Abstract
The involvement of epigenetic processes in the origin and progression of cancer is now widely appreciated. Consequently, targeting the enzymatic machinery that controls the epigenetic regulation of the genome has emerged as an attractive new strategy for therapeutic intervention. The development of epigenetic drugs requires a detailed knowledge of the processes that govern chromatin regulation. Over the recent years, mass spectrometry (MS) has become an indispensable tool in epigenetics research. In this review, we will give an overview of the applications of MS-based proteomics in studying various aspects of chromatin biology. We will focus on the use of MS in the discovery and mapping of histone modifications and how novel proteomic approaches are being utilized to identify and study chromatin-associated proteins and multi-subunit complexes. Finally, we will discuss the application of proteomic methods in the diagnosis and prognosis of cancer based on epigenetic biomarkers and comment on their future impact on cancer epigenetics.
Keywords: Mass spectrometry, quantitative proteomics, chromatin, histone modifications, epigenetic readers, cancer epigenetics
INTRODUCTION
During the development of a multicellular organism, a single cell gives rise to a multitude of differentiated cell types that ultimately form the various tissues and organs of the body. The changes that occur during these cellular differentiation processes are ‘epigenetic’ as the characteristic phenotype of each cell type is brought about without changing the genomic DNA sequence [1]. Instead, the expression of genes encoded in the DNA is regulated by the activity of the underlying chromatin, which is the macro-molecular nucleoprotein complex that governs the structural organization of the genetic material in eukaryotic cells. The basic unit of chromatin is the nucleosome, which consists of 147 bp of DNA wrapped around an octamer made up of two copies of each of the core histones H2A, H2B, H3 and H4 [2]. Nucleosomes are folded into higher order structures by additional proteins, including the linker histone H1, to form chromatin and ultimately the chromosomes. Both DNA and histone proteins carry chemical modifications—DNA is predominantly methylated at cytosines in CpG dinucleotides and histones are subject to a number of post-translational modifications of specific amino acids such as acetylation, methylation, phosphorylation and ubiquitylation—and it has become apparent that these modifications play a major role in regulating the functional state of chromatin [3, 4]. In their entirety, the DNA and histone modifications form the ‘epigenome’ which shapes the flow of information from the genome and defines the identity of a cell.
As chromatin constitutes the primary form of DNA in the nucleus, chromatin modifications influence all DNA-associated processes, such as transcription, replication and DNA repair. These functions are intimately linked to the faithful interpretation and inheritance of the genetic material and are central to inducing and maintaining cell fate decisions. Aberrant epigenetic regulation can, therefore, potentially lead to the accumulation of genetic lesions and the loss of cell identity—events that are associated with the development of cancer. Indeed, abnormal chromatin modification patterns and alterations in the epigenetic machinery are observed in many cancers and these changes are often already apparent in early stage and benign tumors [5]. Therefore, they may precede and thereby facilitate the more severe events associated with malignant transformation, such as mutations in tumor suppressors, activation of proto-oncogenes or genomic instability. The inherent reversibility of epigenetic lesions, unlike genetic mutations, has sparked significant efforts in developing small-molecule inhibitors that target epigenetic processes [6].
The development of these ‘epigenetic therapies’ requires detailed knowledge of the molecular interactions governing chromatin regulation. Next-generation sequencing has enabled the comprehensive genomic mapping of nucleosome positions, higher order chromatin structures, DNA and histone modifications and chromatin-bound factors and is starting to explain various aspects of how the epigenome shapes the expression of the genetic information [7, 8]. But more mechanistic insight is still needed in order to understand how epigenetic modifications effect their biological functions. Mass-spectrometric and proteomic methods are having an increasing impact on epigenetics research. In this review, we will discuss the recent developments and applications of these methods in investigating chromatin biology. Several excellent articles reviewing cancer epigenetics [9, 10], the prospects for epigenetic therapies [6, 11] and the use of proteomic methods in chromatin biology [12–14] have been published recently. We will therefore try to provide an overview over the recent applications of proteomics in epigenetics research with a view to its application in cancer epigenetics and we refer the reader to these reviews for more detailed information.
TECHNOLOGICAL ADVANCEMENTS IN MS-BASED PROTEOMICS FOR EPIGENETIC RESEARCH
Mass spectrometry (MS) has proven to be an indispensable and unbiased tool for the identification and quantification of post-translational modifications (PTMs) on proteins. In this section, we review the ways in which MS has shaped the field of epigenetics by providing a versatile method for the discovery and systematic quantitation of chromatin modifications.
MS for interrogating histone modifications
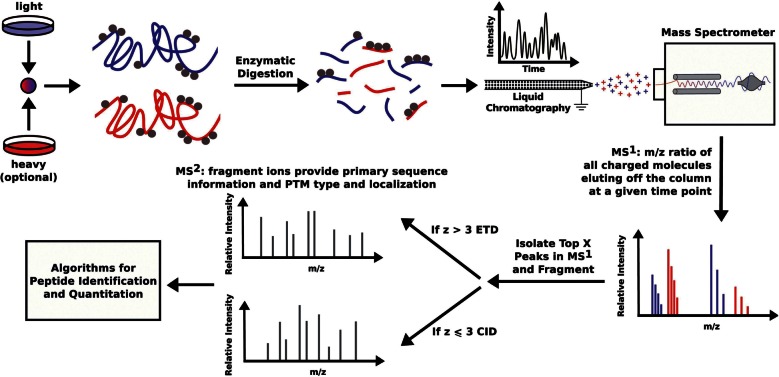
Histones have been notoriously difficult to analyze using conventional proteomics workflows (Figure 1), due to their very basic composition and high frequency and heterogeneity of PTMs. In the collision-induced dissociation (CID) fragmentation of peptides, the occurrence of basic residues (such as lysine, arginine and histidine) tends to inhibit migration of the mobile protein along the protein backbone, resulting in partial and incomplete fragmentation and thus limiting the ability of this technique to localize the sites of PTM. Considering the first 40 residues on the N-terminal tail of histones H2A, H2B, H3 and H4, the composition of basic residues is 30, 38, 38 and 38%, respectively. Furthermore, the majority of these lysine and arginine residues are potential sites of PTM. Although trypsin robustly cleaves the C-terminus of lysine and arginine residues, it is blocked by PTMs such as methylation and acetylation, which are commonly found on histones. Thus, a standard tryptic digestion of differentially modified histones results in overlapping and non-reproducible peptides, which renders analyses such as label-free quantitation extraordinarily difficult. As a result, innovative methods have been and are continuing to be developed for robust analysis of histone PTMs using MS.
