with mRNA, and then either hmtASLMetCAU-Ψ27;f5C34 or the hmtASLMetCAU-Ψ27 was added. Mitochondrial mRNAs tend to lack the 5′-UTR. In order to study the binding of hmtASLMetCAU to the initiation site (P-site), we used a 30-mer oligonucleotide mRNA with either AUG or AUA at the 5′ terminal end followed by the valine codon GUA as the second codon. The binding of the ASLs to AUG in the P-site was detectable, although the level of codon binding was considerably lower than that observed on the E. coli ribosomes. The 5-formyl-group of the f5C34-modified hmtASLMetCAU enhanced the binding of AUG by approximately 40% (Fig. 8a, first two bars). Though the P-site binding of the hmtASLMetCAU-Ψ27;f5C34 to AUA was detectable, the binding of the hmtASLMetCAU-Ψ27 was not (Fig. 8a, second two bars). The presence of f5C34 appears to have enhanced P-site binding. To observe A-site binding, the 55S ribosomes were programmed with a 30-mer mRNA construct consisting of a 5′-leader of twelve uridines, followed by GUA at the P-site and AUG or AUA at the A-site, and ending with another stretch of twelve U residues. The presence of the 5-formyl modification on the ASL enhanced A-site binding to AUG sufficiently for it to be observed (Fig. 8a). However, neither the binding of hmtASLMetCAU-Ψ27 to AUG, nor the binding of either ASL to AUA was observed in the A-site.
Figure 8.
Codon binding by hmtASLMetCAU, the native hmtRNAMetCAU and the unmodified transcript on the bovine mitochondrial ribosome. (a) AUG and AUA codon binding by the hmtASLMetCAU-Ψ27;f5C34 (f5C34) and hmtASLMetCAU-Ψ27 (unm) on the 55S bovine mitochondrial ribosome. The modification f5C34 enhanced P-site binding of the AUG and AUA codons, and A-site binding to the AUG codon. However, A-site binding of the AUA codon by either ASL was not detectable. The binding (pmoles) of hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 are shown under conditions in which the ASLs are saturating. (b) The IF2-facilitated binding of bovine mtRNAMetCAU and its transcript to AUG and AUA in the P-site of the bovine mitochondrial ribosome. The bovine mitochondrial ribosome programmed with AUG was titrated with either mtRNAMetCAU (○) or its transcript ( ). Also, the AUA-programmed ribosome was titrated with either mtRNAMetCAU (◊) or its transcript (
). Also, the AUA-programmed ribosome was titrated with either mtRNAMetCAU (◊) or its transcript ( ).
).
In our mitochondrial 55S ribosome binding assays, only small amounts of the hmtASLMetCAU-Ψ27;f5C34 bound AUG at the A-site and AUA at the P-site. The low level of detectable binding to the P-site, and the inability to detect the binding of either hmtASLMetCAU-Ψ27;f5C34 or the unmodified hmtASLMetCAU to AUA at the A-site may have been due to the very low levels of binding to mitochondrial ribosomes, in general, experienced by us (approximately 0.8 pmol of hmtASLMetCAU-Ψ27;f5C34 bound to AUG at the P-site) and others10,14 in comparison to E. coli 70S ribosomes (4 pmol of hmtASLMetCAU-Ψ27;f5C34 bound to AUG at the P-site10). Also, the codon AUA is not as efficient as AUG in the decoding of methionine in mitochondrial translation. Translation in vitro with a poly(AUG) mRNA resulted in five-fold greater amounts of Met-containing polypeptides being produced than from a poly(AUA) mRNA.14
The lack of full tRNAs, aided by initiation and elongation factors, may also have contribute to the low levels of binding. Therefore, we conducted initiation factor, IF2mt-facilitated, binding of the native bovine mtRNAMetCAU and the full length, but unmodified, transcript to codons at the P-site of the bovine mitochondrial 55S ribosome. Native bovine mtRNAMetCAU was isolated and purified,29 and the unmodified mtRNAMetCAU obtained through transcription in vitro.30 The native tRNA and transcript were aminoacylated and formylated. The IF-2mt-facilitated binding of the native bovine and unmodified transcript of fmet-mtRNAMetCAU to AUG at the P-site were essentially identical at very low levels of input tRNA (Fig. 8b). The affinity of the two tRNAs for AUA at the P-site was considerably lower. However, the absence of modifications on the transcript made no apparent difference in the binding. Thus, the binding of full length native bovine fMet-mtRNAMetCAU and the unmodified transcript to AUG at the P-site of the mitochondrial ribosome was consistent with the results obtained in the binding of the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27. Though the fMet-mtRNAMetCAU and the unmodified transcript comparably bound AUA at the P-site, the extent was diminished to the degree as to not be able to make a reasonable comparison to the binding of the respective mtASLs. The mitochondria uses AUA to initiate translation (AUA at the P-site) only about twenty percent of time, but approximately eighty percent of mitochondrial methionine A-site codons are AUA (twenty percent are AUG).21,31 Thus, the probability of AUA being at the mitochondrial P-site is significantly lower than that of AUG.
