Several single-nucleotide polymorphisms (SNPs) associated with breast cancer risk have been identified through genome-wide association studies. This study investigated the association of eight risk SNPs with breast cancer disease-free survival and overall survival rates. Results suggest that two previously identified breast cancer risk susceptibility loci may influence breast cancer prognosis or comorbid conditions associated with overall survival.
Keywords: Breast cancer, Prognosis, Single-nucleotide polymorphisms, TNRC9, 17q23
Learning Objectives
Describe the results of genome-wide association studies (GWAS) that have identified genetic variants associated with breast cancer risk.
Discuss whether genetic risk variants identified through genome-wide association studies (GWAS) are also associated with breast cancer prognosis.
Describe molecular mechanisms through which germline genetic variants may influence breast cancer survival.
Abstract
Background.
Several single-nucleotide polymorphisms (SNPs) associated with breast cancer risk have been identified through genome-wide association studies (GWAS). We investigated whether eight risk SNPs identified in GWAS were associated with breast cancer disease-free survival (DFS) and overall survival (OS) rates.
Patients and Methods.
A cohort of 739 white women with early-stage breast cancer was genotyped for eight GWAS-identified SNPs (rs2981582, rs1219648 [FGFR2], rs3803662, rs12443621, rs8051542 [TOX3], rs999737 [RAD51L1], rs6504950 [17q23], and rs4973768 [3p24]). Relationships between SNPs and breast cancer outcomes were evaluated using Cox proportional hazard regression models. The cumulative effects of SNPs on breast cancer outcomes were assessed by computing the number of at-risk genotypes.
Results.
At a median follow-up of 121 months (range: 188–231 months) for survivors, 237 deaths (32%) and 186 breast cancer events (25%) were identified among the 739 patients. After adjusting for age, clinical stage, and treatment, rs12443621 (16q12; p = .03) and rs6504950 (17q23; p = .008) were prognostic for OS but not DFS. A higher risk for death was also found in the multivariable analysis of patients harboring three or four at-risk genotypes of the GWAS SNPs compared to patients carrying two or less at-risk genotypes (hazard ratio: 1.60, 95% confidence interval: 1.23–2.24; p = .0008).
Conclusion.
The study results suggest that previously identified breast cancer risk susceptibility loci, rs12443621 (16q12) and rs6504950 (17q23), may influence breast cancer prognosis or comorbid conditions associated with overall survival. The precise molecular mechanisms through which these risk SNPs, as well as others that were not included in the analysis, influence clinical outcomes remain to be determined.
Implications for Practice:
Prior genome-wide association studies (GWAS) have identified rare genetic variations in individual's DNA (SNPs) that increase the risk of breast cancer. However, the role that these GWAS discovered SNPs play in determining survival after a diagnosis of breast cancer is not clear. Here, we found that two GWAS identified SNPs were associated with overall but not breast cancer survival. Replication of these findings in different populations is needed to determine whether GWAS discovered SNPs can be used to develop prognostic and therapeutic approaches for patients with breast cancer.
Introduction
Breast cancer is the most common female malignancy in the United States and is the second leading cause of death from cancer [1]. Despite the excellent 5-year overall survival rate of 83%–92% among women with stage I and II disease [2], the 10-year risk of recurrence is estimated at 20%–40% [3]. Traditional prognostic factors, such as tumor size, grade, and lymph node metastasis status, are still the most important prognostic factors for long-term survival in breast cancer, but less is known about the effect of inherited genetic variation on recurrence risk and overall disease-free survival rates.
It has been estimated that there are 7 million common single-nucleotide polymorphisms (SNPs) in the human genome (with minor allele frequency [MAF] of >5%) [4]. Recently several genome-wide association studies (GWAS) [5–14] have identified multiple breast cancer susceptibility loci—of which the majority are SNPs—that contribute a small effect on breast cancer risk [15]. Some of the GWAS-identified SNPs are found to be at loci containing the genes FGFR2 (rs2981582, rs1219648), LSP1 (rs3817198, rs2771439), MAP3K1 (rs726501, rs889312), TOX3/TOX3 (rs3803662, rs8051542), MRPS30 (rs10941679, rs7705343), COX11 (rs10515083, rs16955329, rs2787487), SLC4A7 (rs4973768), TGFB1 (rs1982073), or ESR1 (rs3020314), or at chromosomes 8p24 (rs13281615, rs283720) and 2q35 (rs13387042, rs10169372). Based on a recent meta-analysis and pooled analysis, which addressed the associations between 145 gene variants and breast cancer, rs2981582 represents the SNP in the FGFR2 gene with the strongest association with breast cancer [16]. What remains unclear is whether these genetic variants continue to influence the biology of the disease after diagnosis and affect breast cancer survival. Although a number of studies have evaluated genetic polymorphisms in relation to prognosis [17–29], no study has examined the relative risks for breast cancer recurrence and death attributable to the recently discovered eight risk SNPs (rs2981582, rs1219648, rs3803662, rs12443621, rs8051542, rs999737, rs6504950, rs4973768).
The list of the GWAS-identified SNPs is growing exponentially. We selected eight risk SNPs that were identified based on a review of the literature [6, 10–11, 23, 30]. We conducted this retrospective analysis to elucidate a possible association between the eight GWAS-identified SNPs and clinical outcome of patients with early-stage breast cancer, adjusting for known clinical and tumor prognostic factors. We hypothesized that SNPs that are associated with increased breast cancer risk could be important genetic markers contributing to poorer breast cancer outcome.