Figure 1:
Overview of MS-based proteomics workflows. Proteins can be labeled with heavy amino acids (heavy) or remain unlabeled (light) by growing cells in appropriate tissue culture media. Isolated proteins are digested into peptides which are then separated by high-performance liquid chromatography (HPLC) and electro-sprayed into the mass spectrometer. The masses of peptides are recorded in a full scan (MS1). Selected peptides are isolated and fragmented and masses determined in a MS/MS spectrum (MS2). The masses of the precursor ions and fragments are used in database searches in order to identify peptide sequences and PTMs that can be assigned to proteins.
Quantitation of histone modifications
Several alternative strategies to conventional tryptic digestion have been proposed in an attempt to overcome the issues associated with generating reproducible histone sequences. Some studies have utilized enzymes with higher specificity, such as Arg-C, which in theory produces only peptides with C-terminal arginines. This approach has successfully been applied in the study of histone H3 variants in Drosophila, where it was found that histone H3.3 possesses marks associated with euchromatin [15] and in the profiling of PTMs on H3.1 and H3.3 during chromatin assembly [16], where specific methylation states of HK9 were discovered to exist on non-nucleosomal variants. Although this approach potentially resolves many of the analytical issues associated with tryptic digests, Arg-C has been reported to be less efficient and specific than trypsin and requires significant protocol optimization for obtaining reproducible digests.
In the spirit of this approach, several chemical derivatization techniques have been developed to create Arg-C-like digests using trypsin. Anhydrides are the most common reagents utilized, as they efficiently react with primary or secondary amines (i.e. unmodified or monomethylated lysine residues) to donate an acyl functional group to the ε-amino group on the lysine side chain. As a result, tryptic activity is blocked at all lysine residues on the histones. The use of deuterated acetic anhydride was the first of such approaches, where it was used over a decade ago in the study of mutant yeast cells [17]. Although a deuterated acetyl group is transferred to the lysine side chains, the corresponding mass shift still results in ambiguity with endogenous acetylation or trimethylation PTMs, which have the same nominal mass of 42 Da. To circumvent this issue, propionic anhydride was used in investigating the effect of G9a and Suv39H knock downs on H3K9-methylation [18]. In contrast to acetic anhydride, which donates an acetyl group to unmodified and monomethylated lysines (C2H2O), propionic anhydride donates a propionyl group (C3H4O), so the mass shift is distinguishable from endogenous acetylation PTMs. Around the same time, a combination of deuterated acetic and propionic anhydride was utilized in the study of drosophila embryos [19], where an inverse relationship between H3K27 or H3K36 hypermethylation was observed on the same peptide, suggesting an exclusive relationship between full occupancy of these repressive and active signals, respectively. The protocol for histone derivatization by propionylation has since been optimized for routine use [20] and extended to include the possibility of incorporating isotopically labeled anhydrides to compare changes in histone marks between different samples in the same LC–MS/MS experiment [21]. This propionylation technique has recently been successfully applied in several studies, such as elucidating the role of macroH2A in melanoma [22], characterizing the effect of deleting the HAT and nucleosome assembly factor necessary for K56ac on other histone modifications in Saccharomyces cerevisiae [23], determining the nucleosomal symmetry of methylation patterns [24], evaluating the specificity of commonly used chromatin immunoprecipitation-grade antibodies [25], identifying novel histone PTMs [26] and mapping the histone code ‘read’ by bromodomain-containing proteins and HP1 [27]. Although clearly a powerful and versatile technique, anhydride esterification of histones is sensitive to water content, thus requiring several drying steps, which can potentially lead to sample loss.
Another important approach for peptide quantitation is ‘stable isotope labeling by amino acids in cell culture’ (SILAC), which facilitates the incorporation of non-radioactive, isotopically labeled amino acids into cultured cells [28]. Standard SILAC experiments involving the incorporation of ‘heavy’ arginine or lysine amino acids have been successfully used for studying histone PTM dynamics across the cell cycle [29, 30], turnover rates of histone marks [31] and nucleosomal partitioning of histones during DNA replication [32]. SILAC has also found an application in probing the dynamics of methylation PTMs [33], in which 13CD3-methionine is introduced into methionine-depleted cell culture media and is metabolically converted into S-adenosyl methionine with a heavy methyl donor group. This technique has been successfully applied to investigate the dynamics of histone methylation within synchronized cell populations [34]. Recent studies have combined the traditional SILAC with this heavy methyl labeling to examine the dynamics of H3K79me throughout the cell cycle [35] and the rates of transition between the different H3K27K36 methylation states [36].
Discovery of novel histone PTMs
In recent years, MS has revealed the existence of several previously unknown histone PTMs. Since 2007 alone, MS has been used to discover novel sites of phosphorylation involved in apoptosis [37], introduced O-GlyNAc as a new histone PTM [38] and revealed a whole arsenal of novel lysine modifications, including propionylation [39, 40], butyrylation [39, 40], formylation [41], succinylation [42], malonylation [42] and crotonylation [26]. The identification of many of these novel histone PTMs has been largely facilitated by recent developments in software that deviate from the conventional approach for unmodified peptide identification based on restricted database searches. For instance, PTMap [43] utilizes sequence alignment to localize sites of PTMs on target proteins while substantially reducing false-positive identifications based on unmatched peaks. While innovative approaches such as these continue to discover novel histone modifications, localization of PTMs on histones remains a challenging bioinformatics problem due to nearly isobaric modifications (e.g. acetylation or trimethylation), isobaric and co-eluting peptides (resulting in mixed tandem mass spectra) [44] and the dynamic range over which these modified forms exist (i.e. low abundance forms often give rise to poor MS/MS fragmentation). The functional implications of many of these modifications are yet to be determined, but these findings highlight the importance of MS in discovering novel marks that may potentially expand the basic set of the ‘combinatorial histone code’ and possess important regulatory roles in diseases such as cancer.