Ability of the 5-formyl-modification to expand codon recognition within the codon box AUN
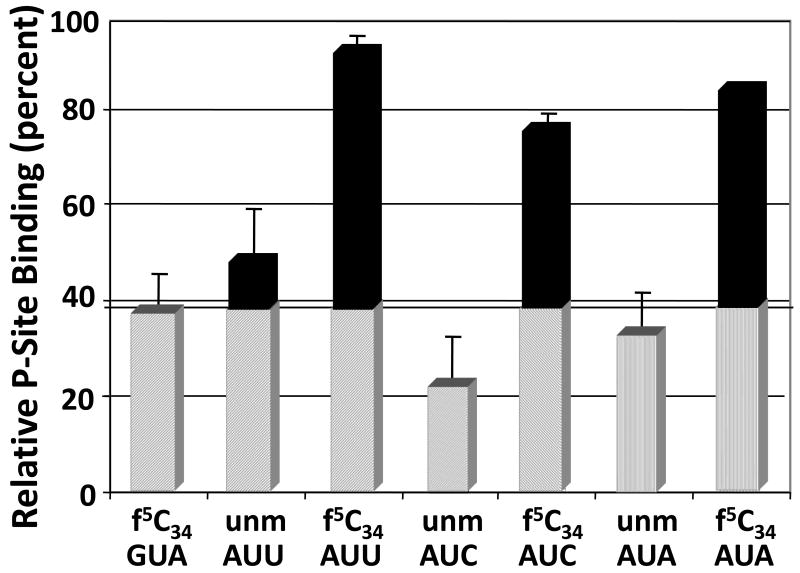
Mitochondria not only initiate translation from AUG and AUA, but also have been found to initiate from the two mitochondrial isoleucine codons AUU and AUC.9,24,32 AUU and AUC are rare initiation codons and may be used to regulate gene expression. The ability of this single and modest modification, f5C34, to enable the one hmtRNAMetCAU to initiate translation with AUA, as well as with AUG, begs the question as to whether f5C34 expands the tRNA's ability to decode the other synonymous codons, AUU and AUG. We found that the hmtASLMetCAU-Ψ27;f5C34 bound to AUU and to AUC with a Kd similar to that for AUA (Fig. 9). Furthermore, as it did in binding to AUA, the modified hmtASLMetCAU-Ψ27;f5C34 bound to AUU and AUC with a dissociation constant that was two- to three-fold less than that of the hmtASLMetCAU-Ψ27 (Fig. 9). The hmtASLMetCAU-Ψ27 was bound to AUU, AUC, and AUA with Kds comparable to, and within error of, that of the negative-control codon for valine GUA (Fig. 9).
Figure 9.
The f5C34-modified hmtASLMetCAU-Ψ27 binds the near-cognate codons AUU and AUC. The modification f5C34 enabled hmtASLMetCAU-Ψ27;f5C34 to bind the near-cognate codons AUU and AUC, as well as AUA, in the P-site of the E. coli ribosome. The relative binding of non-cognate codons by hmtASLMetCAU-Ψ27;f5C34 (f5C34) and hmtASLMetCAU-Ψ27 (unm) are shown under conditions in which the ribosomes are saturated with ASLs, and the results internally normalized (percent). The binding of the valine codon GUA by hmtASLMetCAU-Ψ27;f5C34 (f5C34) is an indication of the degree of non-specific binding in the assay.
Discussion
Determining the structural contributions of f5C34 to the function of mtRNAMetCAU presented a unique challenge because this single RNA has the dual role of participating in initiation and elongation, as well as reading both AUA and AUG. The hmtRNAMetCAU has unique decoding properties because the f5C34 extends codon recognition often to AUA, and to a small extent to AUU and AUC. The hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 exhibited the same overall conformation expected of a hairpin RNA with a stem adopting an A-form RNA structure extended by a non-canonical base pair between C32●C38. The 5′ side of the loop was characterized by the stacking of C32 with the unique C33, and the 3′ side by the stacking of A35, A37 and C38. A sharp turn was observed in the backbone and was characterized by a trans α dihedral angle between the residues C33 and f5C34/C34. This observation is in accordance with the downfield 31P resonance belonging to f5C34/C34 and represents one of the characteristics of the U-turn motif.33,34 Moreover, the “classical” U-turn motif is stabilized by a network of hydrogen bonds between the U332′OH and the N7 of the purine at residue 35 (most often an adenosine), and by a hydrogen bond between the U33H3 exchangeable proton and the phosphate group of residue 36. These hydrogen bonds have been shown to be essential in the formation of the U-turn and to the ribosomal binding properties of the tRNAs.35 Because C33 substitutes for the invariant U33, some of the hydrogen bonds that stabilize the canonical U-turn conformation adopted by cytoplasmic tRNAMetCAU and other tRNAs should not be possible for the hmtRNAMetCAU.33,35 The last hydrogen bond is not possible in the case of a cytosine in position 33, unless the amino group of the cytosine plays the role of hydrogen bond donor. The three dimensional structures of the hmtASLMetCAU-Ψ27;f5C34 and the hmtASLMetCAU-Ψ27 showed that the C33 amino group is directed toward the minor groove and is not involved in direct hydrogen bonding. Nevertheless, distances between the C33N4 and the O2P of residue C32 are on average 4.07 ± 0.72 Å and 3.93 ± 0.55 Å for hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27. If these distances preclude the formation of a direct hydrogen bond, the formation of some indirect hydrogen bonds involving water molecules is highly possible. Water mediated hydrogen bonds can be of importance in the stabilization of the loop conformation and the formation of the observed ersatz U-turn, or “C-turn”. The projection of the U36 residue from the structure and into the solvent is consistent with the NMR data. This is not very surprising when we consider that the uridine is unpaired and between the two stacked purines, A35 and A37. Unpaired uridines have a tendency to bulge from the structure.34 Modifications in position 37 provide stability to the anticodon-codon interaction,36 and are important in driving the conformation of the anticodon loop toward that of the well-known U-turn important to the binding of the codon on the ribosome.37-39 In the anticodon domain structure of the S. pombe initiator tRNAMetCAU, with a uridine in position 33 and the t6A37 modified residue, U36 is also bulged out from the structure,38 whereas in the structure of the unmodified yeast initiator tRNAMet, U36 adopted the C2′-endo sugar conformation and is loosely stacked with A35.41 Direct exposure of U36 to the solvent makes it available for tertiary interactions, in particular with the mitochondrial methionyl-tRNAMet synthetase (mtMRS). mtMRS recognition of the cytoplasmic tRNAMet species includes the anticodon,42,43 and especially residue C34 as a strong identity element.44 Moreover, the residue U36 has been shown to be essential for the tRNAMet – MetRS interaction by being specifically recognized by the MetRS C-terminal extended loop.45