Patients and Methods
The Early-Stage Breast Cancer Repository (ESBCR) is a retrospective cohort of 2,409 women diagnosed with American Joint Committee on Cancer pathologic stage I or II breast cancer and surgically treated (mastectomy or segmental mastectomy) at MD Anderson Cancer Center (MDACC) between 1985 and 2000. Criteria for eligibility and cohort details have been previously described [31]. Briefly, the ESBCR contains detailed information on patient (ethnicity, age, menopausal status) and tumor (clinical and pathological stage, estrogen receptor [ER] and progesterone receptor [PR] status, nuclear grade) characteristics, chemotherapy and endocrine treatment, radiation and surgery type (segmental mastectomy, mastectomy), and epidemiological risk factors, including family history [FH] of breast cancer in at least one first- or second-degree relative. Follow-up information was obtained by direct review of the medical records and linkage to the MDACC Tumor Registry, which mails annual follow-up letters to each registered patient who is known to be alive to determine their clinical status. The MDACC Tumor Registry checks the U.S. Social Security death index and the Texas Bureau of Vital Statistics for the status of patients who fail to respond to the letters.
We selected a subset of the ESBCR population with available formalin-fixed paraffin-embedded normal lymph nodes or blood samples available for DNA extraction for participation in the genotyping project. The subset of the ESBCR population was enriched to include all black (n = 196) and Hispanic patients (n = 208) and a random sample of white patients (n = 986). Because of the small percentage of patients from other ethnic groups, we included only white patients in the study analyses. A total of 247 white patients were excluded from study analysis because of DNA extraction or genotyping failure (n = 251) or insufficient clinical information (n = 6). Patients excluded from the analysis for sample failure were more likely to be diagnosed during 1985–1994 versus 1995–2000 and were also more likely to have stage I versus stage II disease. The final study analysis consisted of 739 patients with breast cancer. The study was approved by the MD Anderson Cancer Center Institutional Review Board.
SNP Selection and Genotyping
We reviewed the literature and selected eight SNPs that were identified as novel breast cancer susceptibility loci by GWAS based on the available literature at the time of genotype analysis [6, 10–11, 23, 30, 32]. All selected SNPs were within genes or linkage disequilibrium (LD) blocks containing genes (10q26-rs2981582, rs1219648 [FGFR2]; 16q12-rs3803662, rs12443621, rs8051542 [TOX3]; 14q24-rs999737 [RAD51L1]; 17q23-rs6504950 [COX11]; 3p24-rs4973768 [SLC4A7]) based on the data from the International HapMap Project (www.hapmap.org, version 23). All SNPs met the following criteria: minor allele frequency (MAF) of ≥0.05, Illumina design score > 0.4, and r2 ≥ 0.8 for binning.
Genotyping of the eight selected SNPs was performed on the Illumina GoldenGate platform (Illumina Inc., San Diego, CA, http://www.illumina.com) as part of a larger array of 1,536 SNPs of other candidate genes. Briefly, genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA, http://www.qiagen.com) according to the manufacturer's protocol [33]. Genomic DNA was extracted from normal formalin-fixed, paraffin-embedded (FFPE) tissue using the appropriate protocol in the PicoPure DNA extraction kit (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) [34]. The call rates for the fixed FFPE samples and non-FFPE samples were 92% and 99%, respectively. Blinded duplicate samples (5%) were included in the array platform, and the duplication concordance was 100%. All genotyping information was analyzed and exported using the GenomeStudio software (Illumina).
Statistical Analysis and Outcome Measures
Disease-free survival (DFS) was defined as the elapsed time between the date of initial treatment until the first date of documented disease recurrence or death by breast cancer. Overall survival (OS) was calculated from the date of initial treatment until the date of death from any cause. Clinical variables included age, stage, ER/PR status, treatment (chemotherapy and/or endocrine therapy), and FH of breast cancer. For each SNP and clinical variable, including age, family history of breast cancer, stage, ER/PR status, nuclear grade, treatment type, year of diagnosis, and surgery type (mastectomy and lumpectomy), univariable Cox proportional hazard regression analysis was used to identify the association with DFS and OS. Because this was an exploratory study, no adjustments for multiple testing were performed [35].
The genetic and clinical variables that were possibly related to survival outcomes by univariable analysis (p ≤ .1) were then included into a multivariable Cox proportional hazard regression model. In the process of fitting the multivariable Cox regression model, a backward selection was used with a p value ≤.05 for the likelihood ratio test. Age at diagnosis was retained in the multivariable model, given its importance in determining survival. Interactions between the SNPs and hormone receptor (ER+ or PR+ versus ER− and PR−) and treatment (none, chemotherapy alone, endocrine therapy alone, chemotherapy, and endocrine therapy) statuses were assessed using the likelihood ratio test (p < .05), comparing the model including an interaction term with the reduced model without the interaction term. Furthermore, the cumulative effects of SNPs were assessed by computing the number of at-risk genotypes based on the results from the univariable analyses. The median survival time was estimated using Kaplan-Meier methods, and the log-rank analysis was used to compare survival between the at-risk genotype groups. Hazard ratios (HRs) and their 95% confidence intervals (CIs) were estimated for all associations between SNPs, clinical variables, and survival outcomes. Finally, we produced a recursive partitioning tree to evaluate higher order interactions between clinical variables and SNPs. Statistical analyses were carried out using SAS version 9.1 (SAS Institute, Cary, NC, http://www.sas.com) and R (http://www.r-project.org).
Results
Patient demographic and clinical characteristics are summarized in Table 1. At a median follow-up of 121 months (range: 188–231 months) for survivors, 237 deaths (32%) and 186 breast cancer events (25%) were identified among the 739 patients. The median OS was 217 months. The majority of patients were postmenopausal (57%) and had stage II disease (72%). Approximately 20% of patients did not receive any systemic treatment for their disease and 25% received endocrine therapy alone. There was minimal missing data for the cohort, ranging from 5.1% for ER/PR status to 0.4% for systemic treatment.
Table 1.