LC–MS/MS approaches for combinatorial histone codes
It is well established that there exists a certain degree of cis- and trans-cross-talk between different sites and types of histone modifications across their N-terminal tails. The methods described in the previous sections based on chemical derivatization and tryptic digestion produce some peptides with multiple potentially modified residues (e.g. H3K9S10K14, H3K27K36), but they abolish long-distance connectivity between modified sites on the same protein (e.g. between H3K4 and H3K27). Thus, to interrogate truly combinatorial histone modifications, one must either analyze intact histones (i.e. top down) or utilize enzymes that produce longer polypeptides (i.e. middle down) containing the modifications of interest, or the N-terminal tails in the case of histones. For instance, the intact N-terminal tails can be generated for middle down analysis by digesting histone H3 with Glu-C to yield the 1–50 N-terminal tail and histone H4 with Asp-N to yield the 1–23 N-terminal tail.
One important advantage to the unusually basic composition of histone N-terminal tails is that they are amenable to fragmentation by electron capture dissociation (ECD) and electron transfer dissociation (ETD). Although the actual mechanisms of dissociation are still not completely understood, ECD and ETD involve the transfer of an unpaired low-energy electron to the peptide for fragmentation and, in contrast to CID, are particularly efficient in the presence of several basic residues. Indeed, studies based on fragmenting the intact N-terminal tails using ECD have been around since 2004 for histone H4 [45] and 2006 for histone H3 [46]. In these early studies, it was demonstrated that off-line chromatography, such as acid–urea cation exchange, could be used to separate histones based upon their degree of acetylation [46]. However, the majority of approaches relied upon standard reversed-phase separation, which results in poor chromatographic resolution between modified forms of the same peptide sequence. As a result, tandem mass spectra are substantially ‘mixed’ in that they contain fragment ions from many isobaric histone isoforms (i.e. modified histone forms with nearly the same number and type of modifications, but distributed to different residues), and it is not possible to confidently identify which concurrent modifications are on the same histone polypeptide.
It was demonstrated in 2004 that off-line fractionation using cation exchange hydrophilic interaction chromatography (HILIC) could be used to substantially reduce the number of modified histone forms present in each ‘mixed’ tandem mass spectrum [47]. This study identified over 150 modified histone H3.3 forms from 30 cation exchange HILIC fractions and elucidated a partial acetyl ordering on histone H3. However, the off-line nature of the fractionation significantly prohibits throughput of this approach, as desalting of each fraction is required prior to analysis. In 2009, a weak cation-exchange hydrophilic interaction chromatography (WCX-HILIC) was presented that enabled the first ‘on-line’ separation and tandem MS acquisition of modified histone isoforms, resulting in the identification and quantitation of several hundred histone H3 modified forms in a single experiment [48]. This approach was extended in 2012 to facilitate the top down analysis of histone H3 [49].
Indeed, these multimodal chromatographic techniques that can resolve hypermodified isoforms of the same protein sequence have advanced our ability to characterize combinatorial histone modifications by MS. Another exciting technology that has only recently been adapted for MS is ion mobility spectrometry (IMS), which has the capacity to further separate analytes based on their gas-phase conformations. In traditional ‘drift-time’ IMS, the ions travel through a buffer gas in a low electric field, resulting in a diffusion-dominant process that separates ions primarily by their collision cross-section, thus providing an orthogonal degree of selectivity to conventional liquid chromatography mass spectrometry (LC–MS) approaches. Several methods for IMS currently exist (for a review see [50]). For instance, differential or field asymmetric waveform IMS separates ions based upon their difference in mobility within a high and low electric field. This technique has recently been demonstrated on modified histone isoforms [51, 52] and can resolve histone H4 acetylation states that are chromatographically similar.
IDENTIFICATION OF PTMS ON CHROMATIN-ASSOCIATED PROTEINS
Despite recent emphasis on the analysis of histone PTMs, it is worth noting that MS has also been useful in identifying PTMs on other chromatin-associating proteins, thus hinting toward the existence of a much broader regulatory capacity for chromatin than what has been proposed by the combinatorial histone code alone.
Several proteins involved in the establishment and maintenance of heterochromatin have been found to be post-translationally modified. For instance, heterochromatin protein 1 has been shown to be hyperphosphorylated [53], where N-terminal phosphorylation promotes chromatin binding [54] and is linked to gene silencing at heterochromatin in other studies [55]. High mobility group proteins have also been shown to possess many PTMs by MS through a combination of bottom up and top down approaches [56]. Several polycomb group proteins have been implicated to be regulated through PTMs [57]. For example, it has been shown that phosphorylation of Ezh2 represses its catalytic activity [58] and enhances its binding to non-coding RNA in a cell-cycle-dependent manner [59].
Regulators of histone acetylation have also been observed to be post-translationally modified. A 2008 study identified 13 phosphorylation sites on the N- and C-terminus of SIRT1, a histone deacetylase (HDAC), using an MS-based approach [60]. A recent review of the PTMs currently known for the sirtuin (SIRT) family of proteins is provided in [61]. Another study in 2011 was also able to identify novel sites of phosphorylation on the CREB-binding protein (CBP) [62], a histone acetyltransferase. Through various other studies, it has been shown that CBP is subject to a range of PTMs, including acetylation, phosphorylation, ubiquitylation and SUMOylation, which regulate its functional interactions, activity and stability [63].