The f5C34 modified residue is one of the most conformationally rigid residues with a C3′-endo sugar conformation that could help in pre-structuring the anticodon for codon interaction.39;46 Yet, UV-monitored studies of thermal stability and circular dichroism studies of stacking interactions of the hmtASLMetCAU-Ψ27 with and without f5C34 indicated that the introduction of f5C34 may enhance the ASL's motional dynamics.15 While the biological function of f5C34 is evident from our work and that of others, the physicochemical contribution of the f5C34 distinguishing hmtASLMetCAU-Ψ27;f5C34 from hmtASLMetCAU-Ψ27 appears ambiguous. However, this apparent contradiction of thermal instability and restricted conformational dynamics is found in other ASLs when the introduction of a modified nucleoside at positions 34 and/or 37 increases the width of the anticodon loop, as does f5C34.47 As with other wobble position-34 modifications,37,48,49 the f5C34 also takes a defined chemical and physical space within the anticodon loop that possibly restricts the conformational dynamics of the other loop nucleosides, in particular A35 and A37, in comparison to that of hmtASLMetCAU-Ψ27 (Fig. 5). In the absence of stable isotope labeling, the restricted dynamics are deduced from a comparison of the family of calculated structures for hmtASLMetCAU-Ψ27;f5C34 with that of hmtASLMetCAU-Ψ27 (Fig. 5) We suggest that the mechanism by which the formyl oxygen of f5C34 may restrict conformational dynamics is similar to that of the C5-oxygen of 5-oxyacetic acid uridine (cmo5U34). Cmo5U34, found in one of the isoaccepting species for each of the E. coli tRNAs for alanine, proline, threonine, serine and valine, enables the tRNA's reading of all four synonymous codons by pre-structuring of the anticodon, and fixing the geometry of the base in the A-site of the ribosome.48;49
Position-37 is not modified in the mtRNAMetCAU. Modifications of the purine at position-37 provide stability to the tRNA/mRNA interactions, and have been shown to be important in driving the conformation of the ASL toward the well-known U-turn structure that accommodates the binding to cognate codons on the ribosome.39,47 The presence of a cytosine in position-33 is quite unusual.16 Of the 21 sequences of tRNAs having a cytosine in position-33, 12 are cytoplasmic initiator tRNAiMetCAU and three are mitochondrial mtRNAMetCAU.16 Surprisingly, the presence of a cytosine residue in position-33 is correlated with the absence of modifications in position-37 in mitochondria, except for that of plants. The structures of the two cytoplasmic tRNAMetCAU species have been studied. Crystal structures have been solved for the entire eukaryotic yeast and E. coli tRNAMetCAU species free,50-53 and in complex with the methionyl-tRNA synthetase (metRS).54 In addition, the anticodon stem and loops of the E. coli tRNAfMetCAU and unmodified tRNAmMetCAU were solved by NMR,41 as well as that of the S. pombe tRNAMetCAU ASL containing the t6A37 modification.38 The anticodon stem and loop domains of yeast, E. coli, human and plant initiator tRNAs have the same primary sequence (residues 29-41) except for position-33 which is a uridine in yeast and E. coli initiator tRNAs, and a cytosine in human and plant initiator tRNAs. This singular difference appears to be responsible for the specific, H2O-mediated hydrolysis of the C34-A35 phosphodiester bond in human and plant initiator tRNAs, and not in yeast and E. coli initiator tRNAs.55 The substitution of U33 with C33 in elongating tRNAs has been shown to decrease codon binding on the ribosome.35,56 Substitution of U33 for C33 in hmtASLMetCAU does not affect P-site binding to either AUG or AUA (not shown). Nevertheless, the S1 nuclease cleavage patterns for the anticodon stem and loop domains of the prokaryotic and eukaryotic initiator tRNAs remain the same whether a cytosine or a uridine is in position-33.57
The A-site anticodon/mRNA interaction is proof-read and stabilized by the 16S rRNA residues G530, A1492 and A1493.58 Crystal structures of the ribosome show that U33 does not make contact with the ribosome, and that the U33N3 is involved in the “classical” U-turn hydrogen bond.18,36 In the NMR structures of the hmtASLMetCAU, C33 lacks that hydrogen bond. Once on the ribosome, the amino group of C33 is available to form tertiary interactions with the rRNA that could stabilize the anticodon/codon binding. Interestingly, the crystal structure mentioned above reveals that the anticodon P-site tRNAPhe wobble base (G34) is stabilized by stacking interactions with the 16S rRNA base C1400 and by packing between the ribose of G34 and the base G966.59 If the same interactions are observed in the case of the hmtRNAMetCAU, the enhanced stacking properties of the f5C34 modified residue could account for stabilization of the hmtASLMet/mRNA interaction. It appears that the C33 may change the conformational dynamic of the ASL loop, and thus could be of importance in stabilizing the f5C●A base pair, as well as that with AUG.