Patient demographic and disease characteristics
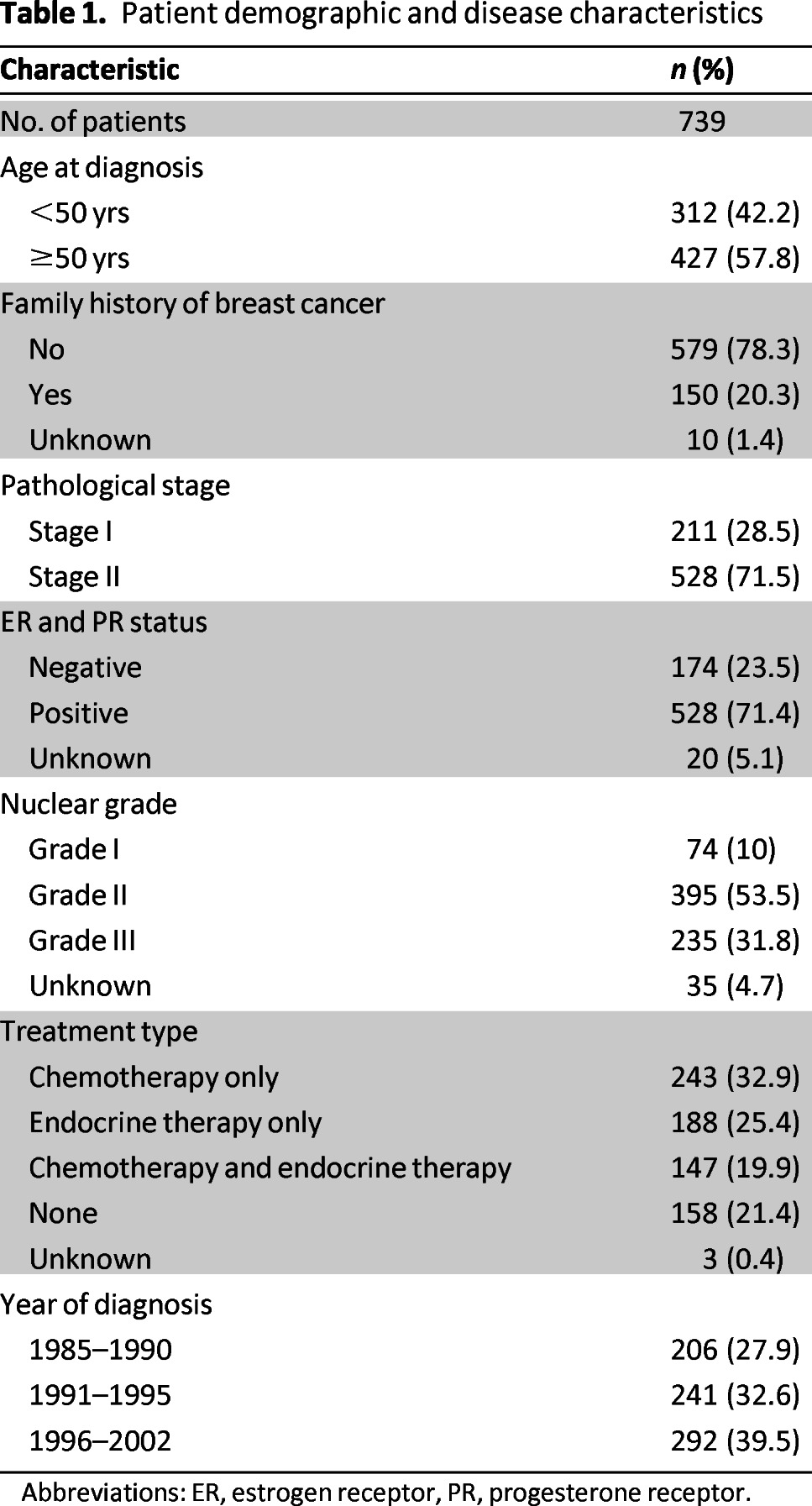
Abbreviations: ER, estrogen receptor, PR, progesterone receptor.
Univariable Associations Between SNPs and Breast Cancer Outcomes
In this cohort of women with a history of early-stage breast cancer, the genotype distributions of eight SNPs were in agreement with Hardy-Weinberg equilibrium. In univariable analysis, none of the SNPs were associated with DFS (Table 2). Significant associations were observed for OS for four SNPs (rs2981582, rs1219648, rs12443621, rs6504950). Homozygous carriers for the minor alleles for rs2981582 and rs1219648 had a decreased risk for death (HR AA vs. GG/AG: 0.6, 95% CI: 0.47–0.98, p = .040; HR GG vs. AA/AG: 0.70, 95% CI: 0.49–1.01). In addition, a decreased risk of death was observed among patients homozygous for the minor alleles for rs121443621 in the dominant model (HR GG vs. AA/AG: 0.70, 95% CI: 0.53–0.95, p = .022). In contrast, OS was worse for homozygous carriers of the minor A allele for rs6504950 both in the codominant model (HR AA vs. GG: 1.70, 95% CI: 1.13–2.61, p = .01) and the dominant model (HR AA vs. GG/AG: 1.70, 95% CI: 1.19–2.68, p = .004). The other investigated SNPs were not associated with OS.
Table 2.