Finally, MS has identified novel PTMs on several proteins involved in ATP-dependent chromatin remodeling. For instance, global phosphoproteomics has revealed novel phosphorylation sites on a number of chromatin remodeling proteins, including hBRM, BRG-1 and SMARCC1 [64]. More targeted MS studies have also identified acetylation on imitation SWI (ISWI) by GCN5 [65].
TECHNOLOGICAL INNOVATION IS THE DRIVER FOR MS-BASED HISTONE PTM ANALYSIS
As detailed in the aforementioned sections, the advancements in our capacity to routinely measure histone PTMs using MS have been largely driven by technological developments. In summary, the major areas that have and will continue to contribute to and shape the future directions of this field are:
(i) Mass spectrometry: mass resolution and sensitivity play an important role in histone PTM analysis. As previously mentioned, acetylation and trimethylation PTMs have very similar masses (42.0106 and 42.0469, respectively) and cannot be distinguished on a low-resolution instrument, such as an ion trap mass spectrometer. About 10 years ago, only few instruments possessed the resolving power to differentiate these two modifications (i.e. fourier transform ion cyclotron resonance (FT-ICR) and quadrupole time-of-flight (Q-TOF) instruments), albeit at the cost of slower scan rates and lower sensitivity. Recent advancements in hybrid mass analyzers (e.g. the Orbitrap Velos) have produced instruments with combined superior scan speed, dynamic range and mass resolution. Continued improvements in these instrument features will further enable mass spectrometry to quantitate histone PTMs across large dynamic ranges in a truly unbiased manner.
(ii) Separation of histone PTMs: an orthogonal yet complementary area to the ongoing enhancements in MS technology is the development of methods that reduce sample complexity on the front end of the mass spectrometer. By reducing the heterogeneity of the population of ions in a given MS scan, one can potentially increase the sensitivity and resolution with which the ion peaks are measured. Thus, recent advancements in chromatography technology (e.g. ultra performance liquid chromatography (UPLC)) and methods (e.g. WCX-HILIC) have directly improved the overall quality of MS data without modifications to the mass analyzer. The generation of higher quality LC–MS data in turn improves the accuracy of subsequent analysis.
(iii) Bioinformatics: the comprehensive characterization of histone PTMs from MS data remains a difficult challenge to date. As was briefly mentioned, specific issues arise from nearly isobaric modifications (acetylation or trimethylation), mixed tandem mass spectra resulting from isobaric and co-eluting forms and missing or poorly fragmented tandem mass spectra due to low abundance modifications. Further developments in software, such as alignment-based methods [43] and incorporation of retention time information into the identifications [44], will further improve the coverage and quality of the readouts provided by these LC–MS platforms for histone PTM analysis.
IDENTIFICATION OF CHROMATIN-ASSOCIATED PROTEINS USING PROTEOMIC METHODS
Global identification of chromatin constituents
In addition to identifying the nature and location of PTMs on histones and other known chromatin-associated proteins, mass spectrometric approaches have been used to identify novel proteins that make up chromatin (Figure 2). The first quantitative analysis of chromatin-bound factors using proteomic approaches was reported in 2003 [66]. For this study, chromatin-enriched fractions were prepared from c-Myc-expressing and non-expressing human B lymphocytes. The isolated proteins were labeled with isotope coded affinity tags (ICAT) [67] and analyzed by MS. This lead to the identification and quantification of 282 differentially expressed proteins, notably mostly transcription factors and chromatin regulators. More recently, SILAC-based quantitative proteomics methods have been used to investigate global changes in chromatin composition after the induction of DNA damage [68, 69] or during DNA replication [70]. In all cases the quantitative filter achieved by combining metabolic labeling with differential treatments of cells allows the identification of changes in protein composition over a background of hundreds of proteins. As demonstrated by a study in 2011, chromatin fractions can also be subjected to limited nuclease digestion and MS analysis, exploiting the higher accessibility of euchromatin over more densely packed heterochromatin, in order to identify proteins associated with these two functional chromatin states [71].
Figure 2:
Strategies for the enrichment of chromatin-bound proteins. Chromatin-bound proteins can be isolated by purifying whole chromosomes, preparing chromatin-enriched fractions or by immuno-affinity purifying specific target proteins using antibodies against DNA- or histone-binding proteins, histone variants or histone PTMs. Specific DNA domains can be isolated via hybridization with specific DNA probes.
In addition to analyzing the protein content of chromatin-enriched fractions, the composition of mitotic chromosomes has been investigated extensively. As mitotic chromosomes are highly condensed structures, they can be biochemically isolated from mitotically arrested cells by sequential centrifugation through sucrose and Percoll gradients [72]. Several studies have reported proteomic analyses of mitotic chromosomes in recent years [73–75], with the most comprehensive characterization so far being performed in 2010 [76]. This study identified 4029 proteins, and the problem of the false-positive identification of ‘hitchhikers’—cytosolic proteins that bind to the charged chromosomes after nuclear envelope breakdown—was overcome by a combination of biochemical purifications with quantitative proteomics and, crucially, sophisticated computational data analyses.
Locus-specific chromatin-interacting proteins
Due to the preparation methods for isolating chromatin fractions and mitotic chromosomes, the aforementioned approaches cannot resolve the exact genomic location of the chromatin-associated factors and are limited to strongly bound proteins. In 2009, this problem was addressed by the development of the proteomics of isolated chromatin segments (PICh) protocol that allows the identification of proteins that are bound to a specific genomic region [77]. The PICh approach entails the hybridization of a locked nucleic acid probe to a complementary genomic sequence and identifying cross-liked and captured proteins by MS. This approach identified many known and novel telomeric proteins, but so far it was only successful on repetitive genomic DNA sequences and with very large cell numbers as starting material. In yeast the copy number problem can be overcome by using multicopy centromere plasmids [78]. The inclusion of tandem repeats of the lactose operator allows the affinity purification of the plasmids via immobilized lac repressors. This system has been used in combination with proteomic techniques to study the composition of kinetochores [79] and the dynamic changes in histone modifications at origins of replication [80]. In 2012, a chromatin affinity purification with mass spectrometry (ChAP–MS) approach was described that utilized a single genomic LexA DNA-binding site and allowed the affinity purification of a small chromatin segment via a LexA ‘affinity handle’ [81]. This system was coupled with SILAC-based proteomics in order to track changes in protein composition at the GAL1 locus upon induction of the gene by galactose. Several transcription-related factors and histone codes were identified.