A pre-structured anticodon is the result of modification-induced, restricted dynamics, and is advantageous for codon binding.39 The three hydrogen bonds between an f5C34 and a guanosine are no different than that of a tRNA's C34●G base pair with the mRNA codon. However, codon recognition and binding of the hmtRNAMetCAU to AUA in the P- or A-sites is problematic because the unconventional f5C34●A interaction would be expected to be quite weak.60 Yet, eighty percent of all methionine codons internal to mitochondrial mRNAs are AUA. We have found that that the f5C34 facilitates the binding of the hmtASLMetCAU-Ψ27;f5C34 at the A-site, and also may effect the kinetics of codon recognition at both the P- and A-sites. In addition, humans, giant pandas and fruit flies have been found to use AUU for initiation with methionine, honey bees use AUC, and mice use both AUU and AUC to initiate translation.24,25 The hmtASLMetCAU-Ψ27;f5C34 read AUU and AUC at the P-site to an extent similar to its binding to AUA, whereas the hmtASLMetCAU-Ψ27 was not able to do so. Thus, the f5C34 appears to have expanded the ability of the tRNA to read all four synonymous codons within the AUN codon box.
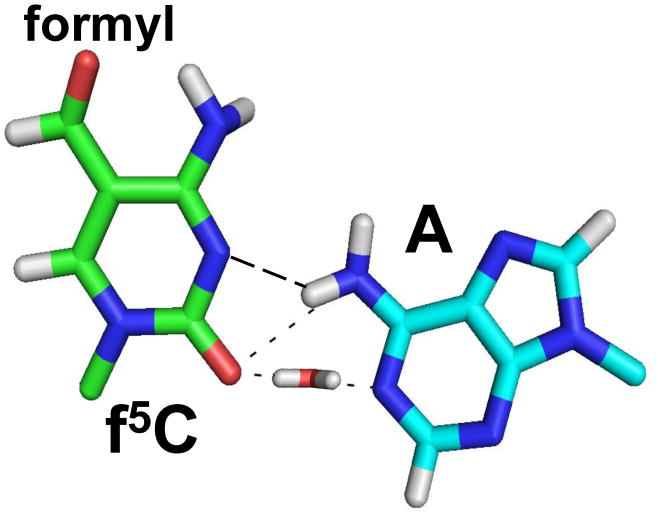
Molecular dynamics simulations of the base pairing of the 5′-monophosphate of f5C34 with that of A under aqueous and neutral conditions61 indicate the potential for novel interactions, including a bridging H2O molecule that stabilizes the interaction (Fig. 10). The f5C34 intranucleotide hydrogen bond that we observed between the f5C34 amino and formyl groups in the NMR-derived structure could increase the stacking interaction between f5C34 and A35 and possibly A37, and thus compensate for the lack of hydrogen bonds in an anticodon f5C●A codon interaction.14 A hydrogen bond between the N4-amino group and the formyl-group oxygen at the position C5 of the f5C34 modification may have altered the electronic properties of the modified residue, and thereby affected the base pairing properties of the f5C34, enabling a f5C34●A mismatch to form on the ribosome, as it has been proposed.14 An additional hydrogen-bond could be formed between f5C34 of the anticodon and A, the third base of the codon, if the adenosine were protonated. This is possible since protonation of an adenosine has been shown to be effective in certain RNA constructs at a pH near 7.62 Nevertheless, an f5C●A+ interaction is still weaker than an f5C●G or C●G interaction. Further studies need to be conducted to reveal the geometry, and stability that the f5C34 modification brings to the interactions of the hmtRNAMet with unconventional codons of mRNA in the A- and P-sites of the ribosome.
Figure 10.
Molecular dynamics simulation of the f5C34●A base pair. The f5C34●A base pair must occur on the mitochondrial ribosome when hmtRNAMetCAU reads the AUA codon in either the A-or P-sites. The base pairing of the 5′-monophosphate of f5C34 with that of A under aqueous and neutral conditions was simulated with molecular dynamics,61 and the lowest energy structure is presented here. The resulting model indicated a possible bridging H2O molecule that stabilizes the interaction.
Materials and Methods
Oligonucleotide Chemical Synthesis and tRNA Preparation
RNAs were chemically synthesized (Thermo Fisher Scientific, Lafayette, CO) using “ACE” chemistry.63 The protected 5-formylcytidine modified nucleotide was chemically synthesized and incorporated at position-34.15 The RNA molecules were deprotected and dialyzed extensively against 20 mM phosphate buffer (10 mM KH2PO4; 10 mM Na2HPO4; pH 6.8; 0.05 mM EDTA) using a membrane with a 3500 Da cutoff. For observation of the exchangeable protons by NMR, the samples were resuspended in 300 μl of 90% H2O/10% D2O. For experiments involving the non-exchangeable protons, the samples were exchanged twice with 99.9% D2O. The NMR sample concentration was between 1.0 and 1.5 mM. Full length, native bovine mtRNAMetCAU and its unmodified transcript were prepared as previously described.30
NMR Spectroscopy
NMR spectra were collected on a DMX Bruker 500 MHz instrument equipped with a triple-resonance 1H, 13C, and 15N probe and three-axis pulsed field gradient capabilities, and on a Varian INOVA 600 MHz instrument equipped with a cryoprobe. NMR spectra were collected with the RNA samples under solution and temperature conditions required and widely used for determination of nucleic acid structures.49,62,64 All NMR data were processed using NMRPIPE.65 Spectra were displayed and analyzed using SPARKY software.66 NOESY spectra in D2O were recorded at 10, 25 and 30° C with mixing times of 50, 150 and 400 ms. TOCSY, COSY and DQF-COSY experiments were acquired at 25 °C. Natural abundance 1H-13C HSQC and 1H-31P HETCOR experiments were recorded in D2O at 25 °C. For the study of exchangeable protons, 1D 1H and 2D NOESY spectra were recorded at different temperatures with various mixing time using the Watergate water suppression sequence. In confirmation of the unimolecular character of the ASLs after purification and dialysis, 1D 1H and 1D-31P spectra were recorded immediately. The 31P NMR spectrum exhibited a highly downfield resonance (characteristic of the canonical U-turn conformation) that we attributed to be P34. This property was confirmed with the 31P resonance assignments using the 31P-1H HECTOR NMR spectrum. Other assignments confirmed that we had a C-turn (a pseudo-like U-turn). For both RNAs (with or without the f5C34 modification), a dipolar connectivity between the protons A35-H8/C33-H1′ was observed within the NOESY spectra also characteristic of the canonical U-turn (Fig. 3). The formation of a C-turn is incompatible with the RNA adopting a duplex conformation. A duplex conformation would probably result in non-canonical, mismatch base pairs between C32-C38, C33-A37 and others. In a duplex conformation, observation of the A35-H8/C33-H1′ dipolar connectivity would require the flipping out of the residue f5C34. This was absolutely not the case as we observed NOE connectivities between the residues f5C34 and A35 (C34/f5C34-H6/A35-H8 and between f5C34 H7–A35 H8) demonstrating that those two residues are stacked.