Univariate association of single-nucleotide polymorphisms with disease-free and overall survival times
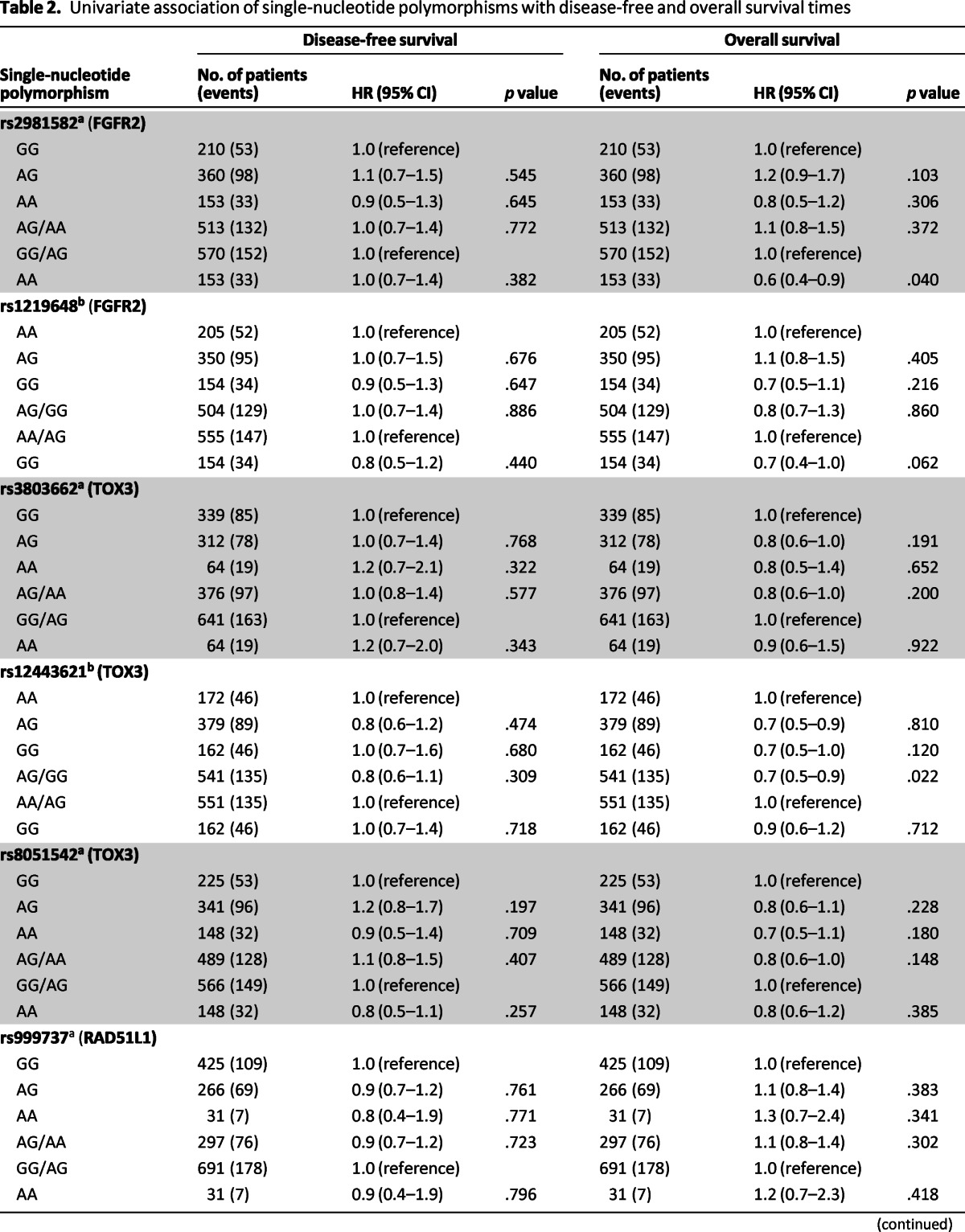
Table 2.
(continued)
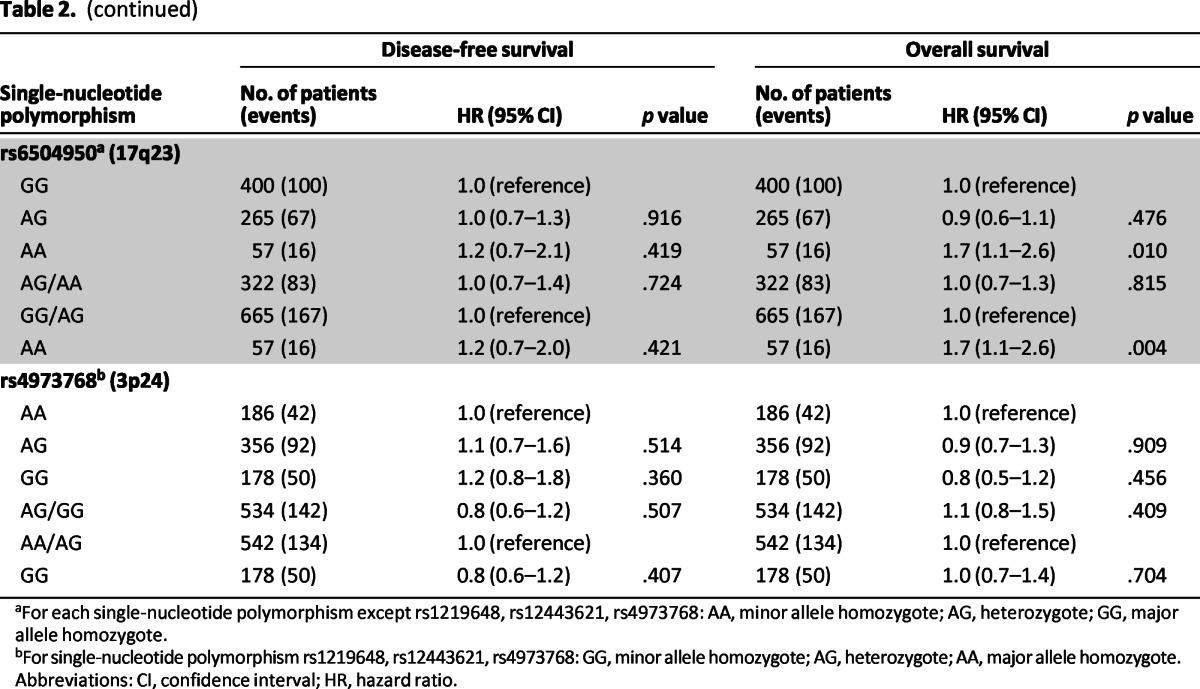
aFor each single-nucleotide polymorphism except rs1219648, rs12443621, rs4973768: AA, minor allele homozygote; AG, heterozygote; GG, major allele homozygote.
bFor single-nucleotide polymorphism rs1219648, rs12443621, rs4973768: GG, minor allele homozygote; AG, heterozygote; AA, major allele homozygote.
Abbreviations: CI, confidence interval; HR, hazard ratio.