IDENTIFICATION OF EPIGENETIC READERS
According to the histone code hypothesis [82, 83], histone modifications form binding sites for the recruitment of epigenetic reader proteins that contain domains that can recognize and bind specific modifications. The identification of modification-binding proteins has been a major focus in chromatin biology in recent years.
Peptide affinity purifications
Most histone modifications are found within the N-terminal histone tails which protrude from the nucleosomes and are thought to be flexible and unstructured. Early studies were based upon the assumption that isolated peptides are a good approximation of histone tails. The first efforts to identify histone modification-binding proteins made use of synthetic peptides resembling the N-terminal tail of histone H3 carrying trimethylated lysines at various positions. In these experiments, immobilized peptides were incubated with nuclear extracts. Bound proteins were eluted, resolved on an SDS–PAGE gel and proteins within visibly stained gel bands were excised and identified by MS. Using this approach, it was shown that the NuRD complex associates with unmodified histone H3 peptides and is prevented from binding by trimethylation at lysine 4 [84]. In a similar approach, the NURF subunit BPTF was identified to bind to H3K4me3-modified histone peptides via its plant homeo domain [85]. In 2007, the peptide pull-down approach was combined with SILAC-based quantitative proteomics in order to achieve a global view of proteins that can interact with H3K4me3-modified histone tails [86]. In 2010, this SILAC peptide pull-down approach was applied to all major lysine trimethylation marks in the histone H3 (K4, K9, K27 and K36) and in histone H4 (K20) tails [87]. Each of these experiments identified around 20–50 modification-interacting proteins, depending on the modified residue.
Chromatin templates
The use of isolated modified histone tail peptides does not take into account that histone modifications are embedded into the chromatin environment. Indeed, certain epigenetic readers might only recognize modifications in the structural context of nucleosomes or larger assemblies of nucleosomes. In chromatin, multivalent interactions may lead to a more selective recognition of precise nucleosomal modification signatures. In order to investigate the cross-talk between DNA and histone modifications within chromatin, a recent study used reconstituted modified nucleosomes as baits in SILAC nucleosome affinity purifications [88]. Post-translationally modified histones were generated using the native chemical ligation technique [89], assembled into nucleosomes together with biotinylated DNA [90] and used in SILAC affinity purifications as described for peptides. By combining CpG-methylated DNA with modified histone H3, the study demonstrated that epigenetic readers can recognize and integrate combinations of epigenetic marks on a nucleosomal template. Furthermore, a recent study reported SILAC affinity purifications using modified nucleosome arrays [91]. Although there is overlap between the data sets obtained with peptide [87], nucleosome [88] and chromatin purifications [91] there are also clear differences. Whether these discrepancies are due to experimental conditions or whether peptides, mono-nucleosomes and nucleosome arrays differ in their capacities to act as binding platforms for epigenetic reader proteins has to be addressed in future studies.
Oligonucleotide-based affinity purifications
SILAC affinity purifications have also been applied to identify methyl CpG-binding proteins [88, 92] and transcription factors that recognize single-nucleotide polymorphisms [93–95] using immobilized DNA oligonucleotides that either included methylated CpG dinucleotides or single base pair exchanges as affinity baits. In addition, oligonucleotide-based SILAC pull-downs have been extended to incorporate RNAs as baits, a development that is of considerable interest for the epigenetics field as it is becoming increasingly apparent that non-coding RNAs play a major role in epigenetic processes [96].
PROBING THE COMPOSITION OF MULTI-SUBUNIT CHROMATIN REGULATORY COMPLEXES
DNA and histone modification maps obtained from numerous genome-wide chromatin immunoprecipitations studies and the identification of histone codes by MS demonstrate that DNA and histone modifications form combinatorial modification patterns within chromatin. It has become apparent that functional chromatin elements such as promoters or enhancers are marked by characteristic modification signatures [8]. Epigenetic effectors must be able to recognize these signatures in order to extract epigenetic information from chromatin. Indeed, a striking number of chromatin regulators contain multiple modification recognition domains and many chromatin regulatory protein complexes contain multiple subunits that can potentially recognize different chromatin modifications [97]. Another aspect of the interpretation of epigenetic information, therefore, is the combinatorial composition of multi-subunit chromatin regulatory complexes that are functionally distinct and that can target their activities to specific chromatin locations depending on their precise subunit composition. Many chromatin-associated complexes have been purified by biochemical techniques or tandem affinity purification (TAP) approaches. The subunits of the purified complexes are usually identified by separating the proteins on a SDS–PAGE gel, ‘in-gel’ digestion of excised protein bands and subsequent analysis by MS. This approach has been successful in several high-throughput studies for the proteome-wide identification of protein complexes in yeast [98–101] and has also been adapted for the identification of chromatin-associated protein complexes [102] using a modified chromatin immunoprecipitation (mChIP) protocol [103]. In 2012, the modular composition of human PRC1 complexes was interrogated by expressing TAP-tagged versions of PRC1 subunits and identifying associated proteins by MS. Six distinct PRC1 complexes were identified, each characterized by a unique PCGF subunit [104]. However, it should be noted that conventional biochemical and affinity purification approaches are prone to false-positive identification of interaction partners. One way to circumvent this problem is to combine affinity purifications of complex subunits with endogenous tagging methods and SILAC labeling techniques [12, 87]. The quantitative readout allows specifically interacting proteins to be distinguished from contaminants and also identifies low-affinity interactions that might not be apparent in conventional purifications. Subunit composition and dynamics in different cell types and developmental stages, however, still constitute challenges that need to be addressed in order to understand the functions of large chromatin regulatory complexes.