NMR restraints generation and structure calculation
NMR data collected at 25° and 30° C were used for the structure calculations. The NOE cross-peaks were integrated using the peak fitting Gaussian function and volume integration in SPARKY software. Using the intensity of the cross peak, and as reference the H5-H6 covalent distance for uridine and cytosine residues, the cross-peaks were qualitatively classified as strong (3.0 1.2 0.0), medium strong (4.0 2.0 0.0), medium (5.0 3.0 0.0), weak (5.0 2.0 0.0) or very weak (6.0 3.0 0.0). No inter-nucleotide restraints were used from the H5′ and H5″ protons. For restraints involving imino protons, (3.0 1.2 0.0) and (5.0 3.0 0.0) intervals were used. A total number of 266 and 275 distance restraints were employed for generating the structures of the hmtASLMetCAU-Ψ27;f5C34 and the hmtASLMetCAU-Ψ27 (Table 2). Consistent with the NMR data, the four stem G●C Watson-Crick base pairs were subjected to six hydrogen-bond restraints (1.8-2.5 Å between the donated hydrogen and the acceptor, and 2.7 - 3.5 Å between the two heavy atoms). No hydrogen bond restraints were used between Ψ27 and A43, despite the observation of an NOE between the Ψ27H3 imino proton and the A42NH2 protons. Dihedral constraints on the ribose ring and backbone were derived from semi-quantitative measurement of the 3JH-H and 3JH-P couplings. Sugar pucker conformations were derived from the observation of the COSY and DQF-COSY spectra. Due to the large U36H1′-H2′ coupling observed, this residue was constrained to the C2′-endo (δ: 160 ± 20°), whereas for the other residues only weak (A43) or very weak H1′-H2′ connectivity was observed on the COSY spectra and these residues were therefore, left unrestrained (A43) or constrained to the C3′-endo conformation (δ: 85 ± 20°). Independent confirmation of the U36 sugar pucker conformation was derived from the 1H-13C HSQC spectra, and the C3′ and C4′ resonances downfield from the main cluster of sugar resonances. The γ torsion angle was derived from the 3JH4′-H5′ and 3JH4′-H5″ peaks in the COSY and DQF-COSY. For residues that lacked H4′-H5′ and H4′-H5″ peaks (J < 5Hz), γ was constrained to the g+ conformation (54 ± 20°). For the other residues, γ was left unrestrained. β dihedral angles were derived from the 3JP-H5′ and 3JP-H5″ couplings observed in the 1H-31P HETCOR NMR spectrum. When the 3JP-H5′ and 3JP-H5″ were clearly absent, β was constrained to the trans conformation (178 ± 20°). When 3JP-H5′ and 3JP-H5″ could be observed, β was left unrestrained. All H3′-P were clearly identified on the 1H-31P HETCOR (3J > 5Hz), so the ε dihedral angle were loosely constraint to avoid the steric forbidden g+ conformation (-120 ± 120°). Dihedral angle restraints for the α and ζ were derived from the observation of the 31P chemical shifts. All of the 31P resonances, except for 31P of the f5C34 and C34 which were found strongly downfield and A37 to a lesser extent, were in a sharp range of chemical shift. The U36 31P chemical shift was upfield to the majority of the 31P resonances. Thus, α and ζ were constraints to avoid the trans conformation resulting in strongly downfield 31P resonances (0 ± 120°). F5C34/C34, A37 and U36 α and ζ dihedral angles were left unrestrained. Each of the intra-aromatic to H1′ NOE connectivities of the different ASL residues had an intensity significantly less than that for the H5-H6 NOE connectivity showing that all the bases adopted an anti glycosidic torsional angle. Thus, the χ glycosidic dihedral angle was constrained to the anti conformation for all residues. NMR restrained molecular modeling of the hmtASLMetCAU-Ψ27;f5C and hmtASLMetCAU-Ψ27 structures was achieved using CNS 1.2.67,68 One hundred structures were calculated using a standard NMR restrained annealing protocol.69 The 10 structures having the lowest total energy, without dihedral or distances restraint violations, were chosen for further analysis. The structure helical parameters were analyzed using X3DNA software.70 The output structures were visualized with MOLMOL and PYMOL.71
Codon Binding