Multivariable Associations Between SNPs and Breast Cancer Outcomes
In multivariable models adjusted for age, ER/PR status, stage, and treatment type, we found two SNPs, rs12443621 and rs6504950, which were independent prognostic markers for OS. Compared with the GG/AG reference group, homozygous carriers of minor A allele for rs6504950 had a higher risk for death (HR: 1.77, 95% CI: 1.15–2.73, p = .008). Compared with the AA reference group, homozygous carriers for the AG/GG for rs12443621 had a decreased risk for death (HR: 0.72, 95% CI: 0.53–0.78, p = .035). In addition, stage II disease was associated with a worse OS (HR: 2.08, 95% CI: 1.45–2.96, p < .001) compared to patients with stage I disease, and patients who received chemotherapy and/or endocrine therapy had a better OS (Table 3) than patients who did not receive systemic treatment. We performed a stratified multivariable analysis to explore the relationship between prognostic SNPs and ER/PR tumor status. The increased risk of death associated with the SNP rs6504950 was seen predominately among patients with ER- or PR-positive tumors (p < .001) compared to patients with ER-negative and PR-negative tumors (p = .30; data not shown). However, the test for interaction between SNPs rs12443621, rs6504950, and ER/PR status on overall survival was not statistically significant. There was no evidence of interaction by treatment status (data not shown).
Table 3.
Multivariate Cox proportional hazards model for overall survival in study cohort
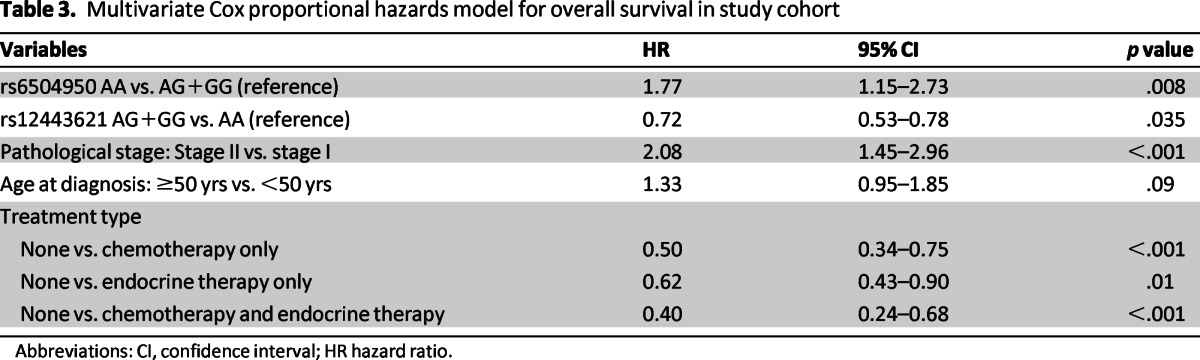
Abbreviations: CI, confidence interval; HR hazard ratio.
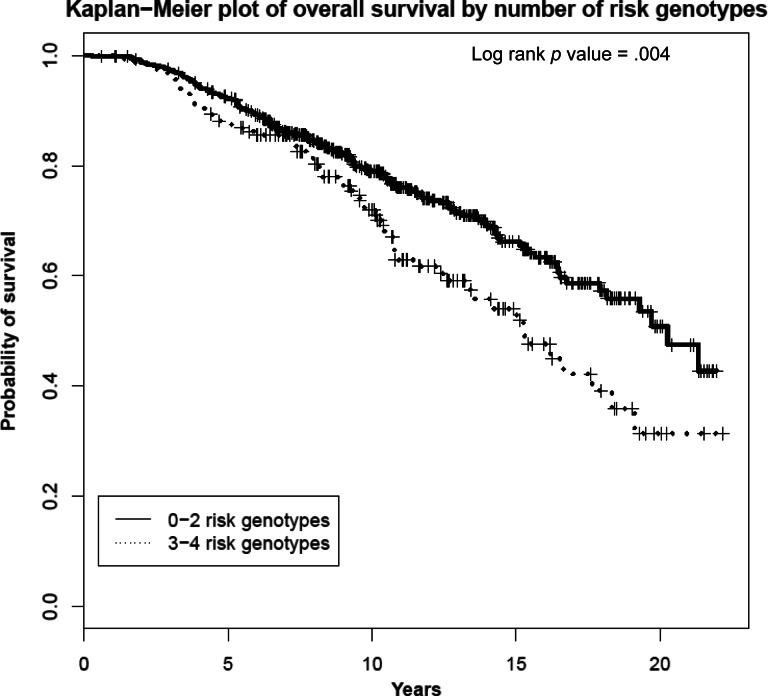
We investigated the cumulative effect of carrying at-risk genotypes for the four SNPs significantly associated with OS in the univariable analysis. We categorized patients into two groups (≤2 vs. 3–4 at-risk genotypes) and used patients carrying ≤2 at-risk genotypes as the reference group. Kaplan-Meier analysis revealed statistical significant differences in OS according to the number of at-risk genotypes (log-rank p = .004; Fig. 1). In multivariable analysis, after adjusting for significant clinical variables, a higher risk for death was found in the subcohort of patients harboring 3–4 at-risk genotypes compared to patients carrying ≤2 at-risk genotypes (HR: 1.60, 95% CI: 1.23–2.24, p = .0008; Table 4).
Figure 1.
Kaplan-Meier plot of overall survival by number of at-risk genotypes.
Table 4.