PROTEOMICS IN CANCER EPIGENETICS
It is becoming widely accepted that perturbations of epigenetic modifications play a pivotal role in the onset, progression and maintenance of cancer. In particular, global DNA hypomethylation and promoter-specific hypermethylation are typical features of cancer cells [105]. Moreover, disruptions in the epigenetic machinery—factors that deposit, remove or recognize modifications or that remodel chromatin—are hallmarks of many cancers [9, 11]. Apart from mutations, deletions or deregulated expression of chromatin regulators, a recurring theme is the aberrant creation of fusion genes through chromosomal translocations. Prominent examples are the fusions of the mixed lineage leukemia (MLL) H3K4-methyltransferase gene with various fusion partners which are found in many aggressive forms of leukemia [106].
Next-generation sequencing applications are providing an increasingly detailed knowledge concerning aberrant DNA methylation in tumors which could be used as biomarker for specific types of cancers [107]. In comparison, the utilization of proteomic methods to measure characteristic changes in histone modifications, epigenetic readers or chromatin-associated factors at the protein level, which could potentially be used for cancer diagnosis and prognosis, is somewhat lagging behind. Recent improvements in the sensitivity and accuracy of mass-spectrometric technologies have driven some advances in the field and several groups have used MS-based proteomics for epigenetic cancer profiling by measuring deregulation of histone modifications in tumor cells. In 2005, the modification patterns of histone H4 in normal human tissues and cancer cells were examined by MS [108] and it was found that tumors show an early global loss of acetyl-H4K16 and trimethyl-H4K20. Moreover, evidence obtained by immunohistochemistry suggests that differences in global histone modification patterns in cancer tissues are predictive of clinical outcomes and associated with the risk of tumor recurrence (reviewed in [109]). In 2011, SILAC-based proteomics was used to quantitatively measure H3 and H4 modifications in several breast cancer cell lines [110]. Significant changes in abundance were observed for several modifications in the N-terminal tails of histones H3 and H4. These results demonstrate that aberrations in global histone modification patterns do occur in cancer tissues. This suggests the existence of ‘histone modification signatures’ with potential use as histone-based prognostic or even diagnostic cancer biomarkers.
As epigenetic changes are not mutations in the DNA sequence, they are in principle reversible. Drugs that target chromatin regulators, therefore, hold great promise for ‘epigenetic therapies’ as a strategy for the treatment of cancer. Inhibitors of DNA methyltransferases and HDACs have already been approved for the treatment of certain cancers [6] and several companies are actively pursuing the development of new epigenetic drugs [111]. Current efforts focus on next-generation HDAC inhibitors and small-molecule inhibitors against histone methyltransferases and histone demethylases. A complementary approach to inhibiting the activities of chromatin modifying enzymes is the targeting of protein–protein interactions. Although targeting protein–protein interactions with small-molecule inhibitors has proven difficult, several recent studies have demonstrated the feasibility of targeting bromodomains in the bromo and extra terminal domain (BET) family of acetyl-lysine recognizing chromatin readers in order to prevent their binding to acetylated histones [112–116]. In one study, an immobilized BET inhibitor was used as bait in a proteomic affinity purification approach to identify the target of the inhibitor and its associated proteins [113]. It was demonstrated that MLL fusion partners are associated with the BET family of proteins and that BET inhibitors are effective against leukemia cell lines driven by these MLL fusions. In addition, BET inhibitors have shown promising results in preclinical leukemia cancer models in vivo demonstrating that targeting interactions is an effective approach for epigenetic cancer therapies.
CONCLUSIONS
Recent and significant advancements in proteomic methodologies have been instrumental in progressing chromatin biology and epigenetics research. We can now identify and quantitate thousands of proteins in complex protein mixtures and we are able to identify entire histone codes on individual histone tails. We have also started to identify readers of chromatin modifications in the context of nucleosomes and chromatin, and genomic manipulation techniques enable the large-scale characterization of protein–protein interactions by genome-wide endogenous tagging of proteins. Although this will potentially enable us to generate system-wide maps of interactions that occur within chromatin, a major challenge is still the identification of proteins at single genomic loci. Elegant approaches that in principle allow the targeted enrichment of a defined genomic region are PICh and ChAP–MS. PICh was successfully applied on telomeric sequences, of which there are 92 in a normal diploid cell and ChAP–MS has suffered from a high level of contaminating proteins so far. It remains to be seen whether these two approaches will be successful in the analysis of proteins at single genomic locations, possibly in combination with further innovations in MS and quantitative proteomics.
Another relatively unexplored area in chromatin biology, which might become increasingly important for cancer epigenetics, is the use of small-molecule inhibitors as baits for the affinity purification of chromatin modifying and demodifying enzymes and epigenetic readers. A seminal study in 1996 identified the first histone deacetylase HDAC1 by using immobilized naturally occurring microbial HDAC inhibitors [117]. The reported application of BET inhibitors as affinity baits to identify interaction partners of the bromodomain-containing BRD proteins using proteomic methods [113] demonstrates the value and applicability of this approach for epigenetics research. As major efforts in developing small-molecule inhibitors against chromatin regulators have only been initiated recently, chemical proteomics holds great promise for understanding inhibitor specificities and to identify and characterize the functions of inhibitor target proteins.
Over the recent years, MS-based proteomics has contributed significantly to our understanding of fundamental epigenetic processes. A desirable application for this technology with respect to cancer epigenetics would be its development into a diagnostic and prognostic tool based on epigenetic biomarkers to indicate treatment options for different types of cancer. Notwithstanding, the anticipated technological advances combined with innovative experimental procedures will greatly improve our abilities to understand the mechanics of chromatin regulation. This knowledge will be indispensable for developing targeted epigenetic therapies for cancer and will ultimately lead to the realization of personalized medicine with improved treatments and less side effects in patients.
Key Points.
Mass spectrometry can be used to identify and quantitate modifications on chromatin-related proteins.
Proteomic tools allow the discovery of novel histone modifications and histone codes on histone tails.
Quantitative proteomics enable large-scale identification of chromatin-bound proteins and epigenetic readers.