In order to observe binding at the P-site, ribosomes were programmed with a 27-mer oligonucleotide mRNA containing a Shine-Dalgarno sequence at the 5′-leader, an AUG or AUA at the P-site, and the valine codon GUA at the A-site. The binding of mRNA was assessed using radiolabeled mRNAs to ensure that the different mRNAs were bound by the ribosome similarly. The binding of the different mRNA sequences to the ribosomes revealed no significant differences (data not shown). Since ASLs bind P-site codons significantly better (∼100-fold) than A-site codons27 and both the hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 bound the valine codon GUA poorly in the A- or P-sites, similar to that of nonspecific ribosome binding (data not shown), the A-site was left empty. Near saturation, the amount of hmtASLMetCAU-Ψ27;f5C34 and hmtASLMetCAU-Ψ27 bound to AUG in the P-site exceeded their binding of AUG in the A-site by many fold (2-4 pmoles bound compared to 0.5-1.0 pmoles). In order to study the binding of the hmtASLs to the A-site codons AUG or AUA, the E. coli 70S ribosome was programmed with a 27-nucleotide mRNA with a 5′-leader with containing the Shine-Dalgarno sequence, the Val codon GUA at the P-site, and either AUG or AUA at the A-site. The P-site was saturated with unmodified ASLValUAC and A-site binding by either hmtASLMetCAU-Ψ27;f5C34 or the hmtASLMetCAU-Ψ27 was tested. Control experiments saturating the GUA-programmed A-site with ASLValUAC after P-site binding revealed similar results as those in the absence of ASLValUAC (data not shown). The 70S ribosomal subunits were prepared from E. coli MRE600.72 Insignificant amounts of the ASLs that had been 5′-endlabeled with 32P using [γ-32P] ATP (MP Biomedicals)15, and were radiochemically detectable (2,000-5,000 CPM), were mixed in a fixed ratio with unlabeled ASLs in a range of concentrations (0-5 μM).49 The filter binding assay (30 min; 25 °C) was performed with activated ribosomes (0.25 μM; 5 pmoles) that had been programmed with mRNA (2.5 μM) and used the ribosomal binding buffer (50 mM HEPES, pH 7.0; 30 mM KCl; 70 mM NH4Cl; 1 mM DTT; 100 μM EDTA; 20 mM MgCl2).49 Data collected from a phosphoimager were measured for radioactive intensity using ImageQuant (Amersham). Nonspecific binding was determined by the binding of ASLs to ribosomes without mRNA and subtracted from the experimental data. In general, the final data is a result of at least three separate experiments, each done with samples in triplicate, ie. minimally nine determinations for each binding with the result in the absence of mRNA subtracted. Data were analyzed using single-site, nonlinear regression (Prism, GraphPad Software, Inc., San Diego, CA).49 Dissociation constants (Kds) were extracted from the binding curves.
Ribosomal Kinetic Assays
The kinetic assays were conducted in a similar fashion as the ribosomal binding assays described above with the exception that only a single low concentration of ASL was used (0.5 μM) and binding was monitored on a time course. Aliquots of ribosomal reactions were filtered after: 0.0, 0.25, 0.5, 1, 5, 10, 15, 20, 30 minutes. The nitrocellulose was washed, dried, and processed as described.
Bovine Mitochondrial 55S Binding Assays
Bovine mitochondrial, 55S, ribosomes were isolated and purified as previously described.10 Bovine 55S mitochondria were programmed with a chemically synthesized 30-mer oligonucleotide. Mammalian mitochondrial mRNAs are leaderless and translation initiation begins at the first nucleoside triplet on the mRNA. Therefore, to study binding at the P-site, we designed the mRNAs (see the following sequences) with an AUG or AUA (in bold) as the first three nucleosides, followed by the valine codon GUA (underlined) at the A-site, and a stretch of poly-Uridines:
5′-AUGGUAUUUUUUUUUUUUUUUUUUUUUUUU-3′
5′-AUAGUAUUUUUUUUUUUUUUUUUUUUUUUU-3′.
The mRNAs of mammalian mitochondria do not consist of a ribosomal binding sequence such as the Shine-Dalgarno sequence found in bacterial mRNAs. However, an mRNA consisting of stretches of poly(U) with the mitochondrial methionine codon AUG has been shown to be successful in translating methionine using an mitochondrial translational system in vitro.10,14 Therefore, this poly(U)-methionine codon mRNA can be incorporated into the mitochondrial 55S ribosome. To study the binding of the hmtASLMetCAU-Ψ27;f5C34 to the A-site, we incorporated the valine codon GUA (intended for the P-site; underlined in the following mRNA sequences), followed by AUG or AUA (in bold) into a string of 24 Uridines:
5′-UUUUUUUUUUUUGUAAUGUUUUUUUUUUUU-3′
5′-UUUUUUUUUUUUGUAAUAUUUUUUUUUUUU-3′.
Full length, native bovine mtRNAMetCAU and its transcript were aminoacylated and formylated.14 The IF2-facilitated binding of the full length tRNAs to AUG and AUA in the P-site was conducted as previously described.73,74
Acknowledgments
The authors would like to thank William Cantara for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health and the National Science Foundation to P.F. A. (2RO1-GM23037 and MCB0548602, respectively), by National Institutes of Health grant GM32734 to L.L.S. and from the United Mitochondrial Disease Foundation to L.L.S, and P.F.A. (05-20), and by Dharmacon-Thermo Fisher Scientific.