Multivariate Cox proportional hazards model for overall survival by number of at-risk genotypes in study cohort
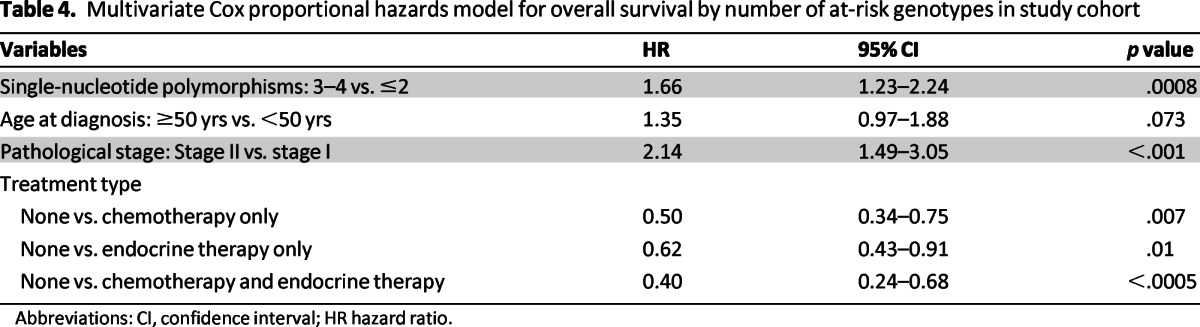
Abbreviations: CI, confidence interval; HR hazard ratio.
Discussion
We evaluated the possible relation between eight GWAS-identified SNPs and breast cancer outcomes in a large cohort of women with early-stage breast cancer. Two GWAS risk SNPs, rs12443621(16q12) and rs6504950 (17q23), were associated with OS, independent of clinical factors. Furthermore, we observed that the risk SNPs (rs2981582, rs1219648, rs12443621, rs6504950) in univariable analysis appeared to act in a codominant (additive) fashion, with mortality risk increasing with the number of at-risk genotypes. To our knowledge, our study is the first to identify the association of these polymorphisms and OS and breast cancer DFS. Replication of our findings in larger study sets and validation is needed before these SNPs can be considered for prognostic risk stratification of patients with breast cancer.
Recent GWAS have identified several SNPs in novel independent loci as markers of breast cancer susceptibility in populations of diverse ethnicity [5, 36]. The rs12443621 prognostic SNP has been identified in an LD block containing the 59 end of the trinucleotide repeat containing 9 (TOX3) gene, located at chromosome 16q12 [5]. The function of the TOX3 gene is unclear; however, it contains a putative high-mobility group box motif, suggesting that it may act as a transcription factor [8]. Of the three polymorphisms identified in the TOX3 gene (rs3803662, rs12443621, and rs8051542) [37], SNP rs3803662 exhibited a stronger association with breast cancer risk in white individuals [5, 32, 38], whereas Chinese [36, 39] studies have showed no association between the rs12443621 or rs3803662 alleles and breast cancer risk [5, 32, 36, 38–39]. Therefore, the clinical relevance of SNP rs12443621 remains unknown. It is not clear why rs12443621 is associated with a decreased OS in this study population. It has been pointed out that genes do not generally act in a simple additive manner but through complex networks involving gene-gene and gene-environment interactions [40]; therefore, it is possible that a minor homozygous genotype or allele associated with increased breast cancer risk may have gene-gene or gene-environment interactions that yield an opposite effect on survival. The other prognostic SNP rs6504950 lies in a 300-kb LD block on 17q23 [11]. Cytochrome C assembly protein 11 (COX11), approximately 10 kb upstream of rs6504950) is the potential causative gene as suggested by the higher levels of COX11 expression in lymphocytes in the HapMap samples (p = .000014) [41]. Of interest, both rs12443621A/G [8, 11, 36] and rs6504950 [42] showed a stronger association with risk of ER-positive than ER-negative breast tumors in those studies. We found that the SNP rs6504950 was associated with a twofold increased risk of death predominately among patients with ER/PR-positive tumors, but the test for interaction was not statistically significant.
There have been an increasing number of epidemiological studies examining the effects of SNPs on cancer outcomes using a candidate gene approach [17–21, 43–44]. These studies have provided insights into the molecular mechanisms through which germline SNPs may influence breast cancer survival [19–20, 22, 24–26, 41, 43, 45–48]. To date, the functional role of the germline polymorphisms in the 16q12 (TOX3) and 17q23 loci is unknown. Smid et al. [49] identified a 69 panel of genes found in tumor DNA relevant to bone metastasis in breast cancer; among those, increased expression of TOX3 was found to be a predictive factor. In prior studies, increased copy numbers of 17q23 chromosomal region have been associated with tumor progression and with poor prognosis in breast cancer [50–52]. Inaki et al. [53] found that 41% (28/69) of breast cancer tumors in their series showed evidence of gene amplification at 17q23 locus. Expression of recurrent fusion gene transcript (RPS6KB1-VMP1) created by a tandem duplication in the 17q23 locus showed a trend toward correlation (p = .06) with poor DFS. In our study, germline SNPs in the 16q12 and 17q23 loci did not influence breast cancer DFS but played a significant role in OS among a favorable prognostic group of older patients with breast cancer who did not receive chemotherapy.
Several limitations of this study should be acknowledged. First, the number of SNPs tested was limited to eight, which is not a comprehensive evaluation of the association between GWAS-identified risk SNPs and breast cancer prognosis. The study was exploratory and we did not adjust for multiple testing; therefore, some of our findings may be due to chance. In addition, the sample size was too small to definitively evaluate for breast cancer outcomes according to ER and PR status or systemic treatment received. In addition, the sample size was too small to definitively evaluate for breast cancer outcomes according to breast tumor subtypes. The Breast Cancer Association Consortium has taken the first step in investigating GWAS-identified susceptibility loci in relation to risk of developing specific breast tumor subtypes; however, information on outcomes is not yet available.
In addition, the ESBCR cohort represents a select group of white women with early-stage disease treated exclusively at a single institution, limiting generalizability of the findings to all patients with breast cancer. Azzatto et al. performed a GWAS of prognosis in patients with breast cancer enrolled in the Nurses' Health Cohort and found no genetic variants associated with breast cancer-specific death. However, that study had few breast cancer deaths (n = 92) and may have been underpowered in the discovery analysis to detect SNPs with modest effects [54]. Similarly, in our study we found no association with breast cancer DFS, but there was an association with OS. The reasons for this specific association are unclear but suggest that any additional biological effect of the genetic variants may be explained by influences on host comorbid conditions, lifestyle factors, or environmental factors that contribute to mortality.