Mass spectrometry-linked affinity purifications can be used to identify chromatin-regulatory protein complexes.
FUNDING
The Medical Research Council (to T.B. and J.B.); Imperial College London (to P.A.D.).
Biographies
Till Bartke is heading the Chromatin Biochemistry Group at the MRC Clinical Sciences Centre where he uses chemical biology and proteomics approaches to study how chromatin regulators interpret DNA and histone modifications.
Julie Borgel is a post-doctoral researcher in the Chromatin Biochemistry Group at the MRC Clinical Sciences Centre and is interested in understanding how chromatin modifications regulate chromatin function.
Peter DiMaggio is a lecturer in the Department of Chemical Engineering at Imperial College London and is interested in the development of customized experimental and computational MS-platforms to characterize the function of chromatin-associating proteins.
References
- 1.Berger SL, Kouzarides T, Shiekhattar R, et al. An operational definition of epigenetics. Genes Dev. 2009;23:781–3. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–9. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 7.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–86. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 9.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11:726. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Eberl HC, Mann M, Vermeulen M. Quantitative proteomics for epigenetics. Chembiochem. 2011;12:224–34. doi: 10.1002/cbic.201000429. [DOI] [PubMed] [Google Scholar]
- 13.Britton LM, Gonzales-Cope M, Zee BM, et al. Breaking the histone code with quantitative mass spectrometry. Expert Rev Proteomics. 2011;8:631–43. doi: 10.1586/epr.11.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert JP, Pawson T, Gingras AC. Mapping physical interactions within chromatin by proteomic approaches. Proteomics. 2012;12:1609–1622. doi: 10.1002/pmic.201100547. [DOI] [PubMed] [Google Scholar]
- 15.McKittrick E, Gaften PR, Ahmad K, et al. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2004;1016:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loyola A, Bonaldi T, Roche D, et al. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–16. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Smith CM, Haimberger ZW, Johnson CO, et al. Heritable chromatin structure: Mapping “memory” in histones H3 and H4. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16454–16461. doi: 10.1073/pnas.182424999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters AHFM, Kubicek S, Mechtler K, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 19.Bonaldi T, Imhof A, Regula JT. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics. 2004;4:1382–96. doi: 10.1002/pmic.200300743. [DOI] [PubMed] [Google Scholar]
- 20.Garcia BA, Mollah S, Ueberheide BM, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933–8. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plazas-Mayorca MD, Zee BM, Young NL, et al. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res. 2009;8:5367–74. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–9. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drogaris P, Wurtele H, Masumoto H, et al. Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure-function relationships. Anal Chem. 2008;80:6698–707. doi: 10.1021/ac800739d. [DOI] [PubMed] [Google Scholar]
- 24.Chen XZ, Xiong J, Xu M, et al. Symmetrical modification within a nucleosome is not required globally for histone lysine methylation. Embo Rep. 2011;12:244–51. doi: 10.1038/embor.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peach SE, Rudomin EL, Udeshi ND, et al. Quantitative assessment of chromatin immunoprecipitation grade antibodies directed against histone modifications reveals patterns of co-occurring marks on histone protein molecules. Mol Cell Proteomics. 2012;11:128–37. doi: 10.1074/mcp.M111.015941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leroy G, Chepelev I, Dimaggio PA, et al. Proteo-genomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 2012;13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–30. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 29.Bonenfant D, Towbin H, Coulot M, et al. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol Cell Proteomics. 2007;6:1917–32. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Pesavento JJ, Yang H, Kelleher NL, et al. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–86. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zee BM, Levin RS, Dimaggio PA, et al. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu M, Long C, Chen X, et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–8. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 33.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–26. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 34.Zee BM, Levin RS, Xu B, et al. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–50. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweet SM, Li M, Thomas PM, et al. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J Biol Chem. 2010;285:32778–86. doi: 10.1074/jbc.M110.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, Sweet SM, Popovic R, et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci USA. 2012;109:13549–54. doi: 10.1073/pnas.1205707109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurd PJ, Bannister AJ, Halls K, et al. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J Biol Chem. 2009;284:16575–83. doi: 10.1074/jbc.M109.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–20. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K, Chen Y, Zhang Z, et al. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J Proteome Res. 2009;8:900–6. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Sprung R, Tang Y, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–9. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wisniewski JR, Zougman A, Mann M. N(epsilon)-Formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36:570–7. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Z, Dai J, Dai L, et al. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–7. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Chen W, Cobb MH, et al. PTMap—a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc Natl Acad Sci USA. 2009;106:761–6. doi: 10.1073/pnas.0811739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiMaggio PA, Jr, Young NL, Baliban RC, et al. A mixed integer linear optimization framework for the identification and quantification of targeted post-translational modifications of highly modified proteins using multiplexed electron transfer dissociation tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2527–43. doi: 10.1074/mcp.M900144-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pesavento JJ, Kim YB, Taylor GK, et al. Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J Am Chem Soc. 2004;126:3386–7. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J Proteome Res. 2006;5:240–7. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 47.Garcia BA, Pesavento JJ, Mizzen CA, et al. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4:487–9. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 48.Young NL, DiMaggio PA, Plazas-Mayorca MD, et al. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–84. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian Z, Tolic N, Zhao R, et al. Enhanced top-down characterization of histone post-translational modifications. Genome Biol. 2012;13:R86. doi: 10.1186/gb-2012-13-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanu AB, Dwivedi P, Tam M, et al. Ion mobility-mass spectrometry. J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 51.Shvartsburg AA, Zheng Y, Smith RD, et al. Separation of variant methylated histone tails by differential ion mobility. Anal Chem. 2012;84:6317–20. doi: 10.1021/ac301541r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shvartsburg AA, Zheng Y, Smith RD, et al. Ion mobility separation of variant histone tails extending to the “middle-down” range. Anal Chem. 2012;84:4271–6. doi: 10.1021/ac300612y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LeRoy G, Weston JT, Zee BM, et al. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–42. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiragami-Hamada K, Shinmyozu K, Hamada D, et al. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol Cell Biol. 2011;31:1186–200. doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimada A, Dohke K, Sadaie M, et al. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 2009;23:18–23. doi: 10.1101/gad.1708009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young NL, Plazas-Mayorca MD, DiMaggio PA, et al. Collective mass spectrometry approaches reveal broad and combinatorial modification of high mobility group protein A1a. J Am Soc Mass Spectrom. 2010;21:960–70. doi: 10.1016/j.jasms.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niessen HE, Demmers JA, Voncken JW. Talking to chromatin: post-translational modulation of polycomb group function. Epigenetics Chromatin. 2009;2:10. doi: 10.1186/1756-8935-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cha TL, Zhou BP, Xia W, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–10. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 59.Kaneko S, Li G, Son J, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasaki T, Maier B, Koclega KD, et al. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flick F, Luscher B. Regulation of sirtuin function by posttranslational modifications. Front Pharmacol. 2012;3:29. doi: 10.3389/fphar.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia Z, Guo M, Ma H. Functional analysis of novel phosphorylation sites of CREB-binding protein using mass spectrometry and mammalian two-hybrid assays. Proteomics. 2011;11:3444–51. doi: 10.1002/pmic.201100121. [DOI] [PubMed] [Google Scholar]
- 63.Ryan CM, Kindle KB, Collins HM, et al. SUMOylation regulates the nuclear mobility of CREB binding protein and its association with nuclear bodies in live cells. Biochem Biophys Res Commun. 2010;391:1136–41. doi: 10.1016/j.bbrc.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 65.Ferreira R, Eberharter A, Bonaldi T, et al. Site-specific acetylation of ISWI by GCN5. BMC Mol Biol. 2007;8:73. doi: 10.1186/1471-2199-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shiio Y, Eisenman RN, Yi EC, et al. Quantitative proteomic analysis of chromatin-associated factors. J Am Soc Mass Spectrom. 2003;14:696–703. doi: 10.1016/S1044-0305(03)00204-6. [DOI] [PubMed] [Google Scholar]
- 67.Gygi SP, Rist B, Gerber SA, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 68.Chou DM, Adamson B, Dephoure NE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi S, Srivas R, Fu KY, et al. Quantitative Proteomics Reveal ATM Kinase-dependent Exchange in DNA Damage Response Complexes. J Proteome Res. 2012;11:4983–91. doi: 10.1021/pr3005524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kubota T, Hiraga S, Yamada K, et al. Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.005561. M110 005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torrente MP, Zee BM, Young NL, et al. Proteomic interrogation of human chromatin. PLoS One. 2011;6:e24747. doi: 10.1371/journal.pone.0024747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29:171–81. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- 73.Morrison C, Henzing AJ, Jensen ON, et al. Proteomic analysis of human metaphase chromosomes reveals topoisomerase II alpha as an Aurora B substrate. Nucleic Acids Res. 2002;30:5318–7. doi: 10.1093/nar/gkf665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gassmann R, Henzing AJ, Earnshaw WC. Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma. 2005;113:385–97. doi: 10.1007/s00412-004-0326-0. [DOI] [PubMed] [Google Scholar]
- 75.Takata H, Uchiyama S, Nakamura N, et al. A comparative proteome analysis of human metaphase chromosomes isolated from two different cell lines reveals a set of conserved chromosome-associated proteins. Genes Cells. 2007;12:269–84. doi: 10.1111/j.1365-2443.2007.01051.x. [DOI] [PubMed] [Google Scholar]
- 76.Ohta S, Bukowski-Wills JC, Sanchez-Pulido L, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–21. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136(1):175–86. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–9. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 79.Akiyoshi B, Nelson CR, Ranish JA, et al. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–99. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17:430–7. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Byrum SD, Raman A, Taverna SD, et al. ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2012;2:198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 83.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 84.Zegerman P, Canas B, Pappin D, et al. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–4. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- 85.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 86.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 87.Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 88.Bartke T, Vermeulen M, Xhemalce B, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–84. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muir TW. Semisynthesis of proteins by expressed protein ligation. Ann Rev Biochem. 2003;72:249–89. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 90.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 91.Nikolov M, Stutzer A, Mosch K, et al. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.005371. M110 005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bartels SJ, Spruijt CG, Brinkman AB, et al. A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein. PLoS One. 2011;6:e25884. doi: 10.1371/journal.pone.0025884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Markljung E, Jiang L, Jaffe JD, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7:e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butter F, Kappei D, Buchholz F, et al. A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep. 2010;11:305–11. doi: 10.1038/embor.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Butter F, Davison L, Viturawong T, et al. Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet. 2012;8:e1002982. doi: 10.1371/journal.pgen.1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Butter F, Scheibe M, Morl M, et al. Unbiased RNA-protein interaction screen by quantitative proteomics. Proc Natl Acad Sci USA. 2009;106:10626–31. doi: 10.1073/pnas.0812099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–6. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gavin AC, Bosche M, Krause R, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 99.Ho Y, Gruhler A, Heilbut A, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 100.Gavin AC, Aloy P, Grandi P, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 101.Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 102.Lambert JP, Fillingham J, Siahbazi M, et al. Defining the budding yeast chromatin-associated interactome. Mol Syst Biol. 2010;6:448. doi: 10.1038/msb.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lambert JP, Mitchell L, Rudner A, et al. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol Cell Proteomics. 2009;8:870–82. doi: 10.1074/mcp.M800447-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao Z, Zhang J, Bonasio R, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45(3):344–56. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 106.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 107.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–92. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 108.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 109.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22:50–5. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 110.Cuomo A, Moretti S, Minucci S, et al. SILAC-based proteomic analysis to dissect the “histone modification signature” of human breast cancer cells. Amino Acids. 2011;41:387–99. doi: 10.1007/s00726-010-0668-2. [DOI] [PubMed] [Google Scholar]
- 111.Katsnelson A. An epic search. Scientist. 2010;24:75–7. [Google Scholar]
- 112.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zuber J, Shi JW, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–11. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]