Abbreviations
- f5C34
5-formylcytidine at position 34
- ASL
anticodon stem and loop
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser spectroscopy
- COSY
correlation spectroscopy
- TOCSY
total correlation spectroscopy
- HSQC
heteronuclear single quantum correlation
- RNA
ribonucleic acids
- tRNA
transfer ribonucleic acids
- mRNA
messenger RNA
- ppm
part per million
- RMSD
root mean square deviation
- NMR
nuclear magnetic resonance
Footnotes
Accession numbers: PDB code:2KRZ (hmtASLMetCAU-Ψ27); 2KRY (hmtASLMetCAU-Ψ27;f5C); BMRB entry: 16655 (hmtASLMetCAU-Ψ27), 16654 (hmtASLMetCAU-Ψ27;f5C)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Aujame L, Freeman KB. Mammalian mitochondrial transfer RNAs: chromatographic properties, size and origin. Nucleic Acids Res. 1979;6:455–469. doi: 10.1093/nar/6.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Söll D, Alfonzo JD. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci USA. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomita K, Ueda T, Ishiwa S, Crain PF, McCloskey JA, Watanabe K. Codon reading patterns in Drosophila melanogaster mitochondria based on their tRNA sequences: a unique wobble rule in animal mitochondria. Nucleic Acids Res. 1999;27:4291–4297. doi: 10.1093/nar/27.21.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos MA, Moura G, Massey SE, Tuite MF. Driving change: the evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- 7.Young IG, Anderson S. The genetic code in bovine mitochondria: sequence of genes for the cytochrome oxidase subunit II and two tRNAs. Gene. 1980;12:257–65. doi: 10.1016/0378-1119(80)90108-0. [DOI] [PubMed] [Google Scholar]
- 8.Anderson S, de Bruijn MH, Coulson AR, Eperon IC, Sanger F, Young IG. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 9.Fearnley IM, Walker JE. Initiation codons in mammalian mitochondria: differences in genetic code in the organelle. Biochemistry. 1987;26:8247–8251. doi: 10.1021/bi00399a034. [DOI] [PubMed] [Google Scholar]
- 10.Takemoto C, Koike T, Yokogawa T, Benkowski L, Spremulli LL, Ueda TA, Nishikawa K, Watanabe K. The ability of bovine mitochondrial transfer RNAMet to decode AUG and AUA codons. Biochimie. 1995;77:104–108. doi: 10.1016/0300-9084(96)88112-0. [DOI] [PubMed] [Google Scholar]
- 11.Crick FH. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 12.Björk GR. Biosynthesis and function of modified nucleosides. In: Söll D, Rajbhandary UL, editors. tRNA: Structure, Biosynthesis, and Function. ASM Press; Washington, D.C: 1995. pp. 165–206. [Google Scholar]
- 13.Moriya J, Yokogawa T, Wakita K, Ueda T, Nishikawa K, Crain PF, Hashizume T, Pomerantz SC, McCloskey JA, Kawai G, Hayashi N, Yokoyama S, Watanabe K. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry. 1994;33:2234–2239. doi: 10.1021/bi00174a033. [DOI] [PubMed] [Google Scholar]
- 14.Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, Watanabe K. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37:1616–1627. doi: 10.1093/nar/gkp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lusic H, Gustilo EM, Vendeix FA, Kaiser R, Delaney MO, Graham WD, Moye VA, Cantara WA, Agris PF, Deiters A. Synthesis and investigation of the 5-formylcytidine modified, anticodon stem and loop of the human mitochondrial tRNAMet. Nucleic Acids Res. 2008;36:6548–6557. doi: 10.1093/nar/gkn703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. Database issue: http://trnadb.bioinf.uni-leipzig.de/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takemoto C, Ueda T, Miura K, Watanabe K. Nucleotide sequences of animal mitochondrial tRNAs(Met) possibly recognizing both AUG and AUA codons. Nucleic Acids Symp Ser. 1999;42:77–78. doi: 10.1093/nass/42.1.77. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe Y, Tsurui H, Ueda T, Furushima R, Takamiya S, Kita K, Nishikawa K, Watanabe K. Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J Biol Chem. 1994;269:22902–22906. [PubMed] [Google Scholar]
- 19.Tomita K, Ueda T, Watanabe K. 5-formylcytidine (f5C) found at the wobble position of the anticodon of squid mitochondrial tRNAMetCAU. Nucleic Acids Symp Ser. 1997:197–198. [PubMed] [Google Scholar]
- 20.Jones CN, Jones CI, Graham WD, Agris PF, Spremulli LL. A disease-causing point mutation in human mitochondrial tRNAMet results in tRNA misfolding leading to defects in translational initiation and elongation. J Biol Chem. 2008;283:34445–34456. doi: 10.1074/jbc.M806992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones CN, Wilkinson KA, Hung KT, Weeks KM, Spremulli LL. Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA. 2008;14:862–871. doi: 10.1261/rna.909208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Montoya J, Ojala D, Attardi G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 24.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 25.Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001;338:371–399. doi: 10.1016/s0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- 27.Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1986;25:3245–3255. doi: 10.1021/bi00359a025. [DOI] [PubMed] [Google Scholar]
- 28.Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi N, Kawakami M, Omori A, Ueda T, Spremulli LL, Watanabe K. Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver. Purification, characterization, and gene structure. J Biol Chem. 1998;273:15085–15090. doi: 10.1074/jbc.273.24.15085. [DOI] [PubMed] [Google Scholar]
- 30.Spencer AC, Heck A, Takeuchi N, Watanabe K, Spremulli LL. Characterization of the human mitochondrial methionyl-tRNA synthetase. Biochemistry. 2004;43:9743–9754. doi: 10.1021/bi049639w. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheffler IE. Mitochondria. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2008. [Google Scholar]