The GWAS have identified several common SNPs that convey breast cancer risk, but it is unclear whether these genetic variants continue to influence the biology of the disease after diagnosis. In our study, we found a significant association between two GWAS-identified SNPs 16q12 (TOX3) and 17q23 loci and OS in a cohort of patients with early-stage breast cancer; however, they were not associated with breast cancer DFS. The reasons for the specific associations are unclear but suggest that any additional biological effect of the genetic variants may be explained by influences on host comorbid conditions, lifestyle factors, or environmental factors that contribute to mortality. These findings provide support for the hypothesis that SNPs in these loci may be biologically important and may be promising targets for diagnostic, prognostic, and therapeutic approaches. These results need confirmation in independent cohorts. Future studies that evaluate the prognostic relationship of these SNPs by breast cancer subtype and type of treatment received may provide additional information that could be used to individualize the care of patients with breast cancer.
This article is available for continuing medical education credit at CME.TheOncologist.com.
Acknowledgments
This work was supported by the National Cancer Institute (grants R01CA089608 to M.L.B. and R03CA123553 to A.M.B.).
Author Contributions
Conception/Design: Patricia A. Thompson, Abenaa M. Brewster
Provision of study material or patients: Patricia A. Thompson, Melissa L. Bondy, Abenaa M. Brewster
Collection and/or assembly of data: Patricia A. Thompson, Aysegul A. Sahin, Melissa L. Bondy, Abenaa M. Brewster
Data analysis and interpretation: Soley Bayraktar, Suk-Young Yoo, Kim-anh Do, Aysegul A. Sahin, Banu K. Arun, Melissa L. Bondy, Abenaa M. Brewster
Manuscript writing: Soley Bayraktar, Suk-Young Yoo, Kim-anh Do, Aysegul A. Sahin, Banu K. Arun, Melissa L. Bondy, Abenaa M. Brewster
Final approval of manuscript: Soley Bayraktar, Patricia A. Thompson, Suk-Young Yoo, Kim-anh Do, Aysegul A. Sahin, Banu K. Arun, Melissa L. Bondy, Abenaa M. Brewster
Disclosures
The authors indicated no financial relationships.
Section editors: Gabriel Hortobágyi: Antigen Express, Galena Biopharma, Novartis, Rockpointe (C/A); Novartis (RF); Taivex (OI); founder and member of the board of directors for Citizen's Oncology Foundation; Kathleen Pritchard: Novartis, Roche, AstraZeneca, Pfizer, Boehringer-Ingelheim, GlaxoSmithKline, Sanofi, Ortho-Biotech, Amgen, Bristol-Myers Squibb (C/A), (H)
Reviewer “A”: None
Reviewer “B”: None
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute. SEER Cancer Statistics Review. [Accessed February 27, 2013]. Available at http://seer.cancer.gov/csr/1975_2009_pops09/index.html.
- 3.Early Breast Cancer Trialists' Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 4.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 5.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han J, Kraft P, Nan H, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 9.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 13.Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 14.Dunning AM, Healey CS, Baynes C, et al. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum Mol Genet. 2009;18:1131–1139. doi: 10.1093/hmg/ddn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pharoah PD, Antoniou A, Bobrow M, et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 16.Peng S, Lu B, Ruan W, et al. Genetic polymorphisms and breast cancer risk: Evidence from meta-analyses, pooled analyses, and genome-wide association studies. Breast Cancer Res Treat. 2011;127:309–324. doi: 10.1007/s10549-011-1459-5. [DOI] [PubMed] [Google Scholar]
- 17.Azzato EM, Lee AJ, Teschendorff A, et al. Common germ-line polymorphism of C1QA and breast cancer survival. Br J Cancer. 102:1294–1299. doi: 10.1038/sj.bjc.6605625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann HS, Otterbach F, Callies R, et al. The AA genotype of the regulatory BCL2 promoter polymorphism (938C>A) is associated with a favorable outcome in lymph node negative invasive breast cancer patients. Clin Cancer Res. 2007;13:5790–5797. doi: 10.1158/1078-0432.CCR-06-2673. [DOI] [PubMed] [Google Scholar]
- 19.Kasimir-Bauer S, Heubner M, Otterbach F, et al. Prognostic relevance of the AQP5–1364C>A polymorphism in primary breast cancer. Mol Med Report. 2009;2:645–650. doi: 10.3892/mmr_00000151. [DOI] [PubMed] [Google Scholar]
- 20.Brendle A, Brandt A, Johansson R, et al. Single nucleotide polymorphisms in chromosomal instability genes and risk and clinical outcome of breast cancer: A Swedish prospective case-control study. Eur J Cancer. 2009;45:435–442. doi: 10.1016/j.ejca.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Gaudet MM, Hunter K, Pharoah P, et al. Genetic variation in SIPA1 in relation to breast cancer risk and survival after breast cancer diagnosis. Int J Cancer. 2009;124:1716–1720. doi: 10.1002/ijc.23919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes S, Agbaje O, Bowen RL, et al. Matrix metalloproteinase single-nucleotide polymorphisms and haplotypes predict breast cancer progression. Clin Cancer Res. 2007;13:6673–6680. doi: 10.1158/1078-0432.CCR-07-0884. [DOI] [PubMed] [Google Scholar]
- 23.Long JR, Kataoka N, Shu XO, et al. Genetic polymorphisms of the CYP19A1 gene and breast cancer survival. Cancer Epidemiol Biomarkers Prev. 2006;15:2115–2122. doi: 10.1158/1055-9965.EPI-06-0464. [DOI] [PubMed] [Google Scholar]
- 24.Otterbach F, Callies R, Frey UH, et al. The T393C polymorphism in the gene GNAS1 of G protein is associated with survival of patients with invasive breast carcinoma. Breast Cancer Res Treat. 2007;105:311–317. doi: 10.1007/s10549-006-9462-y. [DOI] [PubMed] [Google Scholar]