- 33.Moore PB. Structural motifs in RNA. Annu Rev Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 34.Auffinger P, Westhof E. An extended structural signature for the tRNA anticodon loop. RNA. 2001;7:334–341. doi: 10.1017/s1355838201002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashraf SS, Ansari G, Guenther R, Sochacka E, Malkiewicz A, Agris PF. The uridine in “U-turn”: contributions to tRNA-ribosomal binding. RNA. 1999;5:503–511. doi: 10.1017/s1355838299981931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 37.Gustilo EM, Vendeix FAP, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microb. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lescrinier E, Nauwelaerts K, Zanier K, Poesen K, Sattler M, Herdewijn P. The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res. 2006;34:2878–2886. doi: 10.1093/nar/gkl081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agris PF. Bringing order to translation: Contributions of tRNA anticodon domain modifications. EMBO Reports. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zacharias M, Sklenar H. Conformational analysis of single-base bulges in A-form DNA and RNA using a hierarchical approach and energetic evaluation with a continuum solvent model. J Mol Biol. 1999;289:261–275. doi: 10.1006/jmbi.1999.2760. [DOI] [PubMed] [Google Scholar]
- 41.Schweisguth DC, Moore PB. On the conformation of the anticodon loops of initiator and elongator methionine tRNAs. J Mol Biol. 1997;267:505–519. doi: 10.1006/jmbi.1996.0903. [DOI] [PubMed] [Google Scholar]
- 42.Ramesh V, RajBhandary UL. Importance of the anticodon sequence in the aminoacylation of tRNAs by methionyl-tRNA synthetase and by valyl-tRNA synthetase in an Archaebacterium. J Biol Chem. 2001;276:3660–3665. doi: 10.1074/jbc.M008206200. [DOI] [PubMed] [Google Scholar]
- 43.Senger B, Fasiolo F. Yeast tRNAMet recognition by methionyl-tRNA synthetase requires determinants from the primary, secondary and tertiary structure: a review. Biochimie. 1996;78:597–604. doi: 10.1016/s0300-9084(96)80006-x. [DOI] [PubMed] [Google Scholar]
- 44.Pelka H, Schulman LH. Study of the interaction of Escherichia coli methionyl-tRNA synthetase with tRNAfMet using chemical and enzymatic probes. Biochemistry. 1986;25:4450–4456. doi: 10.1021/bi00363a042. [DOI] [PubMed] [Google Scholar]
- 45.Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 46.(16) Kawai G, Yokogawa T, Nishikawa K, Ueda T, Hashizume T, McCloskey JA, Yokoyama S, Watanabe K. Conformational properties of a novel modified nucleoside, 5-formylcytidine, found at the first position of the anticodon of mitochondrial tRNAMet. Nucleosides, Nucleotides and Nucleic acids. 1994;13:1189–1199. [Google Scholar]
- 47.Agris PF, Vendeix FAP, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol. 2006;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 48.Weixlbaumer A, Murphy F, IV, Vendeix FAP, Dziergowska A, Malkiewicz A, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nature Struct Mol Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vendeix FAP, Graham WD, Dziergowska A, Gustilo E, Sproat B, Malkiewicz A, Agris PF. Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry. 2008;47:6117–6129. doi: 10.1021/bi702356j. [DOI] [PubMed] [Google Scholar]
- 50.Barraud P, Schmitt E, Mechulam Y, Dardel F, Tisne C. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res. 2008;36:4894–4901. doi: 10.1093/nar/gkn462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basavappa R, Sigler PB. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schevitz RW, Podjarny AD, Krishnamachari N, Hughes JJ, Sigler PB, Sussman JL. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979;278:188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- 53.Woo NH, Roe BA, Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980;286:346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- 54.Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 55.Perbandt M, Barciszewska MZ, Betzel C, Erdmann VA, Barciszewski J. A critical role of water in the specific cleavage of the anticodon loop of some eukaryotic methionine initiator tRNAs. Mol Biol Rep. 2003;30:27–31. doi: 10.1023/a:1022298519351. [DOI] [PubMed] [Google Scholar]
- 56.Dix DB, Wittenberg WL, Uhlenbeck OC, Thompson RC. Effect of replacing uridine 33 in yeast tRNAPhe on the reaction with ribosomes. J Biol Chem. 1986;261:10112–10118. [PubMed] [Google Scholar]
- 57.Wrede P, Woo NH, Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc Natl Acad Sci USA. 1979;76:3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramakrishnan V. Ribosome Structure and the Mechanism of Translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 59.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 60.Roy A, Panigrahi S, Bhattacharyya M, Bhattacharyya D. Structure, stability, and dynamics of canonical and noncanonical base pairs: Quantum chemical studies. J Phys Chem B. 2008;112:3786–3796. doi: 10.1021/jp076921e. [DOI] [PubMed] [Google Scholar]
- 61.Vendeix FAP, Munoz A, Agris PF. Free energy calculation of modified base-pair formation in explicit solvent: A predictive model. RNA. 2009;15:2278–2287. doi: 10.1261/rna.1734309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cai Z, Tinoco I., Jr Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 63.Scaringe SA. Advanced 5′-silyl-2′-orthoester approach to RNA oligonucleotide synthesis. Methods Enzymol. 2000;317:3–18. doi: 10.1016/s0076-6879(00)17003-x. [DOI] [PubMed] [Google Scholar]
- 64.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons, Inc.; Hoboken, New Jersey: 1986. [Google Scholar]
- 65.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 66.Goddard TD, Kneller D. G SPARKY 3.113. University of California; San Francisco: 2006. http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- 67.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 68.Brünger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 69.Stein EG, Rice LM, Brünger AT. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J Magn Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- 70.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 72.Shapkina T, Lappi S, Franzen S, Wollenzien P. Efficiency and pattern of UV pulse laser-induced RNA-RNA cross-linking in the ribosome. Nucleic Acids Res. 2004;32:1518–1526. doi: 10.1093/nar/gkh320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brooke BE, Haque ME, Spremulli LL. The effect of spermine on the initiation of mitochondrial protein synthesis. Biochem Biophys Res Commn. 2010;391:942–946. doi: 10.1016/j.bbrc.2009.11.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grasso DG, Christian BE, Spencer A, Spremulli LL. Overexpression and purification of mammalian mitochondrial translational initiation factor 2 and initiation factor 3. Methods Enzymol. 2007;430:59–78. doi: 10.1016/S0076-6879(07)30004-9. [DOI] [PubMed] [Google Scholar]