- 25.Papadopoulou E, Tripsianis G, Anagnostopoulos K, et al. Significance of serum tumor necrosis factor-alpha and its combination with HER-2 codon 655 polymorphism in the diagnosis and prognosis of breast cancer. Int J Biol Markers. 2010;25:126–135. doi: 10.1177/172460081002500302. [DOI] [PubMed] [Google Scholar]
- 26.Qu S, Long J, Cai Q, et al. Genetic polymorphisms of metastasis suppressor gene NME1 and breast cancer survival. Clin Cancer Res. 2008;14:4787–4793. doi: 10.1158/1078-0432.CCR-08-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udler MS, Azzato EM, Healey CS, et al. Common germline polymorphisms in COMT, CYP19A1, ESR1, PGR, SULT1E1 and STS and survival after a diagnosis of breast cancer. Int J Cancer. 2009;125:2687–2696. doi: 10.1002/ijc.24678. [DOI] [PubMed] [Google Scholar]
- 28.Varadi V, Bevier M, Grzybowska E, et al. Genetic variation in genes encoding for polymerase zeta subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Res Treat. 2011;129:235–245. doi: 10.1007/s10549-011-1460-z. [DOI] [PubMed] [Google Scholar]
- 29.Shu XO, Long J, Lu W, et al. Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer Res. 2012;72:1182–1189. doi: 10.1158/0008-5472.CAN-11-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebbeck TR, Lynch HT, Neuhausen SL, et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N Engl J Med. 2002;346:1616–1622. doi: 10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- 31.Brewster AM, Do KA, Thompson PA, et al. Relationship between epidemiologic risk factors and breast cancer recurrence. J Clin Oncol. 2007;25:4438–4444. doi: 10.1200/JCO.2007.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen MBWX, Shen W, et al. Association between polymorphisms of trinucleotide repeat containing 9 gene and breast cancer risk: Evidence from 62,005 subjects. Breast Cancer Res Treat. 2011;126:177–183. doi: 10.1007/s10549-010-1114-6. [DOI] [PubMed] [Google Scholar]
- 33.QIAamp DNA Mini and Blood Mini Handbook. [Accessed February 27, 2013]. Available at http://dna.uga.edu/docs/QIAamp_DNA_Mini_and_Blood_Mini_Handbook%20(Qiagen).pdf.
- 34.Applied Biosystems Arcturus PicoPure DNA Extraction Kit. [Accessed April 25, 2013]. Available at: http://www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_086335.pdf.
- 35.Bender R, Lange S. Adjusting for multiple testing: When and how? J Clin Epidemiol. 2001;54:343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- 36.Liang J, Chen P, Hu Z, et al. Genetic variants in trinucleotide repeat-containing 9 (TNRC9) are associated with risk of estrogen receptor positive breast cancer in a Chinese population. Breast Cancer Res Treat. 2010;124:237–241. doi: 10.1007/s10549-010-0809-z. [DOI] [PubMed] [Google Scholar]
- 37.Huijts PE, Vreeswijk MP, Kroeze-Jansema KH, et al. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9:R78. doi: 10.1186/bcr1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reeves GK, Travis RC, Green J, et al. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010;304:426–434. doi: 10.1001/jama.2010.1042. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Zhou X, Huang Z, et al. TNRC9/LOC643714 polymorphisms are not associated with breast cancer risk in Chinese women. Eur J Cancer Prev. 2009;18:285–290. doi: 10.1097/CEJ.0b013e32832bf421. [DOI] [PubMed] [Google Scholar]
- 40.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 41.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broeks A, Schmidt MK, Sherman ME, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: Findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding S, Yu JC, Chen ST, et al. Diverse associations between ESR1 polymorphism and breast cancer development and progression. Clin Cancer Res. 2010;16:3473–3484. doi: 10.1158/1078-0432.CCR-09-3092. [DOI] [PubMed] [Google Scholar]
- 44.Hsieh S, Look MP, Sieuwerts AM, et al. Distinct inherited metastasis susceptibility exists for different breast cancer subtypes: A prognosis study. Breast Cancer Research. 2009;11:R75. doi: 10.1186/bcr2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei H, Hemminki K, Johansson R, et al. PAI-1–675 4G/5G polymorphism as a prognostic biomarker in breast cancer. Breast Cancer Res Treat. 2008;109:165–175. doi: 10.1007/s10549-007-9635-3. [DOI] [PubMed] [Google Scholar]
- 46.Chrisanthar R, Knappskog S, Lokkevik E, et al. Predictive and prognostic impact of TP53 mutations and MDM2 promoter genotype in primary breast cancer patients treated with epirubicin or paclitaxel. PLoS One. 6:e19249. doi: 10.1371/journal.pone.0019249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 48.Colomer R, Monzo M, Tusquets I, et al. A single-nucleotide polymorphism in the aromatase gene is associated with the efficacy of the aromatase inhibitor letrozole in advanced breast carcinoma. Clin Cancer Res. 2008;14:811–816. doi: 10.1158/1078-0432.CCR-07-1923. [DOI] [PubMed] [Google Scholar]
- 49.Smid M, Wang Y, Klijn JG, et al. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24:2261–2267. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 50.Andersen CL, Monni O, Wagner U, et al. High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am J Pathol. 2002;161:73–79. doi: 10.1016/S0002-9440(10)64158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barlund M, Forozan F, Kononen J, et al. Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J Natl Cancer Inst. 2000;92:1252–1259. doi: 10.1093/jnci/92.15.1252. [DOI] [PubMed] [Google Scholar]
- 52.Isola JJ, Kallioniemi OP, Chu LW, et al. Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol. 1995;147:905–911. [PMC free article] [PubMed] [Google Scholar]
- 53.Inaki K, Hillmer AM, Ukil L, et al. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res. 2011;21:676–687. doi: 10.1101/gr.113225.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azzato EM, Pharoah PD, Harrington P, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]